Abstract
In this case-control study, we analyzed the association between eight RegulomeDB-annotated single nucleotide polymorphisms (SNPs) in the MEK1, MEK2, ERK1 and ERK2 genes and polycystic ovarian syndrome (PCOS). Logistic regression analysis demonstrated that MEK1 rs12050732 (OR = 1.29 [95%CI: 1.06-1.58], P = 0.012), ERK2 rs2266966 (OR = 0.81 [95%CI: 0.67-0.99], P = 0.040) and ERK2 rs5999521 (OR = 0.66 [95%CI: 0.51-0.86], P = 0.002) were associated with PCOS risk without adjusting for age and body mass index. Moreover, PCOS risk increased with allele dosage when these three polymorphisms were combined (Ptrend = 0.001). These findings suggest that genetic variants in key MAPK and ERK genes contribute to PCOS risk in Chinese women.
Keywords: variants, MAPK/ERK, RegulomeDB, polycystic ovarian syndrome
INTRODUCTION
Polycystic ovarian syndrome (PCOS) is a common endocrine syndrome associated with oligo-ovulation or anovulation among women of reproductive age [1, 2]. PCOS is characterized by elevated serum luteinizing hormone (LH) and testosterone (T) levels, although the serum follicle-stimulating hormone (FSH) levels are normal [3]. PCOS is also associated with increased risk for metabolic syndromes like insulin resistance, obesity and type 2 diabetes [2, 4, 5]. Both environmental and genetic factors contribute to the complex pathophysiology of PCOS. Candidate gene studies and genome-wide association studies (GWAS) have demonstrated that genetic components play an important role in PCOS susceptibility [6, 7].
MAPK/ERK signaling pathway includes mitogen-activated protein kinases (MAPKs), like extracellular signal-regulated kinases (ERKs), which integrate multiple biochemical and environmental signals through phosphorylation cascades. The MAPK/ERK signaling pathway regulates a wide variety of cellular processes such as proliferation, differentiation, transcriptional regulation and differentiation [8, 9]. The upstream core MAPKs such as MEK1 and MEK2 phosphorylate and activate downstream effector kinases, ERK1 and ERK2 [10]. The MAPK/ERK signaling pathway is associated with steroid biosynthesis in ovarian granulosa cells [11, 12]. Aberrant MAPK/ERK signaling contributes to metabolic signaling defects and excessive ovarian androgen production in women with PCOS [13, 14]. As the above, there is increasing evidence that MAPK/ERK pathway genes are associated with PCOS risk.
Functional annotation of GWAS data has shown that single nucleotide polymorphisms can be useful biomarkers for various human diseases [15, 16]. RegulomeDB is a database that integrates regulatory information from ENCODE and other data sources and is a powerful tool that shows functional SNPs in specific chromosomal regions [17, 18]. It has a scoring system from 1 to 6 for the annotated data on methylation, chromatin structure, protein motifs and binding. The lower score indicates high probability for a variant to be located in a functional region.
Several studies have used RegulomeDB to identify causal polymorphisms associated with human diseases such as lung cancer, colorectal cancer and so on [19–21]. In this case-control study, we analyzed PCOS risk with MAPK/ERK gene polymorphisms that are annotated in RegulomeDB.
RESULTS
Table 1 shows clinical pathological characteristics of PCOS patients and controls in accordance with 2003 Rotterdam PCOS diagnose criteria. PCOS patients showed higher LH and T levels as well as body mass index (BMI) than control subjects.
Table 1. Clinical pathological parameters in PCOS and control subjects.
| Parameters | Case | Control | T-test | P |
|---|---|---|---|---|
| Age | 27.36±5.27 | 30.68±3.66 | 9.030 | <0.001 |
| BMI | 22.84±3.74 | 21.41±3.08 | -5.200 | <0.001 |
| FSH (IU/L) | 6.41±2.67 | 7.33±2.56 | 4.266 | <0.001 |
| LH (IU/L) | 13.27±8.22 | 5.10±3.36 | -14.933 | <0.001 |
| E2 (pg/ml) | 66.94±48.52 | 54.34±39.37 | -3.347 | <0.001 |
| T (ng/ml) | 2.79±7.69 | 1.09±1.01 | -3.428 | <0.001 |
BMI: body mass index; FSH: follicle-stimulating hormone; LH: luteinizing hormone; E2: estradiol; T: total testosterone.
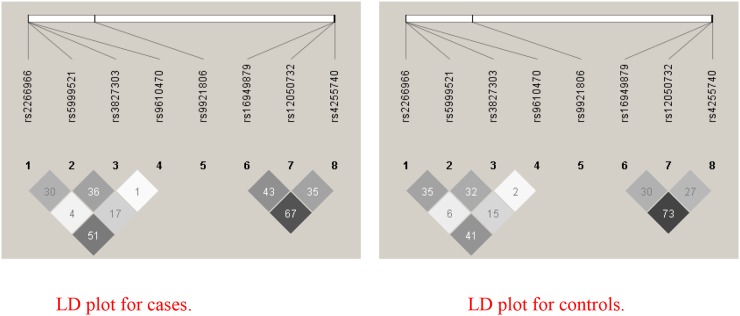
Table 2 shows the genotypes distributions of the 8 SNPs (rs12050732, rs16949879 and rs4255740 in MEK1; rs9921806 in ERK1; rs3827303, rs5999521, rs2266966 and rs9610470 in ERK2) and their association with PCOS. These 8 SNPs showed low pairwise LD each other in cases and controls, respectively (Figure 1). The MEK1 rs12050732 SNP showed association with PCOS in the additive model for unadjusted data (OR=1.29 (95%CI: 1.06-1.58), P = 0.012). The ERK2 rs5999521 SNP showed association with PCOS in the additive model for unadjusted data (OR=0.81 (95%CI: 0.67-0.99), P= 0.040). The ERK2 rs2266966 SNP showed association with PCOS in the additive model for unadjusted data (OR=0.66 (95%CI: 0.51-0.86), P = 0.002). In these all 3 polymorphisms, the data was insignificant when adjusted for age and BMI (P>0.05). The ERK2 rs9610470 SNP did marginal association with PCOS in recessive model (OR=0.16; 95%CI: 0.02-1.37; P = 0.096). The remaining 4 SNPs (rs16949879 and rs4255740 in MEK1; ERK1 rs9921806; rs3827303 in ERK2) showed no association with PCOS risk (Table 2).
Table 2. Association between MEK and ERK polymorphisms and PCOS risk.
| SNP | Genotype | Case | Control | OR (95%CI) | P | OR (95%CI)a | Pa |
|---|---|---|---|---|---|---|---|
| MEK1 rs12050732 | AA | 138 | 159 | 1.00 | 1.000 | 1.00 | 1.000 |
| CA | 188 | 196 | 1.11(0.82-1.50) | 0.518 | 1.06(0.73-1.56) | 0.753 | |
| CC | 79 | 50 | 1.82(1.20-2.77) | 0.005b | 1.54(0.89-2.66) | 0.124 | |
| Dominant | 1.25(0.94-1.67) | 0.126 | 1.16(0.81-1.66) | 0.431 | |||
| Recessive | 1.72(1.17-2.53) | 0.006b | 1.49(0.90-2.46) | 0.124 | |||
| Additive | 1.29(1.06-1.58) | 0.012b | 1.19(0.92-1.55) | 0.179 | |||
| MEK1 rs16949879 | AA | 140 | 142 | 1.00 | 1.000 | 1.00 | 1.000 |
| GA | 173 | 173 | 1.01(0.74-1.39) | 0.930 | 0.99(0.66-1.48) | 0.963 | |
| GG | 75 | 68 | 1.12(0.75-1.67) | 0.585 | 1.31(0.79-2.19) | 0.293 | |
| Dominant | 1.04(0.78-1.40) | 0.775 | 1.08(0.74-1.56) | 0.700 | |||
| Recessive | 1.11(0.77-1.60) | 0.574 | 1.32(0.84-2.09) | 0.233 | |||
| Additive | 1.05(0.86-1.28) | 0.621 | 1.12(0.87-1.44) | 0.366 | |||
| MEK1 rs4255740 | CC | 176 | 174 | 1.00 | 1.000 | 1.00 | 1.000 |
| TC | 189 | 188 | 0.99(0.74-1.33) | 0.967 | 1.01(0.70-1.47) | 0.953 | |
| TT | 39 | 43 | 0.90(0.55-1.45) | 0.657 | 1.18(0.65-2.15) | 0.585 | |
| Dominant Dominant | 0.98(0.74-1.29) | 0.863 | 1.04(0.73-1.48) | 0.824 | |||
| Recessive | 0.90(0.57-1.42) | 0.650 | 1.18(0.67-2.07) | 0.578 | |||
| Additive | 0.96(0.78-1.19) | 0.733 | 1.06(0.81-1.39) | 0.668 | |||
| ERK1 rs9921806 | CC | 261 | 282 | 1.00 | 1.000 | 1.00 | 1.000 |
| TC | 128 | 109 | 1.27(0.93-1.72) | 0.127 | 1.26(0.85-1.86) | 0.255 | |
| TT | 13 | 14 | 1.00(0.46-2.17) | 0.993 | 2.37(0.84-6.67) | 0.103 | |
| Dominant | 1.24(0.92-1.66) | 0.155 | 1.34(0.91-1.95) | 0.134 | |||
| Recessive | 0.93(0.43-2.01) | 0.860 | 2.22(0.79-6.21) | 0.129 | |||
| Additive | 1.16(0.90-1.50) | 0.244 | 1.35(0.97-1.88) | 0.074 | |||
| ERK2 rs3827303 | GG | 258 | 265 | 1.00 | 1.000 | 1.00 | 1.000 |
| CG | 132 | 121 | 1.12(0.83-1.51) | 0.458 | 1.07(0.73-1.58) | 0.737 | |
| CC | 15 | 18 | 0.86(0.42-1.74) | 0.666 | 1.03(0.43-2.44) | 0.949 | |
| Dominant | 1.09(0.81-1.45) | 0.574 | 1.06(0.73-1.54) | 0.745 | |||
| Recessive | 0.82(0.41-1.66) | 0.589 | 1.01(0.43-2.37) | 0.986 | |||
| Additive | 1.04(0.81-1.32) | 0.775 | 1.05(0.77-1.42) | 0.782 | |||
| ERK2 rs5999521 | AA | 162 | 139 | 1.00 | 1.000 | 1.00 | 1.000 |
| GA | 183 | 185 | 0.85(0.63-1.15) | 0.292 | 1.10(0.74-1.63) | 0.638 | |
| GG | 60 | 79 | 0.65(0.43-0.98) | 0.038b | 0.72(0.43-1.20) | 0.203 | |
| Dominant | 0.79(0.59-1.05) | 0.106 | 0.97(0.67-1.41) | 0.886 | |||
| Recessive | 0.71(0.49-1.03) | 0.072 | 0.68(0.43-1.08) | 0.103 | |||
| Additive | 0.81(0.67-0.99) | 0.040b | 0.88(0.69-1.13) | 0.327 | |||
| ERK2 rs2266966 | TT | 279 | 241 | 1.00 | 1.000 | 1.00 | 1.000 |
| CT | 121 | 150 | 0.70(0.52-0.94) | 0.016b | 0.92(0.63-1.33) | 0.643 | |
| CC | 5 | 14 | 0.31(0.11-0.87) | 0.026b | 0.33(0.09-1.14) | 0.079 | |
| Dominant | 0.66(0.50-0.89) | 0.005b | 0.86(0.60-1.23) | 0.408 | |||
| Recessive | 0.35(0.12-0.98) | 0.045b | 0.34(0.10-1.17) | 0.086 | |||
| Additive | 0.66(0.51-0.86) | 0.002 b | 0.81(0.59-1.12) | 0.208 | |||
| ERK2 rs9610470 | TT | 330 | 323 | 1.00 | 1.000 | 1.00 | 1.000 |
| CT | 74 | 76 | 0.95(0.67-1.36) | 0.791 | 1.22(0.79-1.9) | 0.372 | |
| CC | 1 | 6 | 0.16(0.02-1.36) | 0.094 | 0.21(0.02-2.00) | 0.176 | |
| Dominant | 0.90(0.63-1.27) | 0.534 | 1.13(0.73-1.74) | 0.578 | |||
| Recessive | 0.16(0.02-1.37) | 0.096 | 0.21(0.02-1.91) | 0.165 | |||
| Additive | 0.85(0.61-1.17) | 0.319 | 1.03(0.69-1.53) | 0.886 | |||
a Adjusted for age and BMI. b Significant parameters with P < 0.05 are represented in bold.
Figure 1. Linkage disequilibrium (LD) based haploview r2 plot.
We also assessed the cumulative effects of three SNPs (MEK1 rs12050732, ERK2 rs5999521 and ERK2 rs2266966) on PCOS risk (Table 3). We observed allele-dosage association between the number of allele variants and PCOS risk in the additive model for unadjusted data (Ptrend = 0.001) and marginal association when data was adjusted for age and BMI (P-value = 0.09). Individuals with 5-6 risk alleles showed higher risk for PCOS than those with 0–2 risk alleles for unadjusted data (OR = 1.23 (95% CI: 1.08-1.40), P = 0.001). But, the correlation was not significant when the data was adjusted for age and BMI (P = 0.88). Furthermore, we observed no significant association between the three SNPs and PCOS risk in subgroups stratified by age (≤ 30 y and > 30 y) and BMI (≤ 25 kg/m2 and > 25 kg/m2; data not shown).
Table 3. Association between combination of MEK1 rs12050732, ERK2 rs5999521 and ERK2 rs2266966 polymorphisms and PCOS risk.
| No. of risk alleles | case | control | OR (95% CI) | P | OR (95%CI)a | Pa |
|---|---|---|---|---|---|---|
| 0-2 | 82 | 110 | 1 | 1 | 1 | 1 |
| 3 | 73 | 91 | 1.08(0.71-1.64) | 0.73 | 1.38(0.83-2.30) | 0.22 |
| 4 | 113 | 107 | 1.19(0.98-1.45) | 0.08 | 0.92(0.69-1.24) | 0.59 |
| 5-6 | 137 | 98 | 1.23(1.08-1.40) | 0.001b | 1.02(0.83-1.24) | 0.88 |
| Trend | 1.24(1.10-1.41) | 0.001b | 1.14(0.98-1.33) | 0.09 |
a Adjusted for age and BMI. b Significant parameters with P < 0.05 are represented in bold.
DISCUSSION
In this case-control study of Chinese individuals, we evaluated the association of genetic variants identified by RegulomeDB in MAPK/ERK genes with PCOS susceptibility. We found that MEK1 rs12050732, ERK2 rs2266966 and ERK2 rs5999521 were associated with PCOS risk.
The MAPK/ERK pathway plays an important role in the pathogenesis of PCOS. The phospho-ERK1/2 levels are reduced in PCOS granulosa cells [22], whereas the MAPK/ERK signaling pathway is hyperactive in the PCOS endometrium [23]. PCOS patients showed elevated serum T and LH levels [3]. In PCOS patients, MAPK signaling was activated by the LH receptor [24, 25]. Moreover, T levels were elevated when patients were treated with the MAPK kinase inhibitor PD98059 [26, 27]. These data suggest that MAPK/ERK signaling regulates T and LH levels in PCOS patients [24-27].
MEK1 is located on human chromosome 15q22.31 and is a key gene in MAPK pathway [10]. LH consists of α and β subunits, of which the β-subunit synthesis is the rate-limiting step in LH synthesis [28]. Overexpression of constitutively active MEK1 activates the LH β-subunit promoter [24]. MEK1 inhibitor U0126 blocks androgen-induced DNA synthesis, thereby demonstrating the role of MEK1 in androgen metabolism [29]. MEK1 rs12050732 is found in patients with esophageal carcinoma and gastric cancer [30]. MEK1 rs12050732 is a cis-eQTL for DIS3L, which has a RNA exosome subunit with endonucleolytic and 3’-5’exonucleolytic activity that degrades RNA [31].
ERK2 is located on chromosome 22q11.22 and is involved in many critical signal transduction pathways including LH signaling during ovulation [32, 33]. ERK2 deletion in mouse granulosa cells disrupts LH-induced meiotic progression in oocytes, ovulation, and luteinization [34]. ERK2 activation is critical for the normal feedback mechanism of insulin signaling and its aberrant activation contributes to insulin resistance in PCOS [14, 35]. The RegulomeDB score of both the ERK2 SNPs, rs2266966 and rs5999521 was 2b, which indicated binding to many proteins including transcription factors such as CTCF, IRF4, MYC, STAT2, and SP1 [36].
MEK2 is located on chromosome 19p13.3 and phosphorylates MAPK1/ERK2 and MAPK2/ERK3, thereby activating them [37]. RegulomeDB showed only one potentially functional variant of MEK2, which could not be genotyped. ERK1 is located on chromosome 16p11.2 and translocates to the nucleus when activated by upstream kinases [37].
In summary, our study shows that multiple polymorphisms in MAPK/ERK genes are associated with PCOS risk in Chinese women. Further independent studies with large sample sizes are necessary to unravel the molecular mechanisms underlying this association and to further confirm our findings.
MATERIALS AND METHODS
Study population
This study was approved by the Human Research Ethics Committee of Changzhou Maternity and Child Health Care Hospital Affiliated to Nanjing Medical University. We recruited 405 PCOS and control subjects each for this study as described previously [38]. All the study subjects were Chinese women of Han ethnicity. They were recruited from the Department of Obstetrics and Gynecology of the Affiliated Hospital of Medical School of Nanjing University, the Centre of Reproduction Memorial Hospital of Sun Yat-Sen University, and the Department of Obstetrics and Gynecology of Anhui Medical University.
PCOS diagnosis
PCOS was diagnosed according to the revised 2003 consensus on diagnostic criteria and long-term health risks related to PCOS [39]. The criteria for PCOS diagnosis were as follows: (1) oligo ovulation and /or anovulation; (2) clinical and /or biochemical characterization of hyperandrogenism and (3) polycystic ovarian morphology based on ultrasound. At least 2 of the 3 criteria needed to be positive for the case to be classified as PCOS. Moreover, patients with diseases that cause hyperandrogenism such as congenital adrenal hyperplasia, hypothyroidism, Cushing’s syndrome and androgen-secreting tumors were excluded. The study subjects had not received hormone drugs or oral contraceptives three months prior to analysis. All subjects were interviewed face-to-face to collect demographic data and exposure information by trained interviewers. After signing informed consent, peripheral blood was collected during the 3rd, 4th and 5th day of the menstrual cycle and at any time for those who had amenorrhea. Serum levels of FSH, LH, T and estradiol (E2) were measured by RIA assay (Beijing North Institute of Biological Technology of China and the CIS Company of France).
Polymorphism genotyping
We identified all polymorphisms in the MEK1, MEK2, ERK1 and ERK2 genes by Genome Reference Consortium Human Build 37 patch release 13 (GRCh37/hg19) assembly. A lower RegulomeDB score indicated strong probability for a variant to be located in the functional region of a gene. Therefore, a polymorphism with a score of 1 probably affected transcription and expression of a target gene. We selected SNPs that were classified in the 1-2b category (http://regulome.stanford.edu). The selection criteria included minor allele frequency (MAF) ≥ 0.05 and Hardy–Weinberg equilibrium (HWE) ≥ 0.05. Finally, SNPs with R2 values of pairwise linkage disequilibrium (LD) < 0.8 were selected. The genomic DNA isolation method was as described previously [38]. Briefly, genomic DNA was isolated by proteinase K digestion and phenol chloroform extraction from leukocytes isolated from venous blood. Genotyping was performed by Sequenom’s MassARRAY® iPLEX assay according to manufacturer’s instructions without knowing the status of case and control. We included duplicate samples (5%) for internal consistency and three water blanks as negative control in each reaction plate. Eight SNPs were successfully genotyped with call rate > 95% (Table 2).
Statistical analysis
We used χ2 test for categorical variables and Student's t-test for continuous variables to evaluate the differences between the cases and controls. HWE was evaluated using the goodness-of-fit χ2 test among controls. The association between genotypes and PCOS risk with or without adjustments for age and BMI was performed by calculating odds ratios (ORs) and their 95% confidence intervals (CIs) from logistic regression analyses. The statistical analysis was conducted using SPSS Statistics Version 17.0 or PLINK (http://www.cog-genomics.org/plink2/) software.
Abbreviations
- MAP
mitogen-activated protein
- PCOS
polycystic ovarian syndrome
- SNPs
single nucleotide polymorphisms
- BMI
body mass index
- FSH
follicle-stimulating hormone
- LH
luteinizing hormone
- E2
estradiol
- T
total testosterone
- GWAS
genome-wide association studies
- MAPKs
MAP kinases
- ERKs
extracellular signal-regulated kinases
Footnotes
Author contributions
Lingmin Hu and Yiting Zhang performed experiments; Lingmin Hu and Juan Wen analyzed results and prepared figures; Lingmin Hu, Li Chen and Wei Zhou designed the research and wrote the paper; Yong Wang and Juan Wen supervised the study.
CONFLICTS OF INTEREST
The authors declare that no conflicts of interest exist.
FUNDING
This study was supported by the Changzhou Natural Science Foundation (CJ20140021), National Natural Science Foundation of China (81402147, 81600685) Jiangsu Provincial Medical Youth Talent (QNRC2016304), the Natural Science Foundation of Jiangsu Province (BK20160141), and the Medical Science and technology development Foundation of Nanjing Department of Health (YKK16201).
REFERENCES
- 1.Franks S. Polycystic ovary syndrome. Arch Dis Child. 1997;77:89–90. doi: 10.1136/adc.77.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrmann DA. Polycystic ovary syndrome. N Engl J Med. 2005;352:1223–1236. doi: 10.1056/NEJMra041536. [DOI] [PubMed] [Google Scholar]
- 3.Escobar-Morreale HF, Asuncion M, Calvo RM, Sancho J, San Millan JL. Receiver operating characteristic analysis of the performance of basal serum hormone profiles for the diagnosis of polycystic ovary syndrome in epidemiological studies. Eur J Endocrinol. 2001;145:619–624. doi: 10.1530/eje.0.1450619. [DOI] [PubMed] [Google Scholar]
- 4.Heutling D, Schulz H, Randeva H, Dodt C, Lehnert H. [Polycystic ovary syndrome. Prototype of a cardio-metabolic syndrome]. [Article in German] Internist (Berl) 2007;48:144–153. doi: 10.1007/s00108-006-1776-7. [DOI] [PubMed] [Google Scholar]
- 5.Klein J, Craven M, Vuguin PM. Polycystic ovarian syndrome. Adolesc Med State Art Rev. 2015;26:326–342. [PubMed] [Google Scholar]
- 6.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Li Z, You L, Zhao J, Liu J, Liang X, Zhao X, Sun Y, Zhang B, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Zhao H, Cao Y, Yang D, Li Z, Zhang B, Liang X, Li T, Chen J, Shen J, Zhao J, You L, Gao X, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44:1020–1025. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 8.Whitmarsh AJ, Davis RJ. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med (Berl) 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 9.Vossler MR, Yao H, York RD, Pan MG, Rim CS, Stork PJ. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 10.Peti W, Page R. Molecular basis of MAP kinase regulation. Protein Sci. 2013;22:1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S, Maizels ET, DeManno D, St Clair E, Adam SA, Hunzicker-Dunn M. A stimulatory role of cyclic adenosine 3', 5'-monophosphate in follicle-stimulating hormone-activated mitogen-activated protein kinase signaling pathway in rat ovarian granulosa cells. Endocrinology. 1996;137:967–974. doi: 10.1210/endo.137.3.8603610. [DOI] [PubMed] [Google Scholar]
- 12.Tajima K, Dantes A, Yao Z, Sorokina K, Kotsuji F, Seger R, Amsterdam A. Down-regulation of steroidogenic response to gonadotropins in human and rat preovulatory granulosa cells involves mitogen-activated protein kinase activation and modulation of DAX-1 and steroidogenic factor-1. J Clin Endocrinol Metab. 2003;88:2288–2299. doi: 10.1210/jc.2002-020913. [DOI] [PubMed] [Google Scholar]
- 13.Nelson-Degrave VL, Wickenheisser JK, Hendricks KL, Asano T, Fujishiro M, Legro RS, Kimball SR, Strauss JF, 3rd, McAllister JM. Alterations in mitogen-activated protein kinase kinase and extracellular regulated kinase signaling in theca cells contribute to excessive androgen production in polycystic ovary syndrome. Mol Endocrinol. 2005;19:379–390. doi: 10.1210/me.2004-0178. [DOI] [PubMed] [Google Scholar]
- 14.Corbould A, Zhao H, Mirzoeva S, Aird F, Dunaif A. Enhanced mitogenic signaling in skeletal muscle of women with polycystic ovary syndrome. Diabetes. 2006;55:751–759. doi: 10.2337/diabetes.55.03.06.db05-0453. [DOI] [PubMed] [Google Scholar]
- 15.Hong KW, Jeong SW, Chung M, Cho SB. Association between expression quantitative trait loci and metabolic traits in two Korean populations. PLoS One. 2014;9:e114128. doi: 10.1371/journal.pone.0114128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajkumar AP, Christensen JH, Mattheisen M, Jacobsen I, Bache I, Pallesen J, Grove J, Qvist P, McQuillin A, Gurling HM, Tumer Z, Mors O, Borglum AD. Analysis of t(9;17)(q33.2;q25.3) chromosomal breakpoint regions and genetic association reveals novel candidate genes for bipolar disorder. Bipolar Disord. 2015;17:205–211. doi: 10.1111/bdi.12239. [DOI] [PubMed] [Google Scholar]
- 17.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lou J, Gong J, Ke J, Tian J, Zhang Y, Li J, Yang Y, Zhu Y, Gong Y, Li L, Chang J, Zhong R, Miao X. A functional polymorphism located at transcription factor binding sites, rs6695837 near LAMC1 gene, confers risk of colorectal cancer in Chinese populations. Carcinogenesis. 2017;38:177–183. doi: 10.1093/carcin/bgw204. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Akinsanmi I, Arafat D, Cradick TJ, Lee CM, Banskota S, Marigorta UM, Bao G, Gibson G. A burden of rare variants associated with extremes of gene expression in human peripheral blood. Am J Hum Genet. 2016;98:299–309. doi: 10.1016/j.ajhg.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SY, Hong MJ, Jeon HS, Choi YY, Choi JE, Kang HG, Jung DK, Jin C, Do SK, Yoo SS, Seok Y, Lee EB, Shin KM, et al. Functional intronic ERCC1 polymorphism from regulomeDB can predict survival in lung cancer after surgery. Oncotarget. 2015;6:24522–24532. doi: 10.18632/oncotarget.4083. https://doi.org/10.18632/oncotarget.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan CW, Chen MJ, Tai KY, Yu DC, Yang YC, Jan PS, Yang YS, Chen HF, Ho HN. Functional microarray analysis of differentially expressed genes in granulosa cells from women with polycystic ovary syndrome related to MAPK/ERK signaling. Sci Rep. 2015;5:14994. doi: 10.1038/srep14994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song XR, Zhang HY, Zhang YF, Han YK, Li KJ. [Extracellular signal-regulated protein kinase activation in endometrium with polycystic ovary syndrome and its significance]. [Article in Chinese] Zhonghua Fu Chan Ke Za Zhi. 2010;45:767–771. [PubMed] [Google Scholar]
- 24.Yamada Y, Yamamoto H, Yonehara T, Kanasaki H, Nakanishi H, Miyamoto E, Miyazaki K. Differential activation of the luteinizing hormone beta-subunit promoter by activin and gonadotropin-releasing hormone: a role for the mitogen-activated protein kinase signaling pathway in LbetaT2 gonadotrophs. Biol Reprod. 2004;70:236–243. doi: 10.1095/biolreprod.103.019588. [DOI] [PubMed] [Google Scholar]
- 25.Sun C, He M, Ko WK, Wong AO. Mechanisms for luteinizing hormone induction of growth hormone gene transcription in fish model: crosstalk of the cAMP/PKA pathway with MAPK-and PI3K-dependent cascades. Mol Cell Endocrinol. 2014;382:835–850. doi: 10.1016/j.mce.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 26.Patel SS, Beshay VE, Escobar JC, Suzuki T, Carr BR. Molecular mechanism for repression of 17alpha-hydroxylase expression and androstenedione production in granulosa cells. J Clin Endocrinol Metab. 2009;94:5163–5168. doi: 10.1210/jc.2009-1341. [DOI] [PubMed] [Google Scholar]
- 27.Ye DF, Ma HX, Mu WT, Lai MH, Liu H, Zheng YH, Ma WY. [Effective Ingredients of Yangjing Zhongyu Decoction Regulated Androgen Biosyntheses by Mitogen-Activated Protein Kinase Pathway in Porcine Granulose Cells]. [Article in Chinese] Zhongguo Zhong Xi Yi Jie He Za Zhi. 2015;35:847–853. [PubMed] [Google Scholar]
- 28.Matzuk MM, Spangler MM, Camel M, Suganuma N, Boime I. Mutagenesis and chimeric genes define determinants in the beta subunits of human chorionic gonadotropin and lutropin for secretion and assembly. J Cell Biol. 1989;109:1429–1438. doi: 10.1083/jcb.109.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katayama H, Murashima T, Saeki Y, Nishizawa Y. The pure anti-androgen bicalutamide inhibits cyclin A expression both in androgen-dependent and -independent cell lines. Int J Oncol. 2010;36:553–562. doi: 10.3892/ijo_00000529. [DOI] [PubMed] [Google Scholar]
- 30.Li WQ, Hu N, Wang Z, Yu K, Su H, Wang L, Wang C, Chanock SJ, Burdett L, Ding T, Qiao YL, Fan JH, Wang Y, et al. Genetic variants in epidermal growth factor receptor pathway genes and risk of esophageal squamous cell carcinoma and gastric cancer in a Chinese population. PLoS One. 2013;8:e68999. doi: 10.1371/journal.pone.0068999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, Dziembowski A, Jensen TH. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siddappa D, Beaulieu E, Gevry N, Roux PP, Bordignon V, Duggavathi R. Effect of the transient pharmacological inhibition of Mapk3/1 pathway on ovulation in mice. PLoS One. 2015;10:e0119387. doi: 10.1371/journal.pone.0119387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prochazka R, Blaha M. Regulation of mitogen-activated protein kinase 3/1 activity during meiosis resumption in mammals. J Reprod Dev. 2015;61:495–502. doi: 10.1262/jrd.2015-069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM, Richards JS. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science. 2009;324:938–941. doi: 10.1126/science.1171396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corbould A, Kim YB, Youngren JF, Pender C, Kahn BB, Lee A, Dunaif A. Insulin resistance in the skeletal muscle of women with PCOS involves intrinsic and acquired defects in insulin signaling. Am J Physiol Endocrinol Metab. 2005;288:E1047–1054. doi: 10.1152/ajpendo.00361.2004. [DOI] [PubMed] [Google Scholar]
- 36.Anjali G, Kaur S, Lakra R, Taneja J, Kalsey GS, Nagendra A, Shrivastav TG, Devi MG, Malhotra N, Kriplani A, Singh R. FSH stimulates IRS-2 expression in human granulosa cells through cAMP/SP1, an inoperative FSH action in PCOS patients. Cell Signal. 2015;27:2452–2466. doi: 10.1016/j.cellsig.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Guan KL. The mitogen activated protein kinase signal transduction pathway: from the cell surface to the nucleus. Cell Signal. 1994;6:581–589. doi: 10.1016/0898-6568(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhang CW, Zhang XL, Xia YJ, Cao YX, Wang WJ, Xu P, Che YN, Wu XK, Yi L, Gao Q, Wang Y. Association between polymorphisms of the CYP11A1 gene and polycystic ovary syndrome in Chinese women. Mol Biol Rep. 2012;39:8379–8385. doi: 10.1007/s11033-012-1688-7. [DOI] [PubMed] [Google Scholar]
- 39.Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004;19:41–47. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]