Abstract
The microRNAs are a group of 20 nucleotides-long non-coding RNAs. By binding to the 3’UTR region of target mRNA, microRNAs can perform extensive actions mediating gene expression at post-trancriptional stages. It makes microRNAs serve as very crucial regulators in various biological progress including carcinogenesis. Long non-coding RNAs, however, are a subgroup of RNA with the length of 200 nucleotides. Unlike microRNAs, long non-coding RNAs can form secondary of tertiary domain based on their length. With the ability of directly interacting with DNA, RNA, protein, long non-coding RNAs have promoting or inhibitive functions in gene expression regulation. Furthermore, the abnormal expression of certain long non-coding RNAs has roused people’s interest in the role of long non-coding RNAs in tumorigenesis. Although the connection between microRNA/long non-coding RNA and CCA has been a hot field to researchers, the link between molecular mechanism and clinical outcome has been barely built. This review takes a retrospect at the latest researches on the link between microRNA/long non-coding RNA and cholangiocarcinoma and the potential of microRNA/long non-coding RNA serving as distinctive biomarkers for CCA in clinical practice.
Keywords: miRNA, lncRNA, biomarker, cholangiocarcinoma, prediction
INTRODUCTION
Cholangiocarcinoma which can be anatomically divided into intrahepatic cholangiocarcinoma (ICC) and extrahepatic cholangiocarcinoma (ECC) accounts for 10% of primary hepatic tumors [1, 2]. Because of the highly molecular heterogeneity, CCA patients have been linked with bad prognosis and limited therapeutic regimens. Most of CCA are unresectable when discovered since it has progressed into advanced stages. So the improvement in the diagnostic method of CCA is urgent, especially in biomarkers. The classic serum CCA markers such as carbohydrate antigen CA-199 and CA-125 are now regarded as insensitive and unspecific. Since gene expression pattern is greatly changed in CCA background, it is of urgency to identify new epigenetic biomarkers such as miRNA and lncRNA for CCA patients [3].
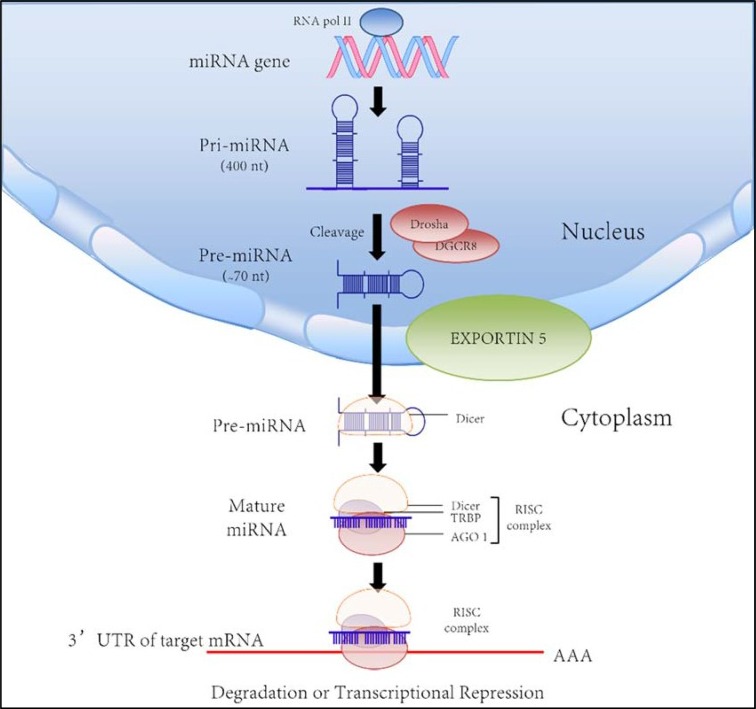
MicroRNAs (miRNAs) refer to a subgroup of small, noncoding RNA that mediate a series of biological events from controlling the growth of insects to deactivating X chromosome in mammals [4, 5]. The first miRNA is found in the nematode Caenorhabditis elegans, but soon researchers discovered that there are thousands of them existing in nearly every speices [6, 7]. However, the exact mechanism of their function is not fully understood yet. Originally, miRNAs derive from long primary miRNAs with several hundred nucleotides in length [8–10]. After cleavage and processing, the small molecule goes through 60–70 nt long precursor and finally becomes 20–25 nt long miRNAs duplexes [11–17]. Then miRNAs will form into RNA-induced silencing complex (RISC). The complex will degrade target mRNA through complementary binding, therefore affecting translational activity [18–21] (Figure 1). Furthermore, a single miRNA has the potential of targeting over hundreds of mRNAs and each mRNA can be targeted by different miRNAs as well which makes the interactive network between miRNA and mRNA even more complex.
Figure 1. Biogenesis and biological function of microRNA [44].
Although previous studies have confirmed that dysregulated miRNAs have a crucial role in tumors [22–26], the underlying mechanisms remain largely unknown. Some miRNAs like miR-21 [27, 28], miR-155 [29, 30] and miR17-92 cluster are recognized as tumor promoter [31, 32], some as tumor suppressors [33]. Although currently the therapeutic application of miRNAs in tumors is still limited, recently researchers have changed their focus to profiling miRNAs in tumor tissues so that miRNAs can be used as predictive, therapeutic or prognostic biomarkers in tumor patients. A research in 2006 first confirmed the biomarker role of miRNAs in CCA patients [34].
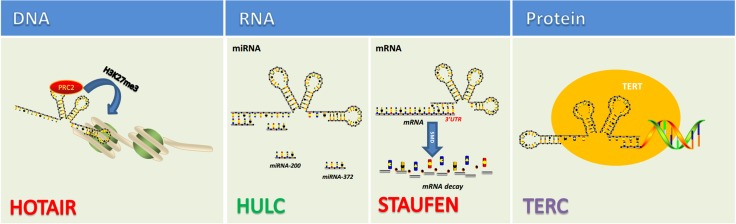
When first discovered, long non-coding RNA (lncRNA) was once called transcriptional noise. However, more and more researches have demonstrated that lncRNAs affect various biological processes including tumorigenic process. Just like mRNA, lncRNAs are transcribed by RNA polymerase II and also have the structural feature of 5′-7-methylguanosine cap and 3′-poly(A) tail [35–38] (Figure 2). The localization and expression of lncRNAs differ in different tumors [39]. In fact, the localization of lncRNA remains ambiguous as some proven existing in nucleus or cytoplasm and some in both [40]. The broad existence of lncRNAs enables them to engage in a wide range of biological processes from chromatin remodeling to translational mediation. To achieve these, lncRNAs have to interact directly with DNA, protein, mRNAs and miRNAs (Figure 3). According to the RNA-seq result of 7 paired ICC and paracancerous tissue, the expression of 230 lncRNAs and 2220 mRNAs are dysregulated in tumor tissues compared with adjacent normal tissues [41]. Among them, 597 mRNAs were targeted by 169 lncRNAs with 219 negatively correlated and 550 positively correlated [41]. Recently, lncRNAs have been found functioning as the downstream molecule of oncogenes and tumor suppressor genes [42]. In this case, lncRNAs can be classified into ‘OncoLncRNA’ and ‘TSLncRNA’ [42]. In a transcriptomic profiling, researchers found that 2148 lncRNAs significantly upregulated, 568 downregulated in ICC tissues which shows big promises to identify certain lncRNAs as biomarkers in ICC patients [43].
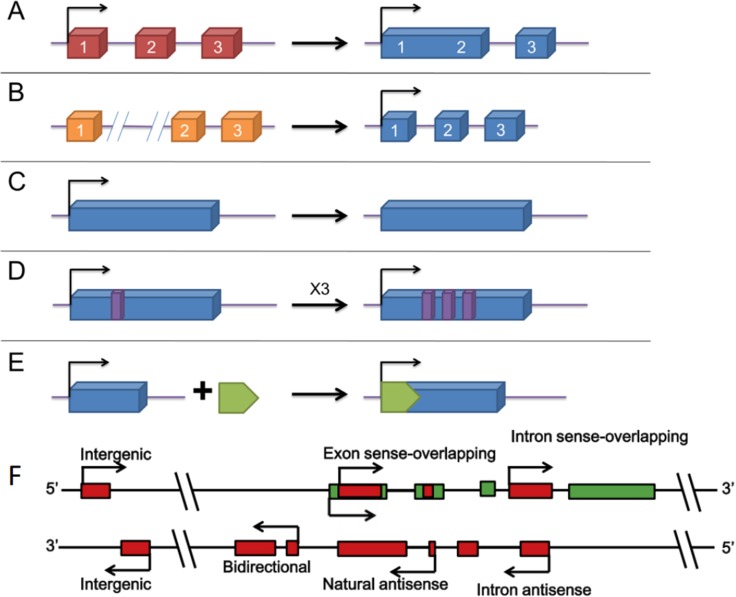
Figure 2. The origins of lncRNAs [87, 88].
(A–E) different modes of lncRNA formation. (A) a lncRNA is transformed from a protein-coding gene with structural damage; (B) two abreast non-transcribed regions generate a lncRNA after chromosomal rearrangement; (C) copy of a noncoding gene by retrotransposition forms a lncRNA without protein-coding ability; (D) a lncRNA with adjacent repeats derives from tandem duplication events; (E) a functional lncRNA with insertion of a transposon. (F) Different kinds of lncRNA genes encode different lncRNAs. Intergenic: a lncRNA gene lies as an independent unit within the genomic interval between two genes. Bidirectional: expression of a lncRNA gene and its neighboring coding transcript on the opposite strand is initiated in close genomic proximity. Intron sense-overlapping: a lncRNA gene lies in the intron of a protein-coding gene on the same strand. Exon sense-overlapping: a lncRNA gene lies in the exons of protein-coding gene on the same strand. Intronic-antisense: a lncRNA lies in the introns of protein-coding gene on the opposite strand in the same region. Natural-antisense: a lncRNA gene lies in the exons of protein-coding gene on the opposite strand.
Figure 3. lncRNAs function via directly interacting with DNA, RNA, protein [89, 90].
miRNA as biomarkers in CCA
Most of previous studies put their focus on the promoting or inhibitive role miRNAs might have in the tumorigenic process of malignant tumors such as CCA. However, more and more researchers have seen the potential of miRNAs serving as biomarkers of diagnosis, prediction, monitoring, therapy and prognosis for CCA patients. Cholangiocarcinoma hold the ability to release small molecules such as miRNAs into bloodstream or bile which further potentiate miRNAs to be the biomarker of CCA (Table 1).
Table 1. microRNAs as potential biomarkers for CCA.
| MicroRNA | Expression | Detectable location | Biomarker category | Sensitivity (%) | Specificity (%) | Target gene | Tumor type |
|---|---|---|---|---|---|---|---|
| 191 [49] | Upregulated | Tissue/serum | Diagnostic/Prognostic | _ | _ | TET1/P53 | ICC |
| 29a [56, 107] | Upregulated | Tissue | Prognostic | _ | _ | HDAC4 | ICC/ECC |
| 21 [34, 57, 108] | Upregulated | Plasm | Diagnostic/Prognostic/Chemoresistance | 87.8 | 90.5 | PTEN, PTPN12, PTPN14 | ICC |
| 221 [57] | Upregulated | Plasm | Diagnostic | _ | _ | DDIT4 | ICC |
| 150-5p [62, 109] | Downregulated | Serum/bile/tissue | Diagnostic | 93.3 | 53.5 | ELK1 | ICC/ECC |
| 122 [66, 110] | Downregulated | Tissue | Prognostic | _ | _ | P53 | ICC/ECC |
| 200a [71, 111] | Downregulated | Tissue | Prognostic | _ | _ | rho-kinase 2/SUZ12 | ICC/ECC combined with hepatolithiasis |
| 203 [72] | Downregulated | Tissue | Prognostic | _ | _ | _ | ICC/ECC |
| 26a [73, 112] | Upregulated | Serum | Diagnostic/Prognostic | 84.8 | 81.8 | Keratin 19 | CCA |
| 29b/205/221 [75] | Downregulated | Tissue | Chemoresistance | _ | _ | PIK3R1/MMP-2(29b) ErbB3/ VEGFA(205) PIK3R1(221) |
ICC/ECC |
| 151-3p/126 [77] | Downregulated(151-3p) Upregulated (126) |
Tissue | Prognostic | _ | _ | _ | Resected ICC/ECC |
| 590-3p [80] | Donwregulated | Serum | Diagnostic/Prognostic | _ | _ | SIP1 | ICC |
| 192/21 (combined) [84, 113] | Both upregulated | Urine | Diagnostic | 63.6(192) 63.6(21) 81.8(combined) |
66.7(192) 71.4(21) 71.4(combined) |
PDCD4 | O.viverrini-infected ICC/ECC |
| 192 [85] | Upregulated | Serum | prognostic | 74 | 72 | _ | O.viverrini-infected ICC/ECC |
| miRNA-483-5p,miRNA-505-3p, miRNA-874,miRNA-885-5p, miRNA-320b, miRNA-92b-3p, miRNA-1275,miRNA-1307-3p(panel)[86] | _ | Plasma | Diagnostic | _ | _ | _ | O.viverrini-infected ICC/ECC |
| 373 [104] | Downregulated | Tissue | Prognostic | _ | _ | MBD2 | ICC/ECC |
| miR-191, miR-486-3p, miR-1274b, miR-16 and miR-484(panel)[114] | _ | Bile | Diagnostic | 67 | 96 | _ | ICC/ECC |
| 34a [115] | Downregulated | Tissue | Prognostic | _ | _ | Smad4 | ECC |
| 224 [116] | Upregulated | Tissue | Prognostic | _ | _ | IL-6 | ICC/ECC |
| 204 [117, 118] | Downregulated | Tissue | Prognostic | _ | _ | Slug | ICC/ECC |
Abbreviations: TET1, ten-eleven translocation 1; HDAC4, Histone deacetylase 4;PTEN, phosphatase and tensin homolog;PTPN, protein tyrosine phosphatase;DDIT4, DNA-damage inducible transcript 4;ELK1, Ets including gene-1; PIK3R1, Phosphoinositide-3-kinase regulatory subunit 1;MMP-2, Matrix metalloproteinase 2;ErbB3, erythroblastic leukemia viral oncogene homolog 3;VEGFA, Vascular endothelial growth factor A;SIP1, Smad-interacting protein 1; PDCD4, programmed cell death 4;MBD2, Methyl CpG Binding Domain protein.
miRNA-191
MiRNA-191 has been found upregulated in various solid malignancies [45–48]. A recent research firstly discovered that the expression of miRNA-191 is increased in ICC tissues and subsequently promotes the proliferation, infiltration and metastasis of tumor cells [49]. According to the research, the miRNA-191-TET1(ten-eleven translocation 1)-P53 axis is held responsible for the pro-tumor role of miRNA-191 in ICC [49]. In a cohort study, miRNA-191 is identified as an independent risk factor of disease-free survival and OS (overall survival) in ICC [49]. It is also found that the high expression of miRNA-191 in tumor tissue is linked with more advanced tumor stage and decreased OS which clearly implies miRNA-191 can be used as a tissue biomarker for predicting the OS of ICC patients [49]. In addition, by using the GEO database (GSE59856), researchers surprisingly discovered that the serum level of miRNA-191 is significantly higher in ICC patients (98) than healthy people (150) which may shed light on using miRNA-191 as a predictive serum biomarker in ICC in the future [49].
miRNA-29a
The expression of miRNA-29a is found upregulated in myeloid leukemia, breast cancer, glioma and nasopharyneal carcinoma [50–52], while downregelated in pancreatic cancer, prostate cancer and gastric cancer [53–55]. The finding indicates that miRNA-29a might have a duel pro/anti-tumor function in human tumors. In CCA tissues, the expression of miRNA-29a is markedly increased [56]. Additionally the increased expression of miRNA-29a is closely associated with the differentiation, clinical stage, metatstasis of CCA [56]. By using statistic method as KaplanMeier survival analysis and Cox regression analysis, the overexpression of miRNA-29a is correlated with patients’ bad prognosis and miRNA-29a is considered as an independent risk factor in CCA [56]. Above all, the results indicate that miRNA-29a might become a prognostic tissue biomarker for CCA patients.
miRNA-21/221
Through RNA sequencing, researchers found that the expression of miRNA-21, miRNA221 is highly increased in ICC [57]. Notably, both of them can be detected in the plasma of ICC patients with a higher concentration than healthy people, indicating their use as diagnostic biomarker in ICC patients [57]. The expression level of miRNA-21 in the plasma of ICC patients is as 3 fold much as healthy people, emphasizing its diagnostic role in ICC [57]. Moreover, the increased expression of miRNA-21 is correlated with more invasive and metastatic behavior in cholangiocarcinoma cells [58]. This finding may indicate that miRNA-21 can also become a novel prognostic plasma biomarker for ICC, however, large scale clinical studies need to be performed to support this hypothesis. Like miRNA-21, the expression of miRNA-221 is significantly increased in ICC patients. Previous studies have shown the pro-tumor function miRNA-221 had in hepatocellularcarcinoma, bladder cancer [59, 60]. Interestingly, a recent research found out the anti-tumor role of miRNA-221 in lung cancer [61].
miRNA-150-5p
Microarray profiling results of normal tissue, PSC (primary sclerosing cholangitis, a major risk factor for CCA) and CCA show that the expression of miRNA-150-5p is significantly downregulated in CCA patients’ serum, bile and tissue [62]. Further experiments proved that the overexpression of miRNA-150-5p could block the proliferation and metastasis of tumor cells while the knockdown of miRNA-150-5p will lead to enhanced proliferation, invasion of tumor cells [62]. The expression of miRNA-150-5p is going through a dynamic change from normal tissue to PSC to CCA which means miRNA-150-5p may be able to inhibit the transition from chronic inflammation to CCA [62]. The increased level of CA19-9 is a clear diagnostic sign for ICC patients [63]. Current data indicates that there is a negative correlation existing between the expression level of miRNA-150-5p and the level of CA19-9 and the pathological stage of CCA [62]. According to this discovery, combined with CA19-9, miRNA-150-5p might serve as a predictive serum/bile/tissue biomarker for CCA in early detection and improve the overall prognosis for CCA patients.
miRNA-122
MiRNA-122 is the most abundant miRNA expressed in liver cells. Its function includes mediating lipid metabolism, hepatitis C virus replication, cell differentiation and hepatic metabolism [64]. The decreased expression of miRNA-122 can lead to dysregulated liver function by disrupting normal mitochondrial functions. It is found that miRNA-122 deficiency may be closely associated with bad prognosis of CCA patients [65]. Coincidentally, the expression of miRNA-122 is markedly lower in cholangiocarcinoma tissues than normal bile duct tissues [66]. Taken together, miRNA-122 may serve as a promising prognostic tissue biomarker for CCA patients in the future.
miRNA-200a
The expression of miRNA-200a is downregulated in a wide range of malignancies and further experiments demonstrated that miRNA-200a has an inhibitory role in many kinds of tumors [67]. Further researches confirmed miRNA-200a deficiency is correlated with the progression of certain types of cancers [67–70]. In ICC patients complicated with hepatolithiasis the expression of miRNA-200a is significantly downregulated in CCA tissues compared with normal bile duct tissues and simple hepatolithiasis tissues [71]. Moreover, the expression level of miRNA-200a is correlated with pathological stage and metastasis of CCA which makes miRNA-200a a potential prognostic tissue biomarker in CCA [71].
miRNA-203
According to a previous study, the expression of miRNA-203 is significantly downregulated in CCA tissues compared with paracancerous tissue [72]. Combined with clinical data, further analysis shows that the expression level of miRNA-203 is closely associated with the size of tumor, differentiation of tumor cells and clinical stage [72]. Hence the miRNA-203 expression level might be a positive factor determining the prognosis of CCA patients [72]. Furthermore, the overall survival rate is much lower in miRNA-203 underexpression group than miRNA-203 overexpression group [72]. According to a multivariate Cox model, miRNA-203 is found to be an independent prognostic factor for CCA patients [72]. In conclusion, these findings suggest that miRNA-203 is a potential prognostic tissue biomarker for CCA.
miRNA-26a
The expression level of miRNA-26a in serum is significantly upregulated in CCA patients than healthy people and closely associated with tumor stage [73]. In order to determine the potential of miRNA-26a in the early diagnosis of CCA, researchers compare the AUC curve of miRNA-26a with that of the classic biomarker CA19-9. Surprisingly, the AUC value of miRNA-26a is higher, suggesting the diagnostic power of miRNA-26a in the early diagnosis of CCA [73]. Furthermore, clinical data demonstrates that there is a clear correlation between miRNA-26a and adverse clinicopathological factors, progression-free and OS rate of CCA patients [73]. Through multivariate analysis, miRNA-26a is identified as an independent prognostic factor in CCA [73]. Taken together, miRNA-26a has the potential of serving as both diagnostic and prognostic serum biomarker in CCA patients.
miRNA-29b/205/221
Gemcitabine (Gem; 2′,2′-difluorodeoxycytidine, dFdC) is a cytidine analogue and currently administered as a chemotherapy drug against cholangiocarcinoma [74]. MiRNA-29b and miRNA-221 are found downregulated in Gem-resistant CCA cell lines and ectopic increase of their expression can restore chemosensitivity of CCA cells towards Gemcitabine [75]. Furthermore, it is found that the overexpression of miRNA205 might grant chemosensitivity towards Gemcitabin to innately Gem-resistant cholangiocarcinoma cells [75]. Taken together, miRNA-29a/205/221 show promises serving as tissue biomarkers of chemosensitivity in CCA patients. Furthermore, trabectedin, an anti-metastatic agent in CCA has been found to inhibit metastasis via downregulating the expression of gene SYK and LGALS1 [76]. These two genes are the direct targets of certain miRNAs such as MiR-21-3p, miR-21-5p and miR-31-3p and miR-1207-5p, miR-1225-5p separately, suggesting the overexpression of these miRNAs is closely associated with the effectiveness of trabectedin [76].
miRNA-151-3p/126
Resection is the only curative treatment for CCA but the post-surgery survival rate is not promising. By analyzing the RNA extracted from resected CCA tissue, 43 miRNA were found significantly dysregulated in tumor tissue than adjacent normal tissue [77]. Of these dysregulated miRNAs, miRNA-151-3p and miRNA-126 were closely associated with post-surgery survival rate though the expression level of them are not correlated with the clinicalpathologic changes [77]. The upregulation of miRNA-151-3p and downregulation of miRNA-126 confer a better prognosis with median survival of 58.7 months than others with median survival of 15.1 months [77]. Above all, miRNA-151-3p and miRNA-126 might serve as a prognostic tissue biomarker for CCA patients who had undergone tumor resection.
miRNA-590-3p
miRNA-590-3p has been found downregulated in various malignancies [78]. In bladder cancer cell lines, the upregulation of miRNA-590-3P has been associated with increased tumor invasiveness [79]. A recent study revealed that miRNA-590-3p is also downregulated in serum, tissue, cell lines of ICC, especially in those with metastatic potential [80]. Further researches confirmed that miRNA-590-3p ameliorated the process of epithelial-to-mesenchymal transition (EMT) by targeting EMT-activator Smad-interacting protein 1(SIP1) [80]. By using AUC curves and Cox proportional hazards mode, the serum level of miRNA-590-3p is identified as an independent diagnostic and prognostic serum biomarker for ICC patients [80]. Intriguingly in another study, miRNA-590-3p is found upregulated in hepatocellular carcinoma (HCC), accelerating tumorigenic process via targeting tumor suppressor gene PDCD4 and PTEN [81].
miRNA and Opisthorchis viverrini-associated ICC
The incidence of ICC is higher in Southeast Asia than any other places in the world, where chronic liver fluke Opisthorchis viverrini (OV) infection has a major contributing role in the disease [82]. Moreover, the miRNA signatures are found unique in the subtypes of ICC with tumorigenic infection of OV [83].
In urine sample, miRNA-192 and miRNA-21 were found significantly higher in O. viverrini-infected patients with periductal fibrosis (PDF) and CCA than healthy people [84]. However, the expression level of miRNA-21 in urine is upregulated in patients with other inflammatory diseases as well [84]. Hence serving as a urine biomarker, miRNA-21 might not be specific enough. For this reason, the detection of miRNA-192 and miRNA-21 are combined together to enhance their diagnostic power. The result shows that the combination markedly increased the sensitivity and specificity in differentiation between O. viverrini-infected patients with PDF and CCA and healthy people [84]. The level of miRNA-192 is found higher in serum samples of OV-associated CCA as well [85]. The high level of miRNA-192 expression is positively correlated with lymph node metastasis and shorter OS [85].
In a 2015 research, OV-induced ICC tumor tissues, distal non-tumor tissues and normal non-tumor tissues from 14 CCA patients and normal tissues from healthy donors were analysed for miRNA expression level by using small RNA-Seq. The profiling result was then confirmed by performing quantitative PCR on paired plasma samples. 67 miRNAs are dysregulated with 35 upregulated and 32 downregulated in ICC tissues compared with distal non-tumor tissues and the dysregulated miRNA number reaches 316 with 144 upregulated and 172 downregulated when compared with normal non-tumor tissues [86]. Since OV-induced ICC shows distinctive challenges in early diagnosis and prognostic prediction, applicable biomarkers for early diagnosis in blood is urgently needed. Interestingly, out of all dysregulated miRNAs 8 miRNAs (miRNA-483-5p, miRNA-505-3p, miRNA-874, miRNA-885-5p, miRNA-320b, miRNA-92b-3p, miRNA-1275, miRNA-1307-3p) are found only detectable in the plasma of ICC patients [86]. This 8 miRNA panel might serve as a novel circulating miRNA-based biomarker for OV-induced ICC patients.
lncRNA as biomarkers in CCA
LncRNAs have been found holding multiple biological functions ranging from cellular proliferation to cellular apoptosis [91]. Moreover, a close connection between dysregulated lncRNA expression and the prognosis of tumor patients has been discovered [92].
LncRNA AFAP1-AS1
LncRNA AFAP1-AS1 is transcribed from the antisense DNA strand of gene AFAP1. Previous studies have demonstrated that AFAP1-AS1 is correlated with malignant behavior such as invasion and metastasis in hepatocellular carcinoma and nasopharyngeal carcinoma [93–95]. A recent study first discovered that the expression level of AFAP1-AS1 is significantly higher in CCA tissue than in paracancerous tissue and the same result was detected in CCA cell lines Hucct and normal biliary tract cells [96]. Further experiments showed that the underexpression of AFAP1-AS1 would restrict the proliferative and metastatic potential of CCA cell lines, Hucct and TFK-1 which confirmed that AFAP1-AS1 has a oncogenic function in cholangiocarcinoma [96]. Taken together, lncRNA AFAP1-AS1 might become a promising diagnostic and prognostic tissue biomarker for CCA.
LncRNA CCAT1
LncRNA CCAT1(colon cancer associated transcript 1) was firstly found overexpressed in colorectal cancer, accelerating the tumorigenic process [97]. It is also demonstrated that the overexpression of CCAT1 is correlated with the occurrence, progression, chemoresistance of tumor cells [98–100]. A recent study showed that the expression level of CCAT1 is markedly higher in cholangiocarcinoma tissue than in paracancerous normal tissue [101]. Furthermore, the upregulation of CCAT1 might lead to bad pathological characteristics, lymph node metastasis and more advanced tumor stage [101]. Statistical data shows that the OS rate of lncRNA CCAT1-overexpressed patients is significantly lower than underexpressed patients and by using multivariate and ROC analysis, lncRNA CCAT1 is found to be an independent prognostic factor for CCA [101]. Taken together, lncRNA CCAT1 can serve as a prognostic tissue biomarker in CCA patients.
LncRNA NEAT-1
BRCA-1 associated protein-1(BAP1) is identified as an anti-tumor factor and found participating in various cellular processes by interacting with other proteins. However, low BAP1 expressing CCA cell lines exhibit higher sensitivity to gemcitabine and cisplatin with lower IC50 [102]. Further studies demonstrated that lncRNA NEAT-1 is a down-stream molecule of BAP1 in treatment response [102]. In NEAT-1 knock-down CCA cell lines, the cytotoxicity of gemcitabine is significantly higher than control group which implies lncRNA NEAT-1 might serve as a chemosensitivity tissue biomarker for CCA.
LncRNA MALAT1
The expression of lncRNA MALAT1 is upregulated in various kinds of cancers such as liver, uterus, lung, breast, prostate, pancreas and cervix [103]. Further studies confirmed that MALAT1 is an independent prognostic factor in some cancers as well [103]. In CCA especially hilar cholangiocarcinoma (HCCA), the expression level of MALAT1 is much higher than paracancerous tissue [104]. In vitro and vivo experiments, MALAT1 exhibits a pro-tumor function in the proliferation, invasion and migration of CCA cells [104]. Furthermore, overexpression of MALAT1 is correlated with lower OS rate, worse TNM stage, larger tumor size and metastasis in HCCA patients [104]. Taken together, lncRNA MALAT1 is a very promising novel prognostic tissue biomarker in HCCA.
LncRNA CPS1-IT1
A 2015 study for the first time discovered that Carbamoyl-phosphate synthase 1 (CPS1) and its lncRNA CPS1 intronic transcript 1 (CPS1-IT1) are co-upregulated at the same time in ICC tissue compared with paracancerous normal tissue [105]. The study suggested that the overexpression of CPS1 and CPS1-IT1 is correlated with increased CA19-9 positivity and lymph node metastasis [105]. Further experiment confirmed that the upregulation of CPS1 and CPS1-IT1 has a negative impact on the OS rate for ICC patients [105]. In conclusion, current study suggests that CPS1-IT1 might serve as a prognostic tissue biomarker for ICC patients.
CONCLUSIONS AND PROSPECTIVE
CCA has a bad prognosis and is still ranked as one of the most lethal malignant diseases. In recent years, many studies have uncovered the connections between miRNA/lncRNA and the cholangiocarcinoma in tumorigenic process and tumor progression, some of which suggested that miRNAs and lncRNAs might serve as novel biomarkers for CCA patients. Although in some studies, the sensitivity and specificity of these novel biomarkers are even better than the most widely used CA19-9, the clinical data supporting these findings are not big enough to be convincing. So in the next step of developing novel biomarkers, researchers should increase the number of their clinical candidates to make their discovery more statistically convincing. In conclusion, in the future miRNAs and lncRNAs may become very promising biomarkers for CCA in early diagnosis, treatment response and prognosis prediction.
ACKNOWLEDGMENTS AND FUNDING
This study was supported by the National Natural Science Foundation of China (81472243 and 81670598), the Key Joint Research Program of Shanghai Health Bureau (2013ZYJB0001).
Footnotes
CONFLICTS OF INTEREST
The authors declared no competing financial interests exist.
REFERENCES
- 1.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366:1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- 2.Hong K, Geschwind JF. Locoregional intra-arterial therapies for unresectable intrahepatic cholangiocarcinoma. Semin Oncol. 2010;37:110–117. doi: 10.1053/j.seminoncol.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Obama K, Ura K, Li M, Katagiri T, Tsunoda T, Nomura A, Satoh S, Nakamura Y, Furukawa Y. Genome-wide analysis of gene expression in human intrahepatic cholangiocarcinoma. Hepatology. 2005;41:1339–1348. doi: 10.1002/hep.20718. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 5.Mlynarczyk SK, Panning B. X inactivation: Tsix and Xist as yin and yang. Curr Biol. 2000;10:R899–903. doi: 10.1016/s0960-9822(00)00847-2. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 13.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 16.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 17.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 24.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 27.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 29.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 32.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 35.Zong X, Tripathi V, Prasanth KV. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, Lam WL. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo X, Gao L, Wang Y, Chiu DK, Wang T, Deng Y. Advances in long noncoding RNAs: identification, structure prediction and function annotation. Brief Funct Genomics. 2016;15:38–46. doi: 10.1093/bfgp/elv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang R, Xia LQ, Lu WW, Zhang J, Zhu JS. LncRNAs and cancer. Oncol Lett. 2016;12:1233–1239. doi: 10.3892/ol.2016.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W, Li Y, Song X, Xu J, Xie J. Genome-wide analysis of long noncoding RNA and mRNA co-expression profile in intrahepatic cholangiocarcinoma tissue by RNA sequencing. Oncotarget. 2017;8:26591–26599. doi: 10.18632/oncotarget.15721. https://doi.org/10.18632/oncotarget.15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao AK, Rajkumar T, Mani S. Perspectives of long non-coding RNAs in cancer. Mol Biol Rep. 2017;44:203–218. doi: 10.1007/s11033-017-4103-6. [DOI] [PubMed] [Google Scholar]
- 43.Lv L, Wei M, Lin P, Chen Z, Gong P, Quan Z, Tang Z. Integrated mRNA and lncRNA expression profiling for exploring metastatic biomarkers of human intrahepatic cholangiocarcinoma. Am J Cancer Res. 2017;7:688–699. [PMC free article] [PubMed] [Google Scholar]
- 44.Pisarello MJ, Loarca L, Ivanics T, Morton L, LaRusso N. MicroRNAs in the Cholangiopathies: Pathogenesis, Diagnosis, and Treatment. J Clin Med. 2015;4:1688–1712. doi: 10.3390/jcm4091688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagpal N, Ahmad HM, Molparia B, Kulshreshtha R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis. 2013;34:1889–1899. doi: 10.1093/carcin/bgt107. [DOI] [PubMed] [Google Scholar]
- 46.Qin S, Zhu Y, Ai F, Li Y, Bai B, Yao W, Dong L. MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma. 2014;61:27–34. [PubMed] [Google Scholar]
- 47.Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, Dov A, Marcusson EG, Bennett CF, Chajut A, Cohen D, Yerushalmi N. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 2010;70:8077–8087. doi: 10.1158/0008-5472.CAN-10-1313. [DOI] [PubMed] [Google Scholar]
- 48.Shen J, DiCioccio R, Odunsi K, Lele SB, Zhao H. Novel genetic variants in miR-191 gene and familial ovarian cancer. BMC Cancer. 2010;10:47. doi: 10.1186/1471-2407-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Zhou ZQ, Yang ZR, Tong DN, Guan J, Shi BJ, Nie J, Ding XT, Li B, Zhou GW, Zhang ZY. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology. 2017;66:136–151. doi: 10.1002/hep.29116. [DOI] [PubMed] [Google Scholar]
- 50.Zhong S, Li W, Chen Z, Xu J, Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene. 2013;531:8–14. doi: 10.1016/j.gene.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 51.Qiu F, Sun R, Deng N, Guo T, Cao Y, Yu Y, Wang X, Zou B, Zhang S, Jing T, Ling T, Xie J, Zhang Q. miR-29a/b enhances cell migration and invasion in nasopharyngeal carcinoma progression by regulating SPARC, COL3A1 gene expression. PLoS One. 2015;10:e0120969. doi: 10.1371/journal.pone.0120969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu C, Wang Y, Kuai W, Sun X, Chen H, Hong Z. Prognostic value of miR-29a expression in pediatric acute myeloid leukemia. Clin Biochem. 2013;46:49–53. doi: 10.1016/j.clinbiochem.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Cui Y, Su WY, Xing J, Wang YC, Wang P, Chen XY, Shen ZY, Cao H, Lu YY, Fang JY. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025872. e25872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trehoux S, Lahdaoui F, Delpu Y, Renaud F, Leteurtre E, Torrisani J, Jonckheere N, Van Seuningen I. Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells. Biochim Biophys Acta. 2015;1853:2392–′2403. doi: 10.1016/j.bbamcr.2015.05.033. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Kong D, Ahmad A, Bao B, Dyson G, Sarkar FH. Epigenetic deregulation of miR-29a and miR-1256 by isoflavone contributes to the inhibition of prostate cancer cell growth and invasion. Epigenetics. 2012;7:940–949. doi: 10.4161/epi.21236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng Y, Chen Y. Increased Expression of miR-29a and Its Prognostic Significance in Patients with Cholangiocarcinoma. Oncol Res Treat. 2017;40:128–132. doi: 10.1159/000455869. [DOI] [PubMed] [Google Scholar]
- 57.Correa-Gallego C, Maddalo D, Doussot A, Kemeny N, Kingham TP, Allen PJ, D’Angelica MI, DeMatteo RP, Betel D, Klimstra D, Jarnagin WR, Ventura A. Circulating Plasma Levels of MicroRNA-21 and MicroRNA-221 Are Potential Diagnostic Markers for Primary Intrahepatic Cholangiocarcinoma. PLoS One. 2016;11:e0163699. doi: 10.1371/journal.pone.0163699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang Q, Liu L, Liu CH, You H, Shao F, Xie F, Lin XS, Hu SY, Zhang CH. MicroRNA-21 regulates the invasion and metastasis in cholangiocarcinoma and may be a potential biomarker for cancer prognosis. Asian Pac J Cancer Prev. 2013;14:829–834. doi: 10.7314/apjcp.2013.14.2.829. [DOI] [PubMed] [Google Scholar]
- 59.Fu B, Wang Y, Zhang X, Lang B, Zhou X, Xu X, Zeng T, Liu W, Guo J, Wang G. MiR-221-induced PUMA silencing mediates immune evasion of bladder cancer cells. Int J Oncol. 2015;46:1169–1180. doi: 10.3892/ijo.2015.2837. [DOI] [PubMed] [Google Scholar]
- 60.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamashita R, Sato M, Kakumu T, Hase T, Yogo N, Maruyama E, Sekido Y, Kondo M, Hasegawa Y. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med. 2015;4:551–564. doi: 10.1002/cam4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu X, Xia M, Chen D, Wu F, Lv Z, Zhan Q, Jiao Y, Wang W, Chen G, An F. Profiling of downregulated blood-circulating miR-150-5p as a novel tumor marker for cholangiocarcinoma. Tumour Biol. 2016;37:15019–15029. doi: 10.1007/s13277-016-5313-6. [DOI] [PubMed] [Google Scholar]
- 63.Patel AH, Harnois DM, Klee GG, LaRusso NF, Gores GJ. The utility of CA 19-9 in the diagnoses of cholangiocarcinoma in patients without primary sclerosing cholangitis. Am J Gastroenterol. 2000;95:204–207. doi: 10.1111/j.1572-0241.2000.01685.x. [DOI] [PubMed] [Google Scholar]
- 64.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, Smith R, Leeson B, Beard R, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu N, Jiang F, He TL, Zhang JK, Zhao J, Wang C, Jiang GX, Cao LP, Kang PC, Zhong XY, Lin TY, Cui YF. The Roles of MicroRNA-122 Overexpression in Inhibiting Proliferation and Invasion and Stimulating Apoptosis of Human Cholangiocarcinoma Cells. Sci Rep. 2015;5:16566. doi: 10.1038/srep16566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472–6498. doi: 10.18632/oncotarget.3052. https://doi.org/10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li MY, Hu XX. Meta-analysis of microRNA expression profiling studies in human cervical cancer. Med Oncol. 2015;32:510. doi: 10.1007/s12032-015-0510-5. [DOI] [PubMed] [Google Scholar]
- 69.Haldrup C, Kosaka N, Ochiya T, Borre M, Hoyer S, Orntoft TF, Sorensen KD. Profiling of circulating microRNAs for prostate cancer biomarker discovery. Drug Deliv Transl Res. 2014;4:19–30. doi: 10.1007/s13346-013-0169-4. [DOI] [PubMed] [Google Scholar]
- 70.Kolesnikoff N, Attema JL, Roslan S, Bert AG, Schwarz QP, Gregory PA, Goodall GJ. Specificity protein 1 (Sp1) maintains basal epithelial expression of the miR-200 family: implications for epithelial-mesenchymal transition. J Biol Chem. 2014;289:11194–11205. doi: 10.1074/jbc.M113.529172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen C, Yang D, Wang Q, Wang X. Expression and Clinical Pathological Significance of miR-200a in Concurrent Cholangiocarcinoma Associated with Hepatolithiasis. Med Sci Monit. 2015;21:3585–3590. doi: 10.12659/MSM.895013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J, Gao B, Huang Z, Duan T, Li D, Zhang S, Zhao Y, Liu L, Wang Q, Chen Z, Cheng K. Prognostic significance of microRNA-203 in cholangiocarcinoma. Int J Clin Exp Pathol. 2015;8:9512–9516. [PMC free article] [PubMed] [Google Scholar]
- 73.Wang LJ, Zhang KL, Zhang N, Ma XW, Yan SW, Cao DH, Shi SJ. Serum miR-26a as a diagnostic and prognostic biomarker in cholangiocarcinoma. Oncotarget. 2015;6:18631–18640. doi: 10.18632/oncotarget.4072. https://doi.org/10.18632/oncotarget.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gesto DS, Cerqueira NM, Fernandes PA, Ramos MJ. Gemcitabine: a critical nucleoside for cancer therapy. Curr Med Chem. 2012;19:1076–1087. doi: 10.2174/092986712799320682. [DOI] [PubMed] [Google Scholar]
- 75.Okamoto K, Miyoshi K, Murawaki Y. miR-29b, miR-205 and miR-221 enhance chemosensitivity to gemcitabine in HuH28 human cholangiocarcinoma cells. PLoS One. 2013;8:e77623. doi: 10.1371/journal.pone.0077623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peraldo Neia C, Cavalloni G, Chiorino G, Ostano P, Aglietta M, Leone F. Gene and microRNA modulation upon trabectedin treatment in a human intrahepatic cholangiocarcinoma paired patient derived xenograft and cell line. Oncotarget. 2016;7:86766–86780. doi: 10.18632/oncotarget.13575. https://doi.org/10.18632/oncotarget.13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McNally ME, Collins A, Wojcik SE, Liu J, Henry JC, Jiang J, Schmittgen T, Bloomston M. Concomitant dysregulation of microRNAs miR-151-3p and miR-126 correlates with improved survival in resected cholangiocarcinoma. HPB (Oxford) 2013;15:260–264. doi: 10.1111/j.1477-2574.2012.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Backes C, Meese E, Lenhof HP, Keller A. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res. 2010;38:4476–4486. doi: 10.1093/nar/gkq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mo M, Peng F, Wang L, Peng L, Lan G, Yu S. Roles of mitochondrial transcription factor A and microRNA-590-3p in the development of bladder cancer. Oncol Lett. 2013;6:617–623. doi: 10.3892/ol.2013.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zu C, Liu S, Cao W, Liu Z, Qiang H, Li Y, Cheng C, Ji L, Li J. MiR-590-3p suppresses epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma by inhibiting SIP1 expression. Oncotarget. 2017;8:34698–34708. doi: 10.18632/oncotarget.16150. https://doi.org/10.18632/oncotarget.16150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang H, Zheng W, Zhao W, Guan C, An J. Roles of miR-590-5p and miR-590-3p in the development of hepatocellular carcinoma. [Article in Chinese] Nan Fang Yi Ke Da Xue Xue Bao. 2013;33:804–811. [PubMed] [Google Scholar]
- 82.Yongvanit P, Pinlaor S, Bartsch H. Oxidative and nitrative DNA damage: key events in opisthorchiasis-induced carcinogenesis. Parasitol Int. 2012;61:130–135. doi: 10.1016/j.parint.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 83.Plieskatt JL, Rinaldi G, Feng Y, Peng J, Yonglitthipagon P, Easley S, Laha T, Pairojkul C, Bhudhisawasdi V, Sripa B, Brindley PJ, Mulvenna JP, Bethony JM. Distinct miRNA signatures associate with subtypes of cholangiocarcinoma from infection with the tumourigenic liver fluke Opisthorchis viverrini. J Hepatol. 2014;61:850–858. doi: 10.1016/j.jhep.2014.05.035. [DOI] [PubMed] [Google Scholar]
- 84.Silakit R, Loilome W, Yongvanit P, Thongchot S, Sithithaworn P, Boonmars T, Koonmee S, Titapun A, Khuntikeo N, Chamadol N, Techasen A, Namwat N. Urinary microRNA-192 and microRNA-21 as potential indicators for liver fluke-associated cholangiocarcinoma risk group. Parasitol Int. 2015;66:479–485. doi: 10.1016/j.parint.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 85.Silakit R, Loilome W, Yongvanit P, Chusorn P, Techasen A, Boonmars T, Khuntikeo N, Chamadol N, Pairojkul C, Namwat N. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: a prospective prognostic indicator. J Hepatobiliary Pancreat Sci. 2014;21:864–872. doi: 10.1002/jhbp.145. [DOI] [PubMed] [Google Scholar]
- 86.Plieskatt J, Rinaldi G, Feng Y, Peng J, Easley S, Jia X, Potriquet J, Pairojkul C, Bhudhisawasdi V, Sripa B, Brindley PJ, Bethony J, Mulvenna J. A microRNA profile associated with Opisthorchis viverrini-induced cholangiocarcinoma in tissue and plasma. BMC Cancer. 2015;15:309. doi: 10.1186/s12885-015-1270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen R, Wang G, Zheng Y, Hua Y, Cai Z. Long non-coding RNAs in osteosarcoma. Oncotarget. 2017;8:20462–20475. doi: 10.18632/oncotarget.14726. https://doi.org/10.18632/oncotarget.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wei MM, Zhou GB. Long Non-coding RNAs and Their Roles in Non-small-cell Lung Cancer. Genomics Proteomics Bioinformatics. 2016;14:280–288. doi: 10.1016/j.gpb.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 90.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feng S, Yao J, Chen Y, Geng P, Zhang H, Ma X, Zhao J, Yu X. Expression and Functional Role of Reprogramming-Related Long Noncoding RNA (lincRNA-ROR) in Glioma. J Mol Neurosci. 2015;56:623–630. doi: 10.1007/s12031-014-0488-z. [DOI] [PubMed] [Google Scholar]
- 92.Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, Xia R, Shu YQ, De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y, Gao W, Zheng S, Zhao X, Chen T, Chen R. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13:137. doi: 10.1186/s12967-015-0490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao L, Zhang S. Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. Tumour Biol. 2016;37:9699–9707. doi: 10.1007/s13277-016-4858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q, Chen P, Shi L, Lian Y, Jing Y, Tang K, Li Z, Zhou Y, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404–20418. doi: 10.18632/oncotarget.4057. https://doi.org/10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi X, Zhang H, Wang M, Xu X, Zhao Y, He R, Zhang M, Zhou M, Li X, Peng F, Shi C, Shen M, Wang X, et al. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget. 2017;8:58394–58404. doi: 10.18632/oncotarget.16880. https://doi.org/10.18632/oncotarget.16880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A, Dayanc BE, Ritter G, Gomceli I, Bostanci EB, Akoglu M, Chen YT, et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130:1598–1606. doi: 10.1002/ijc.26170. [DOI] [PubMed] [Google Scholar]
- 98.Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C, Lu J. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:5427–5434. [PMC free article] [PubMed] [Google Scholar]
- 99.Chen L, Wang W, Cao L, Li Z, Wang X. Long Non-Coding RNA CCAT1 Acts as a Competing Endogenous RNA to Regulate Cell Growth and Differentiation in Acute Myeloid Leukemia. Mol Cells. 2016;39:330–336. doi: 10.14348/molcells.2016.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen J, Zhang K, Song H, Wang R, Chu X, Chen L. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget. 2016;7:62474–62489. doi: 10.18632/oncotarget.11518. https://doi.org/10.18632/oncotarget.11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang XM, Li ZL, Li JL, Zheng WY, Li XH, Cui YF, Sun DJ. LncRNA CCAT1 as the unfavorable prognostic biomarker for cholangiocarcinoma. Eur Rev Med Pharmacol Sci. 2017;21:1242–1247. [PubMed] [Google Scholar]
- 102.Parasramka M, Yan IK, Wang X, Nguyen P, Matsuda A, Maji S, Foye C, Asmann Y, Patel T. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol Cancer. 2017;16:22. doi: 10.1186/s12943-017-0587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 104.Tan X, Huang Z, Li X. Long Non-Coding RNA MALAT1 Interacted with miR-204 to Modulates Human Hilar Cholangiocarcinoma Proliferation, Migration and Invasion by Targeting CXCR4. J Cell Biochem. 2017 doi: 10.1002/jcb.25862. [DOI] [PubMed] [Google Scholar]
- 105.Ma SL, Li AJ, Hu ZY, Shang FS, Wu MC. Coexpression of the carbamoylphosphate synthase 1 gene and its long noncoding RNA correlates with poor prognosis of patients with intrahepatic cholangiocarcinoma. Mol Med Rep. 2015;12:7915–7926. doi: 10.3892/mmr.2015.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang H, Li C, Jian Z, Ou Y, Ou J. TGF-beta1 Reduces miR-29a Expression to Promote Tumorigenicity and Metastasis of Cholangiocarcinoma by Targeting HDAC4. PLoS One. 2015;10:e0136703. doi: 10.1371/journal.pone.0136703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang LJ, He CC, Sui X, Cai MJ, Zhou CY, Ma JL, Wu L, Wang H, Han SX, Zhu Q. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget. 2015;6:5932–5946. doi: 10.18632/oncotarget.3465. https://doi.org/10.18632/oncotarget.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liang Z, Liu X, Zhang Q, Wang C, Zhao Y. Diagnostic value of microRNAs as biomarkers for cholangiocarcinoma. Dig Liver Dis. 2016;48:1227–1232. doi: 10.1016/j.dld.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 109.Wu C, Zhang J, Cao X, Yang Q, Xia D. Effect of Mir-122 on Human Cholangiocarcinoma Proliferation, Invasion, and Apoptosis Through P53 Expression. Med Sci Monit. 2016;22:2685–2690. doi: 10.12659/MSM.896404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peng F, Jiang J, Yu Y, Tian R, Guo X, Li X, Shen M, Xu M, Zhu F, Shi C, Hu J, Wang M, Qin R. Direct targeting of SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma tumourigenesis and metastasis. Br J Cancer. 2013;109:3092–3104. doi: 10.1038/bjc.2013.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang P, Lv L. miR-26a induced the suppression of tumor growth of cholangiocarcinoma via KRT19 approach. Oncotarget. 2016;7:81367–81376. doi: 10.18632/oncotarget.13229. https://doi.org/10.18632/oncotarget.13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chusorn P, Namwat N, Loilome W, Techasen A, Pairojkul C, Khuntikeo N, Dechakhamphu A, Talabnin C, Chan-On W, Ong CK, Teh BT, Yongvanit P. Overexpression of microRNA-21 regulating PDCD4 during tumorigenesis of liver fluke-associated cholangiocarcinoma contributes to tumor growth and metastasis. Tumour Biol. 2013;34:1579–1588. doi: 10.1007/s13277-013-0688-0. [DOI] [PubMed] [Google Scholar]
- 113.Chen YJ, Luo J, Yang GY, Yang K, Wen SQ, Zou SQ. Mutual regulation between microRNA-373 and methyl-CpG-binding domain protein 2 in hilar cholangiocarcinoma. World J Gastroenterol. 2012;18:3849–3861. doi: 10.3748/wjg.v18.i29.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li L, Masica D, Ishida M, Tomuleasa C, Umegaki S, Kalloo AN, Georgiades C, Singh VK, Khashab M, Amateau S, Li Z, Okolo P, Lennon AM, et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014;60:896–907. doi: 10.1002/hep.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qiao P, Li G, Bi W, Yang L, Yao L, Wu D. microRNA-34a inhibits epithelial mesenchymal transition in human cholangiocarcinoma by targeting Smad4 through transforming growth factor-beta/Smad pathway. BMC Cancer. 2015;15:469. doi: 10.1186/s12885-015-1359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang M, Wu X, Cao H, Zhan Q, Xia M, Zhou Q, Cai X, An F. Regulatory role of serum miR-224 in invasiveness and metastasis of cholangiocarcinoma. [Article in Chinese] Zhonghua Gan Zang Bing Za Zhi. 2015;23:748–753. doi: 10.3760/cma.j.issn.1007-3418.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 117.Canu V, Sacconi A, Lorenzon L, Biagioni F, Lo Sardo F, Diodoro MG, Muti P, Garofalo A, Strano S, D’Errico A, Grazi GL, Cioce M, Blandino G. MiR-204 down-regulation elicited perturbation of a gene target signature common to human cholangiocarcinoma and gastric cancer. Oncotarget. 2017;8:29540–29557. doi: 10.18632/oncotarget.15290. https://doi.org/10.18632/oncotarget.15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qiu YH, Wei YP, Shen NJ, Wang ZC, Kan T, Yu WL, Yi B, Zhang YJ. miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol Biochem. 2013;32:1331–1341. doi: 10.1159/000354531. [DOI] [PubMed] [Google Scholar]