Abstract
Production of propionic acid by fermentation of propionibacteria has gained increasing attention in the past few years. However, biomanufacturing of propionic acid cannot compete with the current oxo-petrochemical synthesis process due to its well-established infrastructure, low oil prices and the high downstream purification costs of microbial production. Strain improvement to increase propionic acid yield is the best alternative to reduce downstream purification costs. The recent generation of genome-scale models for a number of Propionibacterium species facilitates the rational design of metabolic engineering strategies and provides a new opportunity to explore the metabolic potential of the Wood-Werkman cycle. Previous strategies for strain improvement have individually targeted acid tolerance, rate of propionate production or minimisation of by-products. Here we used the P. freudenreichii subsp. shermanii and the pan-Propionibacterium genome-scale metabolic models (GEMs) to simultaneously target these combined issues. This was achieved by focussing on strategies which yield higher energies and directly suppress acetate formation. Using P. freudenreichii subsp. shermanii, two strategies were assessed. The first tested the ability to manipulate the redox balance to favour propionate production by over-expressing the first two enzymes of the pentose-phosphate pathway (PPP), Zwf (glucose-6-phosphate 1-dehydrogenase) and Pgl (6-phosphogluconolactonase). Results showed a 4-fold increase in propionate to acetate ratio during the exponential growth phase. Secondly, the ability to enhance the energy yield from propionate production by over-expressing an ATP-dependent phosphoenolpyruvate carboxykinase (PEPCK) and sodium-pumping methylmalonyl-CoA decarboxylase (MMD) was tested, which extended the exponential growth phase. Together, these strategies demonstrate that in silico design strategies are predictive and can be used to reduce by-product formation in Propionibacterium. We also describe the benefit of carbon dioxide to propionibacteria growth, substrate conversion and propionate yield.
Keywords: Propionic acid, Propionibacteria, Metabolic engineering, Genome-scale modelling, Pentose phosphate pathway, Phosphoenolpyruvate carboxykinase overexpression
Highlights
-
•
Genetic engineering strategies were designed using genome-scale models.
-
•
PPP over-expression increased propionate:acetate 4-fold during exponential growth.
-
•
Sparging with CO2 enhanced growth and conversion of glucose to propionate.
-
•
Overexpression of PEPCK and MMD extended the exponential growth phase.
1. Introduction
Propionic acid is a three-carbon compound with a broad range of applications in the food, pharmaceutical and chemical industries. In addition to its use as an intermediate for the synthesis of cellulose fibres, herbicides, perfumes and pharmaceuticals, propionic acid is a strong antimicrobial agent used in animal feed and as a food preservative (Guan et al., 2015b). Propionic acid is mainly produced by petrochemical processes. However, the demand for green chemicals has renewed attention to fermenting propionibacteria for propionic acid production. Propionibacteria are gram-positive, rod-shaped, facultative anaerobes. Dairy propionibacteria are important industrial GRAS (generally recognized as safe) strains used in the food and cheese industry (Meile et al., 2008). Propionibacteria have long been employed as starter and ripening cultures, responsible for flavour development and participating in the formation of cheese eyes (Britz and Riedel, 1994, Thierry and Maillard, 2002). Dairy strains are also used industrially to produce trehalose and vitamin B12 (Kośmider et al., 2012, Ruhal and Choudhury, 2012).
The bioprocess for propionate production still suffers from low productivity, low yield, and expensive downstream purification costs; acetic and succinic acids are produced alongside propionic acid making the downstream separation costly (Liu et al., 2012, Rodriguez et al., 2014). Metabolic engineering strategies aiming to reduce by-products are thus needed. Improved yields can theoretically be achieved by deleting by-product genes, but metabolic manipulation of propionibacteria has developed slowly, and only a few examples exist in the literature engineering the best propionic acid producer, P. acidipropionici. Metabolic engineering strategies have focussed on enhancing flux through the Wood-Werkman cycle, either by directing carbon into the cycle through over-expression of pyruvate and phosphoenolpyruvate carboxylases or the pyruvate:methylmalonyl-CoA carboxyltransferase (Ammar et al., 2014, Liu et al., 2016, Wang et al., 2015b). Other strategies that have been tested include the over-expression of components of the Wood-Werkman cycle itself (Liu et al., 2015, Wang et al., 2015a) or the targeting of acid tolerance associated processes such as trehalose synthesis (Jiang et al., 2015) and the glutamate decarboxylase or arginine deiminase pathways (Guan et al., 2015a). While P. acidipropionici remains resistant to engineering, unique metabolic features of this species were over-expressed in lower producers (Wang et al., 2015b). Few studies have targeted by-product production, and these have ultimately had limited success (Liu et al., 2016, Suwannakham et al., 2006). Ultimately, the limited phenotypic improvements in current studies and limitations in genetic engineering tools necessitate a genome-scale model to further aid design (Wang et al., 2015b).
Here, we utilise a recently developed genome-scale model for P. freudenreichii subsp. shermanii and the pan-Propionibacterium GEM to design genetic modification strategies. Because the production of propionic acid is already growth-coupled, classic growth-coupling strain design algorithms such as OptKnock (Burgard et al., 2003) favour sub-optimal strategies that limit energy generation, leaving cells more susceptible to acid stress. Given the depletion of the pH gradient is regarded as a major mechanism by which weak acids exert their toxicity on cells, we explored strategies favouring propionate production while maintaining high levels of energy generation or even exceeding those of wild-type strains. Strategies probed pathways consistent with the maximum production of energy and propionate in the native metabolism. By simultaneously trying to improve both energy output and propionate production, strains are expected to be readily evolvable to a higher producing phenotype while also improving acid tolerance. Such approaches facilitate the production of high producing mutants with minimum genetic perturbations; a requirement enforced by the few genetic modification tools available. Given the limited success of reducing acetate production in previous studies (Liu et al., 2016, Suwannakham et al., 2006), we over-expressed the PPP to test in silico simulation predictions and show that such a strategy results in lower acetate production without sacrificing growth. We then tested an alternative strategy consisting of over-expressing an ATP-PEPCK and MMD to enhance energy available to the cell and analyse how this affects the phenotype. We observed an extended exponential growth phase in the mutant strain. Because this strategy was reliant on the addition of CO2, the influence of CO2 on the Propionibacterium fermentation was also assessed, revealing undescribed biomass and propionate stimulating effects in propionibacteria.
2. Materials and methods
2.1. Model preparation
Simulations using the pan-Propionibacterium and P. freudenreichii subsp. shermanii models (manuscript in preparation) were performed in MATLAB using the COBRA toolbox (Hyduke et al., 2011). The original excel models were first converted to SBML format and boundary metabolite exchanges were added to allow accumulation using an in-house tool, as previously described (Quek and Nielsen, 2008). Glucose was allowed to be consumed as a carbon source at a rate of 1 mmol/gDW/h, while all other carbon-containing exchange metabolites were set to production only. No external terminal electron acceptors were available, including oxygen, sulphate and nitrate. Ammonium, phosphate, water and hydrogen sulphide fluxes were unconstrained. When required, additional reactions were added from the modelSEED database (Overbeek et al., 2005). Reactions referenced herein have been matched to their unique modelSEED reaction identifiers or by the identifiers given within the original model (Supplementary material Table S1).
2.2. Identification of pathways of interest
Pathways enabling maximum theoretical yield of propionate were hierarchically screened with respect to ATP yield in P. freudenreichii subsp. shermanii. This was performed by changing the objective function to maximise for the propionate boundary exchange reaction with a small weighting for the ATP drain reaction. Subsequent minimisation of total flux was utilised to assist in the identification of pathways that maximise yield. A key reaction in the pathway was then constrained to carry no flux so that alternative pathways could be determined. The procedure was repeated until the maintenance fell below 75% of the wild-type value. The same analysis was also performed on the pan-model. Pathways that could increase propionate production while maintaining a constant maximum energy yield were similarly calculated by maximising for the ATP drain reaction with a small bias for the propionate exchange flux. OptKnock simulations were implemented through the COBRA toolbox (Burgard et al., 2003).
2.3. Pathway yield calculation
Pathway yields were calculated by allowing free flux through central carbon metabolism. Flux through pathways that were identified to enhance propionate production were constrained as appropriate for the relevant scenario by constraining key enzymatic steps. These included citramalate synthase for the citramalate pathway, citrate synthase for the TCA cycle, and formate-tetrahydrofolate ligase for the glycine cleavage pathway. For each strategy, the expected yield of propionate with energy optimisation was calculated by maximising and minimising the propionate yield after maximising for the ATP yield. Additionally, the maximum energy yield associated with maximum propionate production was calculated by maximising the ATP yield with propionate constrained at the maximum theoretical yield of 1.714 moles/mole glucose. In all strategies, the methylmalonyl-CoA carboxyltransferase was set to irreversible (lower bound 0) to prevent infinite energy cycling when the sodium-pumping MMD was added. The relevant constraints for each strategy are detailed as follows: 1. GAPDH replacement of PGK and Bifidobacterium shunt knockout (constrained to 0 flux: phosphoglycerate kinase, phosphoketolase; added to model: GAPDH); 2. Acetate knock-out (constrained to 0 flux: citrate synthase, acetate kinase, cystathionine gamma-synthase, formate-tetrahydrofolate ligase); 3. PPP over-expression (constrained to 0 flux: citrate synthase, citramalate synthase, formate-tetrahydrofolate ligase); 4. Citramalate pathway over-expression (constrained to 0 flux: citrate synthase); 5. TCA cycle over-expression (constrained to 0 flux: citramalate synthase); 6. Glycine cleavage pathway over-expression (added to model: formate dehydrogenase); 7. Linear propionate pathway (added to model: PEPCK, MMD); 8. Combination of strategies 6 and 7 (added to model: formate dehydrogenase, PEPCK, MMD).
2.4. Bacterial strains and growth conditions
E. coli DH5α was grown aerobically in Luria-Bertani medium at 37 °C and supplemented with 50 μg/mL apramycin (Am), 100 μg/mL ampicillin (Ap) or 50 μg/mL kanamycin (Km) when needed. Propionibacterium strains were grown anaerobically at 32 °C on complex medium containing yeast extract 10 g/L, trypticase soy broth 5 g/L, K2HPO4 0.25 g/L, MnSO4 0.05 g/L and glucose 80 g/L to mimic industrial-scale process conditions. Medium components and the carbon source were sterilized separately for 20 min at 121 °C. Sodium lactate broth (NBL) medium containing 10 g/L yeast extract, 10 g/L trypticase soy broth, and sodium lactate 10 g/L (Kiatpapan and Murooka, 2001), were used for the preparation of propionibacteria electrocompetent cells. When needed, media was supplemented with 50 μg/mL of erythromycin (Ery). Propionibacterium strains were kept at −80 °C using glycerol (20%) as cryoprotector. Strains and plasmids used in this study are listed in Table 1.
Table 1.
Bacterial strains, plasmids and primers used in this study.
| Strain, plasmid or primer | Description | Reference |
|---|---|---|
| Strains | ||
| P. acidipropionici | ATCC 4875 | ATCC |
| P. freudenreichii subsp. shermanii | ATCC 9614 | ATCC |
| Pf-ZOP | ATCC 9614 carrying plasmid pPACV76 | This study |
| Pf-EMP | ATCC 9614 carrying plasmid pPACV70 | This study |
| Pf-PAM | ATCC 9614 carrying plasmid pPACV86 | This study |
| E. coli DH5α | E. coli K12 F-lacU169 (φ80lacZ∆M15) endA1 recA1 hsdR17 deoR supE44λ thi-1 gyrA96 relA1 | Life Technologies |
| Plasmids | ||
| pRGO1 | Native plasmid P. acidipropionici ATCC 4875 | (Kiatpapan et al., 2000) |
| pGEM-T | Cloning vector, ApR | Promega |
| pCR-Blunt II-TOPO | Cloning vector, KmR | Invitrogen |
| pSET152 | Cloning vector for conjugal transfer E. coli-Streptomyces, AmR | (Bierman et al., 1992) |
| pPACV16 | pGEM-T derivative carrying orf1, orf2, orf5 and orf6 from pRGO1 | This study |
| pPACV29 | pPACV16 derivative carrying origin for intergeneric transfer and aac(3)IV gene from pSET152, AmR | This study |
| pBluescript II SK (+) | Cloning vector, ApR | Strategene |
| pPACV50 | pPACV29 derivative carrying multiple cloning sites and lacZ alpha fragment from pACYC, AmR | This study |
| pPACV70 | pPACV50 derivative carrying ermE gene from S. erytharea, EryR | This study |
| pPACV76 | pPACV70 derivative carrying zwf, opcA, and pgl genes from P. acidipropionici ATCC 4875, EryR | This study |
| pTOPO-MMD | pCR-Blunt II-TOPO derivative carrying mmd genes from P. acidipropionici ATCC 4875, KmR. | This study |
| pPACV81 | pPACV70 derivative carrying pckA gene from E. coli, EryR | This study |
| pPACV86 | pPACV81 derivative carrying mmd genes from P. acidipropionici ATCC 4875, EryR | This study |
| Primers | ||
| pRGO1_F | ACACCGTGATGCGCCCATGT | |
| pRGO1_R | CTCTACTGCTAGCCATCAGT | |
| pSET152_F | CTGTGCGCAGAGTTGGTAGCTCTTGATCC | |
| pSET152_R | CTGTGCGCAGGATCTTTTCCGCTGCATAA | |
| MCS_F | CTTATCGTTTAAACGCAGTGAGCGCAACGCAATT | |
| MCS_R | GGCGCTGTTTAAACCTAGTCCCATTCGCCATTCAGGC | |
| ermE_F | TTTTCTAGACTACCGCTGCCCGGGTCC | |
| ermE_R | GTGCAGGTACCAGCCCGAC | |
| ZOP_F | TTTCATATGCTCGACACAGGTTCCGC | |
| ZOP_R | TTTTCTAGATCACAGCACGCAGTTGTACC | |
| PermE_F | TTTTCTAGAGGTTCGACTGGCACCGGCACCGCGC | |
| PermE_R | TTTCATATGCGCTGGATCCTACCAACCGG | |
| pckA_F | TTTAAGCTTCATATGCGCGTTAACAATGG | |
| pckA_R | TTTTCTAGAGGTACCTTACAGTTTCGGACCAGCCG | |
| PermE_F1 | TTTAAGCTTGGTACCGGTTCGACTGGCACCGGCACCGCGC | |
| PermE_R1 | TTTTCATATGCGCTGGATCCTACCAACCGG | |
| PAM_F | TTTTGGTACCCATATGCAGGCCGACGGCAGGAC | |
| PAM_R | TTTTTCTAGATTATAACTAGAAACTCCCCATCAGGG | |
| PF_F | CTTCACCCGGGCCATC | |
| PF_R | CCGAGCACGATCCGATG |
2.5. Genomic DNA and plasmid extraction
Genomic DNA of P. freudenreichii subsp. shermanii, P. acidipropionici and Saccharopolyspora erythraea were extracted using Invitrogen PureLink Genomic DNA kit. Plasmid extraction was performed using QIAgen MiniPrep plasmid purification kit. For plasmid extraction from propionibacteria, 5 mL of overnight culture was centrifuged and resuspended in 250 μL of buffer P1 containing 100 μg/mL RNAse and 20 mg/mL of lysozyme (Sigma-Aldrich). Cells were incubated at 37 °C for 30 min and extraction was continued as specified in the plasmid purification kit protocol.
2.6. Construction of expression vector
pRGO1 plasmid extracted from P. acidipropionici ATCC 4875 was used as a template for amplification of a PCR fragment containing regions orf1, orf2, orf5 and orf6 (Kiatpapan et al., 2000), using primers pRGO1_F and pRGO1_R (Table 1). The resulting PCR fragment was cloned into pGEM-T vector obtaining plasmid pPACV16. Plasmid pSET152 was used as a template for amplification of a PCR fragment containing the apramycin resistance gene, aac(3)IV, and the origin of intergeneric transfer (Bierman et al., 1992). Primers pSET152_F and pSET152_R containing FspI restriction sites were used for amplification (Table 1). The resulting fragment was digested with FspI and subsequently ligated to DraI/SspI digested plasmid pPACV16, obtaining plasmid pPACV29. The multiple cloning site and lacZ alpha fragments were amplified from pBluescript II SK (+) using primers MCS_F and MCS_R with PmeI sites (Table 1) and cloned into NheI/Klenow treated plasmid pPACV29 obtaining plasmid pPACV50. The erythromycin resistance gene, ermE, was amplified from Saccharopolyspora erythraea genomic DNA using primers ermE_F and ermE_R with KpnI and XbaI sites, respectively (Table 1). ermE was cloned into digested KpnI/XbaI pPACV50, obtaining plasmid pPACV70. zwf, glucose-6-phosphate 1-dehydrogenase (PACID_22670), opcA, putative allosteric activator of Zwf (PACID_22680) and pgl, 6-phosphogluconolactonase (PACID_22690), genes were amplified as a single fragment using P. acidipropionici ATCC 4875 genomic DNA as a template and primers ZOP_F and ZOP_R with NdeI and XbaI sites, respectively (Table 1). ermE promoter (PermE) was amplified using S. erythraea genomic DNA as a template and primers PermE_F and PermE_R with XbaI and NdeI sites, respectively (Table 1). PACID_22670, PACID_22680 and PACID_22690 and PermE PCR fragments were digested with XbaI/NdeI and ligated to XbaI digested plasmid pPACV70 obtaining plasmid pPACV76.
pckA gene coding for PEPCK from E. coli was amplified from E. coli DH5α genomic DNA as template and primers pckA_F and pckA_R with NdeI and KpnI sites. PermE was amplified using S. erythraea genomic DNA as a template and primers PermE_F1 and PermE_R1 with KpnI and NdeI sites, respectively (Table 1). PEPCK and PermE PCR fragments were digested with KpnI/NdeI and ligated to KpnI digested plasmid pPACV70 obtaining plasmid pPACV81.
PACID_02160, PACID_02170, PACID_02180 and PACID_02190 genes for MMD were amplified as a single fragment using P. acidipropionici ATCC 4875 genomic DNA as a template and primers PAM_F and PAM_R with NdeI and XbaI sites, respectively (Table 1). MMD and PermE, amplified with primers PermE_F and PermE_R, PCR fragments were digested with NdeI/XbaI and ligated to XbaI digested plasmid pCR-Blunt II-TOPO obtaining plasmid pTOPO-MMD. pTOPO-MMD XbaI fragment was posteriorly cloned into XbaI digested pPACV81 obtaining pPACV86.
2.7. Transformation protocol
Transformation of P. freudenreichii subsp. shermanii was performed by electroporation as described in previous studies (Ammar et al., 2013, Kiatpapan et al., 2000). Briefly, cells were grown in NBL medium until OD 0.5–1, cooled on ice for 15 min, and centrifuged at 5000 rpm at 4 °C. Cells were then washed four times with cold glycerol 10% and resuspended in electroporation buffer (1/50 of the original volume). 90 μL of resuspended cells were mixed with 1–2 μg of plasmid DNA in a 0.1 cm cuvette and incubated on ice for 10 min. Electroporation was performed using a MicroPulser Electroporator (Biorad Laboratories, Richmond, CA, USA) at 20 kV/cm and time constants between 4.5 and 5.0 ms. After an electroporation pulse, 1 mL of recovery NBL medium was added and cells were incubated for 3 h at 32 °C. Cells were posteriorly plated on NBL agar supplemented with 50 μg/mL of erythromycin. Colonies appeared on plates after 10–15 days of incubation. Transformants were confirmed by plasmid extraction and restriction profiles. To confirm that the positive transformants were P. freudenreichii subsp. shermanii strains, genomic DNA was extracted and PCR amplification was performed using oligos PF_F and PF_R (Table 1). To confirm segregational stability of plasmids, P. freudenreichii subsp. shermanii transformants were grown for 25 generations without any addition of antibiotics and plated on solid medium with and without erythromycin. No differences were observed between number of colonies in each plate, indicating the plasmids were segregationally stable. Structural stability was also studied by cultivation in selective medium for 25 generations and subsequent plasmid extraction. No changes in the restriction profile were observed, indicating that the plasmids were structurally stable.
2.8. Fermentation conditions
Glycerol stock cultures were revived in 1.5 mL Eppendorf tubes containing 1 mL of complex media inoculated with 0.8% (v/v) of the glycerol stock and grown for 24 h at 32 °C. The culture was transferred to a 15 mL Falcon tube containing 14 mL of complex media and grown for an additional 24 h. 5% (v/v) of this culture was used to inoculate 250 mL serum bottles containing 100 mL of complex media and grown for an additional 24 h. Cells from the serum bottles in mid-exponential phase were used to inoculate the fermenter at an initial OD600nm of 0.3. The Pf-ZOP and Pf-EMP fermentation were performed using a 1 L BIOSTAT® A fermenter (Luna-Flores et al., 2016). The fermenter was equipped with standard probes and controllers, controlling pH, dissolved oxygen, temperature, and agitation. The agitation rate was kept constant at 300 rpm and pH was controlled at 6.4 using 10 M NaOH. The temperature of the culture was maintained at 32 °C using an electric jacket and a recirculation chiller. The exhaust gas condenser was set to 20% to avoid medium evaporation. Prior to inoculation, the fermenter was sparged with N2 for at least 15 min. A constant N2 flow was kept for the entire fermentation at a flow rate of 300 sccm.
Fermentations for strain Pf-PAM and CO2-sparged Pf-EMP were performed in Multifors Infors HT 1 L bioreactors with an operation volume of 0.9 L. The fermenters were equipped with standard probes and controllers, controlling pH, dissolved oxygen, temperature, and agitation. The agitation rate was kept constant at 300 rpm and pH was controlled at 6.4 using 10 M NaOH. The temperature of the culture was maintained at 32 °C. The fermenter was initially sparged with a gas mix Ar (80%) and CO2 (20%) for at least 15 min. A constant gas mix Ar (80%) and CO2 (20%) flow was kept for the entire fermentation at 0.3 VVM (270 sccm). All fermentations were performed in duplicates.
2.9. Analytical methods
The optical density of the culture was measured at 600 nm using a Biochrom Libra S12 UV/Vis. Organic acids and carbohydrates were quantified by ion-exclusion chromatography using an Agilent 1200 HPLC system and an Agilent Hiplex H column (300 × 7.7 mm, PL1170-6830) with a guard column (SecurityGuard Carbo-H, Phenomenex PN: AJO-4490). Sugars and alcohols were monitored using a refractive index detector (Agilent RID, G1362A), set on positive polarity and optical unit temperature of 40oC. Organic acids were monitored at 210 nm (Agilent MWD, G1365B). 30 μL of the sample was injected onto the column using an autosampler (Agilent HiP-ALS, G1367B), and column temperature kept at 40 °C using a thermostatted column compartment (Agilent TCC, G1316A). Analytes were eluted isocratically with 4 mM H2SO4 at 0.4 mL/min for 50 min. Chromatograms were integrated using the software ChemStation.
3. Results
3.1. Strain design algorithms
Initial genetic engineering targets were designed using the OptKnock algorithm. Three key strategies were identified to improve flux towards propionate. The first strategy involved reducing the total energy available to the organism by knocking out the energy yielding steps of glycolysis. Knocking-out phosphoglycerate kinase (pgk) forced the cells to increase organic acid production relative to growth. This can be compensated by the introduction of the NADPH-dependent glyceraldehyde-3-phospate dehydrogenase (gapdh). The final two strategies directly targeted acetate production. The first was the simplistic knockout of acetate, achieved in P. freudenreichii subsp. shermanii by targeting the acetate kinase. The second strategy targeted the redox balance instead to favour propionate production and inhibit acetate production by targeting glucose-6-phosphate isomerase (pgi), forcing flux through the PPP and generating an excess of reduced cofactors. This strategy is superseded by the over-expression of the PPP which can be tailored to maximise for energy production (Fig. 1) and is not commented on further. The knockout strategies result in lower energy yields and are therefore less favourable (Table 2).
Fig. 1.
Phase plane analysis for glucose catabolism by P. freudenreichii subsp. shermanii demonstrating the influence of the propionate yield and PPP expression on the ATP yield with an objective of ATP maximisation. Phosphoglycerate kinase knockout strategy (pgk KO), acetate knockout strategy (acetate KO), TCA or citramalate pathway over-expression strategy (TCA/Cit OE), glycine cleavage based pathway over-expression (Gly cleavage pathway), linear propionic acid pathway strategy (Linear PA pathway) and the combined strategy (Linear PA + Gly cleavage), have been mapped on for comparison. Each point represents optimal over-expression of the given pathway for propionate or energy production or the expected metabolism after knock-out.
Table 2.
Simulated flux distributions under energy maximisation and propionate maximisation with availability of the associated metabolic engineering strategies.
| Strategy |
ATP maximisationa |
PA maximisationb | |||
|---|---|---|---|---|---|
| ATP | PA max | PA min | ATP | ||
| Wild-type | l1-norm solution using central carbon metabolism | 4.00 | 1.33 | 1.33 | 3.43c |
| Knock-out | Energy yielding step of glycolysis (PGK) | 2.00 | 1.33 | 1.33 | 1.71 |
| Acetate kinase (ACK) | 3.43 | 1.71 | 1.71 | 3.43c | |
| Over-expression | Pentose phosphate pathway | 4.00 | 1.33 | 1.33 | 3.43 |
| TCA/citramalate pathways | 4.00 | 1.33 | 1.33 | 3.71 | |
| Glycine cleavage system pathway | 4.00 | 1.6 | 1.33 | 3.43 | |
| Glycine cleavage system pathway with formate dehydrogenase | 4.00 | 1.71 | 1.33 | 4.00 | |
| Linearised PA pathway | 4.33 | 1.33 | 1.33 | 3.86 | |
| Glycine cleavage and linear PA pathways | 4.43 | 1.71 | 1.71 | 4.43 | |
ATP maximisation represents the presumed objective function of metabolism, consistent with wild-type flux distributions. Strategies that enhance the minimum propionate production at maximisation of ATP are growth coupling, while strategies that additionally enhance maximum ATP production likely improve acid tolerance. Non-growth coupled strategies cannot rely on evolution and will require additional interventions to achieve the desired phenotype.
Optimal over-expression of the pathways to achieve the maximum theoretical yield of propionate at the highest ATP yield possible. A higher energy here makes the strategy more favourable.
The wild-type and acetate kinase knock-out pathways are presumed to utilise the pentose phosphate pathway when maximising for propionate production.
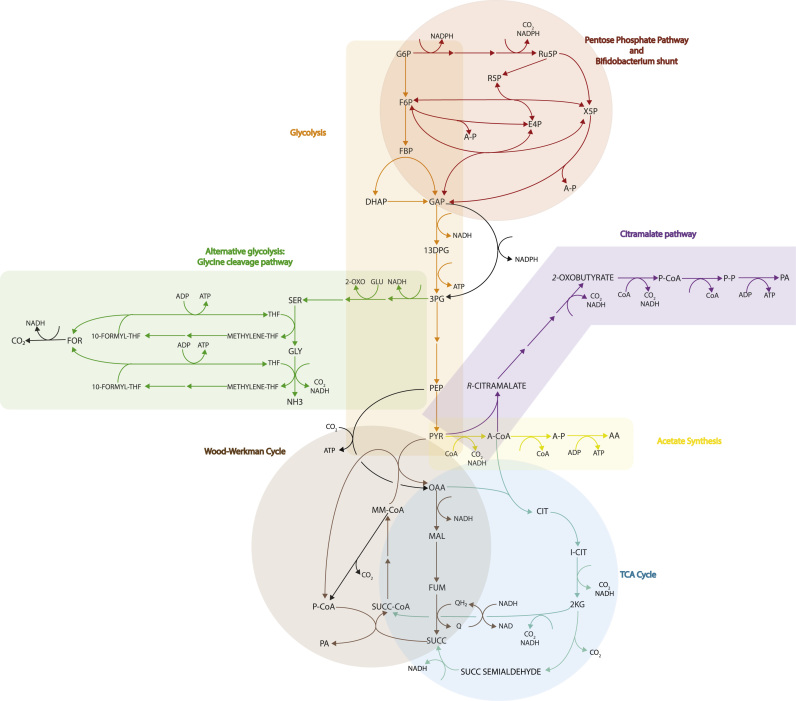
3.2. Native P. freudenreichii subsp. shermanii pathways associated with improved propionic acid yields
The native metabolism of propionate biosynthesis in P. freudenreichii subsp. shermanii was explored using two approaches. Guided by the knowledge that the maximal theoretical yield of propionate is redox limited, we first identified pathways that were capable of achieving the maximum theoretical yield of propionate production. The second objective was to maximise ATP generation. This gave a series of over-expression targets that generate additional reduced cofactors that can shift metabolism towards propionate production under minimal energetic cost. We then studied pathways that could improve propionate production without energy penalty. In all cases, fluxes were redistributed through pathways that produced more reduced cofactors and therefore allowed an increase in propionate titre.
Modelling suggested that at the maximum theoretical yield of propionate production of 1.7 mol/mol glucose, 3.7 mol ATP/glucose can be achieved. This is more than 90% of the maximum energy yield of the native strain (which can achieve 4 ATP/glucose), suggesting that the homofermentative production of propionate suffers only a relatively minor energetic disadvantage. Two pathways were identified that could achieve the maximum propionate yield with this energy yield: the citramalate pathway, which synthesises 2-oxobutyrate from pyruvate and acetyl-CoA that can be further degraded to propionate with a corresponding gain in ATP, and the oxidative branch of the tricarboxylic acid (TCA) cycle (Fig. 2). One final pathway was identified as a potential candidate, the PPP. This pathway suffers a minor energetic penalty compared to the previously mentioned pathways with a yield of 3.4 ATP/glucose at the maximum theoretical yield of propionate (Table 2). The expected propionate and energy yields from this pathway are demonstrated in Fig. 1 at varying levels of expression, with an optimum flux of 86% of the glucose uptake rate to maximise for both energy and propionate.
Fig. 2.
Central carbon metabolism and relevant metabolic pathways of P. freudenreichii subsp. shermanii discussed in this work. Pathways are colour-coded. Upregulation of the glycine cleavage pathway, TCA cycle (via the alpha-ketoglutarate dehydrogenase), the citramalate pathway or the PPP are all possible ways to source additional reduced cofactors for propionate generation, although the ATP generation through the glycine cleavage pathway makes this particularly attractive. Potential knock-out strategies target glycolysis or the acetate synthesis pathway. Non-native enzymes proposed in this work are depicted in black.
The production of propionate and acetate in a 2:1 ratio (4/3 mol propionate/glucose) is expected to correspond to the maximisation of energy for P. freudenreichii subsp. shermanii under the l1-norm flux distribution, which assumes minimisation of total metabolic fluxes. Exploring the maximum range of propionate production while still producing 4 ATP/glucose reveals that propionate can also be produced with formate as an oxidised species rather than acetate. The pathway utilises the synthesis and subsequent degradation of serine via the glycine cleavage system to produce 10-formyltetrahydrofolate, which is then used to regenerate tetrahydrofolate while phosphorylating ADP. This pathway will be referred to hereafter as the glycine cleavage pathway (Fig. 2). The combination of this pathway with glycolysis and the Wood-Werkman cycle enables the production of 1.6 mol propionate/mol glucose and 0.8 moles of formate. Given that formate is another acid it will contribute to both acid stress and process separation costs. Removal is possible by over-expressing a non-native formate dehydrogenase or formate hydrogenlyase. Depending on the conversion efficiency of formate to carbon dioxide, the combination of the glycine cleavage pathway and formate dehydrogenase can achieve a yield of 1.6–1.7 mol propionate/mol glucose, consistent with the maximum theoretical yield, at the highest native energy yield possible of 4 ATP/glucose (Table 2).
3.3. Pan-genome guided design
We probed the diversity of the pan-genome of propionibacteria to look for energetically efficient pathways for propionate production. Most notably, we found P. acidipropionici species contained a sodium-pumping MMD, while P. propionicum contained a PEPCK that uses GTP as a cofactor rather than the typical pyrophosphate-dependent PEPCK of Propionibacterium. The combination of these two enzymes form an alternative linear pathway to the Wood-Werkman cycle for the degradation of PEP to propionate, yielding more energy than the Wood-Werkman cycle itself. According to our model, 4.33 ATP/glucose is expected (Table 2). This scenario does not necessarily alter the ratio of propionate to acetate (Table 2), unless flux through the glycine cleavage system pathway is possible, which may be driven by evolution as this favours generation of ATP.
3.4. Coupling the maximum energy yield with the utmost theoretical yield of propionate
Our results suggest the maximum theoretical yield of propionate can be achieved while achieving the maximum energy yield from glucose through the glycine cleavage system pathway and expression of the formate dehydrogenase. Given that this pathway does not incur an energetic disadvantage to supply reduced cofactors this offers a valuable strategy to increase propionate yield. The introduction of the alternative linear pathway for propionate is advantageous for cells as it enhances the amount of energy that can be achieved through the production of propionate and therefore actively selects for its production, so long as the supply of reduced cofactors does not incur an extra energetic cost. The combination of both strategies marginally improves the maximum energy yield from the fermentation when propionate is the sole product; thus coupling the maximum theoretical yield of propionate to the maximum energy yield while simultaneously improving the maximum energy yield relative to the wild-type strain. Presuming flux through the glycine cleavage based pathway can be coupled to ATP regeneration then a yield of 4.43 ATP/glucose could be achieved with the production of 1.7 moles propionate/mol glucose, which is clearly superior to the other proposed strategies (Fig. 1).
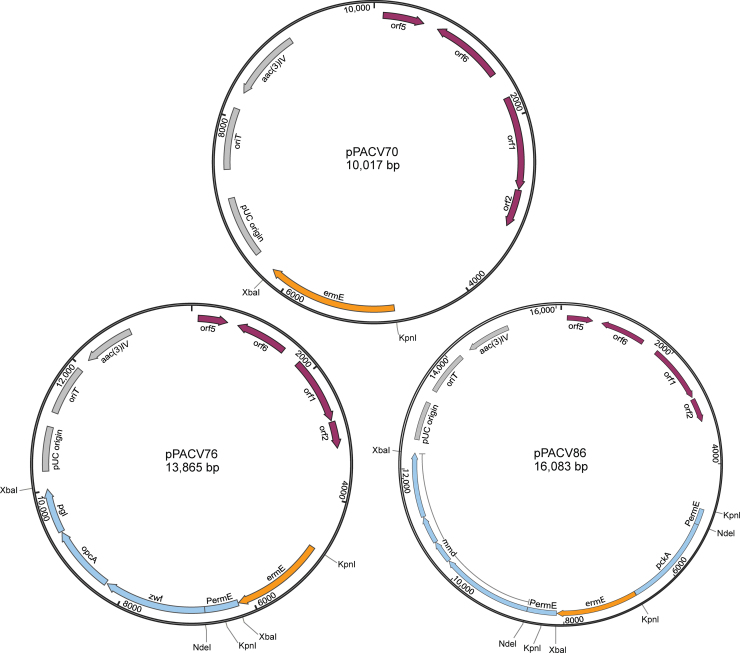
3.5. Construction of expression vectors and transformation of propionibacteria
Over-expression of the PPP was selected as a lead strategy for testing the hypothesis that increasing the availability of reduced cofactors would reduce by-products in propionibacteria. Thus, genes zwf, opcA and pgl, were amplified from P. acidipropionici ATCC 4875 genomic DNA and cloned into pPACV70 under PermE promoter for gene expression, resulting in plasmid pPACV76 (Fig. 3). To improve energy yield from propionate production, a linear pathway for propionate production for P. freudenreichii subsp. shermanii was designed which used the non-native ATP-PEPCK and MMD as well as native components of the Wood-Werkman cycle. To test this pathway, the ATP-PEPCK and MMD were overexpressed.
Fig. 3.
Plasmid maps of pPACV70, pPACV76 and pPACV86. Open reading frames orf1, orf2, orf5 and orf6 from pRGO1 are indicated in burgundy arrows in pPACV70, pPACV76 and pPACV86 maps. zwf, opcA and pgl genes are indicated by blue arrows in pPACV76 map. pckA and mmd genes are indicated by blue arrows in pPACV86 map.
pckA and mmd genes were amplified from E. coli and P. acidipropionici ATCC 4875 genomic DNA, respectively, and cloned into pPACV70 under the control of PermE for expression, resulting in plasmid pPACV86 (Fig. 3).
Plasmids pPACV76, pPACV86 and empty plasmid pACV70 were transformed into P. freudenreichii subsp. shermanii by electroporation obtaining Pf-ZOP, Pf-PAM and Pf-EMP strains, respectively. Plasmid extraction and restriction profile confirmed the presence of each plasmid. Pf-ZOP, Pf-PAM and Pf-EMP strains were confirmed to be P. freudenreichii subsp. shermanii by genomic DNA extraction and PCR amplification.
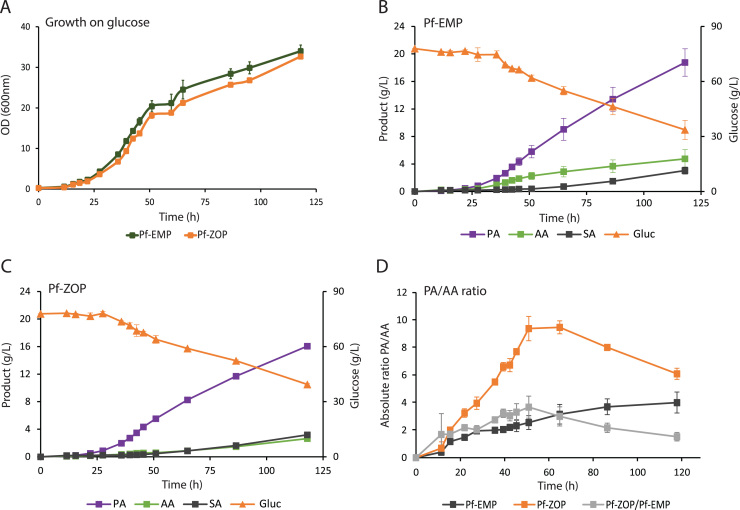
3.6. Effects of zwf, opcA and pgl overexpression on fermentation kinetics
Pf-ZOP and Pf-EMP strains were grown in complex medium with glucose as carbon source to study the effect of expression of zwf, opcA and pgl on cell growth and organic acids production (Fig. 4A, B and C). No difference in growth rates was observed between Pf-ZOP and Pf-EMP strains (Fig. 4A). Glucose consumption and propionate production remained similar between the two strains until the end of the exponential growth phase, at which point the rate of propionate production and glucose consumption was slightly higher in Pf-EMP. In contrast, the rate of acetate production was approximately 3.5 times lower in Pf-ZOP during the exponential growth phase. The ratio of propionate to acetate was improved 4-fold from 2.27 in Pf-EMP to 8.94 in Pf-ZOP (Table 3). During the pseudo-stationary phase, growth occurred at a much reduced rate and acetate production in Pf-ZOP increased to a similar rate as Pf-EMP without a concomitant increase in propionate production (Table 3 and Fig. 4D).
Fig. 4.
Kinetics of batch fermentation of glucose by Pf-EMP and Pf- ZOP strains. A) Cell growth. Production of propionate (PA), acetate (AA), succinate (SA) and glucose consumption (Gluc) in (B) Pf-EMP and (C) Pf-ZOP. D) Absolute PA/AA ratio (g/g). Fermentation of Pf-EMP and Pf-ZOP strains were performed in duplicate. Error bars correspond to the deviation of replicates from the mean.
Table 3.
Fermentation kinetic parameters of Pf-EMP and Pf-ZOP during exponential and stationary growth phase.
|
Exponential phase |
Stationary phase |
||||
|---|---|---|---|---|---|
| Strain | Pf-EMP | Pf-ZOP | Pf-EMP | Pf-ZOP | |
| Growth rate (h−1) | µ | 0.077 ± 0.004 | 0.077 ± 0.004 | 0.006 ± 0.001 | 0.008 ± 0.000 |
| Rates (g/gDW h) | Glucose consumption | 0.198 ± 0.052 | 0.263 ± 0.048 | 0.048 ± 0.012 | 0.048 ± 0.002 |
| Propionate | 0.076 ± 0.005 | 0.086 ± 0.006 | 0.022 ± 0.005 | 0.019 ± 0.001 | |
| Acetate | 0.033 ± 0.003 | 0.009 ± 0.001 | 0.006 ± 0.002 | 0.003 ± 0.000 | |
| Succinate | 0.004 ± 0.001 | 0.001 ± 0.001 | 0.005 ± 0.001 | 0.006 ± 0.001 | |
| Ratios (g/g) | PA/AA | 2.270 ± 0.103 | 8.940 ± 0.433 | 2.900 ± 0.820 | 4.190 ± 0.360 |
| PA/Glc | 0.270 ± 0.057 | 0.280 ± 0.054 | 0.450 ± 0.024 | 0.390 ± 0.027 | |
| Productivity (g/L h) | Glucose consumption | 0.520 ± 0.140 | 0.600 ± 0.097 | 0.400 ± 0.095 | 0.370 ± 0.016 |
| Propionate | 0.200 ± 0.022 | 0.190 ± 0.016 | 0.180 ± 0.042 | 0.150 ± 0.004 | |
| Acetate | 0.083 ± 0.012 | 0.020 ± 0.002 | 0.040 ± 0.024 | 0.034 ± 0.003 | |
| Succinate | 0.009 ± 0.002 | 0.005 ± 0.002 | 0.045 ± 0.007 | 0.044 ± 0.003 | |
*Values corresponding to exponential growth phase (28–43 h), carbon balance 111% Pf-EMP, 90% Pf-ZOP, and to stationary phase (60–120 h), carbon balance 93% Pf-EMP, 88% Pf-ZOP.
3.7. Effects of CO2 sparging and MMD and PEPCK overexpression on fermentation kinetics
Pf-PAM and Pf-EMP strains were grown in complex medium with glucose as carbon source and a 20% carbon dioxide sparge. A CO2 sparge was utilised because preliminary serum bottle trials suggested this was important to achieve a high activity of the PEPCK, observed via enhanced biomass formation (data unpublished). Growth rates remained similar, but the arrest of exponential growth was prolonged slightly in the Pf-PAM strain, resulting in an additional, short growth phase from 50 to 64 h which we will refer to as the transition phase (Fig. 5A). During this transition phase, glucose was rapidly catabolised to propionate and acetate, but the production of organic acids and consumption of sugar remained similar (Fig. 5B, C, Table 5). The production of acetate relative to propionate was increased (Fig. 5D). At the end of this prolonged growth phase, Pf-PAM had achieved a 20% higher biomass than Pf-EMP (Fig. 5D). Throughout the exponential growth phase, glucose was more rapidly converted to propionate, acetate and succinate by Pf-PAM (Table 4). The growth rate and productivities of both acetate and propionate were markedly higher in the transition phase (Table 5) followed by the onset of the pseudo-stationary phase where growth in Pf-PAM was lower and acetate production double compared to Pf-EMP (Table 4).
Fig. 5.
Kinetics of batch fermentation of glucose by Pf-EMP and Pf- PAM strains with a CO2 sparge. A) Cell growth. Production of propionate (PA), acetate (AA), succinate (SA) and glucose consumption (Gluc) in (B) Pf-EMP and (C) Pf-ZOP. D) Absolute PA/AA ratio and gram dry weight ratio (GDW). Fermentation of Pf-EMP and Pf-PAM strains were performed in duplicate. Error bars correspond to the deviation of replicates from the mean and are depicted where replicate data is available.
Table 5.
Fermentation kinetic parameters of Pf-EMP and Pf-PAM during transition phase.
|
Transition phase |
|||
|---|---|---|---|
| Strain | Pf-EMP | Pf- PAM | |
| Growth rate (h−1) | µ | 0.017 ± 0.001 | 0.033 ± 0.003 |
| Rates (g/gDW h) | Glucose consumption | 0.071 ± 0.022 | 0.089 ± 0.008 |
| Propionate | 0.042 ± 0.005 | 0.044 ± 0.004 | |
| Acetate | 0.005 ± 0.001 | 0.008 ± 0.001 | |
| Succinate | 0.004 ± 0.001 | 0.004 ± 0.000 | |
| Ratios (g/g) | PA/AA | 1.019 ± 3.608 | 2.666 ± 3.229 |
| PA/Glc | 0.554 ± 0.108 | 0.498 ± 0.019 | |
| Productivity (g/L h) | Glucose consumption | 0.522 ± 0.154 | 0.699 ± 0.039 |
| Propionate | 0.306 ± 0.028 | 0.349 ± 0.006 | |
| Acetate | 0.040 ± 0.003 | 0.063 ± 0.004 | |
| Succinate | 0.030 ± 0.004 | 0.028 ± 0.000 | |
*Values corresponding to transitional growth phase (50–64 h), carbon balance 113% Pf-EMP, 119% Pf-PAM.
Table 4.
Fermentation kinetic parameters of Pf-EMP and Pf-PAM during exponential and stationary growth phase.
|
Exponential phase |
Stationary phase |
||||
|---|---|---|---|---|---|
| Strain | Pf-EMP | Pf- PAM | Pf-EMP | Pf-PAM | |
| Growth rate (h−1) | µ | 0.083 ± 0.004 | 0.086 ± 0.001 | 0.007 ± 0.001 | 0.005 ± 0.000 |
| Rates (g/gDW h) | Glucose consumption | 0.160 ± 0.026 | 0.205 ± 0.009 | 0.047 ± 0.005 | 0.044 ± 0.004 |
| Propionate | 0.072 ± 0.004 | 0.082 ± 0.004 | 0.021 ± 0.002 | 0.018 ± 0.002 | |
| Acetate | 0.022 ± 0.002 | 0.029 ± 0.001 | 0.003 ± 0.000 | 0.005 ± 0.001 | |
| Succinate | 0.001 ± 0.001 | 0.003 ± 0.000 | 0.005 ± 0.001 | 0.006 ± 0.001 | |
| Ratios (g/g) | PA/AA | 3.259 ± 0.302 | 2.857 ± 0.182 | 7.860 ± 1.536 | 3.373 ± 0.529 |
| PA/Glc | 0.449 ± 0.076 | 0.400 ± 0.025 | 0.442 ± 0.06 | 0.416 ± 0.059 | |
| Productivity (g/L h) | Glucose consumption | 0.406 ± 0.095 | 0.485 ± 0.061 | 0.515 ± 0.017 | 0.529 ± 0.008 |
| Propionate | 0.187 ± 0.022 | 0.192 ± 0.028 | 0.226 ± 0.004 | 0.219 ± 0.007 | |
| Acetate | 0.057 ± 0.009 | 0.068 ± 0.009 | 0.030 ± 0.003 | 0.066 ± 0.003 | |
| Succinate | 0.002 ± 0.003 | 0.006 ± 0.001 | 0.060 ± 0.004 | 0.074 ± 0.003 | |
*Values corresponding to exponential growth phase (24–50 h), carbon balance 130% Pf-EMP, 114% Pf-PAM, and to stationary phase (64–137 h), carbon balance 89% Pf-EMP, 91% Pf-PAM.
4. Discussion
4.1. Feasibility of the proposed genetic engineering strategies
The biological production of propionic acid would be economically feasible if propionibacteria could be engineered to reach near maximum theoretical yields while maintaining high titre and volumetric productivity (Rodriguez et al., 2014). Current strategies in the literature have focussed on directly upregulating enzymatic components of the propionate production pathway (Ammar et al., 2014, Liu et al., 2016, Wang et al., 2015b) or by improving acid tolerance by upregulating the arginine deiminase, glutamate decarboxylase amino acid systems, or trehalose synthesis (Guan et al., 2015a, Jiang et al., 2015). These strategies compete with, rather than suppress, acetate production and have relatively minor improvements in phenotype, or represent a carbon drain and reduce the yield.
Metabolic modelling was therefore used to identify more effective strategies. The classic strain design algorithm, OptKnock (Burgard et al., 2003), was originally used which works on a principle of growth coupling the production of a target product by eliminating alternative, energetically superior pathways. Because propionate is already growth coupled, these strategies inherently either targeted the elimination of other by-products (such as an acetate kinase knockout) or energy generating steps (such as pgk knockout to prevent a net energy yield through glycolysis). Because the fermentation titre is ultimately limited by product inhibition (Blanc and Goma, 1987), primarily thought to be caused by the futile cycling of protons across the membrane, the availability of energy for proton extrusion should dictate acid stress tolerance. Therefore, reducing the energy yield risks trading titre off against fermentation yield and the most successful strategies will enhance propionate production while maintaining, or even improving, the high energy yield of Propionibacterium.
Here, we present a number of engineering strategies, which in silico, achieve higher yield (Table 2). The strategies vary in complexity and the limited capacity to implement these strategies with molecular biology means the testing of any strategy is inherently difficult. A strong focus was therefore placed on ensuring these strategies are robust in their implementation. Each strategy proposed is analysed in detail in the supplementary material Table S2. While P. freudenreichii subsp. shermanii serves as a good propionibacteria model, the ultimate goal is to translate these strategies into strains most suited to industrial production, such as P. acidipropionici, where genetic engineering remains elusive. It is likely that future work needs to address the large variety of restriction modification (RM) systems which have been identified in Propionibacterium, compiled in the REBASE database (Roberts et al., 2015), before this is possible. In other non-model organisms, RM systems have been by-passed by the expression of native methylation components in the cloning host (O'Connell Motherway et al., 2009, Zhang et al., 2012).
In the current study, we explored two (PPP and linear PA) of the strategies identified (Table 2) in the more readily engineered P. freudenreichii subsp. shermanii. The PGK strategy was excluded because it cripples energy production, while the ACK knockout strategy was excluded because P. freudenreichii subsp. shermanii utilises different pathways for acetate production to P. acidipropionici (manuscript in preparation). The TCA and citramalate pathways are energetically advantageous over the PPP but rely on the promiscuity of enzymes associated with acetate metabolism; thus, both pathways would require extensive protein engineering to change enzyme selectivity. Finally, the glycine cleavage pathway is particularly promising on its own or coupled to the linear pathway due to its ability to couple the regeneration of ATP to the production of reduced cofactors. However, it involves at least nine enzymes.
4.2. Influence of PPP over-expression on the metabolism of P. freudenreichii subsp. shermanii
A strategy to manipulate the redox metabolism of Propionibacterium using the PPP is presented. Over-expression of the PPP by upregulating the oxidative branch is a proven strategy (Ahmad et al., 2012) and was performed here by upregulating the first two steps of the oxidative branch of the PPP. The PPP is a favourable engineering target given it is compatible with other engineering strategies already implemented such as upregulation of the Wood-Werkman cycle (Liu et al., 2015, Wang et al., 2015a) and is tractable within the limits of current engineering tools. The PPP has long been thought to be the primary pathway responsible for the increased ratio of propionate to acetate in higher producing propionibacteria (Papoutsakis and Meyer, 1985) but this has never been tested.
Consistent with the modelling, the strategy resulted in a 4-fold improvement in the propionate to acetate ratio and a 13% improvement in the rate of propionate production without increased production of succinate. Our findings demonstrate that over-expression of the PPP is a valid alternative to supplementing the media with glycerol without inhibiting growth, an initial concern of Wang and colleagues (Wang et al., 2015b). This is despite a reduced energy yield from the pathway; suggesting this is partially compensated for by either the increase in NADPH or activity of the PPP which has been shown to combat oxidative stress (Krüger et al., 2011). Importantly, these results show that NADPH can be coupled to the Wood-Werkman cycle, although the exact mechanism remains unclear, and supports the hypothesis that a higher flux through the PPP may be responsible for the divergence in the propionate to acetate ratio from 2 (Papoutsakis and Meyer, 1985).
In Pf-EMP, a molar ratio of propionate to acetate of ~2:1 was observed during exponential growth. This ratio is consistent with the simulated l1-norm flux distribution for the maximisation of energy. At such a ratio, it is expected that flux through the PPP is minimal. In comparison, Pf-ZOP achieved a molar propionate to acetate ratio of ~8. If all additional reduced cofactors required to achieve such a ratio are assumed to come from the PPP, the model predicts that the split ratio of flux between glycolysis and the PPP is approximately 2:3, implying 60% of the glucose uptake was directed towards the PPP. This compares to the 86% required in order to achieve the maximum theoretical yield.
Pf-ZOP appears to experience a partial release of the over-expression phenotype at the onset of the stationary phase. In particular, the propionate production rate reduces with respect to Pf-EMP (Table 3), which ultimately reduces the propionate to acetate ratio from 9.2 (g/g) at the end of the exponential growth phase to 6 (g/g) at the end of the fermentation; although this is still a 55% improvement over Pf-EMP. Conversely, Pf-EMP showed an improvement in the ratio of propionate to acetate at the onset of stationary phase. We postulate this different behaviour may be due to the activity of the Bifidobacterium shunt recently identified in P. freudenreichii subsp. shermanii (manuscript in preparation). The Bifidobacterium shunt catabolises sugars via the PPP and a phosphoketolase enzyme to produce primarily acetate and lactate in Bifidobacterium, where it is over-expressed in response to stress (Sánchez et al., 2005). Expression in response to cold stress (Dalmasso et al., 2012) and apparent activation at the onset of stationary phase suggests it may play a similar role in P. freudenreichii subsp. shermanii.
4.3. Influence of carbon dioxide sparging on the metabolism of P. freudenreichii subsp. shermanii
Fermenters were sparged with CO2 to test the linear propionate pathway strategy. Sparging improved biomass production, which can be observed by comparisons of the strains harbouring the empty plasmid (Fig. 6). While biomass was similar at the end of the exponential growth phase, propionibacteria supplied with CO2 grew for longer, resulting in 25% higher biomass. The rate of acetate production across the exponential growth phase is reduced by one-third and halved in the stationary phase (Table 3, Table 4). This suggests CO2 addition somehow alters metabolism to produce more reduced cofactors and suppresses acetate production.
Fig. 6.
Comparison of fermentations of Pf-EMP with a CO2 sparge (solid line, squares) and without (dotted line, triangles). Sparging with CO2 increased biomass production and allowed increased glucose catabolism.
Our results indicate that in the presence of CO2, glucose is catabolised at a slower rate to all three major fermentation products and propionate is favoured over acetate. Such a phenotype is identical to that observed when acid stress is applied by reduction in the external pH (Feng et al., 2010). Likewise, CO2 can also trigger acid stress (Baez et al., 2009, Sun et al., 2005). Despite the fact that near equal amounts of dissolved CO2 and carbonate will be present in the fermenter at pH 6.4, a lack of homologues for bacterial carbonate transporters (Price, 2011) and a higher permeability of dissolved CO2 (Jones and Greenfield, 1982) leads to cytoplasmic acidification.
Metabolic modelling suggests CO2 fixation does not directly promote propionate production and does not enhance energy. Fixation occurred only when flux was constrained through the native PPi-PEPCK (Siu et al., 1961), resulting in an energetic penalty and the equimolar production of succinate, consistent with experimental results (Wood and Werkman, 1938, Wood and Werkman, 1940). Further conversion to propionate was only viable in the presence of MMD.
The above results lead us to propose a novel mechanism by which CO2 can modify Propionibacterium metabolism by overcoming thermodynamic limitations. Assuming a mechanism of transport by diffusion, intracellular propionate is expected to increase across the course of the fermentation as extracellular propionate accumulates and the pH gradient diminishes. In addition to decoupling the proton gradient, accumulating propionate is expected to inhibit metabolism kinetically and may eventually form a thermodynamic barrier. This is particularly possible given the Wood-Werkman cycle is entirely reversible (Rosner and Schink, 1990). The fixation of CO2 through the PPi-dependent PEPCK would help build the concentrations of key metabolic intermediates in the Wood-Werkman cycle allowing further flux through the pathway and resulting in the increased consumption of substrate observed.
4.4. Influence of the linear pathway for propionate production on the metabolism of P. freudenreichii subsp. shermanii
The linear pathway for propionate production is a promising, energetically beneficial route to propionate identified by studying the pan Propionibacterium genome (manuscript in preparation). The strategy consists of over-expression of the ATP-dependent PEPCK, which improves the growth rate and succinate yield in E. coli strains lacking the PEP carboxylase (Kim et al., 2004) along with the sodium-pumping MMD, the key energy-yielding step in the fermentation of succinate to propionate for organisms such as Propionigenium modestum (Schink and Pfennig, 1982).
Expression of the linear propionate pathway resulted in a continuation of the exponential growth phase at a reduced growth rate. Both glucose catabolism and the production of all fermentation products are stimulated compared to CO2-sparged Pf-EMP. This appears to come at a trade-off for propionate specificity; compared to CO2-sparged Pf-EMP the propionate to acetate yield is lower in the transition and stationary phases. Increased succinate production suggests that the ATP-PEPCK operates in the direction of oxaloacetate synthesis, but suggests MMD activity may be limiting. While an increased growth rate was expected, it is possible that additional energy was captured as polyphosphate and utilised to fuel further growth at the end of the exponential growth phase instead (Clark et al., 1986). A more pronounced phenotype is expected if more PEP is available, such as during the catabolism of disaccharides.
5. Conclusion
In this work, we proposed several genetic engineering strategies for the improvement of propionic acid production in propionibacteria using genome-scale modelling. Two fundamental strategies were identified by exploring pathways encoded within the pan-genome of propionibacteria that lead to the generation of reduced cofactors or enhanced energy production. The first two enzymes of the oxidative branch of the PPP were over-expressed, resulting in a 4-fold increase in the propionate to acetate ratio during the exponential growth phase and demonstrating the ability for NADPH to couple to the Wood-Werkman cycle. Furthermore, our results from the over-expression of the ATP-dependent PEPCK and the sodium-pumping MMD suggest that MMD activity may be limiting and that the ATP-PEPCK operates in the direction of oxaloacetate and ATP synthesis in Propionibacterium. The growth rate of this mutant was higher than that of the empty plasmid strain as predicted by the model. Sparging CO2 into the fermenter was surprisingly beneficial for cell growth and propionate production. These findings suggest a CO2 sparge may be a cheap, novel method to promote the production of propionate, offering new opportunities to influence propionate production in efficient native producers.
Acknowledgments
We thank Manuel Plan for his valuable help with analytics. The authors acknowledge the Queensland node of Metabolomics Australia (MA) at The University of Queensland, a NCRIS initiative under Bioplatforms Australia Pty Ltd. We are grateful to the Australian Research Council Linkage project with Dow Chemicals ARC LP120100517 and The Queensland Government for the Accelerate Fellowship to E.M. T.M. was supported by an Australian Government Research Training Program Scholarship.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2017.11.001.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- Ahmad I., Shim W.Y., Jeon W.Y., Yoon B.H., Kim J.-H. Enhancement of xylitol production in Candida tropicalis by co-expression of two genes involved in pentose phosphate pathway. Bioprocess Biosyst. Eng. 2012;35:199–204. doi: 10.1007/s00449-011-0641-9. [DOI] [PubMed] [Google Scholar]
- Ammar E.M., Jin Y., Wang Z., Yang S.-T. Metabolic engineering of Propionibacterium freudenreichii: effect of expressing phosphoenolpyruvate carboxylase on propionic acid production. Appl. Microbiol. Biotechnol. 2014;98:7761–7772. doi: 10.1007/s00253-014-5836-y. [DOI] [PubMed] [Google Scholar]
- Ammar E.M., Wang Z., Yang S.-T. Metabolic engineering of Propionibacterium freudenreichii for n-propanol production. Appl. Microbiol. Biotechnol. 2013;97:4677–4690. doi: 10.1007/s00253-013-4861-6. [DOI] [PubMed] [Google Scholar]
- Baez A., Flores N., Bolívar F., Ramírez O.T. Metabolic and transcriptional response of recombinant Escherichia coli to elevated dissolved carbon dioxide concentrations. Biotechnol. Bioeng. 2009;104:102–110. doi: 10.1002/bit.22379. [DOI] [PubMed] [Google Scholar]
- Bierman M., Logan R., O'brien K., Seno E., Rao R.N., Schoner B. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Blanc P., Goma G. Kinetics of inhibition in propionic acid fermentation. Bioprocess Biosyst. Eng. 1987;2:175–179. [Google Scholar]
- Britz T., Riedel K.-H. Propionibacterium species diversity in leerdammer cheese. Int. J. Food Microbiol. 1994;22:257–267. doi: 10.1016/0168-1605(94)90177-5. [DOI] [PubMed] [Google Scholar]
- Burgard A.P., Pharkya P., Maranas C.D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Clark J.E., Beegen H., Wood H.G. Isolation of intact chains of polyphosphate from Propionibacterium shermanii grown on glucose or lactate. J. Bacteriol. 1986;168:1212–1219. doi: 10.1128/jb.168.3.1212-1219.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmasso M., Aubert J., Briard-Bion V., Chuat V., Deutsch S., Gilbert J.A. A temporal-omic study of Propionibacterium freudenreichii CIRM-BIA1T adaptation strategies in conditions mimicking cheese ripening in the cold. PLoS One. 2012;7(1):e29083. doi: 10.1371/journal.pone.0029083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Xu H., Yao J., Li S., Zhu H., Ouyang P. Kinetic analysis and pH-shift control strategy for propionic acid production with Propionibacterium freudenreichii CCTCC M207015. Appl. Biochem. Biotechnol. 2010;160:343–349. doi: 10.1007/s12010-008-8300-6. [DOI] [PubMed] [Google Scholar]
- Guan N., Li J., Shin H.D., Du G., Chen J., Liu L. Metabolic engineering of acid resistance elements to improve acid resistance and propionic acid production of Propionibacterium jensenii. Biotechnol. Bioeng. 2015;113:1294–1304. doi: 10.1002/bit.25902. [DOI] [PubMed] [Google Scholar]
- Guan N., Zhuge X., Li J., Shin H.-D., Wu J., Shi Z., Liu L. Engineering propionibacteria as versatile cell factories for the production of industrially important chemicals: advances, challenges, and prospects. Appl. Microbiol. Biotechnol. 2015;99:585–600. doi: 10.1007/s00253-014-6228-z. [DOI] [PubMed] [Google Scholar]
- Hyduke D., Schellenberger J., Que R. COBRA Toolbox 2.0. Protoc. Exch. 2011:1–35. [Google Scholar]
- Jiang L., Cui H., Zhu L., Hu Y., Xu X., Li S., Huang H. Enhanced propionic acid production from whey lactose with immobilized Propionibacterium acidipropionici and the role of trehalose synthesis in acid tolerance. Green Chem. 2015;17:250–259. [Google Scholar]
- Jones R.P., Greenfield P.F. Effect of carbon dioxide on yeast growth and fermentation. Enzyme Microb. Technol. 1982;4:210–223. [Google Scholar]
- Kiatpapan P., Hashimoto Y., Nakamura H., Piao Y.-Z., Ono H., Yamashita M., Murooka Y. Characterization of pRGO1, a plasmid from Propionibacterium acidipropionici, and its use for development of a host-vector system in propionibacteria. Appl. Environ. Microbiol. 2000;66:4688–4695. doi: 10.1128/aem.66.11.4688-4695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiatpapan P., Murooka Y. Construction of an expression vector for propionibacteria and its use in production of 5-aminolevulinic acid by Propionibacterium freudenreichii. Appl. Microbiol. Biotechnol. 2001;56:144–149. doi: 10.1007/s002530100603. [DOI] [PubMed] [Google Scholar]
- Kim P., Laivenieks M., Vieille C., Zeikus J.G. Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl. Environ. Microbiol. 2004;70:1238–1241. doi: 10.1128/AEM.70.2.1238-1241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kośmider A., Białas W., Kubiak P., Drożdżyńska A., Czaczyk K. Vitamin B 12 production from crude glycerol by Propionibacterium freudenreichii ssp. shermanii: optimization of medium composition through statistical experimental designs. Bioresour. Technol. 2012;105:128–133. doi: 10.1016/j.biortech.2011.11.074. [DOI] [PubMed] [Google Scholar]
- Krüger A., Grüning N.-M., Wamelink M.M. The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid. Redox Signal. 2011;15:311–324. doi: 10.1089/ars.2010.3797. [DOI] [PubMed] [Google Scholar]
- Liu L., Guan N., Zhu G., Li J., Shin H.-D., Du G., Chen J. Pathway engineering of Propionibacterium jensenii for improved production of propionic acid. Sci. Rep. 2016;6 doi: 10.1038/srep19963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhu Y., Li J., Wang M., Lee P., Du G., Chen J. Microbial production of propionic acid from propionibacteria: current state, challenges and perspectives. Crit. Rev. Biotechnol. 2012;32:374–381. doi: 10.3109/07388551.2011.651428. [DOI] [PubMed] [Google Scholar]
- Liu L., Zhuge X., Shin H.-D., Chen R.R., Li J., Du G., Chen J. Improved production of propionic acid in Propionibacterium jensenii via combinational overexpression of glycerol dehydrogenase and malate dehydrogenase from Klebsiella pneumoniae. Appl. Environ. Microbiol. 2015;81:2256–2264. doi: 10.1128/AEM.03572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Flores, C.H., Nielsen, L.K., Marcellin, E., 2016. Propionic acid production of Propionibacterium acidipropionici in a BIOSTAT® A.
- Meile L., Le Blay G., Thierry A. Safety assessment of dairy microorganisms: Propionibacterium and Bifidobacterium. Int. J. Food Microbiol. 2008;126:316–320. doi: 10.1016/j.ijfoodmicro.2007.08.019. [DOI] [PubMed] [Google Scholar]
- O'Connell Motherway M., O'Driscoll J., Fitzgerald G.F., Van Sinderen D. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2009;2:321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Begley T., Butler R.M. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoutsakis E.T., Meyer C.L. Fermentation equations for propionic acid bacteria and production of assorted oxychemicals from various sugars. Biotechnol. Bioeng. 1985;27:67–80. doi: 10.1002/bit.260270109. [DOI] [PubMed] [Google Scholar]
- Price G.D. Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth. Res. 2011;109:47–57. doi: 10.1007/s11120-010-9608-y. [DOI] [PubMed] [Google Scholar]
- Quek L.-E., Nielsen L.K. On the reconstruction of the Mus musculus genome scale metabolic network model. Genome Inform. 2008;21:89–100. [PubMed] [Google Scholar]
- Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez B.A., Stowers C.C., Pham V., Cox B.M. The production of propionic acid, propanol and propylene via sugar fermentation: an industrial perspective on the progress, technical challenges and future outlook. Green Chem. 2014;16:1066–1076. [Google Scholar]
- Rosner B., Schink B. Propionate acts as carboxylic group acceptor in aspartate fermentation by Propionibacterium freudenreichii. Arch. Microbiol. 1990;155:46–51. [Google Scholar]
- Ruhal R., Choudhury B. Improved trehalose production from biodiesel waste using parent and osmotically sensitive mutant of Propionibacterium freudenreichii subsp. shermanii under aerobic conditions. J. Ind. Microbiol. Biotechnol. 2012;39:1153–1160. doi: 10.1007/s10295-012-1124-y. [DOI] [PubMed] [Google Scholar]
- Sánchez B., Champomier-Vergès M.-C., Anglade P., Baraige F., Clara G., Margolles A., Zagorec M. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 2005;187:5799–5808. doi: 10.1128/JB.187.16.5799-5808.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B., Pfennig N. Propionigenium modestum gen. nov. sp. nov. a new strictly anaerobic, nonsporing bacterium growing on succinate. Arch. Microbiol. 1982;133:209–216. [Google Scholar]
- Siu P.M., Wood H.G., Stjernholm R.L. Fixation of CO2 by phosphoenolpyruvic carboxytransphosphorylase. J. Biol. Chem. 1961;236:PC21–PC22. [PubMed] [Google Scholar]
- Sun L., Fukamachi T., Saito H., Kobayashi H. Carbon dioxide increases acid resistance in Escherichia coli. Lett. Appl. Microbiol. 2005;40:397–400. doi: 10.1111/j.1472-765X.2005.01714.x. [DOI] [PubMed] [Google Scholar]
- Suwannakham S., Huang Y., Yang S.T. Construction and characterization of ack knockout mutants of Propionibacterium acidipropionici for enhanced propionic acid fermentation. Biotechnol. Bioeng. 2006;94:383–395. doi: 10.1002/bit.20866. [DOI] [PubMed] [Google Scholar]
- Thierry A., Maillard M.-B. Production of cheese flavour compounds derived from amino acid catabolism by Propionibacterium freudenreichii. Le Lait. 2002;82:17–32. [Google Scholar]
- Wang Z., Ammar E.M., Zhang A., Wang L., Lin M., Yang S.-T. Engineering Propionibacterium freudenreichii subsp. shermanii for enhanced propionic acid fermentation: effects of overexpressing propionyl-CoA: succinate CoA transferase. Metab. Eng. 2015;27:46–56. doi: 10.1016/j.ymben.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Wang Z., Lin M., Wang L., Ammar E.M., Yang S.-T. Metabolic engineering of Propionibacterium freudenreichii subsp. shermanii for enhanced propionic acid fermentation: effects of overexpressing three biotin-dependent carboxylases. Process Biochem. 2015;50:194–204. doi: 10.1016/j.ymben.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Wood H.G., Werkman C.H. The utilization of CO2 by the propionic acid bacteria. Biochem. J. 1938;32:1262. doi: 10.1042/bj0321262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H.G., Werkman C.H. The relationship of bacterial utilization of CO2 to succinic acid formation. Biochem. J. 1940;34:129. doi: 10.1042/bj0340129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Wang W., Deng A., Sun Z., Zhang Y., Liang Y., Che Y., Wen T. A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria. PLoS Genet. 2012;8:e1002987. doi: 10.1371/journal.pgen.1002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material