Abstract
BACKGROUND
Studies in rodents provide compelling evidence that microorganisms inhabiting the gut influence neurodevelopment. In particular, experimental manipulations that alter intestinal microbiota impact exploratory and communicative behaviors and cognitive performance. In humans, the first years of life are a dynamic time in gut colonization and brain development, but little is known about the relationship between these two processes.
METHODS
We tested whether microbial composition at 1 year of age is associated with cognitive outcomes using the Mullen Scales of Early Learning and with global and regional brain volumes using structural MRI at 1 and 2 years of age. Fecal samples were collected from 89 typically developing one-year-old infants. 16S rRNA amplicon sequencing was used for identification and relative quantification of bacterial taxa.
RESULTS
Cluster analysis identified 3 groups of infants defined by their bacterial composition. Mullen scores at age 2 differed significantly between clusters. In addition, higher alpha diversity was associated with lower scores on the overall composite score, visual reception scale, and expressive language scale at age 2. Exploratory analyses of neuroimaging data suggest the gut microbiome has minimal effects on regional brain volumes 1 and 2 years of age.
CONCLUSIONS
This is the first study to demonstrate associations between the gut microbiota and cognition in human infants. As such, it represents an essential first step in translating animal data into the clinic.
Keywords: microbiota, infant, cognition, gut, brain, MRI
INTRODUCTION
The gut microbiome is a complex microbial ecosystem which varies between individuals and may be a key modulator of neurodevelopment through the microbiome- gut-brain axis (1; 2). In rodents, experimental manipulations that alter the intestinal microbiota impact anxiety and depression-related behaviors in a multiple well-established paradigms (3–6). Cognitive effects have also been reported (7; 8). Early development appears to be of critical importance as bacterial colonization of germ-free mice does not normalize anxiety phenotypes at 10 weeks of age (9), but does normalize anxiety phenotypes at 3 weeks of age (10). Behavioral changes are accompanied by changes in neurochemistry, gene expression, dendritic morphology, and subcortical brain volumes (5; 10–15). In humans, altered microbial composition of the gut has been reported in children with autism (16–18) and adults with depression (19; 20) and linked to childhood temperament (21) and adult cognition (22), but no studies have addressed when these relationships emerge or directly examined which brain regions may be involved.
The first year of life is the foundational period for microbial colonization of the gut and the most rapid and dynamic phase of postnatal brain development. Gut colonization is initially determined by method of delivery (23). Subsequently, the diversity of species within each individual (alpha diversity) increases rapidly as facultative aerobic Enterobacteriaceae give way to strict anaerobes, which are then displaced by a mixture of Clostridiales (24). Environmental factors influencing colonization include feeding practices (breast versus formula, introduction of solid foods etc.), day care attendance, illness, and exposure to antibiotics (25; 26). The microbiome is also influenced by genetics with 8.8% of taxa showing heritability greater than 0.2 (27). All these factors contribute to significant inter-individual variation (28). While this dynamic progression is transpiring in the gut, several foundational processes are simultaneously occurring in the brain including the proliferation, migration, and apoptosis of glia, myelination of axons, and synaptogenesis (29). These processes are accompanied by dramatic changes in brain tissue volumes (30; 31) and the rapid emergence of various cognitive functions.
A relationship may exist between these two concurrent processes, but this has not been empirically demonstrated. In this prospective, longitudinal study, we tested if the 1- year-old gut microbiome was associated with cognitive outcomes at 1 and 2 years of age. We hypothesized that infant gut microbiome samples would cluster into groups of community similarity and that cognitive ability would differ between clusters. Specifically, we predicted that overall cognitive performance would be highest in clusters with an abundance of putatively beneficial microorganisms such as Lactobacillus (32) or Bacteroides (4). We also predicted that lower alpha diversity (indicating a less mature microbiome) would be correlated with lower cognitive performance given reports that low alpha diversity in infancy precedes negative health outcomes including type 1 diabetes (33) and asthma (34). We also conducted an exploratory analysis to examine whether composition of the gut microbiome at 1 year was associated with global and regional brain volumes assessed through structural magnetic resonance imaging at 1 and 2 years.
METHODS AND MATERIALS
Study Population
We recruited 89 one-year-old infants (twins and singletons) from two prospective longitudinal studies of early brain development at the University of North Carolina at Chapel Hill (30; 31; 35; 36). Exclusion criteria for the parent studies included fetal ultrasound abnormalities and major medical illness of the mother. Informed written consent was obtained from the parent/legal guardian of each subject. This study was approved by the Institutional Review Board of the University of North Carolina at Chapel Hill.
Microbiome Analysis
Parents collected approximately 200 mg of fecal material from a single diaper, placed it in a tube filled with Allprotect reagent (Valencia, CA), and returned it through overnight shipping. Once received, samples were stored at −80°C until analysis. 16S rRNA amplicon sequencing of the V1-V2 gene region (37; 38) was performed on the Illumina MiSeq platform for identification and relative quantification of bacterial taxa as previously described (25; 39). Bioinformatics was performed with Quantitative Insights Into Microbial Ecology (QIIME) software (40). The de novo algorithm was used for OTU picking and chimeric sequences were removed with ChimeraSlayer. Taxonomic assignment, alpha and beta diversity analysis were done via QIIME (41; 42). Clustering analysis was completed with distance metrics applied to the relative genus abundance from the 89 subjects with Partitioning Around Medoids as the clustering algorithm. Cluster scoring methods were used to assess the optimal number of clusters. See Supplementary Methods for details.
Genera Analysis and Co-occurrence Networks
A modified Kruskal Wallis test for zero-inflated data (ZIKW) was applied to identify significant differences in relative genera abundance between clusters (FDR < 5%). Permutation was used to ensure accurate p-values. To better understand the unique community dynamics of each cluster, we used one representative genera from each cluster as a seed to generate co-occurrence networks via Spearman correlations. See Supplementary Methods for details.
Cognitive Testing
The Mullen consists of five separate Scales, with their own age-group standardized normative T-scores and percentiles, and an Early Learning (cognitive) Composite (ELC) similar to an IQ score or the Bayley MDI (43). The 5 Scales measure gross motor, fine motor, visual reception, expressive language, and receptive language skills. The standardized T-scores of 4 scales (gross motor not included) are combined to create the ELC. The Mullen has good standardization and reliability data, with median internal consistency scores ranging between 0.75 and 0.91, and test-retest correlations over 0.82 for 1–25 months. Mullen data were collected over a period of three years by several experienced examiners who were blind to microbiome characteristics. All cognitive data was screened for quality control and no data warranted exclusion from this study (see Supplementary Methods). This provided cognitive data for 86 subjects at 1 year of age and 69 subjects at 2 years of age.
Structural Image Acquisition
T1-weighted images were acquired on a Siemens head-only 3T TIM-Trio scanner (Siemens Medical System, Erlangen, Germany) during unsedated natural sleep (median age at scan 1 = 12.8 months, median age at scan 2 = 25.1 months). Scan parameters at years 1 and 2 were as follows: MP-RAGE time repetition [TR] = 1900 ms, time echo [TE] = 3.74 ms, flip angle = 7°, image resolution = 1×1×1mm. A total of 38 infants at year 1 did not go to sleep or woke up in the scanner (success rate: 57%). A total of 61 infants at year 2 did not go to sleep, woke up in the scanner, or were lost to follow up (success rate: 31%).
Image Analysis
Images were examined to exclude scans with motion and imaging artifacts (year 1 n=3, year 2 n=1). Brain tissue was classified as gray matter, white matter, and cerebrospinal fluid using an atlas-moderated iterative expectation maximization segmentation algorithm with T1 images as previously described (30). Lateral ventricle volume was segmented using ITK-SNAP (44) as previously described (31). Gray matter was parsed into 90 regions by non-linear warping of a neonatal adaptation of the AAL atlas template as previously described (31; 45). With these methods, we obtained measures of intracranial volume, total gray matter, total white matter, total cerebrospinal fluid, lateral ventricle volume, and 90-region gray matter volumes at 1 year of age for 46 subjects and at 2 years of age for 27 subjects.
Association of Cognitive and Brain Outcomes with Cluster Membership and Alpha Diversity
Linear mixed effect models were used to test for effects of cluster membership and alpha diversity on cognitive performance and brain volumes as well as cluster membership on alpha diversity. Subjects with sensory deficits that would impede cognitive testing (n=2) or MRI abnormalities (n=1, gliosis, chronic venous ischemia) were excluded from analyses. Likelihood ratio tests were used to test the significance of the coefficient of each cluster and alpha diversity measure. P-values of primary analyses were uncorrected while secondary analyses were adjusted for multiple comparisons using FDR. Sensitivity analyses were conducted to determine the effect of cluster independent of alpha diversity or alpha diversity independent of cluster by including either cluster membership or alpha diversity metrics as a covariate in the reciprocal analyses. Additional sensitivity analyses looked at change in Mullen scale raw scores from 1 to 2 years as well as 1-year performance in the subset of subjects with data at 2 years. Raw scores are better suited to longitudinal analyses as they are not age-adjusted while t- scores are. Demographic and medical variables associated with cluster membership or alpha diversity and neuroimaging or cognitive outcomes were included as covariates (See Supplementary Methods for details).
Metagenome Prediction from PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to predict metagenome functional content from the 16S rRNA sequencing data (46). The PICRUSt pipeline was used to predict the abundance of Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs by subject at the KO and collapsed KEGG pathway levels. This output was grouped by cluster and analyzed for group differences through Statistical Analysis of Taxonomic and Functional Profiles (STAMP) (47).
RESULTS
Infant microbiota cluster into three groups
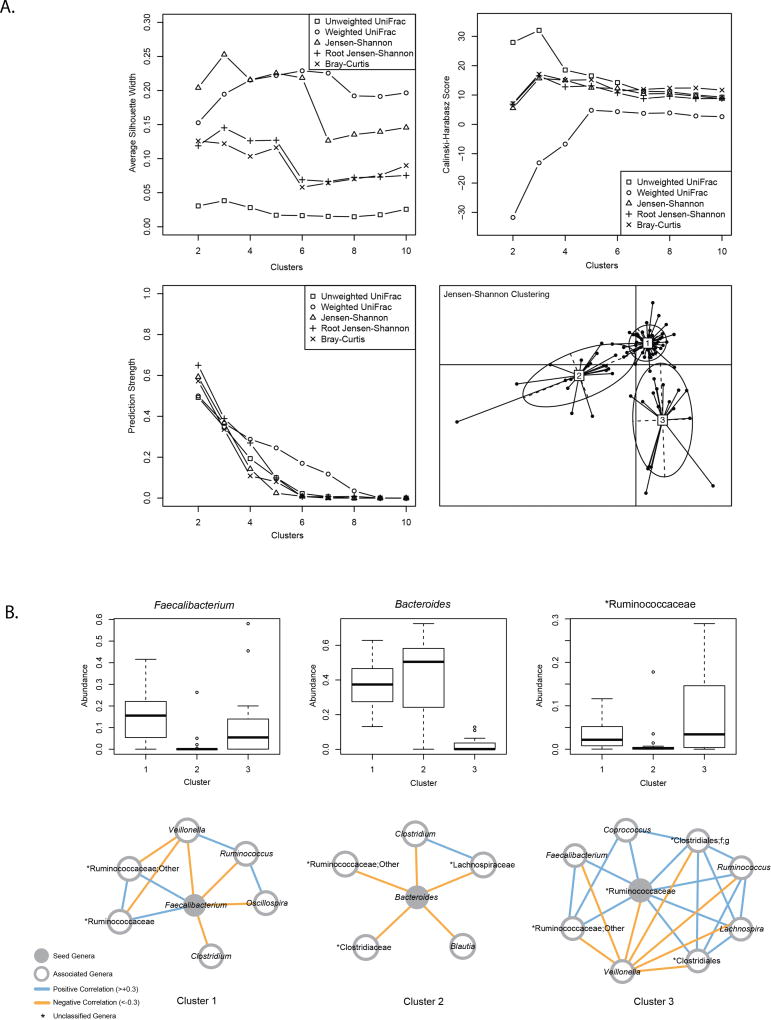
There was modest support for clustering subjects into three groups based on average silhouette width and Calinksi-Harabasz scoring (Figure 1a). The Jensen-Shannon distance metric scored highest for three groups and was used for subsequent analyses. Clusters differed in the abundance of many genera (Supplementary Table 1) with Cluster 1 characterized by relatively high abundance of Faecalibacterium, Cluster 2 by relatively high abundance of Bacteroides, and Cluster 3 by relatively high abundance of an unnamed genus in the family Ruminococcaceae. Co-occurrence networks show positive and negative correlations between different genera (Fig 1b).
Figure 1. Infant gut microbiome at 1 year of age clusters into three groups.
A. Distance metrics (unweighted Unifrac, weighted Unifrac, Jensen-Shannon, Root Jensen-Shannon, Bray-Curtis) were scored with average silhouette width (SW), Calinski-Harabasz (CH), and prediction strength to identify the number of clusters with most support. Modest support for clustering into three groups was supported by SW and CH. B. (Top) Boxplots of genera selected as seed regions for co-occurrence networks. (Bottom) Co-occurrence networks show positive and negative correlations between different genera.
Identification of predictor covariates
Breastfeeding at time of sample collection (1 year), birth method, and paternal ethnicity were significantly different between clusters after FDR-correction (Table 1). Infants in C2 were more likely to be breastfed at 1 year of age and less likely to have been born via cesarean section. Paternal ethnicity in C2 was 90% white compared to C3 and C1 with 71% and 57% white respectively. Having older siblings was associated with increased alpha diversity after FDR correction. Alpha diversity was increased in infants with Black or Asian paternal ethnicity and decreased in infants with Native American paternal ethnicity as compared to white paternal ethnicity (Supplementary Table 1).
Table 1.
Demographic characteristics of group and clusters
| Descriptive Variable | Overall | Cluster 1 | Cluster 2 | Cluster 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | Q-Value | ||
| Income | High | 39 | 43.8 | 22 | 41.5 | 7 | 36.8 | 10 | 58.8 | 0.804 |
| Middle | 23 | 25.8 | 13 | 24.5 | 7 | 36.8 | 3 | 17.6 | ||
| Low | 23 | 25.8 | 15 | 28.3 | 5 | 26.3 | 3 | 17.6 | ||
| Not Available | 4 | 4.5 | 3 | 5.7 | 0 | 0 | 1 | 5.9 | ||
|
| ||||||||||
| Maternal Psychiatric History | Yes | 24 | 27 | 11 | 20.8 | 6 | 31.6 | 7 | 41.2 | 0.432 |
| No | 65 | 73 | 42 | 79.2 | 13 | 68.4 | 10 | 58.8 | ||
|
| ||||||||||
| Paternal Psychiatric History | Yes | 11 | 12.4 | 5 | 9.4 | 4 | 21.1 | 2 | 11.8 | 0.544 |
| No | 78 | 87.6 | 48 | 90.6 | 15 | 78.9 | 15 | 88.2 | ||
|
| ||||||||||
| Cesarean Section | Yes | 44 | 49.4 | 27 | 50.9 | 4 | 21.1 | 13 | 76.5 | 0.035* |
| No | 45 | 50.6 | 26 | 49.1 | 15 | 78.9 | 4 | 23.5 | ||
|
| ||||||||||
| Single or Twin Gestation | Twin | 47 | 52.8 | 30 | 56.6 | 6 | 31.6 | 11 | 64.7 | 0.432 |
| Singleton | 42 | 47.2 | 23 | 43.3 | 13 | 68.4 | 6 | 35.3 | ||
|
| ||||||||||
| Sex | Male | 49 | 55.1 | 28 | 52.8 | 11 | 57.9 | 10 | 58.8 | 0.956 |
| Female | 40 | 44.9 | 25 | 47.2 | 8 | 42.1 | 7 | 41.2 | ||
|
| ||||||||||
| NICU | Yes | 19 | 21.3 | 10 | 18.9 | 4 | 21.1 | 5 | 29.4 | 0.841 |
| No | 70 | 78.7 | 43 | 81.1 | 15 | 78.9 | 12 | 70.6 | ||
|
| ||||||||||
| Maternal Ethnicity | White | 66 | 74.2 | 36 | 67.9 | 18 | 94.7 | 12 | 70.6 | 0.190 |
| Black | 19 | 21.3 | 15 | 28.3 | 1 | 5.3 | 3 | 17.6 | ||
| Asian | 2 | 2.2 | 2 | 3.8 | 0 | 0 | 0 | 0 | ||
| Native American | 2 | 2.2 | 0 | 0 | 0 | 0 | 2 | 11.8 | ||
|
| ||||||||||
| Paternal Ethnicity | White | 60 | 67.4 | 30 | 56.6 | 17 | 89.5 | 13 | 76.5 | 0.049* |
| Black | 23 | 25.8 | 20 | 37.7 | 1 | 5.3 | 2 | 11.8 | ||
| Asian | 5 | 5.6 | 3 | 5.7 | 0 | 0 | 2 | 11.8 | ||
| Native American | 1 | 1.1 | 0 | 0 | 1 | 5.3 | 0 | 0 | ||
|
| ||||||||||
| Surgical Anesthesia | Yes | 5 | 5.6 | 2 | 3.8 | 1 | 5.3 | 2 | 11.8 | 0.544 |
| No | 84 | 94.4 | 51 | 96.2 | 18 | 94.7 | 15 | 88.2 | ||
|
| ||||||||||
| Older Siblings | Yes | 42 | 47.2 | 32 | 60.4 | 4 | 21.1 | 6 | 35.3 | 0.059 |
| No | 47 | 52.8 | 21 | 39.6 | 15 | 78.9 | 11 | 64.7 | ||
|
| ||||||||||
| Currently Breastfed | Yes | 32 | 36 | 14 | 26.4 | 14 | 73.7 | 4 | 23.5 | 0.012* |
| No | 57 | 64 | 39 | 73.6 | 5 | 26.3 | 14 | 76.5 | ||
|
| ||||||||||
| Ever Given Formula | Yes | 64 | 71.9 | 40 | 75.5 | 9 | 47.4 | 15 | 88.2 | 0.071 |
| No | 25 | 28.1 | 13 | 24.5 | 10 | 52.6 | 2 | 11.8 | ||
|
| ||||||||||
| Currently Given Formula | Yes | 19 | 21.3 | 11 | 20.8 | 3 | 15.8 | 5 | 29.4 | 0.558 |
| No | 70 | 78.7 | 42 | 79.2 | 16 | 84.2 | 12 | 70.6 | ||
|
| ||||||||||
| Given Milk Other Than Breastmilk or Formula | Yes | 77 | 86.5 | 47 | 88.7 | 15 | 78.9 | 15 | 88.2 | 0.934 |
| No | 12 | 13.5 | 6 | 11.3 | 4 | 21.1 | 2 | 11.8 | ||
|
| ||||||||||
| Type of Other Milk | Breastmilk Exclusively | 12 | 13.5 | 6 | 11.3 | 4 | 21.1 | 2 | 11.8 | 0.432 |
| Cow's Milk | 72 | 80.9 | 45 | 84.9 | 15 | 78.9 | 12 | 70.6 | ||
| Other (almond, soy) | 5 | 5.6 | 2 | 3.8 | 0 | 0 | 3 | 17.6 | ||
|
| ||||||||||
| Symptoms of Illness in Previous Week | Yes | 43 | 48.3 | 20 | 37.7 | 10 | 52.6 | 13 | 76.5 | 0.059 |
| No | 46 | 51.7 | 33 | 62.3 | 9 | 47.4 | 4 | 23.5 | ||
|
| ||||||||||
| Gastrointestinal Symptoms in Previous Week | Yes | 12 | 13.5 | 6 | 11.3 | 3 | 15.8 | 3 | 17.6 | 0.804 |
| No | 77 | 86.5 | 47 | 88.7 | 16 | 84.2 | 14 | 82.4 | ||
|
| ||||||||||
| Antibiotics within Last Year | Yes | 30 | 33.7 | 15 | 28.3 | 8 | 42.1 | 7 | 41.2 | 0.614 |
| No | 59 | 66.3 | 38 | 71.7 | 11 | 57.9 | 10 | 58.8 | ||
|
| ||||||||||
| Antibiotics During Pregnancy | Yes | 17 | 19.1 | 10 | 18.9 | 3 | 15.8 | 4 | 23.5 | 0.912 |
| No | 72 | 80.9 | 43 | 81.1 | 16 | 84.2 | 13 | 76.5 | ||
| Descriptive Variable |
Overall | Cluster 1 | Cluster 2 | Cluster 3 | |||||
|---|---|---|---|---|---|---|---|---|---|
| N | Mean ± SD | N | Mean ± SD | N | Mean ± SD | N | Mean ± SD | Q-Value | |
| Gestational Age at Birth (days) | 89 | 263.72 ± 16.74 | 53 | 262.94 ± 15.72 | 19 | 268.32 ± 18.4 | 17 | 261.00 ± 17.94 | 0.432 |
|
| |||||||||
| Birth Weight (grams) | 89 | 2917.31 ± 690.21 | 53 | 2885.09 ± 706.73 | 19 | 3129.47 ± 645.65 | 17 | 2780.65 ± 671.66 | 0.432 |
|
| |||||||||
| Apgar at 5 Minutes | 89 | 8.75 ± .79 | 53 | 8.87 ± 0.48 | 19 | 8.47 ± 1.39 | 17 | 8.71 ± 0.59 | 0.316 |
|
| |||||||||
| Maternal Age at Birth (years) | 89 | 31.9 ± 4.93 | 53 | 31.74 ± 5.08 | 19 | 30.95 ± 5.06 | 17 | 33.47 ± 4.17 | 0.432 |
|
| |||||||||
| Maternal Education (years) | 89 | 16.72 ± 2.61 | 53 | 16.62 ± 2.8 | 19 | 17.74 ± 2.33 | 17 | 15.88 ± 1.93 | 0.432 |
|
| |||||||||
| Paternal Age at Birth (years) | 89 | 34.06 ± 5.89 | 53 | 34.13 ± 6.06 | 19 | 33.53 ± 6.95 | 17 | 34.42 ± 4.11 | 0.689 |
|
| |||||||||
| Paternal Education (years) | 89 | 15.61 ± 3.4 | 53 | 15.43 ± 3.49 | 19 | 16.56 ± 3.17 | 17 | 15.09 ± 3.37 | 0.432 |
Cognitive abilities differ between clusters
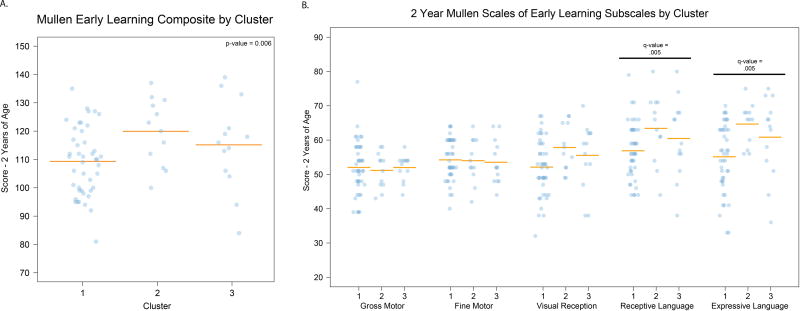
Our primary analysis focused on ELC scores at 1 and/or 2 years of age. Significant differences were observed at 2 years of age (n=69) with C2 showing the highest performance and C1 showing the lowest performance (Figure 2a; p-value = 0.006). In order to determine how differences in ELC at 2 years of age related to specific functional domains, we performed secondary analyses testing whether the five individual Mullen Scales (gross motor, fine motor, visual reception, receptive language, and expressive language) differed between clusters at 2 years of age. Receptive language and expressive language were significantly different between clusters after FDR correction (Figure 2b, q-value = 0.005, q-value = 0.005). No significant differences were observed in Mullen outcomes between clusters at 1 year (n=86).
Figure 2. Performance on Mullen Scales of Early Learning at 2 years of age differs between clusters.
A. Individual value plot showing performance on the Mullen Early Learning Composite by cluster (p-value = 0.006). B. Individual value plot showing secondary analysis of the Mullen Scale performance by cluster (receptive language q-value = 0.005, expressive language q-value = 0.005). Covariates for both analyses: cesarean section, paternal ethnicity, currently breastfeeding, sex, maternal education, paternal age, paternal ethnicity, twin status, income.
Regional gray matter differences between clusters
We tested whether global and regional brain volumes differed between clusters at 1 and 2 years of age (n=46, n=27). At 1 year, the right superior occipital gyrus was largest in C2 and smallest in C3 (covariates: cesarean section, paternal ethnicity, currently breastfeeding, total intracranial volume; q=0.037). At 2 years of age, the left and right caudate nucleus were smallest in C2 and largest in C3 (covariates: cesarean section, paternal ethnicity, currently breastfeeding, total intracranial volume; q=0.001, q=0.003). There were no differences in intracranial volume, total white matter, total gray matter, total cerebrospinal fluid, lateral ventricle volume, or other 90-region parcellation volumes at 1 or 2 years of age.
Predicted metagenome of gut microbiota differs between clusters
Group differences were observed in orthologs and functional categories between clusters at each level of the collapsed KEGG pathways (Supplementary Table 3; q<0.05). As C2 showed the highest performance on the Mullen Scales, we were particularly interested in metabolic differences unique to this group. At level 2 of the KEGG pathways, C2 had increased “Metabolism of Cofactors and Vitamins” as well as decreased “Cell Motility.” Within these categories, genes involved in metabolism of biotin, lipoic acid, folate, and ubiquinone and other terpenoid quinones were increased while genes involved in bacterial chemotaxis, flagellar assembly, bacterial motility proteins, and cytoskeleton proteins were decreased.
Alpha diversity correlates with cognitive performance
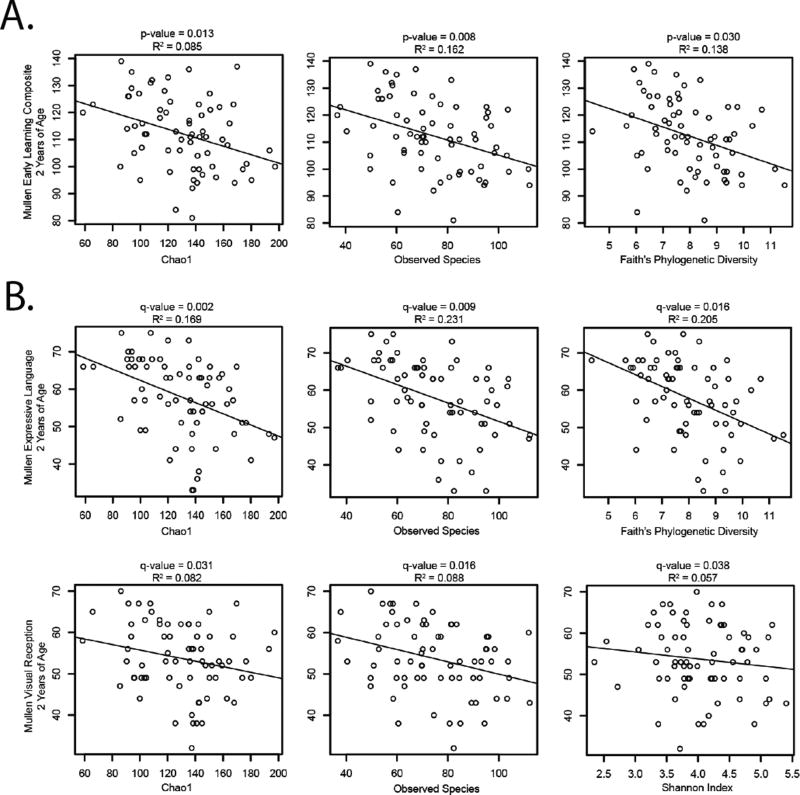
Four measures of alpha diversity, Chao1 (CH1), observed species (OS), Shannon Index (SI), and Faith’s Phylogenetic Diversity (FPD) were tested for associations with the ELC. At 2 years of age, the primary analysis showed a significant negative correlation of ELC with CH1, OS, FPD (Figure 3a; p=0.013, p=0.008, p=0.030). No significant associations were observed at 1 year of age. Secondary analysis of individual Mullen Scales at 2 years of age revealed a significant negative correlation of expressive language with CH1 (q=0.002), OS (q=0.009), and FPD (q=0.016) and negative correlation of visual reception with CH1 (q=0.031), OS (q=0.016), and SI (q=0.038) after FDR correction (Figure 3b). Alpha diversity measures accounted for 5 to 23% of the variance in Mullen scores.
Figure 3. Alpha diversity is negatively correlated with cognitive scores at 2 years of age.
A. Plots of primary analysis, Early Learning Composite versus alpha diversity measures, show negative correlations. Covariates: older siblings, paternal ethnicity, sex, maternal education, paternal age, twin status, income B. Secondary analyses with the separate Mullen Scales show a negative association of expressive language with alpha diversity. Covariates: older siblings, paternal ethnicity, sex, maternal education, paternal age, twin status, income.
Alpha diversity associated with regional gray matter volumes at 2 years of age
Secondary analyses testing the association of alpha diversity metrics with brain volumes at 1 and 2 years of age revealed three regions of significance at age 2 (covariates: older siblings, paternal ethnicity, total intracranial volume). These included the left precentral gyrus (CH1, q=0.025), left amygdala (OS, q=0.046), right angular gyrus (CH1, q=0.025), all with positive directionality. There were no associations with intracranial volume, total gray matter, total white matter, total cerebrospinal fluid, lateral ventricle volume, or other regional volumes at 2 years of age after FDR correction. Similarly, no significant associations of alpha diversity metrics with 1-year brain volumes were observed after FDR correction.
Alpha diversity differs between clusters
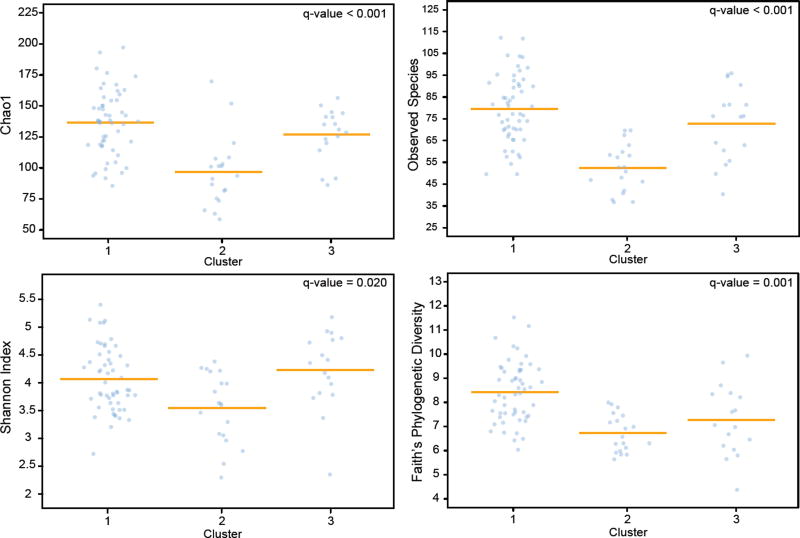
Finally, we tested whether measures of alpha diversity differed between clusters. FPD, SI, OS, and CH1 were all significantly different between clusters with C1 showing the greatest alpha diversity and C2 showing the least (Fig. 4).
Figure 4. Clusters differ by three measures of alpha diversity.
Individual value plots of Faith’s Phylogenetic Diversity, Shannon Index, Observed Species, and Chao1 by cluster. Covariates: cesarean section, paternal ethnicity, currently breastfeeding, older siblings.
Sensitivity Analyses
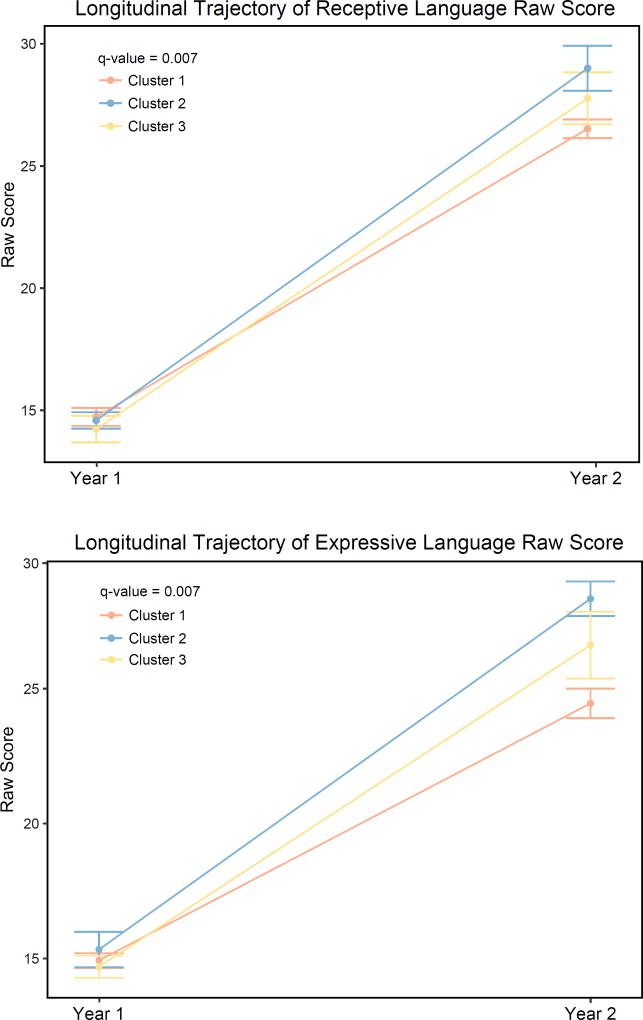
We conducted analyses to determine if cluster membership or alpha diversity had the larger impact on outcomes by including either cluster or individual alpha diversity metrics as a covariate. Most notably, cognitive results of cluster analyses retained significance with CH1, FPD, and SI as covariates, but results were lost with OS (a measurement of richness rather than evenness). ELC at 2 years of age was no longer significantly associated with alpha diversity when cluster was included as a covariate (Supplementary Table 4). A second sensitivity analysis assessed the change in Mullen performance between 1 and 2 years. This was done using raw scores (not age-adjusted) for GM, FM, VR, RL, and EL. Change in performance between 1 and 2 years of age was significantly different between clusters for RL and EL (Figure 5, q=0.007, q=0.007). Finally, we performed an analysis of 1-year cognitive outcome by cluster in a subset of subjects also with data at 2 years (n=69) (Supplementary Figure 1). Results remained null in this group.
Figure 5. Change in receptive language and expressive language scales from 1 to 2 years of age differs between clusters.
Plot of average raw scores by cluster from 1 to 2 years of age. (receptive language q=0.007, expressive language q=0.007) Covariates: sex, age at Mullen testing, maternal education, paternal education, paternal ethnicity, twin status, total household income, maternal psychiatric history
DISCUSSION
There is increasing agreement from preclinical work that the gut microbiota influences brain development during a critical period in early life resulting in long-term changes in behavior. This is the first study to show that variation in the human gut microbiome is associated with cognition in a cohort of typically developing infants during the hypothesized sensitive period. This study does not attempt to address a causative role in the observed relationships, but is an important translational starting point.
Previous work suggests that the human gut microbiome achieves an adult-like state between one to three years of age (48; 49), but to our knowledge this is the first study to test whether distinct groups of subjects, defined by their bacterial composition, are present at this age. In adults, several studies have supported the existence of three clusters (sometimes called enterotypes) characterized by the relative abundance of Bacteroides, Prevotella, and unclassified genera in the family Ruminococcaceae (50; 51) though there is ongoing debate as to whether individual variation is best described in terms of clusters or gradients (52). Our data suggest that at least two of the three canonical adult enterotypes, those characterized by differentially increased abundance of Bacteroides and Ruminococcaceae, may already be present at 1 year of age. Alternatively, the clusters we observed may reflect differences in the developmental rate of the gut microbiome. In particular, the cluster with increased abundance of Bacteroides may reflect a subgroup of children with delayed maturation of the gut microbiome, given recent reports that the first two years of life are characterized by a succession of bacterial taxa in which facultative aerobic Enterobacteriaceae give way to strict anaerobes, including Bacteroides, which are then displaced by a diverse mixture of Clostridiales including Ruminococcaceae, Faecalibacterium, and Lachnospira (24; 53).
The environmental and genetic factors influencing cluster membership were not a primary focus of the current study, but we did investigate a number of potentially important demographic, medical history, and feeding history variables. In keeping with the conceptualization of C2 as a less mature microbiome, children in C2 were more likely to be breastfed at the time of sample collection. Maturation into an adult-like microbiota is primarily determined by cessation of breast-feeding (49). Birth method was significantly different between clusters with C2 (high levels of Bacteroides) less likely to be born via cesarean section. This is in keeping with prior research showing that cesarean section is associated with reduced levels of Bacteroides shortly after birth as well as increased levels of multiple taxa within phyla Firmicutes and Proteobacteria (24). Finally, there was an observed difference in paternal ethnicity between clusters. Maternal race/ethnicity has been previously associated with the infant microbiome (54; 55). This association could reflect both genetic and environmental factors associated with this sociocultural construct. Importantly, the associations we observed between cluster membership and cognitive performance at age 2 were present when controlling for breastfeeding at 1 year, cesarean section, and paternal ethnicity. We did not observe any associations with antibiotic exposure during the first year of life or consumption of formula and other milks. Antibiotic exposure delays microbiota maturation (24), but in the current sample, any taxa suppressed by early antibiotic exposure appear to have rebounded by 1 year. Other factors that might influence cluster membership such as the type and diversity of solid foods and prenatal stress were not measured in the current study, but would represent a valuable addition to future studies.
Clusters at 1 year of age predicted cognitive performance at 2 years of age with C2 (high levels of Bacteroides) showing the highest level of performance (90th percentile) and C1 (high levels of Faecalibacterium) showing the lowest level of performance (72nd percentile), though no individuals scored in the cognitively impaired range (ELC < 70). Examination of the Mullen Scales demonstrated significant differences in receptive language and expressive language, though a similar pattern of C2>C3>C1 was observed for the visual reception scale as well. It is likely that all three metrics contributed to the overall significant difference in ELC. The receptive language and expressive language findings may have relevance to disorders characterized by language impairments and delays. There are interesting parallels between our work and studies in maternal immune activation (MIA) mouse models of neurodevelopmental disorders. MIA mice exhibit altered gut microbiota and decreased ultrasonic vocalizations. These vocalizations were restored through administration of Bacteroides fragilis (4), a member of the genus most prevalent in our high performing expressive language group.
Alpha diversity at 1 year of age also predicted cognitive performance at 2 years of age. Higher diversity was associated with poorer scores on the ELC, visual reception, and expressive language scales. This was somewhat surprising as high alpha diversity in infancy indicates a more mature, adult-like community and low alpha diversity in infancy is associated with negative health outcomes including type 1 diabetes (33) and asthma (34). It has been hypothesized that functional redundancy provided by a highly diverse microbiome may allow individuals to better respond to environmental fluctuations, promoting intestinal homeostasis and maintaining human health (24). However, the notion that alpha diversity is always positive has been challenged recently as high alpha diversity has been found in adult subjects with major depressive disorder (19) and ASD (16). Our data add to this growing body of research which suggests increased diversity is not necessarily beneficial for neurocognitive or neuropsychiatric outcomes. Rather, higher diversity may mean less resources afforded to gut microbiota beneficially impacting neurodevelopment. In addition, delayed maturation of the gut microbiome may be associated with a prolonged period of cortical plasticity and maturation leading to better cognitive performance.
Analysis of the change in Mullen scores for receptive language and expressive language from 1 to 2 years of age were also significantly different between clusters. This result suggests that the gut microbiome may influence the developmental trajectory of cognition. The contribution of the gut microbiome to cognition at 2 years of age may be a downstream consequence of the community’s influence on development at 1 year of age—possibly through changes in systemic immunity, circulating short chain fatty acids and other metabolites, including various neuroactive compounds, or neuromodulation via vagal stimulation, all of which have been described in animal models (1). Alternative explanations for finding associations at 2 years, but not 1 year, include less accurate cognitive assessment at age 1 due to greater reliance on parental report or that scores at 1 year represent individual differences in maturational pace whereas scores at 2 may represent more persistent differences in cognitive ability.
PICRUSt analysis suggested mechanisms that might underlie microbial effects on neurodevelopment. In C2, there was increased predicted prevalence of genes related to the production of vitamins and cofactors (biotin, lipoic acid, folate, ubiquinone, other terpenoid quinones) while genes involved in pathogenicity (bacterial chemotaxis, flagellar assembly, bacterial motility proteins, cytoskeleton proteins) were decreased. The combination of increased neurodevelopmentally important metabolites like folate (56–58) and reduced pathogenic and inflammatory potential from gut commensals may play a role in the higher cognitive performance of C2. However, PICRUSt analysis is limited as it relies on predicted metagenomic information rather than direct measurement of gene expression. Results may be influenced by primer selection, orthology scheme, and reference genome (46).
In exploratory analyses of regional gray matter volumes, we observed several significant associations with microbiome measures. However, these findings were anatomically distributed, mostly unilateral, and varied in direction of effect. Consequently, they should be treated with caution until replicated. Studies incorporating other neuroimaging phenotypes such as cortical thickness and surface area, diffusion tensor imaging (DTI), and functional connectivity or activation may provide additional insights into the neural circuits mediating the microbiome’s impact on cognition.
In conclusion, we have shown that microbial composition of the human gut at 1 year of age predicts cognitive performance at 2 years of age, particularly in the area of communicative behavior. Results may have implications for developmental disorders characterized by cognitive or language delay. Strengths of this study include its prospective design, focus on infancy, and incorporation of structural MRI. Limitations include the following: 1. Microbiota were assessed at a single timepoint when the intake of solid foods varies significantly, potentially resulting in rapid shifts in the gut microbiome (49; 59). Longitudinal studies incorporating multiple measurements of the microbiota would address this limitation and help delineate temporal relationships between microbial colonization and brain development in human infants. 2. This work took advantage of two ongoing prospective longitudinal studies of early brain development (30; 31; 35; 36). Consequently, the behavioral outcomes examined were not selected based on their potential relevance to the microbiome. In rodents, the most consistent behavioral alterations following manipulation of the gut microbiota are in the realm of anxiety and depression-related behaviors. Future studies ought to directly probe these domains. 3. Functional differences between clusters were inferred from marker gene data. Future studies using broad-spectrum metabolomics or transcriptomics would yield more insight into mechanisms underlying the relationships we observed. Ultimately, with further research, it may become possible to guide the development of the gut microbiome through targeted interventions, thereby supporting cognitive development.
Supplementary Material
IN THIS ISSUE.
Infancy is a period when the human brain undergoes rapid changes. It is also the foundational period for microbial colonization of the gut. Little is known about the relationship between these processes. In a preliminary study of 89 infants, we demonstrate that microbial composition at 1 year of age is associated with cognition at 2 years of age. Results deepen our understanding of the human microbiome-gut-brain axis and may have implications for improving cognitive outcomes.
Acknowledgments
Jennifer Prater served as lead study coordinator with additional assistance from Dianne Evans and Wendy Neuheimer. Maggie Fox, Haley Parrish, Margo Williams, Mallory Turner, Emma Brink, Monica Ferenz, and Kassidy Jezierski at the FPG Child Development Institute conducted Mullen Scales of Early Learning assessments. Joe Blocher & Rachel Steiner at the Neuro Image Research and Analysis Laboratories provided image analysis. We thank the families for their participation in this study. This study was supported by NIH Grants T32 NS007432, P30 DK34987, R01 HD053000, U01 MH070890, R33 MH104330, and the Foundation of Hope for Research & Treatment of Mental Illness.
Footnotes
Author contributions: Conceptualization, J.H.G., X.G., A.L.T., and R.C.K.; Statistical Analysis, K.X., M.A., and M.A.A.; Image analysis, M.A.S.; Behavior analysis: B.D.G.; Microbiome analysis, M.A.A.; Data Curation, A.L.C.; Writing – Original Draft, A.L.C.; Writing – Review & Editing, R.C.K., A.L.C., M.A.A., K.X., M.A., M.A.S., J.H.G., X.G., A.L.T. and B.D.G.; Visualization, A.L.C. and K.X.; Supervision, J.H.G. and R.C.K.; Project Administration, A.L.C., and R.C.K.; Funding Acquisition, X.G., J.H.G., and R.C.K.
FINANCIAL DISCLOSURES
R.C.K is a co-investigator on a grant from Nestle/Wyeth entitled “Interrelationships of Nutrition, Gut Microbiota, as well as Brain & Cognitive Development in Early Life”. The other authors declare no biomedical financial interests or potential conflicts of interest.
References
- 1.Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 3.De Palma G, Blennerhassett P, Lu J, Deng Y, Park aJ, Green W, et al. Microbiota and host determinants of behavioural phenotype in maternally separated mice. Nat Commun. 2015;6:7735. doi: 10.1038/ncomms8735. [DOI] [PubMed] [Google Scholar]
- 4.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Pnas. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016:1–11. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 7.Fröhlich EE, Farzi A, Mayerhofer R, Reichmann F, Jačan A, Wagner B, et al. Cognitive impairment by antibiotic-induced gut dysbiosis: Analysis of gut microbiota-brain communication. Brain Behav Immun. 2016 doi: 10.1016/j.bbi.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savignac HM, Tramullas M, Kiely B, Dinan TG, Cryan JF. Bifidobacteria modulate cognitive processes in an anxious mouse strain. Behav Brain Res. 2015;287:59–72. doi: 10.1016/j.bbr.2015.02.044. [DOI] [PubMed] [Google Scholar]
- 9.Neufeld K-AM, Kang N, Bienenstock J, Foster JA. Effects of intestinal microbiota on anxiety-like behavior. Commun Integr Biol. 2011;4:492–4. doi: 10.4161/cib.4.4.15702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2012;18:1–8. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 11.Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. The probiotic Bifidobacteria infantis: An assessment of potential antidepressant properties in the rat. J Psychiatr Res. 2008;43:164–174. doi: 10.1016/j.jpsychires.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255–265. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 13.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, et al. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J Physiol. 2004;5581:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 15.Luczynski P, Whelan SO, O’Sullivan C, Clarke G, Shanahan F, Dinan TG, Cryan JF. Adult microbiota-deficient mice have distinct dendritic morphological changes: Differential effects in the amygdala and hippocampus. Eur J Neurosci. 2016:1–13. doi: 10.1111/ejn.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon Ma. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism. 2013;4:42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomova A, Husarova V, Lakatosova S, Bakos J, Vlkova B, Babinska K, Ostatnikova D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol Behav. 2015;138:179–187. doi: 10.1016/j.physbeh.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Kleiman SC, Watson HJ, Bulik-Sullivan EC, Huh EY, Tarantino LM, Bulik CM, Carroll IM. The Intestinal Microbiota in Acute Anorexia Nervosa and During Renourishment: Relationship to Depression, Anxiety, and Eating Disorder Psychopathology. Psychosom Med. 2015:969–981. doi: 10.1097/PSY.0000000000000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christian LM, Galley JD, Hade EM, Schoppe-Sullivan S, Kamp Dush C, Bailey MT. Gut microbiome composition is associated with temperament during early childhood. Brain Behav Immun. 2015;45:118–127. doi: 10.1016/j.bbi.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Real JM, Serino M, Blasco G, Puig J, Daunis-I-Estadella J, Ricart W, et al. Gut microbiota interacts with brain microstructure and function. J Clin Endocrinol Metab. 2015;100:4505–4513. doi: 10.1210/jc.2015-3076. [DOI] [PubMed] [Google Scholar]
- 23.Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, et al. Association of Cesarean Delivery and Formula Supplementation With the Intestinal Microbiome of 6-Week-Old Infants. JAMA Pediatr. 2016;170:1–8. doi: 10.1001/jamapediatrics.2015.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:1–14. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson AL, Monteagudo-Mera A, Cadenas MB, Lampl ML, Azcarate-Peril MA. Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front Cell Infect Microbiol. 2015;5:3. doi: 10.3389/fcimb.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrich JK, Davenport ER, Beaumont M, Bell JT, Clark AG, Ley RE. Genetic Determinants of the Gut Microbiome in UK Twins Correspondence. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dogra S, Sakwinska O, Soh S-E, Ngom-Bru C, Brück WM, Berger B, et al. Rate of establishing the gut microbiota in infancy has consequences for future health. Gut Microbes. 2015;6:321–5. doi: 10.1080/19490976.2015.1078051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knickmeyer RC, Gouttard S, Kang C, Evans D, Wilber K, Smith JK, et al. A structural MRI study of human brain development from birth to 2 years. J Neurosci. 2008;28:12176–82. doi: 10.1523/JNEUROSCI.3479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilmore JH, Shi F, Woolson SL, Knickmeyer RC, Short SJ, Lin W, et al. Longitudinal development of cortical and subcortical gray matter from birth to 2 years. Cereb Cortex. 2012;22:2478–2485. doi: 10.1093/cercor/bhr327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–5. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostic ADD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A-M, et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allerg. 2014;44:842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 35.Gilmore JH, Lin W, Prastawa MW, Looney CB, Vetsa YSK, Knickmeyer RC, et al. Regional Gray Matter Growth, Sexual Dimorphism, and Cerebral Asymmetry in the Neonatal Brain. J Neurosci. 2007;27:1255–1260. doi: 10.1523/JNEUROSCI.3339-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilmore JH, Schmitt JE, Knickmeyer RC, Smith JK, Lin W, Styner M, et al. Genetic and environmental contributions to neonatal brain structure: A twin study. Hum Brain Mapp. 2010;31:1174–1182. doi: 10.1002/hbm.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–9. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romano-Keeler J, Azcarate-Peril MA, Weitkamp J-H, Slaughter JC, McDonald WH, Meng S, et al. Oral colostrum priming shortens hospitalization without changing the immunomicrobial milieu. J Perinatol. 2016:1–6. doi: 10.1038/jp.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. correspondence QIIME allows analysis of high- throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat Publ Gr. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lozupone C, Knight R. UniFrac : a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozupone C, Hamady M, Knight R. UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullen E. Mullen Scales of Early Learning: AGS Edition. AGS. Circle Pines: American Guidance Service Inc; 1995. [Google Scholar]
- 44.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, Shen D. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langille M, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes J, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2009;457:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nat May. 2011;12:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science (80-) 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 52.Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, Knight R. Rethinking enterotypes. Cell Host Microbe. 2014;16:433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature. 2016;534:263–6. doi: 10.1038/nature17940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levin A, Sitarik A, Havstad S, Fujimura K, Wegienka G, Andrea C-B, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Reports. 2016;6:31775. doi: 10.1038/srep31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stearns JC, Zulyniak MA, de Souza RJ, Campbell NC, Fontes M, Shaikh M, et al. Ethnic and diet-related differences in the healthy infant microbiome. Genome Med. 2017;9:32. doi: 10.1186/s13073-017-0421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatzi L, Papadopoulou E, Koutra K, Roumeliotaki T, Georgiou V, Stratakis N, et al. Effect of high doses of folic acid supplementation in early pregnancy on child neurodevelopment at 18 months of age: the mother–child cohort “Rhea” study in Crete, Greece. Public Health Nutr. 2012;15:1728–1736. doi: 10.1017/S1368980012000067. [DOI] [PubMed] [Google Scholar]
- 57.Julvez J, Fortuny J, Mendez M, Torrent M, Ribas-Fito N, Sunyer J. Maternal use of folic acid supplements during pregnancy and four-year-old neurodevelopment in a population-based birth cohort. Paediatr Perinat Epidemiol. 2009;23:199–206. doi: 10.1111/j.1365-3016.2009.01032.x. [DOI] [PubMed] [Google Scholar]
- 58.Roza SJ, van Batenburg-Eddes T, Steegers EaP, Jaddoe VWV, Mackenbach JP, Hofman A, et al. Maternal folic acid supplement use in early pregnancy and child behavioural problems: The Generation R Study. Br J Nutr. 2010;103:445–452. doi: 10.1017/S0007114509991954. [DOI] [PubMed] [Google Scholar]
- 59.De Meij TGJ, Budding AE, De Groot EFJ, Jansen FM, Kneepkens CMF, Benninga MA, et al. Composition and stability of intestinal microbiota of healthy children within a Dutch population. FASEB J. 2016;30:1512–1522. doi: 10.1096/fj.15-278622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.