Abstract
The tumor suppressor PTEN is frequently inactivated in glioblastoma. PTEN-L is a long form of PTEN produced by translation from an alternate upstream start codon. Unlike PTEN, PTEN-L has a signal sequence and a tract of six arginine residues that allow PTEN-L to be secreted from cells and be taken up by neighboring cells. This suggests that PTEN-L could be used as a therapeutic to restore PTEN activity. However, effective delivery of therapeutic proteins to treat CNS cancers such as glioblastoma is challenging. One method under evaluation is cell-mediated therapy, where cells with tumor-homing abilities such as neural stem cells are genetically modified to express a therapeutic protein. Here, we have developed a version of PTEN-L that is engineered for enhanced cell-mediated delivery. This was accomplished by replacement of the native leader sequence of PTEN-L with a leader sequence from human light-chain immunoglobulin G (IgG). This version of PTEN-L showed increased secretion and an increased ability to transfer to neighboring cells. Neural stem cells derived from human fibroblasts could be modified to express this version of PTEN-L and were able to deliver catalytically active light-chain leader PTEN-L (lclPTEN-L) to neighboring glioblastoma cells.
Keywords: PTEN, PTEN-L, glioblastoma, cell-mediated therapy, signal sequence, neural stem cell
Introduction
The tumor suppressor PTEN is lost in many cancer types.1 In glioblastoma, about 80% of patients show loss of one copy of PTEN, and about 40% of patients show complete loss of PTEN expression.2, 3 Bioinformatics analyses suggest that partial PTEN loss is an early event in glioblastoma formation,4 with the consequence that most or all glioblastoma cells within a patient will have this genetic aberration. Mouse models indicate that both complete and partial loss of PTEN are able to promote glioblastoma formation.5 Cell culture studies have shown that restoration of PTEN in glioblastoma cell lines can lead to reduced migration and proliferation, along with increased susceptibility to apoptosis.6 As well, studies in several mouse models of cancer have shown that reactivation of PTEN expression in established cancers can cause regression.7, 8 These observations suggest that pharmacologic strategies to reverse the consequences of PTEN loss may be effective in glioblastoma.
The best described function of PTEN is its ability to dephosphorylate and inactivate the second messenger phosphatidylinositol 3,4,5 trisphosphate (PIP3).1 PIP3 is generated by the class I phosphoinositide 3-kinases (PI 3-kinases). Thus, one strategy to pharmacologically reverse the effects of PTEN loss in cancer cells is to antagonize the action of the PI 3-kinases.9 Alternatively, kinases activated downstream of PIP3 may be valid targets. However, the phosphatase activity of PTEN is subject to a complex set of regulatory interactions,10 and PTEN has additional functions beyond breakdown of PIP3.11, 12 These activities will not be restored by inhibition of PI 3-kinase or downstream kinases.
Direct replacement of tumor suppressors such as PTEN in cells has generally been viewed as pharmacologically unfeasible. However, a recent publication suggests that this may not be the case. Hopkins et al.13 have described a version of PTEN, known as PTEN long (PTEN-L), that is secreted by cells and taken up by neighboring cells.14 They produced recombinant PTEN-L and showed that it could be used in subcutaneous animal models of cancer to effectively reduce tumor growth. However, there are numerous issues with treating cancer effectively with protein therapeutics, including the relatively poor penetration of therapeutic proteins into tissues and tumors. This is an especially significant problem in glioblastoma, where therapeutic proteins need to cross the blood-brain barrier if administered systemically. Direct administration of therapeutic proteins within the CNS does not solve this problem, because perfusion of brain tissue is generally poor. Strategies such as convection-enhanced delivery15 attempt to overcome this issue but still result in limited perfusion.16, 17 As well, they generally do not deliver proteins in a sustained fashion,18 which may be desirable for therapeutic efficacy. Cell-mediated delivery may be one strategy to overcome this. Various cell types, including monocyte/macrophages,19 mesenchymal stem cells,20 and neural stem cells,21 appear to have some ability to home to cancer cells. If these cells are engineered to express a therapeutic protein, it may be possible to exploit the cancer cell homing ability of these cells to deliver therapeutic proteins in a sustained manner and at high local concentrations.
PTEN-L is produced as a result of the use of an alternate upstream start codon.14, 22 This adds an additional 173 amino acids to the amino terminus of normal PTEN. This additional sequence contains a polyarginine sequence that promotes uptake by other cells.14 It also contains a polyalanine sequence that appears to function as a signal sequence to direct secretion via the classical secretion pathway (although a mitochondrial location for PTEN-L has been proposed by another group22). Here, we have constructed an engineered version of PTEN-L that contains an optimized leader sequence and assessed the consequences of this modification on PTEN-L secretion from cells and uptake by neighboring cells.
Results
Phylogenetic Analysis of PTEN-L
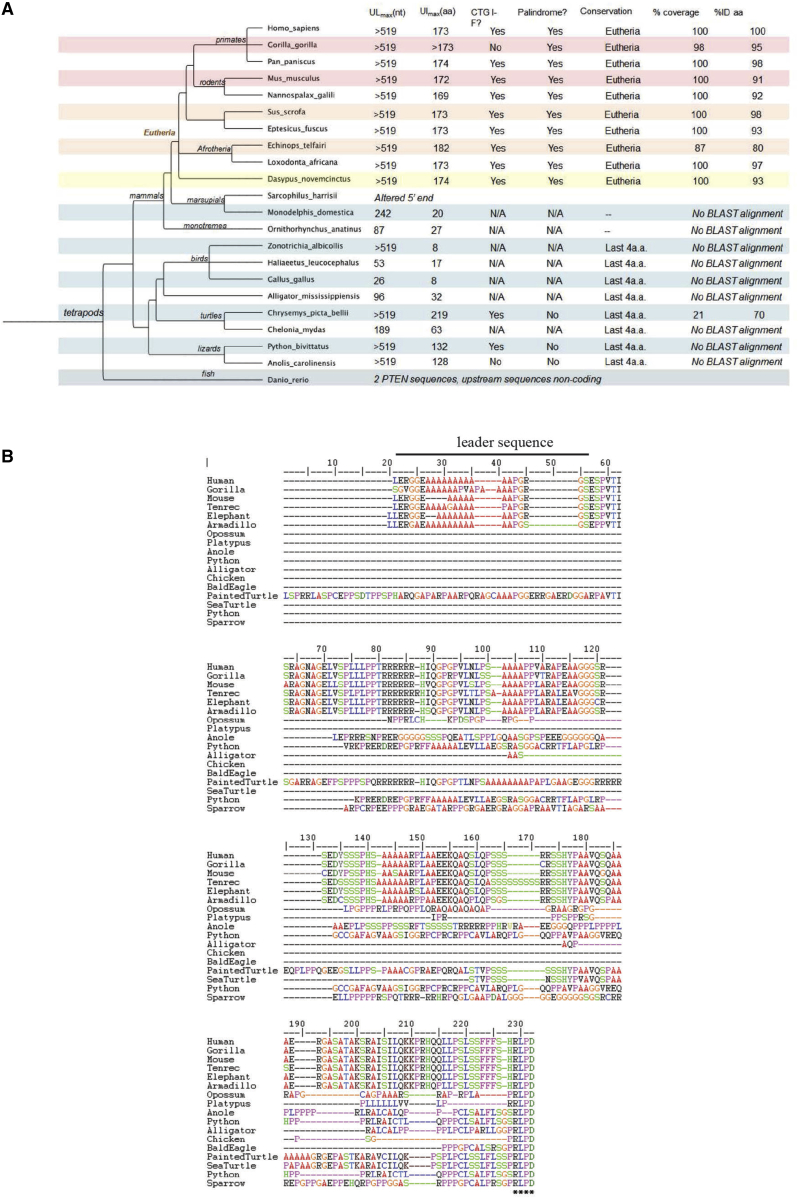
An analysis of the additional sequence in PTEN-L for homology with different species was performed, because this might suggest alternate designs for PTEN-L. Compared with a previous analysis of PTEN-L sequence conservation,14 this analysis used a local alignment strategy rather than a global one and used a greater number of non-mammalian sequences in the comparison. Upstream nucleotide sequences from several tetrapod species’ pten genes were extracted and translated to amino acids using DAMBE.23 The translated upstream sequences were aligned to human PTEN-L using BLASTp24 and evaluated for percent coding length conservation and identity. This analysis showed that PTEN-L arose relatively recently in evolution and is only evident in Eutheria, a clade of the class Mammalia that primarily consists of placental mammals (Figure 1A). A multiple sequence alignment of all amino acid sequences was produced using MUSCLE25 to evaluate sequence similarity. The multiple sequence alignment was then back-translated to codon sequences using DAMBE to assess the nucleotide identity of aligned sections and the presence of an in-frame, palindromic alternative start site (Figure 1B). The six-arginine tract required for transduction is perfectly preserved across six Eutheria species except for Tenrec, which has seven arginines (Figure 1B). The putative leader sequence is also conserved within this group, containing polyalanine tracts of varying length (Figure 1B). This is unusual in that the leader sequences of highly secreted proteins typically contain more hydrophobic amino acids. The polyalanine and polyarginine tracts are encoded by frameshifted variants of the same nucleotide sequence (polyalanine by tandem repeats of the CGG trinucleotide and polyarginine by tandem repeats of GGC) (Figure 2). The nucleotide repeats represent microsatellites, simple repetitive elements that are very abundant in eukaryotic genomes. They frequently occur within the coding regions of genes including 17% of human genes.26 Microsatellites are thought to arise fortuitously through replicative slippage, but they can affect the function of genes and their products, and their accumulation can be driven by natural selection.27 The polyamino acid repeats in PTEN-L may have arisen through microsatellite repeat expansion at a recent stage of vertebrate evolution and may have been retained in Eutheria either because they impart some selective advantage or because they have yet to be eliminated through negative selection or genetic drift. This suggests that the cell-to-cell transfer ability of PTEN-L may be limited, and that it could be improved through rational design.
Figure 1.
Species Tree for PTEN-L and Alignment of PTEN-L Sequences
(A) Species tree for PTEN-L. The species tree was generated as described in the Results. ULmax(nt) and UImax(aa) indicate numbers of conserved nucleotides and amino acids, respectively. The column labeled CTG-IF indicates whether the CTG initiation codon for endogenous PTEN-L is present. The column labeled Palindrome indicates whether the palindromic sequence identified by Liang et al.22 as being required for initiation of PTEN-L expression is present. (B) Alignment of PTEN-L sequences. Alignments were generated as described in the Results. The black bar indicates the proposed leader sequence in PTEN-L.
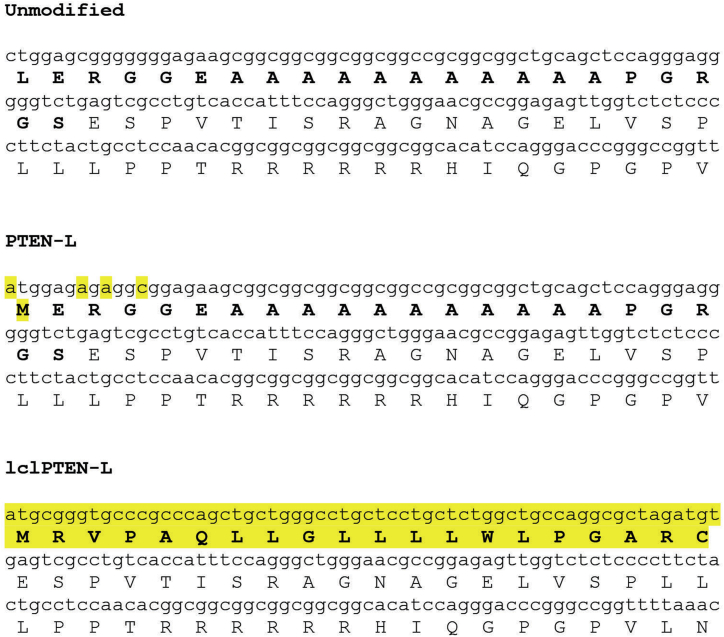
Figure 2.
PTEN-L and lclPTEN-L Designs
The top sequence shows the naturally occurring sequence coding for PTEN-L. The middle panel shows the sequence changes made to this naturally occurring PTEN-L sequence to generate the construct for high-level expression of PTEN-L. The methionine codon at the start of normal PTEN was also changed to a glycine codon in this construct to suppress expression of normal PTEN. The bottom sequence shows the sequence of lclPTEN-L in which the putative leader sequence is replaced by a human light-chain IgG leader sequence. Changed amino acids and bases with respect to the naturally occurring PTEN-L sequence are highlighted in yellow. Amino acids of the PTEN-L putative leader sequence and the human light-chain IgG leader sequence are in boldface.
PTEN-L Constructs
To evaluate leader sequence function in PTEN-L, we made expression vectors for normal PTEN-L and PTEN-L with an optimal leader sequence from a highly secreted protein. The PTEN-L expression construct was made by cloning the 519 base pair 5′ sequence unique to PTEN-L from T47D cells and then joining this to cDNA encoding normal PTEN using PCR splicing.28 As part of this splice, the start CTG codon of PTEN-L was changed to ATG and the start codon of normal PTEN was changed to a glycine codon (Figure 2). In addition, silent mutations were introduced into the string of seven G nucleotides downstream of the PTENL start codon to avoid issues with G quadruplex formation that could potentially interfere with translation.29 Rounds of PCR were used to replace the cDNA for the 22-amino acid PTEN-L leader sequence with a synthetic sequence encoding the leader sequence of a human immunoglobulin Gκ (IgGκ) light chain30 (Figure 2). The protein encoded by this construct was designated lclPTEN-L (“lcl” is an acronym for light-chain leader). Each cDNA was subcloned into the doxycycline-inducible lentiviral vector pLVX-Tight-puro, and replication-incompetent lentiviruses were generated using a four-plasmid transfection system into 293T cells as described previously.31
Expression and Activity of PTEN-L and lclPTEN-L
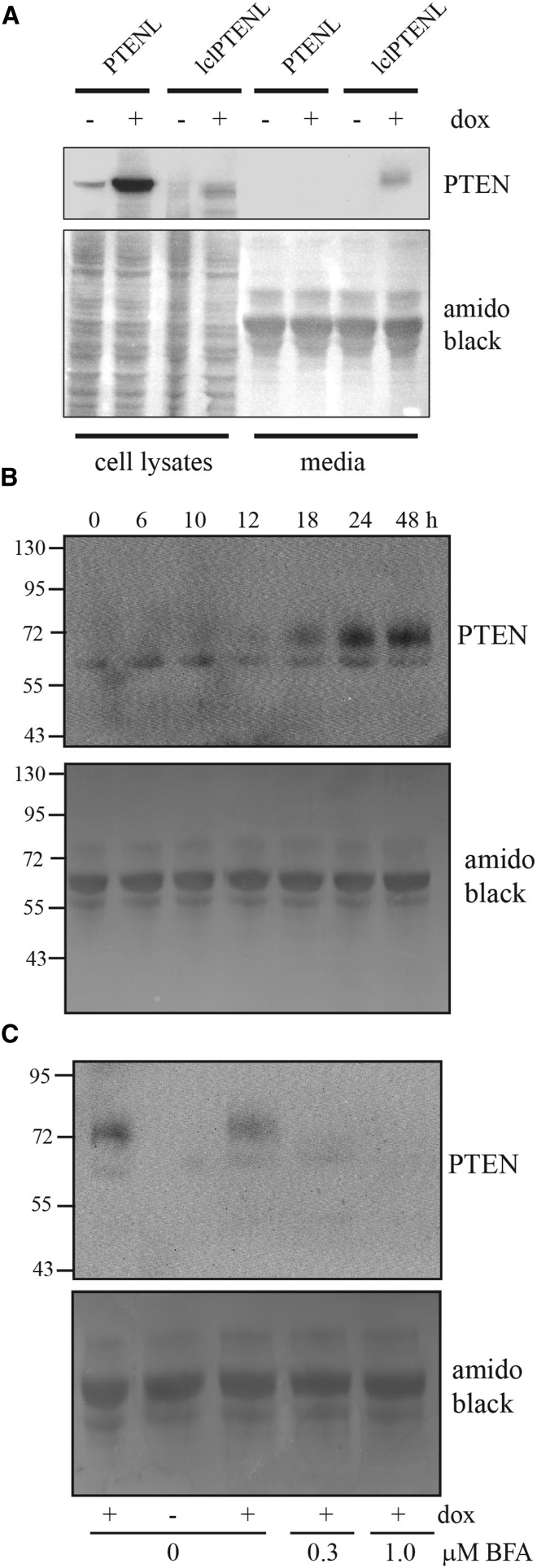
Expression of PTEN-L and lclPTEN-L was first assessed in the U87MG human glioblastoma cell line. These cells were chosen because they are PTEN null32 and effects of PTEN restoration in U87MG cells have been studied previously by multiple groups (e.g., Wick et al.,6 Tamura et al.,33 and Raftopoulou et al.34). Figure 3A shows a western blot analysis of cell lysates and conditioned media before and after doxycycline induction. With U87MG/PTEN-L cells, a single band of the expected size for PTEN-L (65 kDa) is observed in cell lysates from doxycycline-treated cells (Figure 3A). For lclPTENL, a much fainter band is seen in cell lysates that comigrates with the PTEN-L band (Figure 3A). A slightly lower band of similar intensity is also detected. This may represent lclPTEN-L with a cleaved leader sequence. In conditioned media, no PTEN-L was detected with doxycycline-treated U87MG/PTENL cells (Figure 3A). This is consistent with the initial description of PTEN-L, where it was only detected in conditioned media after concentration.14 However, with U87MG/lclPTEN-L cells, a band of the same size as PTEN-L could be detected in conditioned media (Figure 3A). Figure 3B shows a time course for the detection of this band after doxycycline induction. lclPTEN-L could be detected 18 hr after doxycycline induction and continued to accumulate in media up to 48 hr, the longest time point tested. Brefeldin A is a natural product that blocks protein transport from the endoplasmic reticulum to the Golgi during the classical pathway for protein secretion.35 Brefeldin A effectively blocked the secretion of lclPTEN-L into media, consistent with its secretion via this pathway (Figure 3C).
Figure 3.
Expression of PTEN-L and lclPTENL
(A) U87MG cells engineered for inducible expression of PTEN-L or lclPTEN-L were treated with 1 μM doxycycline for 48 hr. U87MG cells were grown in media containing 0.5% FBS to reduce interference of the BSA band in western blots of conditioned media. Total cell lysates and conditioned media were collected and analyzed by western blot. The top panel shows the blot probed with PTEN antibody. The bottom panel shows that same blot stained with amido black to show equal loading of total protein in each lane. (B) U87MG cells expressing lclPTEN-L were treated with doxycycline for the indicated period of time. Conditioned media were then analyzed by western blotting with antibody to PTEN. (C) U87MG cells expressing lclPTEN-L were treated without or with doxycycline for 24 hr in the presence of the indicated concentrations of brefeldin A. Conditioned media were then analyzed by western blotting using antibody to PTEN.
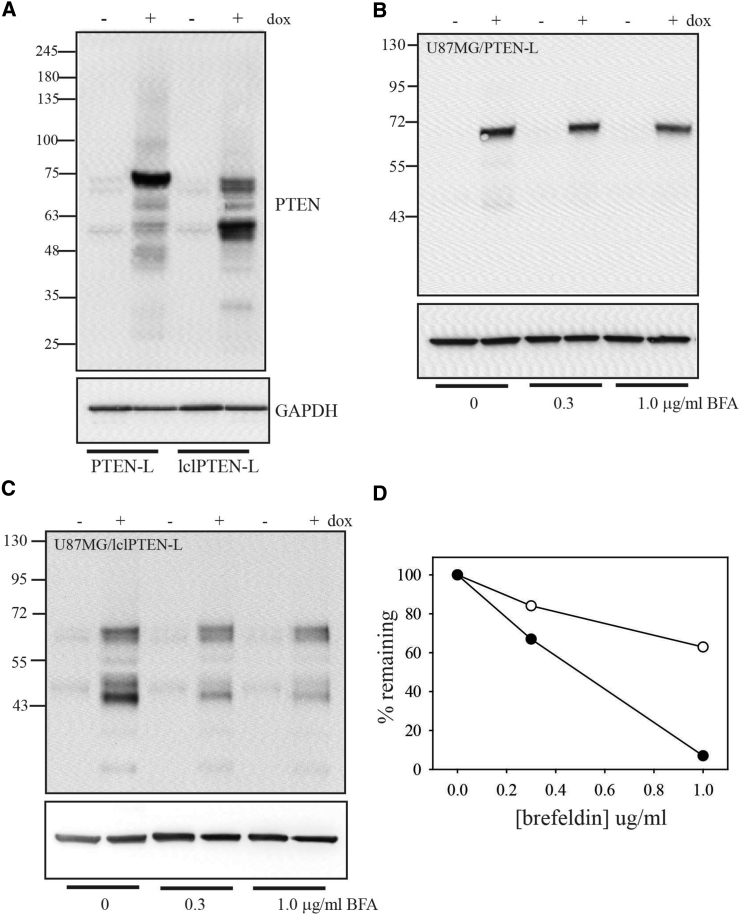
Along with the full-length band, total cell lysates of U87MG/lclPTEN-L cells also showed a lower molecular weight band (Figure 4A). This was absent in U87MG/PTEN-L cells. This band presumably represents a proteolytic product of lclPTEN-L. The PTEN antibody used here was raised against a PTEN C-terminal peptide. This proteolytic product therefore likely represents lclPTEN-L that has been proteolytically cleaved at a site close to the start of normal PTEN protein sequence (normal PTEN is 47 kDa). Intracellular PTEN-L and lclPTEN-L could in theory arise either from cytoplasmic synthesis or from reuptake of protein secreted from the cell. As an initial step to distinguish between these possibilities, cells were treated with brefeldin A. Blockade of the classical secretion pathway with brefeldin A typically results in the degradation of affected proteins via the endoplasmic reticulum-associated protein degradation (ERAD) pathway.36 Treatment of U87MG/PTEN-L cells with brefeldin A did not affect intracellular levels of PTEN-L (Figure 4B). Treatment of U87MG/lclPTEN-L cells with brefeldin A caused a reduction in intracellular levels of lclPTEN-L. This was particularly evident with the lower molecular weight lclPTEN-L band, which decreased to less than 10% of its levels in untreated cells (Figures 4C and 4D). This suggests that most of the intracellular lclPTEN-L is due to reuptake of secreted protein.
Figure 4.
Proteolysis of lclPTENL
(A) U87MG cells expressing either PTEN-L or lclPTEN-L were treated without or with doxycycline for 48 hr. Total cell lysates were analyzed by western blot with antibody to PTEN. (B and C) U87MG cells expressing either PTEN-L (B) or lclPTEN-L (C) were treated for 24 hr with doxycycline and/or brefeldin A as indicated. Total cell lysates were analyzed by western blot using antibody to PTEN. (D) Quantitation of changes in intracellular lclPTEN-L with brefeldin A treatment. Open circles show quantitation of the high molecular weight band, and filled circles show quantitation of the low molecular weight band.
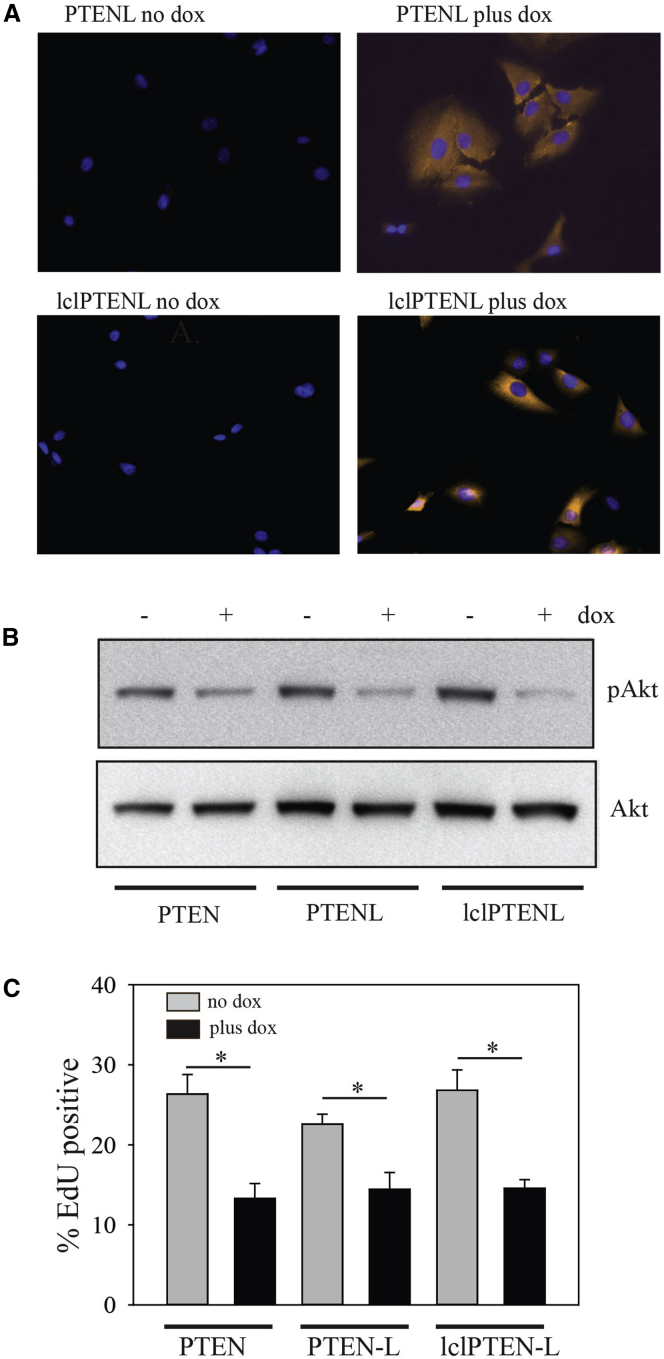
Immunofluorescence showed that PTEN-L and lclPTEN-L exhibited a punctate cytosolic subcellular distribution by immunofluorescence, similar to that described by Naguib et al.37 for normal PTEN, without any nuclear staining evident (Figure 5A). PTEN-L and lclPTEN-L both suppressed Akt phosphorylation to a similar extent to that seen with PTEN (Figure 5B; Figure S1). They also suppressed U87MG cell proliferation to a similar extent to that seen with PTEN (Figure 5C).
Figure 5.
Subcellular Distribution and Catalytic Activity of PTEN-L and lclPTENL
(A) Immunofluorescence was performed on U87MG cells expressing either PTEN-L or lclPTEN-L that were treated without or with doxycycline for 48 hr. PTEN immunofluorescence is shown pseudocolored in orange. Nuclei are stained with DAPI. Immunofluorescence for the top and bottom panels was performed at different times and is for comparison of subcellular distribution rather than expression levels. (B) Total cell lysates from U87MG cells expressing PTEN, PTEN-L, or lclPTEN-L, without or with 48 hr doxycycline treatment, were probed with antibodies to phosphorylated and total Akt. A representative blot from three independent experiments is shown. Quantitation of data from the three experiments is shown in Figure S1. (C) U87MG cells expressing doxycycline-inducible PTEN, PTEN-L, or lclPTEN-L were treated with or without doxycycline for 48 hr. For the last 2 hr of the 48-hr induction, EdU was added to the media. Cells were then fixed and proliferation was assessed by determining the percentage of EdU+ cells in 10 randomly chosen fields of view (between 500 and 1,000 cells total). Error bars show the mean ± SE. *p < 0.05 by the Mann-Whitney rank-sum test.
Cell-to-Cell Transfer of PTEN, PTEN-L, and lclPTEN-L
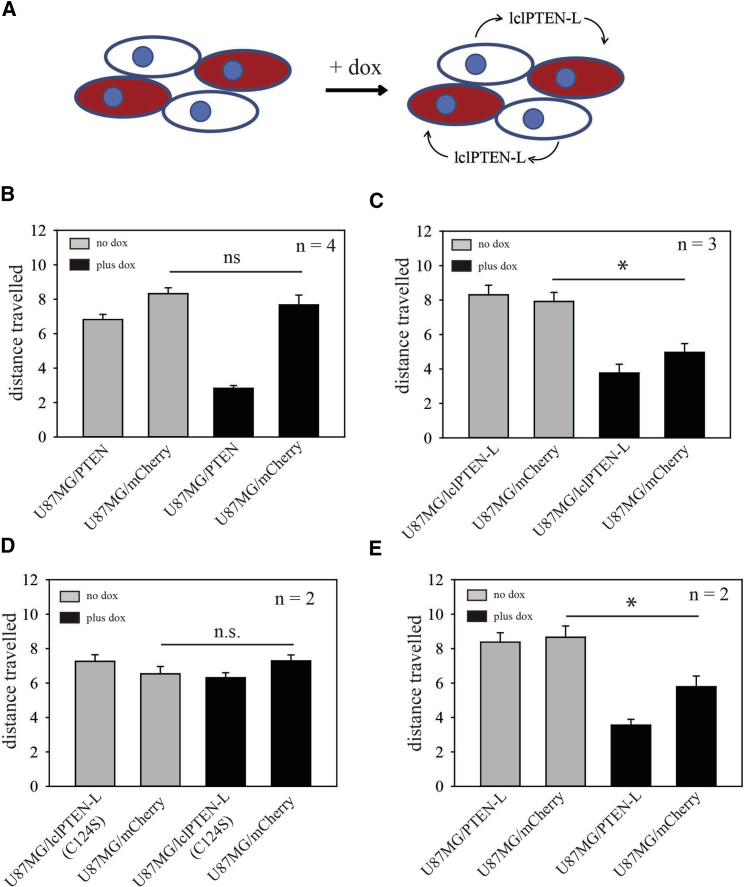
In order to test the ability of PTEN, PTEN-L, and lclPTENL to transfer from one cell to another cell in an active state, coculture experiments were performed with “donor” cells (U87MG cells expressing the different versions of PTEN) and “recipient” cells (U87MG cells expressing mCherry fluorescent protein) (Figure 6A). Effects on cell motility of U87MG/mCherry recipient cells can then be used as a readout for transfer of biologically active PTEN. Motility was assessed using videomicroscopy, which allowed separate assessment of donor and recipient cells. In these coculture experiments, induction of normal PTEN with doxycycline suppressed the motility of U87MG/PTEN donor cells as expected,33 but had no effect on the motility of U87MG/mCherry recipient cells (Figure 6B). In cocultures of U87MG/lclPTEN-L cells with U87MG/mCherry recipient cells, motility of U87MG/lclPTEN-L cells was also suppressed upon induction of expression (Figure 6C). In contrast with the results with U87MG/PTEN cells, motility of cocultured U87MG/mCherry recipient cells was also significantly suppressed, consistent with cell-to-cell transfer of lclPTEN-L (Figure 6C). As an additional control, a lentiviral vector for expression of lclPTEN-L in which PTEN was rendered catalytically inactive with a C124S mutation was made.38 Although lclPTEN-L with the C124S mutation was expressed and secreted similar to lclPTEN-L (Figure S2), it was unable to suppress motility in either expressing cells or co-cultured non-expressing cells (Figure 6D). In cocultures of U87MG/PTEN-L cells with U87MG/mCherry cells, motility of U87MG/PTEN-L cells was suppressed upon induction with doxycycline (Figure 6E). The motility of U87MG/mCherry recipient cells was also suppressed (Figure 6E). This suppression was slightly less than that observed when U87MG/lclPTEN-L cells were used as the donor cells. The above experiments were done using a 1:1 ratio of donor and recipient cells. When the ratio of donor cells to recipient cells was changed to 1:3, U87MG/lclPTEN-L cells were still able to repress U87MG/mCherry cell recipient cell motility (Figure 7A), whereas U87MG/PTENL were not able to do this (Figure 7B). As a second readout of lclPTEN-L transfer, the proliferation of co-cultured U87MG/mCherry recipient cells was also assessed using 5-ethynyl-2’-deoxyuridine (EdU) incorporation. U87MG cells expressing lclPTEN-L also significantly repressed U87MG/mCherry cell recipient cell proliferation (Figure 7C).
Figure 6.
Assay for Cell-to-Cell Transfer of Catalytically Active PTEN
(A) Schematic of experiment. U87MG cells engineered for inducible lclPTENL were cocultured with U87MG cells expressing mCherry fluorescent protein. Cells with inducible lclPTENL are shown in white; cells expressing mCherry are shown in red. The motility of both populations is determined by videomicroscopy. (B–E) Cells expressing PTEN (B), lclPTEN-L (C), lclPTEN-L(C124S) (D), and PTEN-L (E) were cocultured with U87MG cells labeled with mCherry. Videomicroscopy was used to determine distance traveled for both cell populations in the cocultures in the absence and presence of doxycycline. Bar graphs show the mean ± SEM for distance traveled (in pixels). Data shown are pooled from multiple experiments (approximately 30–50 cells analyzed in total), with number of experiments (n) shown in the top right corner of each bar graph. *p < 0.05 by the Mann-Whitney rank-sum test.
Figure 7.
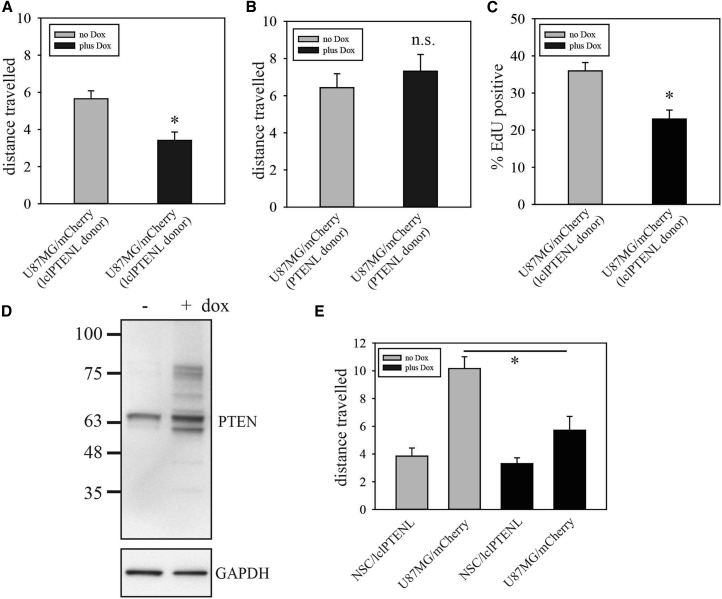
lclPTEN-L Cell-to-Cell Transfer at Lower Donor/Recipient Cell Ratios and with Neural Stem Cell Donor Cells
(A) Analysis of lclPTENL cell-to-cell transfer was performed as described in Figure 6C, except that the ratio of donor U87MG/lclPTENL cells to U87MG/mCherry cells was approximately 1:3, not 1:1. (B) Analysis of PTENL cell-to-cell transfer with a donor to recipient cell ratio of 1:3. (C) U87MG/lclPTEN-L cells were cocultured with U87MG/mCherry cells at a 1:1 ratio and treated without or with doxycycline for 72 hr. Cells were then labeled with EdU for 2 hr and fixed. mCherry/EdU double-positive cells and total mCherry+ cells were then counted under immunofluorescence microscopy for 10 randomly selected fields of view. (D) Neural stem cells were generated from iPSCs as described in Materials and Methods. These were transduced with lentiviruses for Tet activator expression and expression of lclPTEN-L, and selected with G418 and puromycin. Cells were then treated without or with doxycycline for 48 hr, and total cell lysates were analyzed by western blot with antibody to PTEN. (E) Neural stem cells engineered for inducible expression of lclPTEN-L were cocultured in neural stem cell media with U87MG cells labeled with mCherry. Videomicroscopy was used to determine distance traveled for both cell populations in the cocultures in the absence and presence of doxycycline. (A–D) Bar graphs show the mean ± SEM for distance traveled (in pixels). *p < 0.05 by the Mann-Whitney rank-sum test.
Expression of lclPTEN-L in Human Neural Stem Cells
Neural stem cells are being evaluated as vehicles for protein delivery in glioblastoma.21, 39, 40, 41 To determine whether these could be engineered for cell-to-cell transfer of lclPTEN-L, we derived neural stem cells from human fibroblasts via an induced pluripotent stem cell (iPSC) reprogramming and dual SMAD inhibition approach.42 These were positive for nestin, Sox2, and Pax6, while negative for Oct4 and β-III-tubulin, confirming their neural stem cell phenotype (Figure S3). Neural stem cells were transduced with a lentiviral vector expressing Tet activator followed by transduction with a lentiviral vector encoding inducible lclPTEN-L. This was done following the same protocol used for U87MG cells, except that higher virus concentrations were used so that a high density of neural stem cells was maintained during drug selections. This is necessary to maintain healthy populations of these cells in culture. Figure 7D shows that neural stem cells could be engineered for lclPTEN-L expression. LclPTEN-L behaved in a similar fashion to lclPTEN-L expressed in U87MG cells in that the predominant form on western blots was a proteolytic product that migrated slightly below the band for endogenous, wild-type PTEN. Under the culture conditions used, neural stem cells showed little motility, and this was unaffected by induction of lclPTEN-L expression (Figure 7E). They were able to repress the motility of cocultured mCherry-labeled U87MG cells upon lclPTEN-L induction, showing that they could transfer active lclPTEN-L to neighboring cells (Figure 7E).
Discussion
In order to assess the transfer of catalytically active versions of PTEN between cells, we developed a coculture assay that uses inhibition of glioblastoma cell motility as a readout of transfer of PTEN catalytic activity. We chose an assay with a biological activity readout rather than assaying for transfer of PTEN protein from cell to cell, because the latter type of assay gives no indication of activity and is therefore largely meaningless. U87MG glioblastoma cells were used to develop this assay because they are PTEN null32 and are well-known to show reduced motility upon reintroduction of PTEN.33, 34 Two controls were used to validate this assay. First, expression of normal PTEN effectively repressed motility of PTEN-expressing U87MG cells, but had no effect on cocultured non-PTEN-expressing U87MG cells. Thus, PTEN expression does not induce decreased motility on neighboring cells via indirect signaling mechanisms. Second, although lclPTEN-L was able to repress motility in neighboring cells, a catalytically inactive version of lclPTEN-L had no effect on the motility of neighboring cells. Thus, inhibition of motility is not due to non-specific effects of secretion of a protein with a polyarginine tract.
Using this assay, we do observe cell-to-cell transfer of PTEN-L in an active form, in agreement with the initial description of PTEN-L.14 However, our data suggest that the PTEN-L polyalanine tract may function relatively weakly as a signal sequence for secretion via the classical secretion pathway. Replacement of the polyarginine tract in PTEN-L with a bona fide signal sequence resulted in marked changes in PTEN-L behavior. lclPTEN-L, but not PTEN-L, could be detected in conditioned media without concentration. Intracellular levels of lclPTEN-L were sensitive to brefeldin A treatment, consistent with it being due to reuptake of secreted lclPTEN-L. Intracellular PTEN-L expression was unaffected by brefeldin A treatment, suggesting that intracellular PTEN-L levels may be due in part to inefficient secretion, rather than reuptake.
Our findings with lclPTEN-L show that PTEN-L can be engineered for efficient secretion via the classical secretion pathway in a form that is effectively taken up by neighboring cells. Repression of motility in non-expressing cells by lclPTEN-L was more effective than that seen with PTEN-L, consistent with the observed higher secretion levels. It may be possible to further improve the efficacy of lclPTEN-L by replacing the PTEN sequence in lclPTEN-L with the version of PTEN (ePTEN) with enhanced activity described by Nguyen et al.43
When expressed in either U87MG cells or neural stem cells, lclPTEN-L, but not PTEN-L, underwent proteolysis to a form that was slightly shorter than normal PTEN. This proteolysed form was not detected in conditioned media, where only the full-length form was observed. This suggests that proteolysis occurs upon reuptake of lclPTEN-L into cells, rather than in the endoplasmic reticulum or Golgi. Although the mechanism by which polyarginine tracts allow uptake of exogenous proteins into cells is not known, a possible mechanism is a partial unfolding/refolding mechanism occurring at the cell membrane. This could potentially render lclPTEN-L transiently susceptible to proteolysis. The shorter, proteolysed version of lclPTEN-L is only slightly smaller than normal PTEN and may therefore retain full activity.
Genetically modified neural stem cells are being evaluated for glioblastoma therapy, and the results of a recent first-in-human study are encouraging.44 We were able to engineer fibroblast-derived human neural stem cells to express lclPTEN-L. When these were cocultured with U87MG cells, they were able to repress the motility of glioblastoma cells, indicating that they could transfer lclPTEN-L catalytic activity to neighboring cells. Previous cell-mediated protein delivery strategies have been limited to delivering proteins that act on the cell surface of neighboring cells. This study expands the potential of cell-mediated protein delivery, showing that it can be used to deliver a protein that acts intracellularly on neighboring cells.
Materials and Methods
Antibodies and Inhibitors
PTEN rabbit monoclonal antibody 138G6, pAkt rabbit polyclonal antibody, and Oct4 2890S rabbit antibody were from Cell Signaling Technologies (Danvers, MA, USA). Total Akt goat polyclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Nestin MAB5326 mouse monoclonal and Sox2 AB5603 rabbit antibody were from Millipore (Etobicoke, ON, Canada). Pax6 901301 rabbit antibody and βIII-tubulin 801201 mouse monoclonal antibody were from BioLegend (San Diego, CA, USA). Brefeldin A was from Sigma-Aldrich (Oakville ON, Canada).
DNA Constructs
The following primers were used: PTENLA: 5′-AGGGAAGGTGGAAGCCGTGG-3′; PTENLB: 5′-GGCTGTCATGTCTGGGAGCC-3′; PTENL5: 5′-gctcAtggagAgAggcggagaagcggcggcgg-3′; PTENL3: 5′-GATGGCTGTGCCGTCTGGGAGCCTGTGGCTG-3′; PTENLx: 5′-GGATCCgctcAtggagAgAggcggagaagc-3′; PTENLY: 5′-GCTAACGATCTCTTTGATGATGGCTGTGCCGTCTGGG-3′; PTENlz: 5′-cccagacggcacagccatcatcaaagagatcgttagc 3′; PTENlentiREV: 5′–GAATTCTCAGACTTTTGTAATTTGTGTATGC-3′; LDR1: 5′-GGCTGCCAGGCGCTAGATGTGAGTCGCCTGTCACCATTTCC-3′; LDR2: 5′-GCTGGGCCTGCTCCTGCTCTGGCTGCCAGGCGCTAGATGTG-3′; LDR3: 5′-GGATCCATGCGGGTGCCCGCCCAGCTGCTGGGCCTGCTCCTGCTCTG-3′.
cDNA was prepared from T47D breast cancer cells using the Applied Biosystems High-Capacity cDNA Reverse Transcription kit (Foster City, CA, USA). The sequence encoding the 173 amino acids unique to PTEN-L was then PCR amplified using the primers PTENLA and PTENLB. This was ligated into the plasmid pDrive and fully sequenced. PTEN-L and lclPTEN-L constructs were made by PCR splicing using the DNA polymerase mix from the QIAGEN Long-Range PCR Kit (QIAGEN, Montreal, QC, Canada). To make cDNA for full-length PTEN-L with changes to enhance expression, we amplified cDNA from above with the primers PTENL5 and PTENL3, which introduced the sequence changes shown in Figure 1. The PTEN-L sequence was then amplified with the primers PTENLX and PTENLY. PTEN cDNA was amplified with the primers PTENLZ and PTENlentiRev. These two PCR products were then fused by PCR splicing using the primers PTENLX and PTENLentiRev. The final PCR product was ligated into pDRIVE and fully sequenced. cDNAs with the correct sequence were then subcloned into pLVX-Tight-Puro lentiviral expression vector (Clontech, Mountain View CA, USA).
To make cDNA for lclPTEN-L, we performed three rounds of PCR with the primers LDR1/PTENL3, LDR2/PTENL3, and LDR3 and PTENL3. This was then fused to PTEN cDNA as above, substituting the primer LDR3 for PTENLX. The PTEN C124S mutant was produced by site-directed mutagenesis using a QuikChange XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). cDNA for PTEN(C124S) was PCR spliced to the lclPTEN-L upstream sequence and subcloned into pLVX-Tight-Puro as above.
Cell Culture
U87MG cell culture was done as described previously.31 The identity of these cells was confirmed by short tandem repeat (STR) analysis. Cells were routinely confirmed to be free of mycoplasma contamination by PCR analysis with the VenorGeM Mycoplasma detection kit (Sigma-Aldrich).
Lentiviral Particle Generation and Transductions
Lentiviral particle generation was performed by four plasmid transfection into 293T cells as described previously.31 Transduction of U87MG cells was also done as described previously.31 Induction of protein expression was done using 1 μg/mL doxycycline.
Western Blotting, Immunofluorescence, and Proliferation Assays
Western blotting was done as described previously.31 Chemiluminescence from horseradish peroxidase (HRP)-conjugated secondary antibodies was detected with the Alpha Innotech FluorChem system (Santa Clara, CA, USA) and quantitated using Alphaview software. Amido black staining of the blot was used as a control for protein loading. Immunofluorescence was also done as described previously.31 Proliferation assays were performed using Click-iT EdU Alexa Fluor 488 Imaging Kits from Thermo Fisher Scientific (Waltham, MA, USA).
Videomicroscopy
Lentivirus with cDNA for mCherry fluorescent protein was made as above using the plasmid pLV-mCherry (plasmid 36084; Addgene). U87MG cells transduced with this lentivirus were cultured at a 1:1 ratio with U87MG cells expressing different versions of PTEN-L. Videomicroscopy was performed as described previously.45 Quantitation of the motility of the two cell populations was determined using the MtrackJ plugin46 in ImageJ software (NIH, Bethesda, MD, USA), also as described previously.45
Human Neural Stem Cell Generation and Lentiviral Transduction
Human iPSCs were generated from fibroblasts following the method described by Okita et al.47 iPSC-derived neural stem cells were generated using a modified dual-SMAD inhibition/neural induction protocol.42 iPSCs were dissociated and plated onto Matrigel-coated dishes at a density of 3.5 × 104 cells/cm2. Once wells were ∼80% confluent, differentiation was initiated by culturing cells in varying ratios of knockout serum replacement (KSR) media and N2 media (days 1–4, 100% KSR media; days 5–6, 3:1 ratio KSR:N2 media; days 7–8, 1:1 ratio KSR:N2 media; days 9–10, 1:3 ratio KSR:N2 media; days 11–12, 100% N2 media). TGF-β inhibitor SB-431542 (10 μM) was added for the first 4 days and the BMP inhibitor LDN-193189 (0.5 μM) for all 12 days.48 At day 12, cells were harvested with Accutase and are plated in the presence of 10 μM Rock inhibitor Y-27632 at a density of 1 × 105 cells/cm2 for selection and expansion of neural stem cells. Neural stem cells were maintained and passaged at high density on Matrigel-coated dishes in neural stem cell media (Neurobasal A medium supplemented with B27, N2, EGF, and FGF2). Lentiviral transductions were done overnight with undiluted media from four plasmid-transfected 293T cells. Cocultures with U87MG cells for videomicroscopy were done in neural stem cell media on either Matrigel- or laminin-coated plates (results were similar on both substrates).
Statistics
Statistical analyses were performed using SigmaPlot12 software. Data from videomicroscopy analyses were not normally distributed as determined by the Shapiro-Wilk test, and statistical significance was determined using the Mann-Whitney rank-sum test. A p value less than 0.05 was considered significant.
Author Contributions
S.J.L. made the constructs for expression of PTEN-L and modified versions of PTEN-L. S.J.L., A.G., and D.J. performed the experiments on the expression and cell-to-cell transfer of PTEN-L and modified versions of PTEN-L. C.V. and D.A.G. performed the analysis of PTEN-L evolutionary conservation. L.M.J. and W.L.S. generated the human neural stem cells. I.A.J.L. designed the experiments and wrote the initial manuscript. All authors reviewed the manuscript and provided comments that were incorporated in the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by operating grant MOP-136793 from the Canadian Institutes of Health Research (to I.A.J.L.), funds from the Ottawa Regional Cancer Foundation and Ottawa Hospital Foundation (to I.A.J.L.), and a catalyst grant by the Heart and Stroke Foundation Canadian Partnership for Stroke Recovery (to L.M.J. and W.L.S.). A.G. was supported by an Ontario Graduate Scholarship, and L.M.J. was supported by an Ontario Institute for Regenerative Medicine postdoctoral fellowship.
Footnotes
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.omtm.2017.11.006.
Supplemental Information
References
- 1.Chalhoub N., Baker S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozawa T., Riester M., Cheng Y.K., Huse J.T., Squatrito M., Helmy K., Charles N., Michor F., Holland E.C. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rankin S.L., Zhu G., Baker S.J. Review: insights gained from modelling high-grade glioma in the mouse. Neuropathol. Appl. Neurobiol. 2012;38:254–270. doi: 10.1111/j.1365-2990.2011.01231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wick W., Furnari F.B., Naumann U., Cavenee W.K., Weller M. PTEN gene transfer in human malignant glioma: sensitization to irradiation and CD95L-induced apoptosis. Oncogene. 1999;18:3936–3943. doi: 10.1038/sj.onc.1202774. [DOI] [PubMed] [Google Scholar]
- 7.Saborowski M., Saborowski A., Morris J.P., 4th, Bosbach B., Dow L.E., Pelletier J., Klimstra D.S., Lowe S.W. A modular and flexible ESC-based mouse model of pancreatic cancer. Genes Dev. 2014;28:85–97. doi: 10.1101/gad.232082.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miething C., Scuoppo C., Bosbach B., Appelmann I., Nakitandwe J., Ma J., Wu G., Lintault L., Auer M., Premsrirut P.K. PTEN action in leukaemia dictated by the tissue microenvironment. Nature. 2014;510:402–406. doi: 10.1038/nature13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., Abraham R.T. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y., Paluch B.E., Wang X., Jiang X. PTEN at a glance. J. Cell Sci. 2012;125:4687–4692. doi: 10.1242/jcs.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mense S.M., Barrows D., Hodakoski C., Steinbach N., Schoenfeld D., Su W., Hopkins B.D., Su T., Fine B., Hibshoosh H., Parsons R. PTEN inhibits PREX2-catalyzed activation of RAC1 to restrain tumor cell invasion. Sci. Signal. 2015;8:ra32. doi: 10.1126/scisignal.2005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassi C., Ho J., Srikumar T., Dowling R.J., Gorrini C., Miller S.J., Mak T.W., Neel B.G., Raught B., Stambolic V. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341:395–399. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulido R., Baker S.J., Barata J.T., Carracedo A., Cid V.J., Chin-Sang I.D., Davé V., den Hertog J., Devreotes P., Eickholt B.J. A unified nomenclature and amino acid numbering for human PTEN. Sci. Signal. 2014;7:pe15. doi: 10.1126/scisignal.2005560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins B.D., Fine B., Steinbach N., Dendy M., Rapp Z., Shaw J., Pappas K., Yu J.S., Hodakoski C., Mense S. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science. 2013;341:399–402. doi: 10.1126/science.1234907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lonser R.R., Sarntinoranont M., Morrison P.F., Oldfield E.H. Convection-enhanced delivery to the central nervous system. J. Neurosurg. 2015;122:697–706. doi: 10.3171/2014.10.JNS14229. [DOI] [PubMed] [Google Scholar]
- 16.Voges J., Reszka R., Gossmann A., Dittmar C., Richter R., Garlip G., Kracht L., Coenen H.H., Sturm V., Wienhard K. Imaging-guided convection-enhanced delivery and gene therapy of glioblastoma. Ann. Neurol. 2003;54:479–487. doi: 10.1002/ana.10688. [DOI] [PubMed] [Google Scholar]
- 17.Mastorakos P., Song E., Zhang C., Berry S., Park H.W., Kim Y.E., Park J.S., Lee S., Suk J.S., Hanes J. Biodegradable DNA nanoparticles that provide widespread gene delivery in the brain. Small. 2016;12:678–685. doi: 10.1002/smll.201502554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta A.M., Sonabend A.M., Bruce J.N. Convection-enhanced delivery. Neurotherapeutics. 2017;14:358–371. doi: 10.1007/s13311-017-0520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biglari A., Southgate T.D., Fairbairn L.J., Gilham D.E. Human monocytes expressing a CEA-specific chimeric CD64 receptor specifically target CEA-expressing tumour cells in vitro and in vivo. Gene Ther. 2006;13:602–610. doi: 10.1038/sj.gt.3302706. [DOI] [PubMed] [Google Scholar]
- 20.Chan J.K., Lam P.Y. Human mesenchymal stem cells and their paracrine factors for the treatment of brain tumors. Cancer Gene Ther. 2013;20:539–543. doi: 10.1038/cgt.2013.59. [DOI] [PubMed] [Google Scholar]
- 21.Bagó J.R., Sheets K.T., Hingtgen S.D. Neural stem cell therapy for cancer. Methods. 2016;99:37–43. doi: 10.1016/j.ymeth.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang H., He S., Yang J., Jia X., Wang P., Chen X., Zhang Z., Zou X., McNutt M.A., Shen W.H., Yin Y. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 2014;19:836–848. doi: 10.1016/j.cmet.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia X. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013;30:1720–1728. doi: 10.1093/molbev/mst064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gemayel R., Vinces M.D., Legendre M., Verstrepen K.J. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 27.Mularoni L., Ledda A., Toll-Riera M., Albà M.M. Natural selection drives the accumulation of amino acid tandem repeats in human proteins. Genome Res. 2010;20:745–754. doi: 10.1101/gr.101261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thornton J.A. Splicing by overlap extension PCR to obtain hybrid DNA products. Methods Mol. Biol. 2016;1373:43–49. doi: 10.1007/7651_2014_182. [DOI] [PubMed] [Google Scholar]
- 29.Bugaut A., Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabat E.A., Wu T.T., Foeller C., Perry H.M., Gottesman K.S. NIH Publication No. 91-3242; 1991. Sequences of Proteins of Immunological Interest, Fifth Edition. [Google Scholar]
- 31.Gont A., Hanson J.E., Lavictoire S.J., Parolin D.A., Daneshmand M., Restall I.J., Soucie M., Nicholas G., Woulfe J., Kassam A. PTEN loss represses glioblastoma tumor initiating cell differentiation via inactivation of Lgl1. Oncotarget. 2013;4:1266–1279. doi: 10.18632/oncotarget.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishii N., Maier D., Merlo A., Tada M., Sawamura Y., Diserens A.C., Van Meir E.G. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura M., Gu J., Takino T., Yamada K.M. Tumor suppressor PTEN inhibition of cell invasion, migration, and growth: differential involvement of focal adhesion kinase and p130Cas. Cancer Res. 1999;59:442–449. [PubMed] [Google Scholar]
- 34.Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 35.Niu T.K., Pfeifer A.C., Lippincott-Schwartz J., Jackson C.L. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith M.H., Ploegh H.L., Weissman J.S. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naguib A., Bencze G., Cho H., Zheng W., Tocilj A., Elkayam E., Faehnle C.R., Jaber N., Pratt C.P., Chen M. PTEN functions by recruitment to cytoplasmic vesicles. Mol. Cell. 2015;58:255–268. doi: 10.1016/j.molcel.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maehama T., Dixon J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 39.Aboody K.S., Brown A., Rainov N.G., Bower K.A., Liu S., Yang W., Small J.E., Herrlinger U., Ourednik V., Black P.M. Neural stem cells display extensive tropism for pathology in adult brain: evidence from intracranial gliomas. Proc. Natl. Acad. Sci. USA. 2000;97:12846–12851. doi: 10.1073/pnas.97.23.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metz M.Z., Gutova M., Lacey S.F., Abramyants Y., Vo T., Gilchrist M., Tirughana R., Ghoda L.Y., Barish M.E., Brown C.E. Neural stem cell-mediated delivery of irinotecan-activating carboxylesterases to glioma: implications for clinical use. Stem Cells Transl. Med. 2013;2:983–992. doi: 10.5966/sctm.2012-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagó J.R., Alfonso-Pecchio A., Okolie O., Dumitru R., Rinkenbaugh A., Baldwin A.S., Miller C.R., Magness S.T., Hingtgen S.D. Therapeutically engineered induced neural stem cells are tumour-homing and inhibit progression of glioblastoma. Nat. Commun. 2016;7:10593. doi: 10.1038/ncomms10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen H.N., Yang J.M., Afkari Y., Park B.H., Sesaki H., Devreotes P.N., Iijima M. Engineering ePTEN, an enhanced PTEN with increased tumor suppressor activities. Proc. Natl. Acad. Sci. USA. 2014;111:E2684–E2693. doi: 10.1073/pnas.1409433111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Portnow J., Synold T.W., Badie B., Tirughana R., Lacey S.F., D’Apuzzo M., Metz M.Z., Najbauer J., Bedell V., Vo T. Neural stem cell-based anticancer gene therapy: a first-in-human study in recurrent high-grade glioma patients. Clin. Cancer Res. 2017;23:2951–2960. doi: 10.1158/1078-0432.CCR-16-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gont A., Hanson J.E., Lavictoire S.J., Daneshmand M., Nicholas G., Woulfe J., Kassam A., Da Silva V.F., Lorimer I.A. Inhibition of glioblastoma malignancy by Lgl1. Oncotarget. 2014;5:11541–11551. doi: 10.18632/oncotarget.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meijering E., Dzyubachyk O., Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 47.Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 48.Mica Y., Lee G., Chambers S.M., Tomishima M.J., Studer L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013;3:1140–1152. doi: 10.1016/j.celrep.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.