Abstract
Microglia and macrophage cells are the primary producers of cytokines in response to neuroinflammatory processes. But these cytokines are also produced by other glial cells, endothelial cells, and neurons. It is essential to identify the cells that produce these cytokines to target their different levels of activation. We used dual RNAscope® fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) techniques to visualize the mRNA expression pattern of pro- and anti-inflammatory cytokines in microglia/macrophages cells. Using these methods, we can associate one mRNA to specific cell types when combining with different cellular markers by immunofluorescence. Results from RNAscope® probes IL-1β, TNFα, TGFβ, IL-10 or Arg1, showed colocalization with antibodies for microglia/macrophage cells. These target probes showed adequate sensitivity and specificity to detect mRNA expression. New FISH detection techniques combined with immunohistochemical techniques will help to jointly determine the protein and mRNA localization, as well as provide reliable quantification of the mRNA expression levels.
Keywords: in situ hybridization, RNAscope, Traumatic brain injury (TBI), Macrophages, Immunofluorescence, Microglia cells, Neuroinflammation, Cytokines
Background
The mRNA in situ hybridization technique is a useful tool that allows the specific and selective labeling of RNA sequences in brain slices in a cell-dependent manner (Grabinski et al., 2015). Furthermore, the use of antibodies against these specific cytokines can produce variable results due to the detection limits of the technique. Namely, because these cytokines are expressed in low abundance, the detection limit becomes the limiting factor for the use of antibodies. Lastly, fluorescence in situ hybridization (FISH) combined with immunohistochemistry (ISH) allows the examination of cytokine mRNA profiles in distinct cells with high selectivity and specificity, thus allowing us to identify the precise cellular source of cytokine production following TBI. This protocol describes how the combination of FISH and immunofluorescence imaging can bridge the gap between mRNA and protein analysis. We can identify the target mRNA being produced by microglia/macrophage cells. Analysis of both RNA and protein expression in the same tissue allows differentiating between cell specific production of microglia/macrophage cells and other cell types. There are limited studies on the effects of sex on inflammation profile following traumatic brain injury (TBI). In our recent publication (Villapol et al., 2017), we used RNAscope® technology combined with immunofluorescence to determine pro-inflammatory (e.g., IL1β and TNFα) and anti-inflammatory (e.g., TGFβ and Arg1) cytokine mRNA expression profiles in microglia/macrophages in the injured brains of male and female mice. Our data demonstrate that a mixed pattern of both pro-inflammatory and anti-inflammatory cytokine expression occurred in microglia/macrophages in the first week after TBI. Also, we have previously shown that IL-10 levels are significantly increased in microglia/macrophages in the injured cortex of NOX2-/- mice (Barrett et al., 2017).
In summary, the use of FISH improves specificity and sensitivity when examining cytokine mRNAs in distinct cells, confirmed by antibody co-immunostaining for microglia/macrophages. This method allows us to identify the cellular source of cytokine production following brain injury with much-improved confidence.
Materials and Reagents
Thick Whatman paper (Fischerbrand® Chromatography Paper) (Fisher Scientific, catalog number: 05-714-4)
Gelatin-coated glass slides (Superfrost Plus) (Fisher Scientific, catalog number: 12-550-15)
Microscope cover glass (24 x 50 mm) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3422ERI)
ImmEdge hydrophobic barrier pen (Advanced Cell Diagnostics, catalog number: 310018)
Kimwipes (KCWW, Kimberly-Clark, catalog number: 34133)
Foil paper
Mice (C57BL/6 from THE JACKSON LABORATORY, catalog number: 000664)
RNAscope® positive control probe-Mm-Ppib (peptidylprolyl isomerase B) (Advanced Cell Diagnostics, catalog number: 313911)
RNAscope® negative control probe-DapB (Advanced Cell Diagnostics, catalog number: 310043)
RNAscope® probe-Mm-Tgfβ1 (Transforming growth factor beta 1) (Advanced Cell Diagnostics, catalog number: 407751)
RNAscope® probe-Mm-IL1β (interleukin 1 beta) (Advanced Cell Diagnostics, catalog number: 316891)
RNAscope® probe-Mm-TNFα (tumor necrosis factor alpha) (Advanced Cell Diagnostics, catalog number: 311081)
RNAscope® probe-Mm-Arg1 (Arginase 1) (Advanced Cell Diagnostics, catalog number: 403431)
Phosphate-buffered saline (PBS), 10x solution (Fisher Scientific, catalog number: BP39920)
Paraformaldehyde (PFA) stock to prepare 4% (w/v) PFA, PRILLS (Electron Microscopy Sciences, catalog number: 19200)
Sucrose Crystalline stock (Fisher Scientific, catalog number: S5) to prepare 30% (w/v) solution diluted in ultra-pure water
Ultra-pure water (e.g., Milli-Q)
Freshly prepared 50% (v/v) ethanol in ultra-pure water (e.g., Milli-Q water) solution
Freshly prepared 70% (v/v) ethanol in ultra-pure water (e.g., Milli-Q water) solution
Absolute ethanol (200-proof ethanol) (Fisher Scientific, catalog number: BP28184)
RNAscope® H2O2 & protease plus reagents (Advanced Cell Diagnostics, catalog number: 322330)
RNAscope® 2.5 HD detection reagents-RED (Advanced Cell Diagnostics, catalog number: 322360) (read Note 1)
RNAscope® target retrieval reagents (Advanced Cell Diagnostics, catalog number: 322000)–previously known as pretreatment 2 solution
Normal goat serum (NGS) blocking solution (Vector Laboratories, catalog number: S-1000)
Triton X-100
Polyclonal anti-rabbit Iba-1 (ionized calcium binding adaptor molecule-1) antibody (Wako Pure Chemical Industries, catalog number: 019-19741)
Polyclonal anti-rabbit P2Y12 antibody (AnaSpec, catalog number: AS-55042A)
Polyclonal anti-rat F4/80 antibody (R&D systems, catalog number: MAB5580)
Goat anti-rabbit Alexa Fluor® 488 secondary antibody (Thermo Fisher Scientific, catalog number: A-11034)
Goat anti-rat Alexa Fluor® 488 secondary antibody (Thermo Fisher Scientific, catalog number: A-11006)
Fluoro-gel (with Tris buffer) mounting medium, 20 ml (Electron Microscopy Science, catalog number: 17985-10)
Glycerol
Ethylene glycol
RNAscope® Wash Buffer Reagents (4 x 60 ml) (Advanced Cell Diagnostics, catalog number: 310091)
4’,6-Diamidino-2-phenylindole (DAPI) solution (Sigma-Aldrich, catalog number: D9542)
Antifreeze solution (see Recipes)
1x wash buffer (see Recipes)
1x antigen retrieval solution (see Recipes)
DAPI solution (see Recipes)
Equipment
GilsonTM PIPETMAN ClassicTM Pipets (Gilson, models: P20, P200, P1000, catalog numbers: F123600, F123601, F123602)
2 L beaker
Extra-long forceps (Thermo Fisher Scientific, FisherbrandTM, catalog number: 10-316A)
Timer, TraceableTM NanoTM (Fisher Scientific, FisherbrandTM, catalog number: 14-649-83)
Slide Holder Handle, 24 slides (Electron Microscopy Sciences, catalog number: 62543-06)
Sliding microtome, MicromHM 430 (Thermo Fisher Scientific, Thermo ScientificTM, model: HM 430, catalog number: 910010)
HybEzTM hybridization system (Advanced Cell Diagnostics, catalog number: 310010)
ACD HybEZTM humidity control tray with lid (Advanced Cell Diagnostics, catalog number: 310012)
Tissue Tek Slide Stain Set with EasyDipTM slide staining (Electron Microscopy Sciences, catalog number: 62540-01)
ACD EZ-slide holder/rack (Advanced Cell Diagnostics, catalog number: 310017)
Thermix® Hot plate with magnetic stirrer, Model 210T (Fisher Scientific, model: Model 210T, catalog number: 11-493-210T)
Fluorescent images were acquired on an Axioplan 2 microscope (Carl Zeiss, model: Axioplan 2, catalog number: 451485) with a Photometrics camera (CoolSNAP, fx, Axioskop, Roper Scientific, serial number: A02M86017)
Leica SP8 confocal microscope (Leica Microsystems, model: Leica TCS SP8)
Software
GraphPad Prism software V. 5.0 (GraphPad Software, Inc., La Jolla, CA)
Adobe Photoshop CS5 V. 12.0 (Adobe Photoshop, CC)
AxionVision 4V, 4.8.2.0., licensed to: 3018897 (Carl Zeiss MicroImaging, GmbH)
ImageJ64 software (National Institute of Health)
Procedure
Fluorescent in situ hybridization was performed using RNAscope® Technology 2.5 Red fluorescent kit for fresh frozen tissue, with some modifications.
All target probes consist of 20 short double-Z oligonucleotide probe pairs that are gene specific and were obtained from ACD.
RNAscope® Probe-Mm-Arg1 (accession number NM_007482.3, target region 2-1114)
RNAscope® Probe-Mm-TNFα (accession number NM_013693.2, target region 41-1587)
RNAscope® Probe-Mm-Tgfβ1 (accession number NM_011577.1, target region 588-1913)
RNAscope® Probe-Mm-IL1β (accession number NM_008361.3, target region 2-950)
RNAscope® Positive Control Probe-Mm-Ppib (accession number NM_011149.2, target region 98-856)
RNAscope® Negative Control Probe-DapB (targets a bacterial gene)
-
Tissue
Euthanize mice with CO2 and transcardially perfuse with ice-cold phosphate buffered saline (PBS) as was previously described (Villapol et al., 2017).
Post fix the whole brains with 4% (v/v) PFA overnight.
Transfer the mice brains into a 30% (v/v) sucrose solution in PBS for dehydration for 48 h at 4 °C, or until the brain sinks completely to the bottom of the tube.
Section the mice brains at 20 μm-thick in coronal orientation with a sliding microtome.
Cryoprotect brains in an antifreeze solution (see Recipes) and store at -20 °C.
-
Before starting
Prepare 1.5 L of 1x wash buffer (see Recipes).
Set HybEZTM hybridization system to 40 °C.
Cut Whatman paper and place at center on the bottom of the HybEZTM humidity tray and add approximately 50 ml of ultra-pure water.
Warm HybEZTM humidity control tray containing wet Whatman paper for 30 min before use.
Allow ‘Pretreat 1’ solution (hydrogen peroxide solution) to equilibrate at room temperature (RT) before use.
Equilibrate amplification reagents (AMP solution 1-6, obtained from ACD) at RT.
Warm controls and target probes for 10 min at 40 °C and cool at RT before use.
-
RNAscope® Technology for Fluorescent in situ hybridization (FISH)
Mount coronal brain sections (20 μm-thick) on gelatin-coated slides and store at -80 °C until use. Attempt to locate brain sections on the bottom of the slide to avoid using an excessive amount of buffer during washes.
Place slides on a vertical plastic staining rack and immerse in 50% freshly made ethanol solution for 5 min.
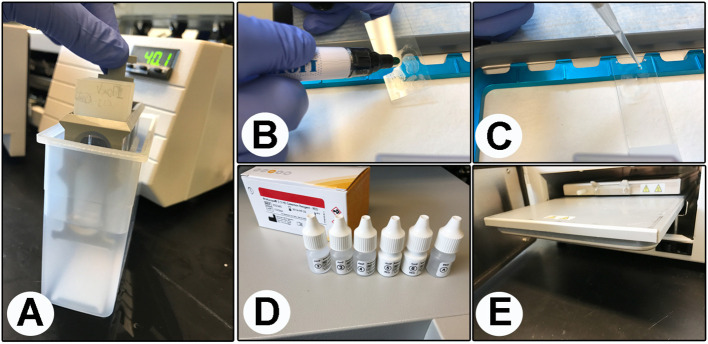
Transfer the vertical plastic staining rack containing the slides into an EasyDipTM slide staining system containing 70% ethanol solution for 5 min (Figure 1A and Video 1).
Transfer the vertical plastic staining rack containing the slides into an EasyDipTM slide staining system containing 100% ethanol solution for 5 min.
Transfer the vertical plastic staining rack containing the slides into an EasyDipTM slide staining system containing a second 100% ethanol solution for 5 min (read Note 1).
Remove slides from 100% EtOH, and let them air dry completely at RT for 30 min in a slide holder.
Draw a hydrophobic barrier around each section using an immEdge hydrophobic barrier pen (Figure 1B and Video 1).
From this step forward, do not allow the brain sections to dry out completely.
-
Pretreatment 1 (hydrogen peroxide treatment)
Place slides on ACD EZ-slide holder/rack and add 60-100 μl per brain section of pretreat solution 1 to each section (Figure 1C and Video 1, read Note 2).
Incubate the brain section in pretreat solution 1 for 10 min at RT. Start the timer once you add the pretreat solution 1 to the first slide.
Decant the pretreat solution 1 by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe if necessary.
Place slides into the vertical plastic staining rack submerged in ultra-pure water.
Wash slides by moving the staining rack up and down several times.
-
Pretreatment 2 (antigen retrieval solution)
Bring 350 ml 1x antigen retrieval solution (pretreatment solution 2) (see Recipes) to a boil using a hot plate. (It usually takes 40 min to reach boiling.) It’s recommendable to cover the container with foil paper.
Check to ensure the temperature is between 100-104 °C.
Slowly submerge the vertical plastic staining rack with slides using long curved forceps into the boiling antigen retrieval solution.
Check to ensure the temperature is between 100-104 °C.
Boil slides for 2 to10 min (it depends on the quality of tissue).
Rinse slides in ultra-pure water (repeat once more).
-
Pretreatment 3 (protease digestion)
Decant the ultra-pure water by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe if necessary.
Place slides on the ACD EZ-slide holder/rack.
Add about 60-100 μl per brain section of pretreatment solution 3 (protease solution) to cover entire tissue section (Figure 1D and Video 1, read Note 2).
Place the ACD EZ-slide holder/rack into the pre-warmed HybEZTM Oven with lid and incubate for 30 min at 40 °C.
Remove the ACD EZ-slide holder/rack, and place the ACD HybEZTM humidity control tray back into the HybEZTM Oven hybridization system (Figure 1E and Video 1).
Decant the pretreatment solution 3 by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe if necessary.
Place slides into the vertical plastic staining rack submerged in ultra-pure water.
Wash slides by moving the slide rack up and down several times.
-
Target probe hybridization
Decant the ultra-pure water by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe if necessary.
Place slides on the ACD EZ-slide holder/rack (read Note 3).
Add 60-100 μl per brain section of target probes or control probes to cover entire tissue section (read Note 2).
Place the ACD EZ-slide holder/rack into the pre-warmed ACD HybEZTM humidity control tray and incubate for 2 h at 40 °C in the HybEZTM hybridization system.
Remove the ACD EZ-slide holder/rack, and place the ACD HybEZTM humidity control tray back into the HybEZTM hybridization system.
Decant the control and target probes by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe.
Place slides into the vertical plastic staining rack submerged in 1x wash buffer.
Wash slides by moving the slide rack up and down several times for 2 min.
-
Amplification steps (AMP 1-AMP 6)
Decant the 1x wash buffer by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe.
Place slides on ACD EZ-slide holder/rack, do not let the tissue sections dry out completely.
Add 60-100 μl per brain section of AMP solution to each tissue section (read Note 2).
Place the ACD EZ-slide holder/rack into the ACD HybEZTM humidity control tray and incubate according to Table 1.
Decant AMP solution by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe.
Place slides on a vertical plastic staining rack submerged into 1x wash buffer.
Wash slides by moving the slide rack up and down several times for 2 min.
-
Detect the signal (10 min at RT)
Mix 60 μl of Red-A and 1 μl of Red-B solutions per slide. Protect from light.
Decant the 1x wash buffer by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe.
Place slides on ACD EZ-slide holder/rack, do not let the tissue sections dry out completely.
Add 60 μl of detection solution to each tissue section (read Note 2). Make sure to start with the positive and negative control slides.
Incubate for 10 min at RT inside the ACD HybEZTM humidity control tray with lid. Protect from light. Start the timer immediately after adding the detection solution to the first slide.
Check every two minutes the progression of the hybridization signal by comparing the positive and negative controls. The positive control should be reddish in color, whereas the negative control should show little to no background.
Decant the detection solution by flicking the slides into a waste container or by tapping the side of the slides on a Kimwipe.
Place slides on a vertical plastic staining rack submerged into ultra-pure water.
Wash slides by moving the slide rack up and down several times for 2 min.
Before proceeding with immunohistochemistry, check the hybridization signal under the microscope. Positive hybridization signal consists of several punctate signals (red dots) representing the mRNA transcript (Wang et al., 2012) (Figure 2).
-
Immunofluorescence
Transfer the vertical plastic staining rack containing the slides into an EasyDipTM slide staining system containing PBS.
Wash slides by moving the slide rack up and down several times. Repeat three times.
Block in 5% (v/v) normal goat serum (NGS) in PBS supplemented with 0.3% (v/v) Triton X-100 (PBST) for 1 h at RT (or overnight at 4 °C) inside the ACD HybEZTM humidity control tray with lid.
-
Incubate in primary antibody in 5% (v/v) NGS in PBST overnight at 4 °C at the following dilutions:
polyclonal anti-rabbit Iba-1 (1:500)
polyclonal anti-rabbit P2Y12 (1:1,000)
polyclonal anti-rat F4/80 (1:200)
Place slides on a vertical plastic staining rack submerged into PBS. Wash slides by moving the slide rack up and down several times. Repeat three times for 5 min each.
Incubate with corresponding Alexa Fluor® 488 secondary antibodies (1:1,000 in 5% (v/v) NGS in PBST) for 1-2 h at RT inside the ACD HybEZTM humidity control tray with lid. Protect from light.
Place slides on a vertical plastic staining rack submerged into PBS. Wash slides by moving the slide rack up and down several times. Repeat three times for 5 min each. Protect from light.
Check the immunofluorescence signal under the microscope. Positive immunofluorescence signal consists of a bright green detection of the antigen (Figure 3).
Counterstain nuclei with DAPI solution (see Recipes) for 10 min. Protect from light.
Place slides on a vertical plastic staining rack submerged into PBS. Wash slides by moving the slide rack up and down several times. Repeat three times for 5 min each. Protect from light.
Rinse with Milli-Q water and let dry.
Coverslip brain sections with Fluoro-gel with Tris buffer mounting medium. Let dry.
Figure 1. Components of the RNAscope protocol.
A. Washes in distilled water; B. Draw a hydrophobic barrier around each section; C. Add pretreatment solutions; D. Amplification solutions; E. Holder/rack.
Video 1. Critical steps of the Fluorescent in situ hybridization (FISH) protocol.

We show the critical steps for the FISH technique described in this protocol as follows: boil the slides with the antigen retrieval solution, place slides on ACD EZ-slide holder/rack into the HybEZTM Oven, create a barrier around tissue sections, add 60-100 μl per probe or amplification solution to each section, move the slide rack up and down, and acquire representative confocal images.
Table 1. Amplification solutions and their corresponding incubation times and temperatures.
| Amplification solution | Incubation time (min) | Temperature (°C) |
| *AMP 1 | 30 | 40 |
| *AMP 2 | 15 | 40 |
| *AMP 3 | 30 | 40 |
| *AMP 4 | 15 | 40 |
| AMP 5 | 30 | RT |
| AMP 6 | 15 | RT |
| *For amplification steps 1-4, read Note 3. | ||
Figure 2. Representative images of positive control probes and nuclei (DAPI, blue) on selected brain sections.
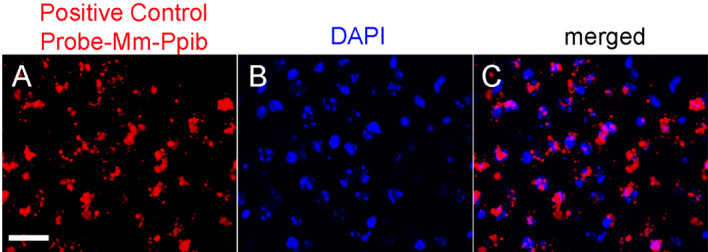
Scale bar = 50 μm.
Figure 3. Representative confocal images of RNAscope.
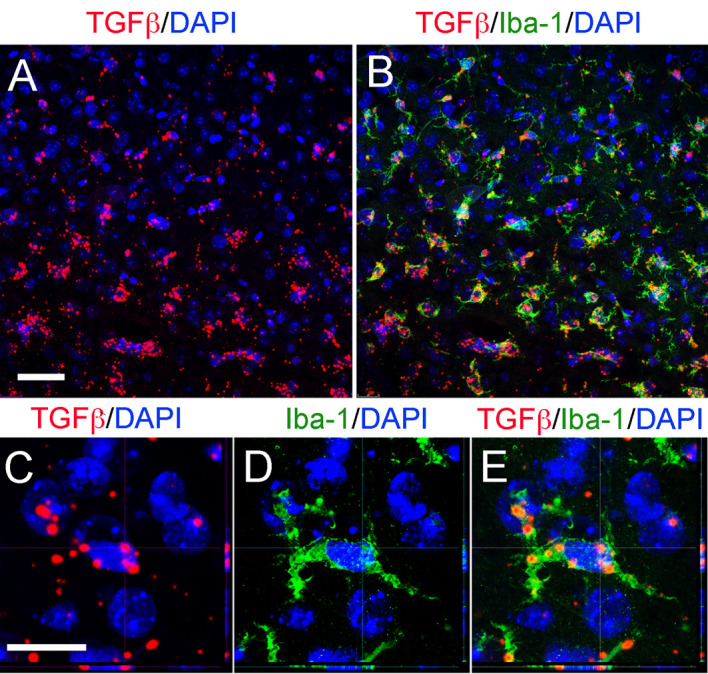
®fluorescent assay using a specific probe to detect TGFβ combined (red) with DAPI staining for nuclei (blue) and immunostaining for microglia/macrophages cells (Iba-1) in the cortex after TBI. Orthogonal projections confocal images and scale bars for A and B (50 μm) and C-E (20 μm).
Data analysis
Details regarding quantification of immunofluorescence and in situ mRNA hybridization has been provided by (Allen et al., 2016; Barrett et al., 2017; Villapol et al., 2017). Briefly:
Positive in situ mRNA hybridization signal consists of a punctate signal representing a single mRNA transcript (Wang et al., 2012).
Take images using a Leica SP8 confocal microscope at 10x and 20x magnification. Take 3-6 microscopic fields within the brain area of interest. All microscope and camera settings (i.e., light level, exposure, gain, etc.) should be identical for all images.
Assign the color label to far red (excitation 647 nm, emission 690 ± 10 nm) for mRNA hybridization signal. Assign the color label to green for the antigen of interest.
Analyze the images with AxionVision software and quantify the hybridization signal with ImageJ64 software (National Institute of Health).
Manually count the number of mRNA-positive cells that colocalize with DAPI nuclei that represent the number of positive cells detected with the antigen of interest. The negative probe used as a control should not contain any stained cells.
Crop and resize images using Adobe Photoshop CS5 if necessary.
Apply changes in brightness and contrast equally to the entire image, and all corresponding images within the same dataset.
Analyze data with a two-way analysis of variance (ANOVA) with Bonferroni post hoc test using GraphPad Prism software v. 5.0 (GraphPad Software, Inc., La Jolla, CA). The two independent variables were sex (female vs. male) and time-point (e.g., 4 h, 1, 3, 7 and 30 days after TBI). A P value < 0.05 was considered statistically significant. Express all figures and tables as mean ± SEM.
Notes
Mounted brain sections can be stored in 100% EtOH following the serial dehydration procedure. At this stage, slides can be stored at -20 °C for a week.
In every pretreatment and amplification step make sure the brain section is covered in solution. We work with mouse brain sections; therefore, bigger tissue might need more volume of solution to cover the section. Not doing so will result in higher noise signal.
Keep the humidity tray warm during the assay.
Because it is important for the tissue to remain wet or humid during the in situ hybridization procedure, it is recommended to work in sets.
At all times check that the integrity of the hydrophobic barrier is not compromised.
All reagents were stored as recommended by the manufacturers.
Recipes
-
Antifreeze solution
30% (v/v) glycerol
30% (v/v) ethylene glycol
40% (v/v) 0.01 M PBS
-
1x wash buffer
1.47 L ultra-pure water (e.g., Milli-Q water)
30 ml of 50x wash buffer
Mix well
-
1x antigen retrieval solution (previously known as pretreatment solution 2)
35 ml of 10x antigen retrieval solution
315 ml of ultra-pure water
Mix well and bring to boil (20 min to reach boiling)
-
DAPI solution
Dilute DAPI in PBS to a 1:50,000 solution
Vortex thoroughly. Failing to do so, will result in blue precipitate on the tissue
Acknowledgments
This work was supported by NIH grants R03NS095038 (S. Villapol), R01NS067417 (M.P. Burns), and the Dean of Biomedical Research (Toulmin pilot project) provided by the Georgetown University Medical Center (I. Mocchetti). We would like to say thanks to Dr. Kathy Maguire-Zeiss for letting us use her fluorescence microscope, and to Lucas Djavaherian for his help recording the video. This protocol has been adapted from (Barrett et al., 2017, Villapol et al., 2017). The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Allen M., Ghosh S., Ahern G. P., Villapol S., Maguire-Zeiss K. A. and Conant K.(2016). Protease induced plasticity: matrix metalloproteinase-1 promotes neurostructural changes through activation of protease activated receptor 1. Sci Rep 6: 35497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett J. P., Henry R. J., Villapol S., Stoica B. A., Kumar A., Burns M. P., Faden A. I. and Loane D. J.(2017). NOX2 deficiency alters macrophage phenotype through an IL-10/STAT3 dependent mechanism: implications for traumatic brain injury. J Neuroinflammation 14(1): 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grabinski T. M., Kneynsberg A., Manfredsson F. P. and Kanaan N. M.(2015). A method for combining RNAscope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS One 10(3): e0120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villapol S., Loane D. J. and Burns M. P.(2017). Sexual dimorphism in the inflammatory response to traumatic brain injury. Glia 65(9): 1423-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F., Flanagan J., Su N., Wang L. C., Bui S., Nielson A., Wu X., Vo H. T., Ma X. J. and Luo Y.(2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14(1): 22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]