Abstract
Directed evolution of orthogonal aminoacyl-tRNA synthetases (AARSs) enables site-specific installation of non-canonical amino acids (ncAAs) into proteins. Traditional evolution techniques typically produce AARSs with greatly reduced activity and selectivity compared to their wild-type counterparts. We designed phage-assisted continuous evolution (PACE) selections to rapidly produce highly active and selective orthogonal AARSs through hundreds of generations of evolution. PACE of a chimeric Methanosarcina spp. pyrrolysyl-tRNA synthetase (PylRS) improved its enzymatic efficiency (kcat/KMtRNA) 45-fold compared to the parent enzyme. Transplantation of the evolved mutations into other PylRS-derived synthetases improved yields of proteins containing non-canonical residues up to 9.7-fold. Simultaneous positive and negative selection PACE over 48 h greatly improved the selectivity of a promiscuous Methanocaldococcus jannaschii tyrosyl-tRNA synthetase variant for site-specific incorporation of p-iodo-L-phenylalanine. These findings offer new AARSs that increase the utility of orthogonal translation systems and establish the capability of PACE to efficiently evolve orthogonal AARSs with high activity and amino acid specificity.
Orthogonal translation systems (OTSs) allow non-canonical amino acids (ncAAs) to be site-specifically incorporated into recombinant proteins, enabling researchers to expand the genetic code. Over 200 ncAAs have been installed into designer proteins using OTSs1,2 in prokaryotes, eukaryotic cells, and even in whole animals3. The most common strategy for genetic code expansion in vivo requires three key components. An unused or rare codon (typically the TAG nonsense codon) is placed into a coding sequence at the position(s) of desired ncAA incorporation. An orthogonal tRNA (o-tRNA) that is not recognized by host endogenous aminoacyl-tRNA synthetases (AARSs) decodes the nonsense codon during translation. Lastly, an orthogonal AARS is required, which is typically evolved by researchers to selectively aminoacylate the o-tRNA, but not endogenous tRNAs, with the target ncAA (Supplementary Results, Supplementary Fig. 1). This AARS component must be generated for each different ncAA of interest, and evolving a tailor-made orthogonal AARS is the most challenging and labor-intensive requirement of this strategy.
Although researchers have evolved many AARSs to incorporate ncAAs into proteins, outstanding challenges limit their utility and generality. Laboratory evolution of AARSs with altered amino acid specificity typically relies on three to five rounds of sequential positive and negative selections from a library containing randomized residues in the amino acid-binding pocket. The limited number of rounds of selection conducted reflects the effort required to complete each round of evolution, which is typically on the order of one week or longer. A consequence of conducting relatively few rounds of selection on focused libraries is that AARSs routinely emerge with suboptimal properties, including ∼1,000-fold reduced activity (kcat/KM) compared to their wild-type counterparts4, and modest selectivity for the target ncAA over endogenous amino acids. The diminished enzymatic efficiency and selectivity of many laboratory-evolved AARSs are longstanding challenges that limit the production and purity of expressed proteins containing ncAAs5.
Here we developed phage-assisted continuous evolution (PACE) selections that enable rapid laboratory evolution of orthogonal AARSs over hundreds of generations of mutation, selection, and replication. We evolved pyrrolysyl-tRNA synthetase (PylRS) variants with up to 45-fold improved enzymatic efficiency (kcat/KMtRNA), and the evolved mutations also improved activity in previously reported PylRS-derived variants without further evolution, resulting in up to 9.7-fold higher expression of ncAA-containing protein. Mutations also emerged that split PylRS into mutually dependent N- and C-terminal fragments that retained high activity and specificity. In addition, in 48 h we evolved a promiscuous mutant Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (MjTyrRS) into a variant with >23-fold higher selectivity for p-iodo-L-phenylalanine over undesired substrates. These results establish a rapid and effective approach to improve catalytic efficiency and amino acid specificity in AARS enzymes.
Results
Development of positive selections for PACE of AARSs
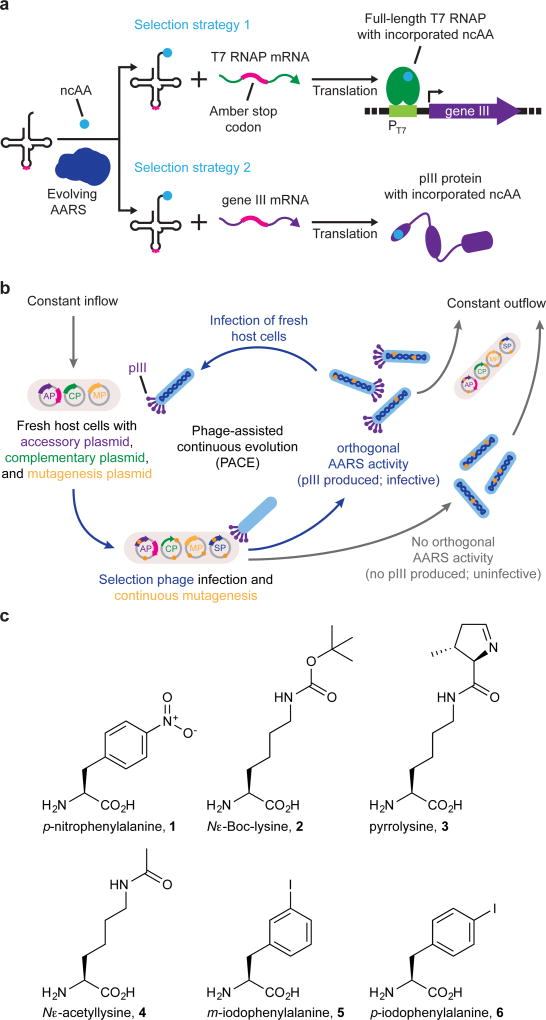
PACE enables the rapid laboratory evolution of diverse classes of proteins including polymerases6–8, proteases9, genome-editing agents10, and insecticidal proteins11. Although PACE has not previously been used to evolve aminoacylation activity, we hypothesized that it should be possible to establish a PACE selection for AARS activity by linking tRNA aminoacylation to the expression of gene III, which encodes a filamentous bacteriophage protein, pIII, that is required for phage infectivity12. We envisioned two strategies by which aminoacylation of an orthogonal amber suppressor tRNA would induce pIII production through amber suppression. In the first strategy, amber suppression of premature stop codons in the T7 RNA polymerase (T7 RNAP) gene allows translation of full-length T7 RNAP, which transcribes gene III from an upstream T7 promoter. This approach results in pIII production in an amplified manner since each amber suppression event can give rise to many gene III transcripts. In the second, more stringent strategy, amber suppression of premature stop codons in gene III results in direct translation of full-length pIII without amplification (Fig. 1a,b and Supplementary Fig. 2).
Figure 1. Overview of PACE positive selections for the continuous evolution of AARS activity and the noncanonical amino acids used in this study.
(a) Strategies for linking AARS activity to the expression of gene III, which encodes the pIII protein required for phage to be infectious. In strategy 1, AARS-catalyzed aminoacylation of an amber suppressor tRNA enables translation of full-length T7 RNAP from a transcript containing a premature amber stop codon. T7 RNAP subsequently drives expression of gene III from the T7 promoter (PT7). In strategy 2, orthogonal aminoacylation permits full-length translation of pIII from gene III mRNA containing a premature amber stop codon. (b) Diagram of PACE with selection strategy 1 plasmids shown. The accessory plasmid (AP) encodes gene III and the amber suppressor tRNA. The complementary plasmid (CP) encodes T7 RNAP. The mutagenesis plasmid (MP) increases the rate of evolution during PACE through arabinose-induced production of mutagenic proteins. The selection phage encodes all phage genes except gene III, which is replaced by the evolving AARS gene. SPs capable of catalyzing aminoacylation of the amber suppressor tRNA in the host E. coli result in production of pIII protein. Under continuous dilution in the fixed-volume vessel (the “lagoon”), phage that trigger the production of pIII propagate faster than the rate of dilution, resulting in the continuous enrichment of SPs encoding active AARS variants. (c) Non-canonical amino acids used in this study.
To implement the first selection strategy, we identified permissive residues in T7 RNAP that would not inhibit enzymatic activity when mutated to a wide variety of amino acids, and we determined how many amber codons are needed in the T7 RNAP gene to make full-length translation of the polymerase completely dependent on orthogonal translation. We installed amber mutations in the T7 RNAP gene at Ser12, Ser203, Tyr250, Tyr312, and Ser527, positions predicted from the crystal structure13 that avoid perturbation of RNA polymerization or DNA binding. Suppression with p-nitro-L-phenylalanine (p-NF; 1) (Fig. 1c) at combinations of these sites using a previously evolved MjTyrRS variant (p-NFRS)14,15 revealed that a minimum of two amber stop codons were required for transcriptional activation by T7 RNAP to become fully dependent on both the AARS and the ncAA substrate (Supplementary Fig. 3a,b). Similar results were observed with site-specific installation of Nɛ-Boc-L-lysine (BocK; 2)16 using a chimeric PylRS (chPylRS), comprising residues 1–149 of Methanosarcina barkeri PylRS (MbPylRS) and residues 185–454 of Methanosarcina mazei PylRS (MmPylRS) (Supplementary Fig. 3c).
To test the ability of this selection to support phage propagation, selection phage (SP) expressing either p-NFRS (SP-p-NFRS) or chPylRS (SP-chPylRS) were propagated non-continuously in cultures of host E. coli cells harboring an accessory plasmid (AP) and complementary plasmid (CP) that together expressed the requisite amber suppressor tRNA, T7 RNAP(S12TAG, S203TAG), and gene III downstream of a T7 promoter. We observed that SP propagation in these cultures was dependent on the presence of a matched ncAA substrate (Supplementary Fig. 4a,b). Together, these results validate the PACE selection strategy based on amplified expression of gene III through amber suppression of two or more stop codons in T7 RNAP.
To implement the second, more stringent selection strategy based on direct amber suppression of premature stop codons in gene III, we installed amber mutations at positions Pro29, Pro83, Thr177, or Tyr184 of gene III. These residues were chosen because they are predicted to be uninvolved in pIII binding to the host cell TolA protein or to the host cell F pilus17–19. The N-terminal signal peptide of pIII, which spans residues 1–18, was not targeted for amber suppression, as this region is required for insertion of pIII into the host inner membrane20–22. We tested the ability of this selection to support phage propagation by challenging selection phage expressing either p-NFRS (SP-p-NFRS) or chPylRS (SP-chPylRS) to propagate non-continuously in cultures of host E. coli cells harboring an accessory plasmid that expressed the requisite amber suppressor tRNA and gene III containing one or more premature stop codons. We found that each of the positions we mutated in pIII were permissive to ncAA incorporation, and the presence of a single premature stop codon in the coding sequence of pIII was sufficient to make robust phage propagation dependent on AARS activity from SP-p-NFRS or SP-chPylRS (Supplementary Fig. 4c,d). Collectively, these developments identify positions in T7 RNAP and pIII that tolerate a range of amino acid side chains, and thereby establish two strategies to link AARS activity to phage infectivity through amber suppression of premature stop codons in T7 RNAP or in gene III.
Next, we sought to demonstrate that the PACE positive selection for aminoacylation based on amber suppression of stop codons in T7 RNAP could support activity-dependent phage propagation in the continuous flow format of PACE. We conducted 48-h mock PACE selections and found that SP-p-NFRS propagated at high phage titer levels in a lagoon supplemented with p-NF substrate without further adaptation. In a separate control lagoon, SP expressing a kanamycin resistance gene rather than an AARS were unable to propagate in the positive selection and rapidly washed out (Supplementary Fig. 5a). We also demonstrated that the PACE positive selection based on direct amber suppression of stop codons in gene III supported activity-dependent phage propagation in continuous flow. In a single, 30-h mock PACE using this selection strategy, active SP-p-NFRS was highly enriched starting from a 1:1 input mixture of SP-p-NFRS and an SP expressing an unrelated gene (Supplementary Fig. 5b). Together, these results confirmed that both selection strategies were capable of supporting phage propagation in PACE in a manner dependent on orthogonal AARS activity.
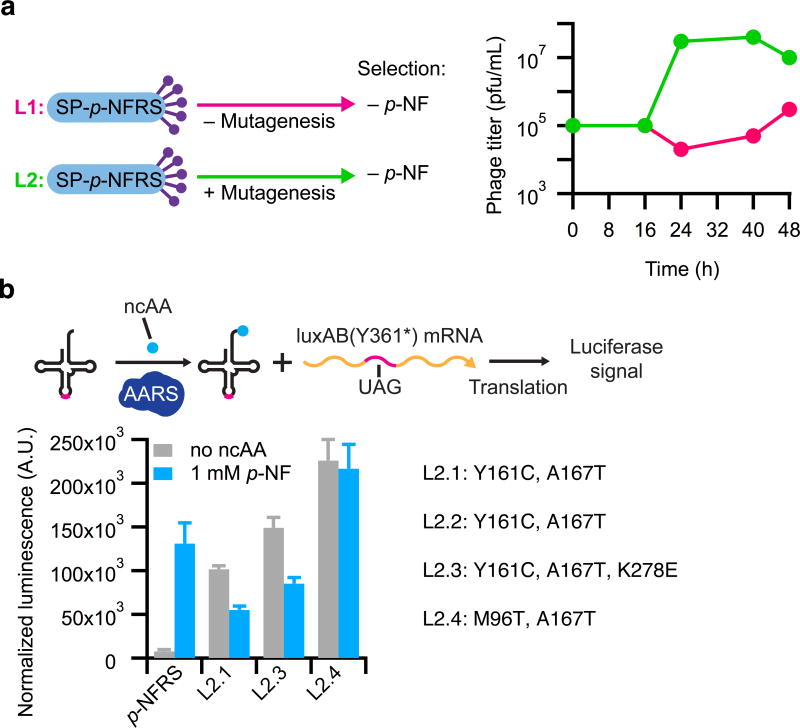
In an additional mock PACE experiment, we demonstrated that positive selection could evolve greatly increased activity from an AARS with little starting activity on a target amino acid. Using the selection requiring amber suppression of stop codons in T7 RNAP, SP-p-NFRS was challenged to evolve acceptance of endogenous amino acids by propagating the phage in Davis rich media (DRM)8 that was not supplemented with p-NF. Under these conditions, high titers of SP-p-NFRS were dependent on induction of a mutagenesis plasmid (MP), MP4, that enhances the rate of mutagenesis in the host E. coli23 (Fig. 2a). This observation suggests that mutation of the AARS was required in order for SP to propagate when the cognate amino acid substrate was unavailable. Sanger sequencing analysis of clonal phage from the experiments confirmed that more mutations accumulated in the gene encoding p-NFRS when MP4 was induced. Additionally, the evolved mutants, but not the starting p-NFRS, displayed strong aminoacylation activity in the absence of p-NF using a luciferase reporter of amber suppression (Fig. 2b), confirming that the AARS evolved to accept one or more canonical amino acids during PACE. Together, results of these experiments validated the positive selection for continuously evolving an orthogonal AARS.
Figure 2. Evolution of AARS activity during mock PACE.
(a) p-NFRS was challenged to aminoacylate the amber suppressor tRNA in the absence of its cognate ncAA substrate, p-NF, over 48 h of positive selection PACE conducted in two separate lagoons (L1 and L2). Enhanced mutagenesis from the MP was supplied in L2 only. Phage titers of L2 (green) rapidly increased after 16 h, while titers in L1 (magenta) were relatively stable throughout the evolution. (b) Mutations in PACE-evolved clones and the relative amino acid substrate specificities of clones from L2. Relative aminoacylation activity was compared in the PACE host strain by measuring the luminescence signal resulting from amber suppression of a premature stop codon at position 361 of a luciferase gene (luxAB). More coding mutations were obtained in phage isolates from L2, in which the MP provided enhanced mutagenesis, and every characterized L2 mutant emerged from PACE with increased activity on endogenous amino acids (no ncAA) compared to the progenitor enzyme, p-NFRS. Values and error bars in b reflect the mean and s.d. of at least three independent biological replicates.
Continuous evolution of enhanced PylRS variants
PylRS from archaebacteria are preferred evolutionary starting points for genetic code expansion efforts due to their tRNA orthogonality in a range of hosts. Wild-type PylRS variants, however, are hampered by poor catalytic efficiency24, which is typically further diminished as an undesired consequence of traditional laboratory evolution5. We hypothesized that our newly validated PACE selections would improve PylRS catalytic efficiency and might provide an improved evolutionary starting point for future AARS evolution efforts.
We evolved chPylRS—the chimera of residues 1–149 of MbPylRS and residues 185–454 of MmPylRS—to have improved aminoacylation activity over 497 h of PACE in three segments (Fig. 3a). The chimera was initially created by exchanging the C-terminal region of MbPylRS with that of the more active MmPylRS25, resulting in an enzyme that has higher specific activity for L-pyrrolysine (3) than MmPylRS, and improved solubility compared to the wild type enzymes. To further improve chPylRS in PACE, we used the non-proteinogenic substrate analog, BocK, rather than the natural cognate substrate, L-pyrrolysine, which is not readily available26. We reasoned that an extended evolution on the substrate analog would select for activity improvements without affecting substrate specificity.
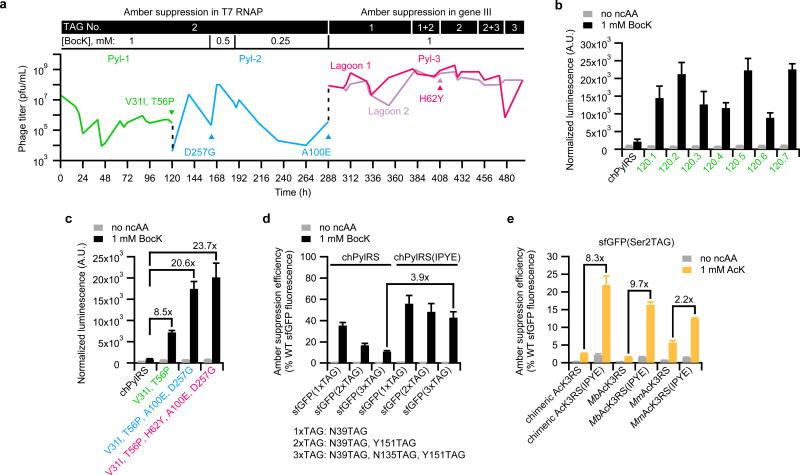
Figure 3. Continuous evolution and characterization of chimeric pyrrolysyl-tRNA synthetase (chPylRS) variants with enhanced aminoacylation activity.
(a) PACE was performed in three segments designed to gradually increase selection stringency. The first two segments (Pyl-1 and Pyl-2) used the selection requiring amber suppression of two stop codons in T7 RNAP, and the final segment (Pyl-3) used the selection requiring direct amber suppression of stop codons in gene III. The number of stop codons in the gene required for each selection and the concentration of BocK substrate are shown above the phage titer graph. Dotted lines (black) indicate transfer of evolved phage from the end of each PACE segment into the subsequent segment. Colored triangles indicate convergence toward the specified mutations. (b,c) The relative expression of luciferase containing BocK at position 361 resulting from aminoacylation by progenitor enzyme, chPylRS, compared to evolved variants from the end of PACE segment Pyl-1 (b) or compared to variants containing only the consensus mutations from the end of each PACE segment (c). Label colors correspond to PACE segments in a. (d) The relative efficiency of multisite BocK incorporation into sfGFP resulting from aminoacylation by chPylRS variants with or without beneficial mutations discovered in PACE (V31I, T56P, H62Y, and A100E; IPYE). (e) The relative efficiency of AcK incorporation at position 2 of sfGFP resulting from aminoacylation by AcK3RS variants with or without transplanted mutations from PACE. Values and error bars in b–e reflect the mean and s.d. of at least three independent biological replicates.
In the first two segments of PACE (Pyl-1 and Pyl-2), SP-chPylRS was evolved using the less stringent selection requiring amber suppression of T7 RNAP. During Pyl-1, the flow rate was modulated to increase selection stringency. The pool of phage surviving Pyl-1 was further evolved in Pyl-2, which challenged SP-chPylRS to propagate as the ncAA substrate concentration was incrementally reduced. The final PACE segment, Pyl-3, was conducted in two lagoons using the more stringent selection strategy, and the number of amber stop codons in gene III was incrementally increased from one to three to increase demands on PylRS efficiency. This approach gradually increased selection stringency over the 497-h evolution of chPylRS, and emerging variants had survived on average 268 generations of mutation, selection, and replication6.
Clonal SP isolates from the 120-h endpoint of Pyl-1 were sequenced, which revealed mutations throughout the PylRS gene with strong convergence toward a pair of mutations in PylRS: V31I and T56P. Sequencing of clonal isolates from the second PACE segment (Pyl-2) revealed strong convergence toward D257G at 162 h, and full convergence on A100E by the end of Pyl-2 (288 h). The additionally stringent conditions of the Pyl-3 segment selected for complete convergence toward mutation H62Y in all sequenced clones from 408 h of PACE (Supplementary Tables 1 –4).
Each of the Pyl-1 variants exhibited improved aminoacylation activity in a luciferase reporter of amber suppression with BocK (Fig. 3b). Comparison of the consensus mutations acquired in each segment of PACE showed that the two combined mutations from Pyl-1 increased the luciferase signal 8.5-fold compared to the progenitor chPylRS, and the additional two mutations from Pyl-2 improved the amber suppression signal 21-fold. The variant containing all consensus mutations from the three segments of PACE provided 24-fold improved amber suppression signal compared to chPylRS while maintaining substrate specificity (Fig. 3c). Further analysis of the consensus mutations acquired in the first two segments of PACE demonstrated that D257G did not substantially contribute to enhance the activity of chPylRS (Supplementary Fig. 6a). Therefore, the tetramutant comprising V31I, T56P, H62Y, and A100E was responsible for the large improvement in apparent activity.
We incorporated BocK at up to three positions in sfGFP to compare the relative activity of chPylRS to the tetramutant variant, chPylRS(IPYE), containing the activity-enhancing mutations from PACE (Fig. 3d and Supplementary Fig. 7a–d). Expression of sfGFP containing three BocK residues was improved nearly 4-fold by chPylRS(IPYE) compared to chPylRS. Biochemical characterization of chPylRS(IPYE) using BocK confirmed that the kcat improved 8.7-fold, and the KM for tRNAPyl substrate improved 5.7-fold, such that the catalytic efficiency (kcat/KMtRNA) of the evolved variant was enhanced 45-fold compared to chPylRS (Table 1). These findings indicate that the increased apparent activity of the tetramutant results from catalytic enhancement of chPylRS, rather than solely from non-catalytic improvements such as enhanced protein expression or stability. The outcome of these experiments demonstrates that PACE positive selection is highly effective at improving the activity of an AARS commonly used for genetic code expansion.
Table 1.
Kinetic parameters of chPylRS variants containing mutations from PACE.
| PylRS variant | kcat, s−1 × 10−3 | KM ATP, µM | KM tRNA, µM | KMBocK, mM |
kcat/KMtRNA, µM−1· s−1 × 10−3 |
Relative catalytic efficiency |
|---|---|---|---|---|---|---|
| chPylRS | 11.88 ± 0.18 | 2.54 ± 0.16 | 0.26 ± 0.07 | 1.03 ± 0.05 | 45.69 | 1 |
| V31I, T56P | 73.15 ± 1.01 | 5.74 ± 0.20 | 0.10 ± 0.02 | 0.82 ± 0.18 | 731.50 | 15.9 |
| V31I, T56P, A100E | 110.23 ± 4.65 | 3.45 ± 1.19 | 0.13 ± 0.03 | 0.91 ± 0.08 | 847.92 | 18.4 |
| V31I, T56P, H62Y, A100E | 103.87 ± 2.37 | 3.96 ± 0.52 | 0.05 ± 0.01 | 1.13 ± 0.23 | 2,077.40 | 45.2 |
The mean values and standard errors were calculated from three replicates
None of the activity-enhancing coding mutations discovered through PACE were in the active site of the enzyme, but they were instead localized exclusively in the N-terminal domain of chPylRS, which is involved in tRNA binding27,28 and is typically not targeted for mutagenesis in laboratory evolution efforts. Since these changes occur at conserved residues in MbPylRS and MmPylRS, we speculated that the PACE mutations may generally improve the activity of other natural and engineered PylRS homologs. Amber suppression assays using several different reporters demonstrated that the activity of the MbPylRS(IPYE) variant was dramatically enhanced while the MmPylRS(IPYE) variant was also improved, albeit more modestly (Supplementary Fig. 6b–d). To test the possibility that the beneficial mutations would also enhance evolved PylRS enzymes, which are also conserved at these locations, we transplanted the four PACE-derived mutations into AcK3RS, which was previously evolved to accept Nɛ-acetyl-L-lysine (AcK; 4)29,30. MbAcK3RS, MmAcK3RS, and chimeric AcK3RS (chAcK3RS) variants containing the four mutations each exhibited increased expression of reporter proteins up to 9.7-fold compared to their unmodified PylRS counterparts, without sacrificing amino acid selectivity (Fig. 3e and Supplementary Figs. 6e and 8). Reporter expression was also enhanced more than 5-fold when the mutations were transplanted into the PylRS-derived IFRS, which was previously evolved to charge m-iodo-L-phenylalanine (5)24,31 (Supplementary Fig. 6f). Collectively, these results suggest that the beneficial mutations discovered exclusively in the N-terminal domain of chPylRS substantially enhance activity in all six additional PylRS variants tested.
Unexpected Evolution of Split PylRS Enzymes
Although there was no strong convergence toward new beneficial coding mutations between the 408-h and 497-h time points, 13 of 16 (81%) of the sequenced SP isolates from the two lagoons of Pyl-3 acquired a surprising frameshift in their coding sequences by 497 h. Of the 13 affected clones, 12 of these contained a single frameshift at one of four different locations in chPylRS (Supplementary Tables 3 and 4). In each case, the shifted reading frame in the chPylRS gene produced a premature ochre (TAA) or opal (TGA) stop codon resulting in a truncated protein of 93, 99, or 102 residues. In addition, one of the 13 affected isolates from the 497-h time point contained an in-frame ochre stop codon at position Lys-90, resulting in a truncated protein of 89 residues. Downstream of the premature stop codon in every case was a Met codon at canonical position 107 of chPylRS. We hypothesized that protein synthesis may reinitiate32–34 from Met-107 resulting in a split chPylRS. In assays of amber suppression, the split chPylRS(IPYE) variants exhibited comparable apparent activity as the full-length chPylRS(IPYE) enzyme. Activity of the split variants was strictly dependent on the presence of both fragments (Supplementary Fig. 9a,c and Supplementary Table 6). In contrast, split chPylRS variants lacking the PACE-evolved coding mutations in their N-terminal fragment had substantially lower activity (Supplementary Fig. 9b). Results from western blot analysis and ESI-MS analysis of split variants confirmed that translational reinitiation occurred at Met-107 (Supplementary Figs. 11 and 12). The prevalence of the split PylRS variants suggest a fitness advantage to the split constructs during PACE, although the molecular basis of this potential advantage is currently unknown.
Split PylRS variants also appear in nature, as PylRS homologs in certain bacteria are expressed from two separate genes (pylSc and pylSn). The Desulfitobacterium hafniense pylSn encodes a 110-residue polypeptide that is homologous to the N-terminal region of archaeal PylRS35, and an alignment of PylSn to the N-terminal split PylRS evolved in PACE shows that they terminate near the same location (Supplementary Fig. 13). These observations together demonstrate the ability of PACE to evolve unexpected changes in protein topology.
Development and validation of AARS negative selections
While positive selection PACE greatly increased the activity of PylRS, the evolution of AARSs to recognize non-cognate substrates requires negative selections to minimize activity on endogenous amino acids. We therefore developed a PACE negative selection that links tRNA aminoacylation to the inhibition of phage propagation. We previously used a dominant-negative variant of pIII (pIII-neg) as the basis of a PACE negative selection for RNA polymerase activity and DNA binding activity.8,10 Because pIII-neg poisons the infectivity of emergent phage, variants possessing undesired activity are unable to effectively propagate and are gradually washed out from the evolving pool of SP under constant dilution.
In the PACE negative selection for aminoacylation, amber suppression of two stop codons in T7 RNAP(S12TAG, S203TAG) allows transcriptional activation of the gene encoding pIII-neg. Amber suppression in this context thus results in expression of pIII-neg and reduced progeny phage infectivity (Fig. 4a and Supplementary Fig. 14a,b). Mock PACE negative selections with SP-p-NFRS confirmed negative selection against AARS activity. SP-p-NFRS in the presence of p-NF quickly washed out of PACE lagoons under negative selection, whereas an SP lacking any AARS activity propagated robustly under the same conditions (Supplementary Fig. 15a–c). These findings established a PACE negative selection against undesired aminoacylation activity.
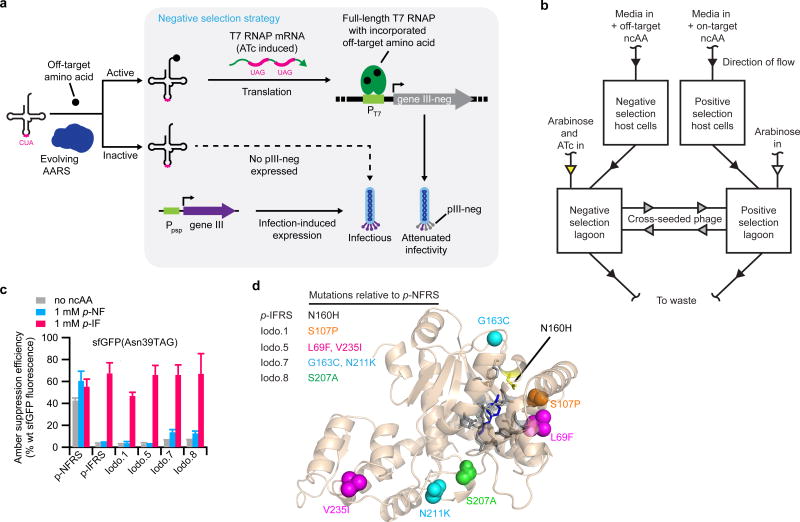
Figure 4. Evolution of AARS variants from dual positive- and negative-selection PACE with greatly improved amino acid specificity.
(a) Strategy for linking undesired aminoacylation to gene III-neg expression, which encodes the pIII-neg protein. When undesired aminoacylation occurs in the negative selection, pIII-neg is produced, impeding progeny phage infectivity. In the absence of undesired aminoacylation, only pIII is produced, resulting in infectious phage progeny. Negative-selection stringency is modulated by ATc concentration. (b) Diagram of dual-selection PACE using simultaneous positive and negative selections. Evolving phage are continuously cross-seeded between positive and negative selection lagoons at a 50-fold dilution. (c) The relative site-specific incorporation efficiency of either endogenous amino acids (no ncAA), p-NF, or p-IF at position 39 of sfGFP resulting from aminoacylation by p-NFRS, p-IFRS, or evolved variants from PACE (Iodo.1, Iodo.5, Iodo.7, and Iodo.8). (d) Predicted position of mutations evolved during dual-selection PACE. The shown crystal structure is the p-NFRS protein sequence aligned to PDB 2AG6 (ref. 37), which is the crystal structure of an AARS that has the identical protein sequence of p-IFRS and is bound to the ncAA substrate, p-bromo-L-phenylalanine (dark blue). The colored spheres in the crystal structure correspond to the colored mutations in the table to the left. Active-site residues within a 5 Å radius around the ncAA substrate are colored gray. Values and error bar in c reflect the mean and s.d. of at least three independent biological replicates.
Continuous evolution to improve amino acid selectivity
Homogeneity of ncAA incorporation is often crucial for downstream applications, as it is usually impractical or impossible to purify proteins containing the desired ncAA substitution from mixtures containing undesired amino acids at the position(s) of interest. The amino acid selectivity of evolved AARSs is therefore a critical determinant of their utility. The laboratory-evolved MjTyrRS variant, p-NFRS, selectively charges p-NF in minimal media14,15, but overnight expression in Luria-Bertani (LB) media demonstrated that p-NFRS also efficiently charges Phe in the presence or in the absence of 1 mM p-NF (Supplementary Fig. 16a–c). Additionally, p-NFRS is a polyspecific enzyme, as it efficiently charges p-iodo-L-phenylalanine (p-IF; 6) in addition to p-NF (Supplementary Fig. 16d). We examined the ability of coupled PACE positive and negative selections to generate a highly specific AARS by evolving p-NFRS to charge p-IF selectively.
The current design of the positive and negative PACE selections for aminoacylation necessitates that they be performed in separate vessels either in the presence or in the absence of desired ncAA, respectively. We hypothesized, however, that the opposing selections could be coupled continuously by constantly exchanging small volumes of material from opposing PACE lagoons, which would allow the pool of AARS variants to be evolved in both selections simultaneously, rather than performing iterative counterselections (Fig. 4b). This strategy’s effectiveness relies on (i) the only actively replicating element in the selection lagoons is the SP, (ii) the comparatively small number of host cells that are diverted into the opposing selection should not greatly affect either selection due to the much larger population of correct host cells being continuously infused, and (iii) any contaminating ncAA diverted into the opposing selection would be diluted to a very low concentration that would be insufficient to support effective aminoacylation. We hoped that coupling the opposing selections lagoons would provide an opportunity for SP variants capable of propagating in both selections—i.e., those AARS variants that evolved high amino acid selectivity—to outcompete variants able to propagate exclusively in one of the opposing selections.
SP-p-NFRS was evolved for 24 h of positive selection PACE toward p-IF followed by 24 h of coupled positive selection with negative selection against the undesired ncAA, p-NF (Supplementary Fig. 17a,b). SPs that acquired preferential activity toward p-IF in PACE were isolated from the evolved pool using a single round of non-continuous counterselections. To enrich variants possessing little to no activity on the undesired ncAA, endpoint SPs from the PACE negative selection were challenged to propagate non-continuously on negative-selection host cells in media containing 4 mM p-NF. The resulting SPs were then challenged with positive-selection host cells in the presence of the desired substrate, 1 mM p-IF, for their ability to promote formation of activity-dependent plaques (the result of phage propagation in semi-solid media) and eight of the resulting plaques were sequenced (Supplementary Fig. 18a,b).
Of the eight sequenced phage isolates, four acquired no new mutations in the AARS gene, but instead emerged from PACE with weakened ribosome binding sites driving AARS expression. Each of the remaining four SP variants contained one or more coding mutations and demonstrated a strong preference for charging p-IF over p-NF (Fig. 4c,d and Supplementary Table 7). The best-performing PACE-evolved variant, Iodo.5, which contained mutations L69F and V235I with respect to p-NFRS, matched the amino acid specificity of a previously reported MjTyrRS variant, p-IFRS, that was evolved to charge p-IF through positive and negative selection on agar plates36,37. Based on our limit of detection in the assay, expression of sfGFP(N39TAG) using variant Iodo.5 was >23-fold higher with p-IF than with p-NF.
The protein sequences of p-IFRS and p-NFRS differ by only a single amino acid; p-NFRS contains Asn160 and p-IFRS contains His160. It is possible that His160 also emerged in PACE but was not isolated. We further tested Iodo.5 by expressing the sfGFP reporter in LB media containing both 1 mM p-NF and 1 mM p-IF in a single culture. Intact protein mass spectrometry of the resulting purified protein revealed the desired mass corresponding to incorporation of p-IF with only trace p-NF incorporation and no detectable incorporation of Phe at the site of interest (Supplementary Fig. 19a,b). These results establish that PACE can rapidly evolve a highly selective AARS from a polyspecific variant in 48 h with no library cloning. This method also eliminates the need for repeated plasmid isolation and retransformation, and it performs positive and negative selection simultaneously and continuously, dramatically increasing the total rounds of evolution performed on the AARS.
Discussion
This study establishes the ability to perform continuous directed evolution of AARSs, dramatically increasing the number of generations of evolution that are possible in practical timescales compared to stepwise laboratory evolution techniques. Using PACE, we evolved PylRS and MjTyrRS variants with greatly improved activities and specificities without cloning mutagenized libraries or performing time-intensive selections one round at a time.
PACE of chPylRS improved enzymatic activity exclusively through N-terminal domain mutations. Wild-type PylRS exhibits poor solubility due to its hydrophobic N-terminal domain28, preventing crystallization of the full-length enzyme25,38. Thus, it would be difficult to correctly predict N-terminal domain residues that would benefit from optimization a priori. Mutations emerging from PACE were transplantable into the conserved positions of PylRS orthologs, and these mutations improved expression of ncAA-containing proteins by all four previously evolved PylRS variants tested, suggesting activity was broadly improved without further evolution of these variants. Surprisingly, we also discovered highly active split-PylRS variants arising from frameshift mutations or an in-frame mutation to a stop codon.
Other recent efforts have also begun to address the long-known inefficiencies of OTSs. Improvements in multisite incorporation of an ncAA into a single polypeptide chain are achievable using multiplex automated genome engineering39 to evolve chromosomally incorporated MjTyrRS variants in genetically recoded E. coli in which release factor 1 is deleted40,41. Ectopic expression of an engineered release factor improves multi-site incorporation by a PylRS:tRNAPyl pair in HEK293T cells42. The efficiency of OTSs can also be improved through optimization of the o-tRNA43–45. The N-terminal mutations of chPylRS from PACE improved multisite incorporation of an ncAA, and these mutations are likely to provide further enhancement when utilized in conjunction with previous methods39–45 that improve OTS efficiency.
Simultaneous positive and negative PACE selections obviated the need for iterated plasmid isolation and retransformation, which is traditionally a rate-limiting step in protein evolution. We used these continuously coupled selections to evolve, in 48 h, the polyspecific p-NFRS into an AARS that selectively charges p-IF, excluding the undesired incorporation of Phe or p-NF. Additional improvements to the negative selection and dual-selection strategy may further expand AARS evolution capabilities. For example, a nitrilase-activatable ncAA tagging (NANCAT)46 strategy may allow the opposing selections to be conducted in the same lagoon by decaging the ncAA exclusively in positive selection host cells. Enabling rapid evolution of AARS variants with entirely new activity on desirable ncAAs is also a future goal, which might be accomplished by initiating PACE from an SP-AARS library containing mutagenized active site residues. We envision that the capabilities established in this study will enable efficient production of additional highly evolved AARS variants that have generally improved utility in genetic code expansion efforts.
Online Methods
General methods
PCR and all cloning steps were performed in HyClone water (GE Healthcare Life Sciences). In all other experiments, water was purified by a MilliQ purification system (EMD Millipore). PCR was performed with Q5 Hot Start High-Fidelity DNA polymerase (New England Biolabs) when unmodified primers were used, and Phusion U Hot Start DNA polymerase (Thermo Fisher Scientific) was used when deoxyuridine-containing primers were required for USER cloning. Plasmids and selection phage were prepared using isothermal assembly with Gibson Assembly 2x Master Mix (New England Biolabs), USER cloning with USER enzyme (New England Biolabs), or ligation cycling reaction with Ampligase (Epicentre). Genes were either synthesized from gBlock gene fragments (Integrated DNA Technologies) or PCR amplified from native sources. Chimeric PylRS, MbPylRS, and MmPylRS were obtained from pTECH plasmid sources from previous studies. Premature stop codons and single point mutations were placed into genes using the Q5 Site-Directed Mutagenesis kit (New England Biolabs). The gene encoding p-NFRS was PCR amplified from the pEVOL plasmid, which was generously provided to us by P. Schultz of the Scripps Research Institute. DNA vector amplification was performed using TOP10, Mach1 (Thermo Fisher Scientific) or NEB 5-alpha F’ Iq (New England Biolabs) cells. All Sanger sequencing of plasmids and SPs was performed from DNA samples that had been amplified using the Illustra Templiphi 100 Amplification Kit (GE Healthcare Life Sciences). All ncAAs used in these studies were purchased from Chem-Impex International (>99.0% purity) except for 4-nitro-L-phenylalanine (Nanjing Pharmatechs, 97% purity) and 4-iodo-L-phenylalanine (Astatech, Inc., 95% purity).
Non-continuous phage propagation
S10308 cells (25 µL) were electroporated with the accessory plasmid of interest and a complementary plasmid, when required (Supplementary Table 8). Transformed cells recovered 1 h in SOC media (New England Biolabs) at 37 °C while shaking. Recovered cells were plated on LB agar (United States Biologicals) containing the antibiotics required for plasmid maintenance and grew 20 h at 37 °C. Single colonies of the transformed cells were picked and grown for 16 h in a 37 °C shaker at 230 rpm using 3 mL of Davis rich media (DRM)8 containing antibiotics. The saturated cultures were diluted 1,000-fold into 3 mL of identical media or media supplemented with 1 mM ncAA where noted. The diluted cultures were grown at 37 °C while shaking to mid-log phase (absorbance at 600 nm (A600) = 0.5–0.7). Once the desired cell density was reached, the cultures were inoculated with SP to provide a desired starting titer of ∼1×105 pfu/mL. A dilution reference was also prepared by diluting an identical volume of SP into media containing no cells. All cultures and dilution references were shaken for 16 h at 37 °C. The resulting saturated cultures were centrifuged 8 min at 3,000g, and the supernatant was filtered using a 0.22 µm cellulose acetate, Spin-X centrifuge tube filter (Costar), and the samples were stored at 4 °C.
Plaque assay
S1030 cells transformed with the appropriate plasmids were grown in 2xYT liquid media (United States Biologicals) supplemented with antibiotics required for plasmid maintenance to A600 = 0.6–0.8. Phage supernatant was serially diluted at 10-fold or 100-fold increments yielding either eight or four total samples, respectively, including undiluted sample. For each phage sample, 100 µL of cells were combined with 10 µL of phage. Within 2 min from phage infection, 950 µL of 55 °C top agar (7 g/L bacteriological agar in 2xYT; no antibiotics) was added and mixed with the phage-infected cells by gentle pipetting once up and down while avoiding formation of bubbles. The final mixtures were plated onto quartered Petri plates that had been previously poured with 1.5 mL of bottom agar (15 g/L bacteriological agar in 2xYT; no antibiotics). Once the overlaid agar congealed, the plates were incubated 16 h at 37 °C to allow plaque formation. When plaque formation was dependent on orthogonal AARS activity, 1 mM ncAA was also added to all liquid and solid media when denoted. When clonal-phage isolates were required, well separated plaques were picked from plates and grown individually at 37 °C while shaking in 3 mL of DRM supplemented with 1 mM ncAA of interest where required. The resulting saturated cultures were pelleted at 3,000g for 8 min, and the phage supernatant was sterile filtered and stored at 4 °C for further analysis.
Phage-assisted continuous evolution of aminoacyl-tRNA synthetases
In general, the PACE apparatus—including host-cell strains, lagoons, chemostats, and media—was used as previously described9. All liquid and solid media contained antibiotics required for plasmid maintenance unless indicated otherwise. To prepare each PACE strain, the accessory plasmid (AP), complementary plasmid (CP), and MP or drift plasmid (DP) of interest were cotransformed into electrocompetent S1030 cells, which recovered for 1 h in SOC medium without antibiotics (New England Biolabs). The recovered transformants were plated onto 2xYT agar containing 0.4% glucose to prevent induction of mutagenesis prior to PACE, and colonies were grown for 16–20 h in a 37 °C incubator. Three colonies were picked and resuspended in DRM. A portion of each suspension was tested for arabinose sensitivity as previously described11, and the remainder was used to inoculate liquid cultures in DRM, which were subsequently grown for 16 h in a 230 rpm shaker at 37 °C.
Each PACE chemostat was prepared by diluting an arabinose-sensitive overnight culture into 40 mL or 80 mL of DRM, which was supplemented with ncAA where noted, and the chemostats grew at 37 °C while stirring with a magnetic stir bar. Once the culture reached an approximate cell density of A600 = 1.0, fresh DRM (supplemented with ncAA where noted) was used to continuously dilute the chemostat culture at a dilution rate of 1.6 chemostat volumes per h while maintaining a constant culture volume as previously described.6
Lagoons flowing from the chemostats were continuously diluted using the indicated flow rates while maintaining a 25-mL constant volume by adjusting the height of the needle drawing waste out of each lagoon. All lagoons were supplemented with 25 mM arabinose from a syringe pump to induce mutagenesis from the MP or DP, unless otherwise indicated. Arabinose supplementation began at least two hours prior to phage infection to insure cells were maximally induced at the start of each experiment. Lagoons were also supplemented with anhydrotetracycline (ATc), where noted, to induce either genetic drift (mutagenesis under weak or no selective pressure) or negative selection depending on the nature of the host-cell plasmids.
Samples of evolving SP pools were taken periodically at indicated time points from the waste line of each lagoon. Collected samples were centrifuged at 10,000g for 2 min, and the supernatant was passed through a 0.22 µm filter and stored at 4 °C for subsequent analysis. Phage titers were determined by plaque assays using S1059 cells (containing the phage-responsive pJC175e8 plasmid to report total phage titer) and untransformed S1030 cells (reporting cheaters from unwanted recombination of gene III into the SP) for all collected samples. Activity-dependent plaque assays were performed for mock selection PACE experiments, using S1030 cells cotransformed with the AP and CP used in the host cells of the corresponding experiment. Mock selections were also monitored by PCR performed on phage aliquots using primers DB212 (5’-CAAGCCTCAGCGACCGAATA) and DB213 (5’-GGAAACCGAGGAAACGCAA), which anneal to regions of the phage backbone flanking the gene of interest.
Evolution of chPylRS (Pyl-1)
Host cells cotransformed with pDB021CH(+), pDB023f, and DP4 were maintained in an 80 mL chemostat using media containing 1 mM BocK. At the beginning of PACE, genetic drift was induced (200 ng/mL ATc) in a lagoon that was flowing from the chemostat at 1 lagoon volume per h. The lagoon was infected with 108 pfu of SP-chPylRS to start the experiment. ATc supplementation was adjusted to 20 ng/mL at 16 h of PACE to slowly reduce the amount of genetic drift, and ATc supplementation was stopped at 24 h. The lagoon flow rate was increased to 2 volumes per h at 40 h of PACE to increase selection stringency for the remainder of the experiment, which ended at 120 h.
Continuation of chPylRS evolution (Pyl-2)
Three preparations of media were used, which contained different concentrations of BocK (DRM-A: 1 mM BocK; DRM-B: 0.5 mM BocK; DRM-C: 0.25 mM BocK). Host cells cotransformed with pDB021CH(+), pDB023f, and MP4 were maintained in a 40 mL chemostat containing DRM-A at the start of the experiment. A single lagoon was flowed from the chemostat at 1 lagoon volume per h, and the experiment was initiated by infecting the lagoon with 100 µL (2×104 pfu) of the evolved pool of SP collected from the 120-h endpoint of Pyl-1. To increase the selection stringency during the experiment, the media being pumped into the chemostat was changed to DRM-B at 42 h of PACE and was changed to DRM-C at 69 h. The experiment ended at 168 h.
Continuation of chPylRS evolution (Pyl-3)
PACE was conducted in two separate lagoons, L1 and L2, and a concentration of 1 mM BocK was maintained throughout the experiment. Selection stringency was increased during the experiment by modulating the lagoon flow rate and altering the ratio of host cells in the lagoons to increase the number of amber suppression events required to produce full-length pIII during translation (Host-A: pDB038 and DP6; Host-B: pDB038a and DP6; Host-C: pDB038b and DP6). Host cells were maintained separately in three, 40 mL chemostats (C1–C3, respectively), and each chemostat was individually prepared and coupled to both lagoons, as needed, over the course of the experiment to minimize media waste and to minimize the total growth time of each chemostat culture.
At the start of the experiment L1 and L2 were continuously diluted with Host-A from C1 at a rate of 0.5 lagoon volumes per h, and genetic drift was induced only in L1 (100 ng/mL ATc). Each lagoon was infected with 108 pfu of clonal-phage isolate SP-Pyl2.288-2, which was isolated from the Pyl-2 segment (Supplementary Table 2). The flow rate from C1 through each lagoon was increased to 1 lagoon volume per h at 41 h of PACE. At the 91-h mark of the experiment, L1 and L2 were fed a 1:1 mixture of Host-A:Host-B supplied from C1 and C2, respectively, and the flow through each lagoon was maintained at 1 lagoon volume per h. At the 120-h mark, 100% Host-B was flowed to each lagoon at 0.5 lagoon volumes per h, and the flow rate was later doubled to 1 lagoon volume per h at 136 h of PACE. At the 162 h mark, L1 and L2 were fed a 1:1 mixture of Host-B:Host-C supplied from C2 and C3, respectively, and the flow through each lagoon was maintained at 1 lagoon volume per h. ATc supplementation to L1 was stopped at 184 h of PACE to end genetic drift. At the 190-h time point of PACE, 100% Host-C was flowed to each lagoon at 0.5 lagoon volumes per h for the remainder of the experiment, which was stopped at 209 h.
Evolution of p-NFRS with dual selection
The media of the positive selection contained 1 mM p-IF, and media of the negative selection contained 4 mM p-NF. Three host-cell strains (Host-A: pDB007(+), pDB023f1, and DP4; Host-B: pDB007(+), pDB023f1, and MP4; Host-C: pDB007(+)ns2a, pDB016, and MP4) were used during the experiment, which were maintained separately in three, 80 mL chemostats (C1–C3), respectively. Host-A was pumped into a positive-selection lagoon (L1-pos) at a flow rate of 1 lagoon volume per h, and genetic drift was induced by supplementation with 200 ng/mL ATc to the lagoon. L1-pos was infected with 108 pfu of SP-p-NFRS to initiate the experiment, and supplementation of ATc was stopped at 24 h to end genetic drift. Concomitant with the end of genetic drift, C1 was disconnected from L1-pos, and the lagoon was connected to C2 (containing Host-B), which was pumped into L1-pos at 1 lagoon volume per h. Also at this time, L1-pos was cross coupled to a second lagoon (L2-neg), which was being continuously flowed with negative-selection Host-C from C3. The maximum level of negative-selection stringency from Host-C was maintained by supplementing L2-neg with 30 ng/mL ATc. Cross coupling of the opposing selection lagoons was accomplished using two lines of Masterflex Microbore two-stop tubes (silicone; platinum cured; 0.89 mm ID) (Cole-Parmer), which each had a dead volume of 1 mL. One of the tubes was used to transfer small volumes of culture from L1-pos into L2-neg, and the second tube transferred material in the opposing direction. Material was peristaltically pumped through the cross-coupling lines with a Masterflex L/S Standard Digital Drive (Cole-Parmer) equipped with a Masterflex L/S 8-channel multichannel pump head for microbore tubing (Cole-Parmer). The flow rate through each cross-coupling line was initially set to 0.5 mL/h to maintain a 50-fold dilution of the transferred material into the opposing lagoons. The flow rate through L1-pos and L2-neg was doubled to 2 lagoon volumes per h at 28 h, and flow through the cross-coupling lines was adjusted to 1 mL/h to maintain 50-fold dilution of transferred material in each direction. The experiment ended at 48 h.
Luciferase assay
S1030 cells (25 µL) were electroporated with the appropriate plasmid(s) and recovered in SOC media (New England Biolabs) for 1 h while shaking at 37 °C. Transformed cells were plated and grown overnight at 37 °C on LB agar containing the antibiotics required for plasmid maintenance. Single colonies were used to inoculate 2–3 mL of DRM containing antibiotics and were grown overnight at 37 °C while shaking at 230 rpm. The saturated overnight cultures were diluted 100-fold in a 96-well deep well plate using 1 mL of DRM containing the required antibiotic and supplemented with 1 mM ncAA where denoted. The plate was shaken at 37 °C for 2 h at 230 rpm and then supplemented with the indicated concentration of isopropyl-β-D-thiogalactosidase (IPTG), anhydrotetracycline (ATc), or 1 mM arabinose—depending on the nature of the plasmids—to induce protein expression. The plate continued to incubate with shaking at 37 °C for an additional 2–3 h until maximum luminescence signal was observed. Each luminescence measurement was taken on 150 µL of each culture, which had been transferred to a 96-well black wall, clear bottom plate (Costar). The A600 and luminescence measurements from each well were taken using an Infinite M1000 Pro microplate reader (Tecan). Background A600 measurements were taken on wells containing media only. The raw luminescence value from each well was divided by the background-subtracted A600 value of the corresponding well to provide the luminescence value normalized to cell density. All variants were assayed in at least biological triplicate, and error bars represent the standard deviation of the independent measurements.
sfGFP assay
In assays of MjTyrRS variants (p-NFRS, p-IFRS, and PACE-evolved), a pDB070 plasmid containing the AARS of interest and a pET28b(+) containing the sfGFP of interest were cotransformed into chemically competent BL21 Star (DE3) cells (Thermo Fisher Scientific). The transformed cells recovered in SOC (New England Biolabs) for 1 h while shaking at 37 °C and were then plated and grown overnight at 37 °C on LB agar containing 50 µg/mL kanamycin and 25 µg/mL chloramphenicol. Single colonies were used to inoculate 2 mL of LB media (United States Biologicals) containing antibiotics and were grown overnight at 37 °C while shaking at 230 rpm. The saturated overnight cultures were diluted 100-fold in a 96-well deep well plate using 600 µL of LB media containing the required antibiotic and were grown at 37 °C to a cell density of A600 = 0.3 while shaking at 230 rpm. AARS expression was induced by addition of LB media (200 µL) containing antibiotics and the additional components to provide each well with a final concentration of 200 ng/mL anhydrotetracycline (ATc) and 1 mM ncAA where indicated. Incubation continued until cultures reached a cell density of A600 = 0.5. Each well was then supplemented with 1 mM isopropyl-β-D-thiogalactosidase (IPTG) to induce sfGFP expression.
In assays of PylRS variants (including AcK3RS variants), a pTECH plasmid containing the AARS of interest and a pBAD plasmid containing the sfGFP of interest were cotransformed into chemically competent TOP10 cells (Thermo Fisher Scientific). The transformed cells recovered in SOC (New England Biolabs) for 1 h while shaking at 37 °C and were then plated and grown overnight at 37 °C on LB agar containing 100 µg/mL carbenicillin and 25 µg/mL chloramphenicol. Single colonies were used to inoculate 2–3 mL of LB media (United States Biologicals) containing antibiotics and were grown overnight at 37 °C while shaking at 230 rpm. The saturated overnight cultures were diluted 100-fold in a 96-well deep well plate using 500 µL of LB media containing the required antibiotic. The plate was shaken at 37 °C for 3 h at 230 rpm and an additional 500 µL of LB was added containing antibiotics and additional components to provide each well with a final concentration of 1 mM ncAA where denoted and 1.5 mM arabinose to induce expression of sfGFP.
For all experiments, the cultures incubated with shaking at 37 °C for an additional 16 h after induction of sfGFP, and 150 µL of each culture was transferred to a 96-well black wall, clear bottom plate (Costar). The A600 and fluorescence (excitation = 485 nm; emission = 510 nm; bandwidth of excitation and emission = 5 nm) readings from each well were taken using an Infinite M1000 Pro microplate reader (Tecan). Background A600 and background fluorescence measurements were taken on wells containing LB media only. The background-subtracted fluorescence value from each well was divided by the background-subtracted A600 value of the same well to provide the fluorescence value normalized to cell density. All variants were assayed in at least biological triplicate, and error bars represent the standard deviation of the independent measurements.
Protein expression and purification of sfGFP
Expression of His-tagged sfGFP was performed using the plasmids, cell strains, and antibiotic concentrations described in the methods for sfGFP assays. Saturated overnight cultures were prepared from single colonies of cotransformed cells, which were diluted 1,000-fold into 300 mL of LB media containing 1 mM ncAA where denoted and were grown while shaking at 230 rpm at 37 °C. Once the cultures utilizing a pDB070 plasmid grew to a cell density of A600 = 0.3, AARS expression was induced by supplementing with anhydrotetracycline (ATc) to a final concentration of 200 ng/mL, and incubation was continued. Cultures utilizing a pTECH plasmid did not require this step as the AARS was expressed constitutively. Once cultures grew to a cell density of A600 = 0.5, sfGFP expression was induced by supplementation with a final concentration of 1 mM isopropyl-β-D-thiogalactosidase (IPTG) for cultures utilizing a pET28b(+) plasmid or a final concentration of 1 mM arabinose for cultures utilizing a pBAD plasmid. Incubation with shaking at 37 °C continued for an additional 16 h after induction of sfGFP expression. Cells were harvested by centrifugation at 5,000g for 10 min at 4 °C, and the resulting pellets were resuspended in B-PER II Bacterial Protein Extraction Reagent (Thermo Fisher Scientific) containing EDTA-free protease inhibitor cocktail (Roche). The soluble fractions of the cell lysates were diluted by an equal volume of equilibration buffer (20 mM Tris (pH 7.4), 10 mM imidazole, 300 mM NaCl) and were separately loaded onto a column containing 2 mL of HisPur Ni-NTA resin (Thermo Fisher Scientific) that had been pre-washed with two bed-volumes of equilibration buffer. The resin was washed with two bed-volumes of wash buffer (20 mM Tris (pH 7.4), 25 mM imidazole, 300 mM NaCl) and protein was then eluted in 3 mL of elution buffer (20 mM Tris (pH 7.4), 250 mM imidazole, 300 mM NaCl). The purified protein was dialyzed against 20 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1 mM 2-mercaptoethanol (BME), and 10% glycerol. Purified protein was stored at −80 °C until analysis.
AARS variant expression and purification for aminoacylation assays
The genes of chPylRS variants and MjTyrRS variants were cloned into pET15a and transformed into BL21(DE3) (New England Biolabs) cells for expression. Cells were grown in 500 mL of LB media supplemented with 100 µg/mL ampicillin at 37 °C to an A600 of 0.6–0.8, and protein expression was induced by addition of 0.5 mM IPTG (chPylRS variants) or 1 mM IPTG (MjTyrRS variants). Cells were incubated at 30°C for an additional 6 h (chPylRS variants) or 4 h (MjTyrRS variants) and harvested by centrifugation at 5,000g for 10 min at 4 °C. The cell pellet was resuspended in 15 mL of lysis buffer (50 mM Tris (pH 7.5), 300 mM NaCl, 20 mM imidazole), and cells were lysed by sonication. The crude extract was centrifuged at 20,000g for 30 min at 4 °C. The soluble fraction was loaded onto a column containing 2 mL of Ni-NTA resin (Qiagen) previously equilibrated with 20 mL of lysis buffer. The column was washed with 20 mL of lysis buffer. The bound protein was then eluted with 2 mL of 50 mM Tris (pH 7.5), 300 mM NaCl, 300 mM imidazole. The purified protein was dialyzed with 50 mM HEPES-KOH (pH 7.5), 50 mM KCl, 1 mM DTT and 50% glycerol and stored at −80 °C for further studies.
Purification of c-Myc-chPylRS-6xHis variants
The chPylRS variants were cloned into the pTech plasmid using insertion primers that incorporate the N-terminal c-Myc sequence (MEQKLISEEDL-) and the C-terminal 6xHis sequence (−GSHHHHHH). BL21 star (DE3) cells (Thermo Fisher Scientific) transformed with the appropriate pTech plasmids were grown in LB media (United States Biologicals) supplemented with 25 µg/mL chloramphenicol. For each variant, a saturated overnight culture was prepared from a single colony, and a 1:100 dilution of culture was made into 5 mL of fresh LB media containing chloramphenicol. The starter culture grew at 37 °C while shaking at 230 rpm until the cell density reached A600 = 0.3. The starter culture was then used to inoculate a 1 L culture of LB media containing chloramphenicol, which continued to incubate while shaking for an additional 16 h. Cells were harvested by centrifugation at 5,000g for 10 min at 4 °C, and cell pellets were resuspended in lysis buffer (20 mM Tris (pH 7.4), 300 mM NaCl, 10 mM imidazole, and EDTA-free protease inhibitor cocktail (Roche)). The cells were lysed by sonication on ice, and the crude extract was centrifuged at 15,000g for 15 min at 4 °C. Lysates were loaded onto columns containing 2 mL of HisPur Ni-NTA resin (Thermo Fisher Scientific) that had been pre-washed with two bed-volumes of equilibration buffer. The resin was washed with 10 bed-volumes of wash buffer (20 mM Tris (pH 7.4), 25 mM imidazole, 300 mM NaCl) and protein was then eluted in 3 mL of elution buffer (20 mM Tris (pH 7.4), 250 mM imidazole, 300 mM NaCl). The purified protein was dialyzed against 20 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol. Purified protein was stored in 20% glycerol at −80 °C until analysis.
Western blot analysis of c-Myc-chPylRS-6xHis variants
Cell lysates (30 µL) of expressed protein were combined with 25 µL of XT Sample Buffer (Bio-Rad), 5 µL of 2-mercaptoethanol, and 40 µL water. The samples were heated at 70 °C for 10 min and 7.5 µL of prepared sample was loaded per well of a Bolt Bis-Tris Plus Gel (Thermo Fisher Scientific). Precision Plus Protein Dual Color Standard (4 µL) Bio-Rad was used as the reference ladder. The loaded gel was run at 200V for 22 min in 1x Bolt MES SDS running buffer (Thermo Fisher Scientific). The gel was transferred to a PVDF membrane using the iBlot 2 Gel Transfer Device (Thermo Fisher Scientific). The membrane was blocked for 1 h at room temperature in 50% Odyssey blocking buffer (PBS) (Li-Cor) and was then soaked 4 times for 5 min in PBS containing 0.1% Tween-20 (PBST). The blocked membrane was soaked with primary antibodies (rabbit anti-6xHis (1:1,000 dilution) (Abcam, ab9108) and mouse anti-c-Myc (1:7,000 dilution) (Sigma-Aldrich, M4439)) in 50% Odyssey buffer (PBS) containing 0.2% Tween-20 for 4 h at room temperature. The membrane was washed four times in PBST, and then soaked for 1 h in the dark at room temperature with secondary antibodies (IRDye donkey anti-mouse 800CW IgG (1:20,000 dilution) (Li-Cor, 926-32212) and IRDye goat anti-rabbit 680RD IgG (1:20,000 dilution) (Li-Cor, 926-68071)) in Odyssey buffer containing 0.01% SDS, 0.2% Tween-20. The membrane was washed 4 times in PBST and finally rinsed with PBS. The membrane was scanned using an Odyssey Imaging System (Li-Cor).
LCMS analysis of intact purified proteins
Purified protein samples were diluted to 10 µM in dialysis buffer lacking reducing agent or glycerol prior to analysis on an Agilent 6220 ESI-TOF mass spectrometer equipped with an Agilent 1260 HPLC. Separation and desalting was performed on an Agilent PLRP-S Column (1,000A, 4.6 × 50 mm, 5 µm). Mobile the phase A was 0.1% formic acid in water and mobile phase B was acetonitrile with 0.1% formic acid. A constant flow rate of 0.250 mL/min was used. Ten microliters of the protein solution were injected and washed on the column for the first 3 min at 5% B, diverting non-retained materials to waste. The protein was then eluted using a linear gradient from 5% B to 100% B over 7 min. The mobile phase composition was maintained at 100% B for 5 min and then returned to 5% B over 1 minute. The column was then re-equilibrated at 5%B for the next 4 min. Data was analyzed using Agilent MassHunter Qualitative Analysis software (B.06.00, Build 6.0.633.0 with Bioconfirm). The charge state distribution for the protein produced by electrospray ionization was deconvoluted to neutral charge state using Bioconfirm’s implementation of MaxEnt algorithm, giving a measurement of average molecular weight. The average molecular weight of the proteins were predicted using ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/), and each calculation was adjusted for chromophore maturation in sfGFP and any ncAA substitutions.
Amber suppressor tRNA preparation
Template plasmid containing the tRNAPyl or tRNACUATyr/Opt gene was purified with the plasmid maxi kit (Qiagen). The plasmid containing tRNAPyl (100 µg) was digested with BstNI (New England Biolabs). The tRNACUATyr/Opt gene was amplified by PCR. The BstNI digested template DNA or PCR product was purified by phenol chloroform extraction, followed by ethanol precipitation and dissolved in double distilled water. A His-tagged T7 RNA polymerase was purified over a column of Ni-NTA resin according to manufacturer’s instructions (Qiagen). The transcription reaction (40 mM Tris (pH 8); 4 mM each of UTP, CTP, GTP, and ATP at pH 7.0; 22 mM MgCl2; 2 mM spermidine; 10 mM DTT; 6 µg pyrophosphatase (Roche Applied Science); 60 µg/mL of DNA template, approximately 0.2 mg/mL T7 RNA polymerase) was performed in 10 mL reactions overnight at 37 °C. The tRNA was purified on 12% denaturing polyacrylamide gel containing 8 M urea and TBE buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA). UV shadowing was used to illuminate the pure tRNA band, which was excised and extracted three times with 1M sodium acetate pH 5.3 at 4 °C. The tRNA extractions were then ethanol precipitated, dissolved in RNase-free distilled water, pooled, and finally desalted using a Biospin 30 column (BioRad). The tRNA was refolded by heating to 100 °C for 5 min and slow cooling to room temperature. At 65 °C, MgCl2 was added to a final concentration of 10 mM to aid folding. A His-tagged CCA adding enzyme was purified over column of Ni-NTA resin according to manufacturer’s instructions (Qiagen). 16 µM refolded tRNA in 50 mM Tris (pH 8.0), 20 mM MgCl2, 5 mM DTT, and 50 µM NaPPi was incubated at room temperature for 1 h with approximately 0.2 mg/mL CCA-adding enzyme and 1.6 µCi/µL of (α-32P)-labeled ATP (PerkinElmer). The sample was phenol/chloroform extracted and then passed over a Bio-spin 30 column (Bio-Rad) to remove excess ATP.
Aminoacylation assay
A 20 µL aminoacylation reaction contained the following components for chPylRS variants: 50 mM HEPES-KOH (pH 7.2), 25 mM KCl, 10 mM MgCl2, 5 mM DTT, 10 mM ATP, 25 µg/mL pyrophosphatase (Roche Applied Science), 10 mM amino acids, 500 nM PylRS variants, 5 µM unlabeled tRNAPyl, and 100 nM 32P-labeled tRNAPyl. A 20 µL aminoacylation reaction contained the following components for MjTyrRS variants: 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 10 mM MgCl2, 10 mM ATP, 20 µM unlabeled tRNACUATyr/Opt, 3 µM 32P-labeled tRNACUATyr/Opt, 2 µM MjTyrRS variants. Various concentrations of ATP (1–100 µM), BocK (0.1–10 mM), Pyl (5–500 µM), Phe (0.1–3.2 mM), p-NF (1–32 mM), p-IF (1–32 mM), and tRNA (0.05–5 µM) were used to determine KM values for corresponding substrates. Time points were taken at 5 min, 20 min and 60 min by removing 2 µL aliquots from the reaction and immediately quenching the reaction into an ice-cold 3 µL quench solution (0.66 µg/µL nuclease P1 (Sigma) in 100 mM sodium citrate (pH 5.0)). For each reaction, 2 µL of blank reaction mixture (containing no enzyme) was added to the quench solution as the start time point. The nuclease P1 mixture was then incubated at room temperature for 30 min and 1 µL aliquots were spotted on PEI-cellulose plates (Merck) and developed in running buffer containing 5% acetic acid and 100 mM ammonium acetate. Radioactive spots for AMP and AA-AMP (representing free tRNA and aminoacyl-tRNA, respectively) were separated and then visualized and quantified by phosphorimaging using a Molecular Dynamics Storm 860 phosphorimager (Amersham Biosciences). The ratio of aminoacylated tRNA to total tRNA was determined to monitor reaction progress.
Data Availability
Selection plasmids used in this study will be available through Addgene. Other materials are available upon reasonable request.
Methods-Only References
All references cited in the Online Methods appear in the main text.
Supplementary Material
Acknowledgments
The authors thank Sunia Trauger at the Small Molecule Mass Spectrometry Laboratory at Harvard University for providing expertise with intact protein mass spectrometry analysis. This work was supported by the Defense Advanced Research Projects Agency N66001-12-C-4207 (D.R.L.), the U.S. National Institutes of Health (NIH) R01 EB022376 (D.R.L.), R35 GM118062 (D.R.L.), R21 AI119813 (C.F.), R01 GM022854 (D.S.), and R35 GM122560 (D.S.), the Department of Energy FG02-98ER2031 (D.S.), and the Howard Hughes Medical Institute (D.R.L). D.I.B is supported by a Ruth L. Kirschstein National Research Service Award (F32 GM106621).
Footnotes
Author Contributions
D.I.B. designed the research, performed experiments, analyzed data, and wrote the manuscript. D.R.L. designed and supervised the research and wrote the manuscript. D.S. designed and supervised the research. C.F. performed protein purification, in vitro aminoacylation assays, aided with in vivo amber suppression assays, and analyzed data. L.T.G designed the chimeric chPylRS variant for evolution in PACE, performed protein purification, performed in vitro aminoacylation assays, and analyzed data. C.M. aided in mutation analysis of evolved chPylRS variants from PACE. All authors contributed to editing the manuscript.
Competing Financial Interests
The authors have filed a provisional patent application on the PACE system and related improvements.
References
- 1.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 2.Wan W, Tharp JM, Liu WR. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta. 2014;1844:1059–1070. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 4.Umehara T, et al. N-acetyl lysyl-tRNA synthetases evolved by a CcdB-based selection possess N-acetyl lysine specificity in vitro and in vivo. FEBS Lett. 2012;586:729–733. doi: 10.1016/j.febslet.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 5.O’Donoghue P, Ling J, Wang Y-S, Söll D. Upgrading protein synthesis for synthetic biology. Nat. Chem. Biol. 2013;9:594–598. doi: 10.1038/nchembio.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leconte AM, et al. A population-based experimental model for protein evolution: effects of mutation rate and selection stringency on evolutionary outcomes. Biochemistry. 2013;52:1490–1499. doi: 10.1021/bi3016185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson JC, Badran AH, Guggiana-Nilo DA, Liu DR. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 2014;10:216–222. doi: 10.1038/nchembio.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickinson BC, Packer MS, Badran AH, Liu DR. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Nat. Commun. 2014;5:5352. doi: 10.1038/ncomms6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubbard BP, et al. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat. Methods. 2015;12:939–942. doi: 10.1038/nmeth.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badran AH, et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature. 2016;533:58–63. doi: 10.1038/nature17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riechmann L, Holliger P. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell. 1997;90:351–360. doi: 10.1016/s0092-8674(00)80342-6. [DOI] [PubMed] [Google Scholar]
- 13.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 14.Tsao ML, Summerer D, Ryu Y, Schultz PG. The genetic incorporation of a distance probe into proteins in Escherichia coli. J. Am. Chem. Soc. 2006;128:4572–4573. doi: 10.1021/ja058262u. [DOI] [PubMed] [Google Scholar]
- 15.Grunewald J, et al. Immunochemical termination of self-tolerance. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11276–11280. doi: 10.1073/pnas.0804157105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanagisawa T, et al. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Lubkowski J, Hennecke F, Pluckthun A, Wlodawer A. The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p. Nat. Struct. Biol. 1998;5:140–147. doi: 10.1038/nsb0298-140. [DOI] [PubMed] [Google Scholar]
- 18.Lubkowski J, Hennecke F, Pluckthun A, Wlodawer A. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure. 1999;7:711–722. doi: 10.1016/s0969-2126(99)80092-6. [DOI] [PubMed] [Google Scholar]
- 19.Deng LW, Perham RN. Delineating the site of interaction on the pIII protein of filamentous bacteriophage fd with the F-pilus of Escherichia coli. J. Mol. Biol. 2002;319:603–614. doi: 10.1016/S0022-2836(02)00260-7. [DOI] [PubMed] [Google Scholar]
- 20.Boeke JD, Model P. A prokaryotic membrane anchor sequence: carboxyl terminus of bacteriophage f1 gene III protein retains it in the membrane. Proc. Natl. Acad. Sci. U. S. A. 1982;79:5200–5204. doi: 10.1073/pnas.79.17.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boeke JD, Model P, Zinder ND. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol. Gen. Genet. 1982;186:185–192. doi: 10.1007/BF00331849. [DOI] [PubMed] [Google Scholar]
- 22.Rapoza MP, Webster RE. The filamentous bacteriophage assembly proteins require the bacterial SecA protein for correct localization to the membrane. J. Bacteriol. 1993;175:1856–1859. doi: 10.1128/jb.175.6.1856-1859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Badran AH, Liu DR. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat. Commun. 2015;6:8425. doi: 10.1038/ncomms9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo LT, et al. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16724–16729. doi: 10.1073/pnas.1419737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavran JM, et al. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong ML, Guzei IA, Kiessling LL. An asymmetric synthesis of L-pyrrolysine. Org. Lett. 2012;14:1378–1381. doi: 10.1021/ol300045c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang R, Krzycki JA. PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine. J. Biol. Chem. 2012;287:32738–32746. doi: 10.1074/jbc.M112.396754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herring S, et al. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007;581:3197–3203. doi: 10.1016/j.febslet.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 31.Ho JM, et al. Efficient reassignment of a frequent serine codon in wild-type Escherichia coli. ACS Synth. Biol. 2016;5:163–171. doi: 10.1021/acssynbio.5b00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 33.Andre A, et al. Reinitiation of protein synthesis in Escherichia coli can be induced by mRNA cis-elements unrelated to canonical translation initiation signals. FEBS Lett. 2000;468:73–78. doi: 10.1016/s0014-5793(00)01198-4. [DOI] [PubMed] [Google Scholar]
- 34.Cabantous S, Rogers Y, Terwilliger TC, Waldo GS. New molecular reporters for rapid protein folding assays. PLoS One. 2008;3:e2387. doi: 10.1371/journal.pone.0002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nozawa K, et al. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature. 2009;457:1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie J, et al. The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat. Biotechnol. 2004;22:1297–1301. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- 37.Turner JM, Graziano J, Spraggon G, Schultz PG. Structural plasticity of an aminoacyl-tRNA synthetase active site. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6483–6488. doi: 10.1073/pnas.0601756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanagisawa T, et al. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J. Mol. Biol. 2008;378:634–652. doi: 10.1016/j.jmb.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 39.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amiram M, et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol. 2015;33:1272–1279. doi: 10.1038/nbt.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmied WH, Elsasser SJ, Uttamapinant C, Chin JW. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc. 2014;136:15577–15583. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan C, Xiong H, Reynolds NM, Söll D. Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic Acids Res. 2015;43:e156. doi: 10.1093/nar/gkv800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 45.Guo J, Melancon CE, 3rd, Lee HS, Groff D, Schultz PG. Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids. Angew. Chem. Int. Ed. Engl. 2009;48:9148–9151. doi: 10.1002/anie.200904035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, et al. Nitrilase-activatable noncanonical amino acid precursors for cell-selective metabolic labeling of proteomes. ACS Chem. Biol. 2016;11:3273–3277. doi: 10.1021/acschembio.6b00765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Selection plasmids used in this study will be available through Addgene. Other materials are available upon reasonable request.