Abstract
Reactive sulfur species (RSS) such as H2S, HS•, H2Sn, (n = 2–7) and HS2•- are chemically similar to H2O and the reactive oxygen species (ROS) HO•, H2O2, O2•- and act on common biological effectors. RSS were present in evolution long before ROS, and because both are metabolized by catalase it has been suggested that “antioxidant” enzymes originally evolved to regulate RSS and may continue to do so today. Here we examined RSS metabolism by Cu/Zn superoxide dismutase (SOD) using amperometric electrodes for dissolved H2S, a polysulfide-specific fluorescent probe (SSP4), and mass spectrometry to identify specific polysulfides (H2S2-H2S5). H2S was concentration- and oxygen-dependently oxidized by 1 μM SOD to polysulfides (mainly H2S2, and to a lesser extent H2S3 and H2S5) with an EC50 of approximately 380 μM H2S. H2S concentrations > 750 μM inhibited SOD oxidation (IC50 = 1.25 mM) with complete inhibition when H2S > 1.75 mM. Polysulfides were not metabolized by SOD. SOD oxidation preferred dissolved H2S over hydrosulfide anion (HS-), whereas HS- inhibited polysulfide production. In hypoxia, other possible electron donors such as nitrate, nitrite, sulfite, sulfate, thiosulfate and metabisulfite were ineffective. Manganese SOD also catalyzed H2S oxidation to form polysulfides, but did not metabolize polysulfides indicating common attributes of these SODs. These experiments suggest that, unlike the well-known SOD-mediated dismutation of two O2•- to form H2O2 and O2, SOD catalyzes a reaction using H2S and O2 to form persulfide. These can then combine in various ways to form polysulfides and sulfur oxides. It is also possible that H2S (or polysulfides) interact/react with SOD cysteines to affect catalytic activity or to directly contribute to sulfide metabolism. Our studies suggest that H2S metabolism by SOD may have been an ancient mechanism to detoxify sulfide or to regulate RSS and along with catalase may continue to do so in contemporary organisms.
Keywords: Antioxidants, Oxidants, Redox, Reactive oxygen species, Superoxide, Hydrogen peroxide, Reactive Species Interactome
Highlights
-
•
Polysulfides are reactive sulfide species (RSS) and are similar to reactive oxygen species (ROS).
-
•
RSS may be the antecedent of redox regulatory and stress-related modalities.
-
•
RSS likely persist in modern-day organisms and are regulated by SOD.
1. Introduction
Sequential one-electron reduction of molecular oxygen produces three reactive oxygen species (ROS), superoxide (O2•-), hydrogen peroxide (H2O2) and hydroxyl radical (HO•) before terminating as water;
| O2 -e- → O2•-, O2•- -e- → H2O2, H2O2 -e- → HO•, HO• -e- → H2O· | (1) |
In addition to their toxicity, H2O2, and arguably O2•-, are considered to be important regulatory molecules [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17] necessitating careful regulation of their titers. Superoxide dismutase (SOD) and catalase (Cat) are well known antioxidant enzymes, the former catalyzes the dismutation of superoxide to oxygen and peroxide (Eq. (2)) while the latter catalyzes peroxide dismutation to oxygen and water (Eq. (3)).
| 2O2•- +2H+ → O2 + H2O2, | (2) |
| 2H2O2 → O2 + H2O· | (3) |
We [18] recently pointed out a number of chemical and biological similarities between ROS produced from oxygen (Eq. (1)) and reactive sulfide species (RSS) produced from the one-electron oxidation of hydrogen sulfide (H2S). These produce the thiyl radical (HS•), hydrogen persulfide (H2S2) and persulfide radical anion, ‘supersulfide’ (HS2•-) before terminating in elemental sulfur (S2) which may ultimately cyclize to S8 (Eq. (4); note for clarity this is not balanced for H+, or S).1
| H2S +e- → HS•, HS• +e- → H2S2, H2S2 +e- → HS2•-, HS2•- +e- → S2(8). | (4) |
A number of recent observations directly and anecdotally suggest RSS may play a far greater role in redox biology than previously appreciated. First, oxygen and sulfur have six valence electrons and both exert much of their signaling through interaction with cysteine sulfur (Cys-SH) in regulatory proteins, i.e., Cys-SH peroxidation produces sulfenyls (Cys-SOH) while persulfidation (sulfhydration) produces cysteine persulfides, Cys-S-SH [19], [20], [21], [22]. Not surprisingly, a number of regulatory systems have been shown to be regulated by either H2O2 or H2S2 (or H2Sn where n = 2–5) and the effector responses appear to be identical [20], [21], [22], [23], [24], [25], [26]. However, H2S can also reduce protein disulfide bonds and affect enzyme activity [27], which at neutral pH H2O cannot. Second, we [28] have shown that many of the methods used to measure ROS that are based on fluorescent probes or amperometric electrodes are also sensitive to RSS and quite often more so. The inability to discriminate between ROS and RSS when using these methods suggests that RSS may be more biologically relevant than previously appreciated. Third, we [29] have also demonstrated that catalase is an effective sulfide/sulfur oxidoreductase that uses oxygen to oxidize both H2S and H2Sn. It can also use H2O2 to oxidize H2S and in the absence of oxygen, catalase can generate H2S from thiorexodin and NADPH. Fourth, catalase effectively oxidizes the probe dichlorofluorescein (DCF), which is often used to detect ROS. This reaction can further confound studies on ROS production as it will give the appearance of ROS activity when there is none.
However, evolution may provide the most compelling argument for the involvement of RSS in biological systems and the significance of “antioxidant” systems in their regulation. Life began in an anoxic and reducing environment around 3.8 billion years ago (bya) and likely depended upon reducing equivalents, mainly in the form of H2S, to provide the energy and catalytic capacity to reduce CO2 and generate organic carbon and primitive amino acids, i.e., the “iron-sulfur world” proposed by Wächtershäuser [30]. These conditions generally persisted until the advent of oxidative photosynthesis in cyanobacteria which increased atmospheric oxygen to 0.5–1% and heralded in the “great oxidation event” (GOE) around 2.3 bya [31], [32], [33]. It is thought that in the absence of O2 (and, therefore, O2•-) prior to this period, there was either no need for SOD enzymes to evolve [34], or they acquired importance only after O2 became prevalent [35]. However, oxidation of H2S concomitant with CO2 reduction would have generated RSS, which, in turn, needed to be regulated. It is also possible that other electron acceptors were employed before O2 became prevalent. The fact that “antioxidant” enzymes including catalase, SOD, thioredoxin and peroxiredoxin all appear to have evolved long before the GOE [34], [35], [36], [37] suggests that they performed important biochemical functions on substrates other than ROS. We proposed these substrates were RSS [18] and our initial studies on catalase [29] appears to bear this out.
In the present study we further explore the hypothesis that antioxidant enzymes were originally involved in RSS metabolism and that remnants of these activities persist to the present day. Specifically, we show that SOD oxidizes H2S to form persulfides. These results suggest that SOD may have contributed to the metabolism of RSS in early forms of life albeit utilizing different electron acceptors and may still function in this regard in extant biological systems where O2 is prevalent.
2. Materials and methods
2.1. Chemicals
SSP4 (3′, 6′-Di(O-thiosalicyl)fluorescein) was purchased from Dojindo molecular Technologies Inc. (Rockville, MD). Sodium di-, tri- and tetrasulfide (Na2S2, Na2S3 and Na2S4). were kindly provided by Dojindo Laboratories. GYY 4137 was generously provided by Matt Whiteman University of Exeter. Sodium sulfide (Na2S), potassium polysulfide (K2S2) and superoxide dismutase (SOD, 3700 units/mg) were obtained from Sigma. Carbon monoxide (CO, 1 mM), carbonyl sulfide (COS, 20 mM) and sulfur dioxide (SO2, 1.4 M) solutions were prepared by bubbling pure gases through a sintered glass aerator into buffer for 20–30 min. All other chemicals were purchased from either Sigma-Aldrich (St. Louis, MO) or ThermoFisher Scientific (Grand Island, NY).
Phosphate buffer (PBS; in mM): 137 NaCl, 2.7, KCl, 8 Na2HPO4, 2 NaH2PO4. pH was adjusted with 10 mM HCl or NaOH to 7.4 (all but pH experiments) or 6.0, 7.0 or 8.0 (pH experiments). HEPES buffer (in mM): 145 NaCl, 3, KCl, 0.57 MgSO4·7H2O, 2 CaCl2·2H2O, 5 glucose, 3 HEPES acid, 7 HEPES sodium salt, pH 7.4. The ratio of HEPES acid to sodium salt was adjusted to produce pH 6, 7 or 8 as needed.
2.2. Polysulfide measurement
The polysulfide-specific fluorescent probe, SSP4 was used to measure polysulfides. Samples and test compounds were aliquoted into black 96-well plates in a darkened room and fluorescence was measured in normoxic conditions on a SpectraMax M5e plate reader (Molecular Devices, Sunnyvale, CA). Typically, fluorescence was measured every 10 min over 90 min.
2.3. Hypoxia
The contribution of oxygen to SOD catalysis of sulfur molecules was examined by sparging buffer with 100% N2, CO, COS or SO2 for 30 min and then placing the sealed buffer in a hypoxia chamber (model 856-HYPO hypoxia chamber, Plas Labs, Inc. Lansing, MI) under 100% N2. This lowered the ambient O2 to less than 0.35%, which under normal barometric conditions (~747 ± 2 mmHg) produced an O2 concentration less than 3.8 μM. Dry chemicals were stored in the chamber prior to use. The compounds of interest were then dissolved in the buffer, and placed in the well plates, covered, and allowed to react for 90 min, the same period as samples in normoxia. The plates were then removed from the chamber and fluorescence measured for an additional 90 min. Time 0 fluorescence was assumed to represent 90 min incubation in hypoxia. A parafilm liner was placed inside the plate cover to help reduce diffusion of gases.
2.4. Amperometric measurement of O2, H2O2 and H2S
Amperometric O2 and H2O2 sensors, ISO-OXY-2 and ISO-HPO-2, respectively, were purchased from WPI (World Precision Instruments, Sarasota, FL). They are designed for tissue culture with 2 mm dia replaceable membrane sleeves and a reported detection limit of 0.1% (ISO-OXY-2) and < 100 nM (ISO-HPO-2). It should be noted that the ISO-HPO-2H2O2 sensor cannot be used when H2S is present as it is >24 times more sensitive to H2S than it is to H2O2 [28].
H2S amperometric sensors with a sensitivity of 14 nM H2S gas (~100 nM total sulfide) were constructed in-house as described previously [38]. The sensors were connected to a WPI TBR 4100 Free Radical Analyzer and data was archived on a laptop PC with software provided by the manufacturer and exported into Microsoft Excel. The H2S sensor was calibrated periodically throughout each day with fresh standards made up in anoxic phosphate buffer (pH 7.4). The H2S sensor does not respond to polysulfides or other oxidized forms of sulfur.
A reaction chamber with a side ports for the H2S and O2 sensors and a 1-cm wide by 2 cm deep central well was purchased from WPI (NOCHM-4). A polycarbonate stopper with a hole in the stopper permitted venting the head space air when the stopper was lowered into the chamber and provided an access port for sample injection with a Hamilton microliter syringe. The chamber was placed on a magnetic stirrer and stirred with a Teflon micro stir bar. Compounds of interest were injected through the stopper and the reactions monitored for 10–30 min or longer if necessary.
2.5. Mass spectrometry confirmation of polysulfide formation by superoxide dismutase
Ultrahigh performance liquid chromatography, tandem mass spectrometry (UPLC-MS/MS) detection was used to further identify and quantify the polysulfides formed from the reaction of SOD with H2S. Polysulfide formation at each time point was captured using the derivatisation agent iodoacetamide (IAM) which also facilitated detection by UPLC-MS/MS.
The derivatised polysulfide compositions were analyzed by UPLC-MS/MS on a Waters Aquity UPLC system with a Xevo TQ-S detector. Solvent A was H2O + 5 mM ammonium acetate; solvent B was 95% acetonitrile + 5% H2O + 5 mM ammonium acetate. An Aquity UPLC CSH C18 (1.7 µm), 2.1 × 100 mm column was used for the separation with the gradient used described in Table 1, the injection volume was 5 μl and the multiple reaction monitoring conditions used are listed in Table 2.
Pure sodium tetrasulfide was used to quantify the polysulfides. In order to do so a known amount of tetrasulfide powder was weighed out and derivatised with an excess of IAM in pH 7.4 ammonium phosphate buffer. During this process the tetrasulfide speciates, forming hydrosulfide, disulfide, trisulfide, tetrasulfide and pentasulfide. The proportion of each of these species is taken as the concentration of that species in proportion to the initial tetrasulfide that was weighed out. For example, if the initial concentration of tetrasulfide would be 100 μM, and the peak for trisulfide represents 10% of the total peak areas for all of the polysulfide species seen then the concentration of trisulfide would be taken as 10 μM.
Three separate conditions were used, 1) 1 μM SOD + 1 mM H2S (Na2S) in normoxia; 2) 1 μM SOD + 1 mM H2S in hypoxia (150 min) followed by normoxia; 3) 1 mM H2S without SOD in normoxia. All reaction mixtures were made up in pH 7.4 ammonium phosphate buffer. Between 0 and 90 min a 50 μl aliquot of each reaction mixture was taken and added to 50 μl 100 mM IAM and mixed. Two additional time points for each set of conditions were also taken at 150 min and 180 min (condition 1). A rough standard curve was produced using pure sodium tetrasulfide salt (Na2S4) derivatized with excess IAM.
2.6. Data analysis
Data was analyzed and graphed using QuatroPro (Corel Corporation, Ottawa Ont, Canada) and SigmaPlot 13.0 (Systat Software, Inc., San Jose, CA). Statistical significance was determined using one-way ANOVA and the Holm-Sidak test (SigmaPlot 13.0). Results are given as mean + or ± SE; significance was assumed when p ≤ 0.05.
3. Results
3.1. Amperometric measurements of H2S consumption (Fig. 1A-C)
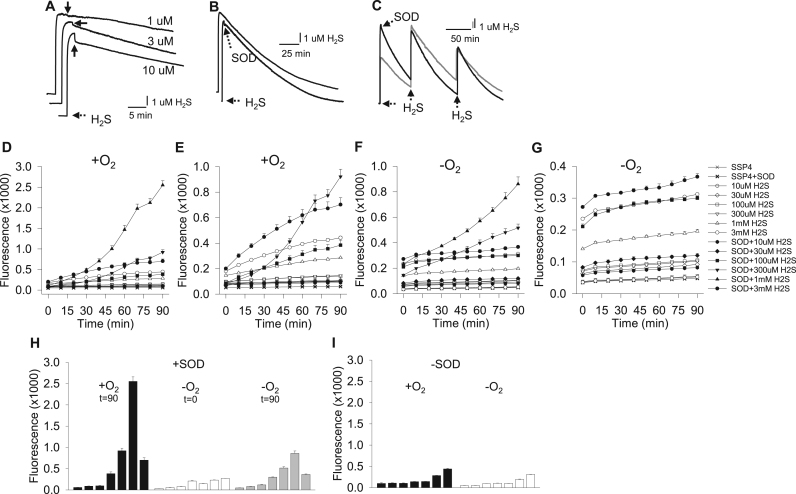
Fig. 1.
Amperometric traces of H2S metabolism (A-C) and fluorometric measurement of polysulfide production (SSP4 fluorescence) from SOD metabolism of H2S (D-I). (A) Traces showing the effects of increasing SOD concentration on 10 μM H2S. Addition of SOD (arrows) produces a rapid drop in H2S that becomes more pronounced at higher SOD concentrations. (B) Decrease in 10 μM H2S with or without 3 μM SOD for 150 min (C) H2S decreases faster after three consecutive 10 μM H2S injections with SOD (black line) than without SOD (gray line). Decrease in H2S without SOD in (A-C) likely due to inherent volatility, H2S consumption by sensor and possible unidentified reaction(s) in chamber. (D, E) polysulfide (SSP4 fluorescence) formation in normoxia from H2S in the absence (open symbols) or presence (solid symbols) of 1 μM SOD (D, full scale, E, expanded scale with 300 μM and 1 mM H2S omitted). (F, G) polysulfide formation in hypoxia (G, expanded scale with 300 μM and 1 mM H2S omitted). Time 0 min represents sample incubation in 100% N2 for 90 min prior to analysis (equivalent to 90 min in normoxia in D and E). (H) Summary of D and F with SOD after 90 min in normoxia (black bars), 90 min in hypoxia (white bars, t = 0 in plate reader), or 90 min after removal from hypoxia (gray bars). (I) summary of D and F without SOD after 90 min in normoxia (black bars) or hypoxia (white bars). SOD concentration-dependently increased SSP4 fluorescence between 100 μM and 1 mM H2S, whereas at 3 mM H2S fluorescence was inhibited, no inhibition was observed when SOD was absent. Time 0 min samples in normoxia and hypoxia were essentially similar indicating no appreciable polysulfide production after 90 min in hypoxia. All fluorescence values are mean +SE, n = 4 replicates).
H2S concentration was monitored in real time amperometrically to identify interactions of SOD with H2S. Aerobic addition of SOD to 10 μM H2S produced a rapid decrease in H2S that was proportional to the SOD concentration (Fig. 1A); 1, 3 and 10 μM SOD significantly decreased H2S by 0.17 ± 0.02, 0.35 ± 0.02 and 1.01 ± 0.5 μM H2S, respectively (p < 0.001, n = 3 for all). H2S concentration decreased at a slow and steady rate in the absence of SOD (32.1 ± 2.6% decrease; n = 9) and significantly (p = 0.014) faster in the presence of 3 μM SOD (43.4 ± 3.45 decrease, n = 7) after the initial rapid effect.
The rate of SOD-mediated H2S oxidation has been reported to increase as much as 600 fold after 1 h [39] (see also Discussion). Although H2S concentration slowly decreases over time in our chamber we were able to examine this reaction using the amperometric sensor to track H2S concentration in real-time for up to 250 min under our experimental conditions. As shown in Fig. 1B, when 3 μM SOD was added to 10 μM H2S there was a monoexponential decline in H2S concentration for 150 min with an overall rate constant of 0.62 h−1 (curve-fit, p < 0.001). Because it is possible that sufficient H2S was consumed in this reaction we then repeated this experiment with two additional 10 μM H2S doses after the initial 10 μM H2S-3 μM SOD reaction (Fig. 1C). Again, there was no obvious indication of an increased rate of metabolism over the 250 min experimental period; the rate constants for the three H2S treatments were 0.72, 0.73 and 0.76 h−1 (curve-fit p < 0.001 for all). Rate constants for H2S in the absence of SOD (Fig. 1B,C) were correspondingly lower; single H2S, 0.43 h−1; and three consecutive H2S, 0.48, 0.43, 0.46 h−1. These experiments show that SOD accelerates the rate of H2S disappearance and there is no obvious further increase in rate over 4 h.
3.2. Evidence for polysulfide formation by SOD metabolism of H2S (Fig. 1D-I, Fig. 2A-C)
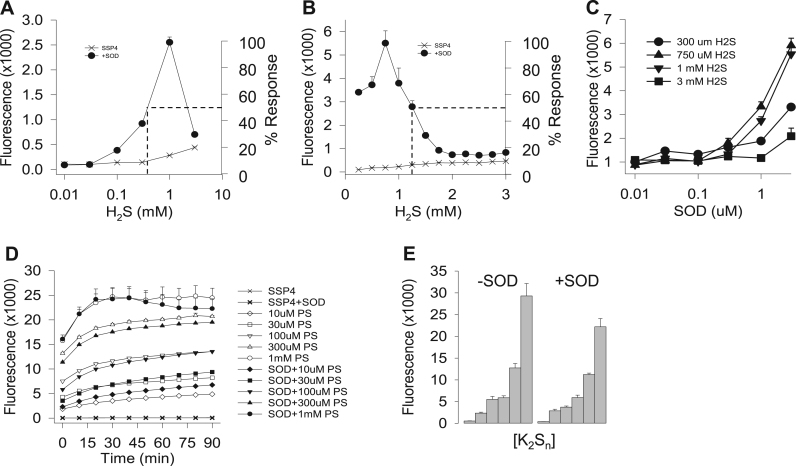
Fig. 2.
SOD metabolism of H2S and polysulfides. (A, B) Effects of increasing H2S concentration on polysulfide production (SSP4 fluorescence) after 90 min in normoxia with or without 1 μM SOD (values in A from Fig. 1D). Note: log H2S scale in A and linear, 250 μM H2S increments, in B. Dashed lines indicate approximate EC50 (380 μM H2S, A) and IC50 (1.25 mM H2S, B). (C) SOD concentration-dependently increases polysulfide production. (D) Concentration-dependent SSP4 fluorescence from K2Sn is independent of the absence (open symbols) or presence (solid symbols) of 1 μM SOD in normoxia. (E) After 90 min in hypoxia, SSP4 fluorescence produced by K2Sn alone (-SOD, left panel) is essentially identical to that produced in the presence of 1 μM SOD (+SOD, right panel). All points are mean +SE, n = 4 replicates.
We then used the polysulfide-specific fluorescent probe SSP4 to determine if polysulfides were formed by SOD metabolism of H2S. As shown in Fig. 1D-F, this was indeed the case, H2S from 100 μM to 1 mM concentration-dependently increased SSP4 (polysulfide) fluorescence in normoxia in the presence of 1 μM SOD. However, H2S concentrations below 100 μM did not appear to produce PS, and fluorescence was decreased with 3 mM H2S. As a control in the absence of SOD, H2S concentration-dependently increased SSP4 fluorescence, however, there was no inhibitory effect at 3 mM H2S and the maximum fluorescence (polysulfide concentration) produced by 3 mM H2S was only 17% of that produced by 1 mM H2S in the presence of SOD. The increase in fluorescence from SOD-mediated polysulfide production also appeared to be delayed at the outset and then increase exponentially starting around 20 min; this delay was not evident in the absence of SOD.
The above experiment was then repeated in hypoxia to examine the contribution of oxygen to H2S metabolism by SOD. SSP4 fluorescence after 90 min in hypoxia (t = 0 min in Fig. 1G-I) was unchanged and essentially the same as the initial fluorescence in normoxia (t = 0 min in Fig. 1D-F). In addition, the SSP4 fluorescence during the ensuing 90 min in normoxia increased in a H2S concentration-dependent and SOD-dependent pattern similar to that observed in normoxia over the same period, although the magnitude of increase was considerably less in the hypoxia-treated plates. These studies suggest that O2 is required for much of the polysulfide production from H2S in the presence of SOD. SSP4 fluorescence in the hypoxia-treated plates during the subsequent 90 min in the plate reader (in normoxia) was less than fluorescence in normoxic plates over the same period, which is likely the result of a delay in O2 diffusion into the previously hypoxic wells.
To determine if polysulfides could be produced by proteins without a catalytic core, but with an exposed cysteine, such as albumin [40], we incubated 0, 1, 10, 100 and 1000 μM H2S with 0, 1, 10, 100 and 1000 μM albumin and monitored SSP4 fluorescence. Although H2S produced a slight increase in SSP4 fluorescence this was not affected by albumin (Fig. S1). This suggests that a redox reactive cysteine alone is not sufficient to generate polysulfides from H2S.
The H2S-mediated concentration-dependent increase in polysulfide production after 90 min incubation in normoxia with or without 1 μM SOD (from Fig. 1D) is shown in Fig. 2A. The apparent EC50 for SOD-mediated polysulfide production at 90 min, assuming 1 mM H2S was the maximum (100%) response, was ~380 μM H2S. Polysulfide production in the absence of SOD continued to increase with increasing H2S, and the EC50 was not determined. The relatively sharp peak in SOD-mediated SSP4 fluorescence between 300 μM H2S and 3 mM H2S was examined further with 250 μM incremental increases in H2S. As shown in Fig. 2B, peak polysulfide production occurred at 750 μM H2S and this was nearly thirty-fold greater than polysulfide production with 750 μM H2S in the absence of SOD. There was no significant difference in fluorescence between 750 μM and 1 mM H2S although 1 mM appeared to be part of a downward trend. Fluorescence sharply and concentration-dependently decreased when H2S concentration exceeded 1 mM and complete inhibition was evident by 1.75 mM. The apparent IC50 was approximately 1.5 mM H2S (Fig. 2B). There was no H2S-dependent inhibition of polysulfide production in the absence of SOD.
Polysulfide production was also dependent on SOD concentration over a range of H2S concentrations (Fig. 2C). SOD from 0.01 to 3 μM concentration-dependently increased polysulfide production at 0.3, 0.75, 1.0 and 3 mM H2S. Fluorescence significantly (p < 0.05) increased between 0.1 and 0.3 μM SOD with 300 and 750 μM H2S but not with 1 or 3 mM H2S. With 1 mM H2S fluorescence did not significantly increase until SOD concentration exceeded 0.3 μM and with 3 mM H2S fluorescence did not increase until the SOD concentration was 3 μM. There was no significant difference in fluorescence between 750 μM and 1 mM H2S at any SOD concentration. These results suggest that the threshold SOD concentration for polysulfide production is around 0.3 μM and they confirm that SOD is inhibited when H2S concentration is 3 mM.
3.3. SOD does not metabolize polysulfides (H2Sn; Fig. 2D,E))
We then examined whether or not SOD could also metabolize polysulfides using the mixed polysulfide, K2Sn. As shown in Fig. 2D, K2Sn concentration-dependently increased SSP4 fluorescence in both the presence and absence of SOD and this was similar to the effects of carrying out this reaction for 90 min in hypoxia (Fig. 2E). Therefore, SOD does not appear to metabolize inorganic polysulfides either in the presence or absence of oxygen.
3.4. MS detection of polysulfides (Fig. 3A-D)
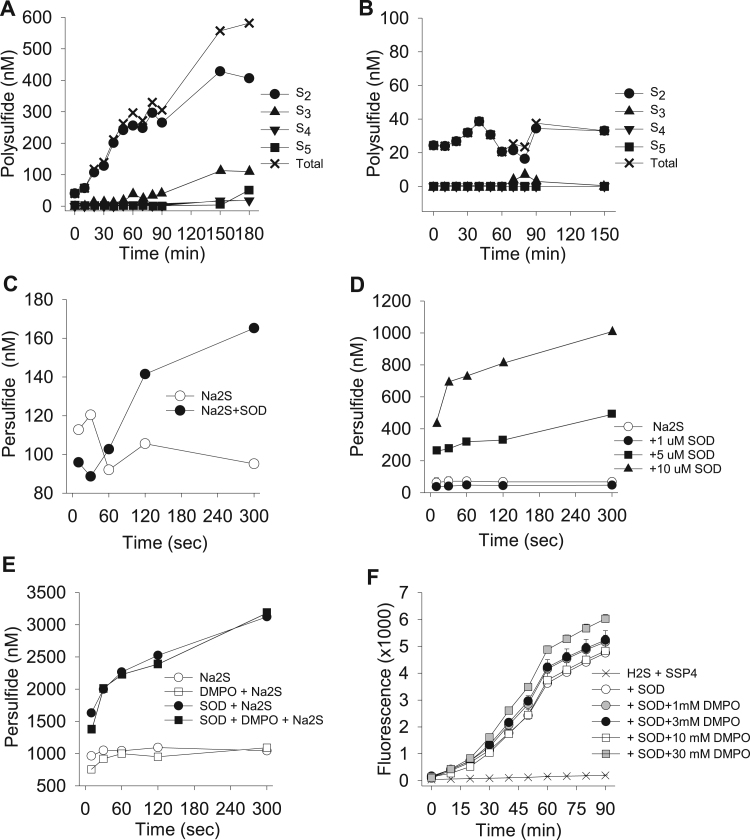
Fig. 3.
Mass spectrometric identification of polysulfides produced by SOD metabolism of Na2S (A-E). (A, B) Production over extended period from 1 mM Na2S under normoxic conditions with (A) or without (B) 1 μM SOD. (A) Persulfide (H2S2) is predominately produced by SOD, whereas other polysulfides appear later. (B) A small amount of persulfide initially appears, likely as a contaminant of Na2S, but no additional polysulfide production is evident (note expanded scale in B). (C-E) short-term (5 min) measurements of persulfide production from 1 mM Na2S. (C) Persulfide is produced from 1 mM Na2S and 1 μM SOD within 2 min and is further increased at 5 min. (D) SOD concentration-dependently increases persulfide production from1 mM Na2S. Note rapid initial rate of persulfide production at elevated SOD concentrations. (E) The radical scavenger DMPO (2 mM) does not affect persulfide production by 10 μM SOD and 1 mM Na2S. (F) Confirmation of the inability of 1–30 mM DMPO to affect polysulfide production (SSP4 fluorescence) by 3 μM SOD metabolism of 750 μM Na2S. A-E, n = 1, F, mean +SE, n = 4.
Detection of polysulfides with mass spectrometry-based methodology allowed us to then identify specific polysulfides that were produced by SOD metabolism of H2S as Na2S (Fig. 3A-D). Under normoxic conditions 1 μM SOD increased the rate of production of persulfide (H2S2) from 1 mM H2S (Fig. 3A). Longer-chain polysulfides, namely H2S3 and H2S5 were formed later in the reaction which could have been the result of recombination of H2S2. Polysulfides were not produced when SOD was incubated with H2S in hypoxia (not shown). A small amount of H2S2 contamination was initially evident in 1 mM Na2S in the absence of SOD (Fig. 3B), but this did not increase over time, and the amount of this background signal was minimal compared to the H2S2 produced by SOD.
Persulfide formation was evident within 2 min after addition of 1 μM SOD to 1 mM H2S (Fig. 3C). SOD concentration-dependently increased persulfide formation from 1 mM H2S (Fig. 3D) and at 5 and 10 μM SOD there appeared to be a very rapid rate of persulfide production that occurred within the first 10 s (faster than the samples could be processed) followed by a slower, steady rate of production. Both fast and slow processes appeared to be dependent on SOD concentration.
3.5. Effect of DMPO on SOD metabolism of H2S (Fig. 3E,F)
In order to determine if persulfides were produced by SOD-catalyzed one-electron reduction of two H2S molecules to form two thiyl radicals which would subsequently combine to form the persulfide, we added the spin trap radical scavenger 5,5-Dimethyl-1-Pyrroline-N-Oxide (DMPO, 2 mM) to 10 μM SOD prior to addition of 1 mM H2S and subsequently monitored persulfide formation by mass-spectrometry (Fig. 3E,F). As shown in Fig. 3E, DMPO did neither affect rate nor magnitude of persulfide formation. To confirm the inability of DMPO to affect this reaction, we added 1, 3, 10, or 30 mM DMPO to 3 μM SOD and 750 μM Na2S and measured polysulfide production with SSP4 (Fig. 3F). Again, there was no appreciable effect of DMPO. There are number of possible explanations for this, 1) persulfide formation may be achieved without thiyl radical formation via a two-electron oxidation of one H2S prior to combining with a second H2S molecule, 2) the rate of reaction of two thiyl radicals with one another out competes the reaction with DMPO, or 3) DMPO has limited access to the catalytic site of SOD. Clearly, additional studies are necessary to identify the mechanisms involved.
3.6. Relative importance of dissolved H2S or HS- in SOD oxidation (Fig. 4)
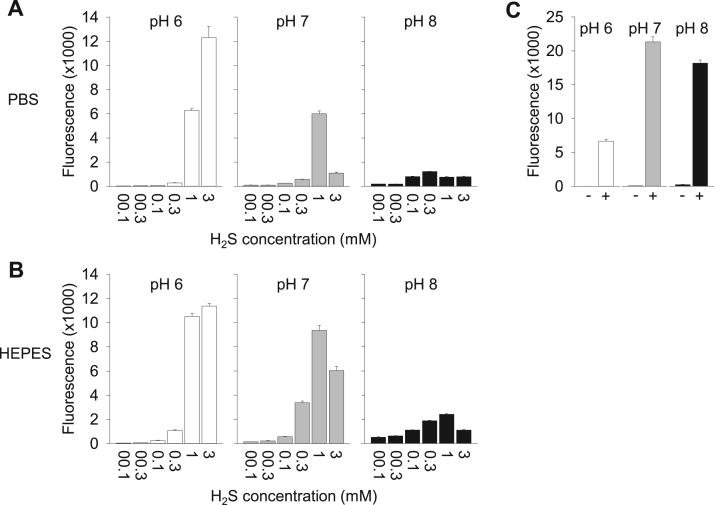
Fig. 4.
Relative importance of dissolved H2S or HS- anion in SOD oxidation. (A) The effects of increasing H2S concentration on SOD (1 μM) oxidation and formation of polysulfides (SSP4 fluorescence) was determined at pH 6, 7, and 8 in PBS buffer. As pH increased, polysulfide formation decreased and the maximum fluorescence was progressively shifted to lower H2S concentrations. (B) Effects of increasing H2S concentration on SOD (1 μM) oxidation and formation of polysulfides (SSP4 fluorescence) in HEPES buffer are similar to those in PBS. (C) pH-dependence of SSP4 fluorescence with (+) or without (-) 300 μM polysulfides in PBS. Fluorescence of SSP4 plus polysulfides is dramatically decreased at pH 6.0 indicating that polysulfide production from SOD and H2S is underestimated at pH 6.0 and that pH has negligible effect on SSP4 alone. All points are mean +SE, n = 4 replicates.
H2S is a weak acid in equilibrium: H2S ↔ HS- ↔ S2-. The pKa1 of this reaction at 20°C is 6.98 and the pKa2 is >12 [41]. Thus, within the physiological range of pH (6.0–8.0) we could adjust the H2S gas to hydrosulfide anion (HS-) ratio from 10:1 to 1:10, a one hundred-fold difference. Because SOD activity is unaffected over this range of pH [34] this allowed us to determine if dissolved H2S or HS- was the preferred substrate for SOD. As shown in Fig. 4A, the H2S concentration-dependent increase in SSP4 fluorescence, indicative of H2S oxidation to polysulfides, decreased dramatically as pH was increased from 6 to 8 in PBS buffer. A similar response was observed when the experiment was carried out in HEPES buffer (Fig. 4B). This effect was not due to a pH effect on SSP4 because SSP4 fluorescence with mixed polysulfides (H2Sn, presumably the end-product of SOD metabolism) in PBS was actually lower (approximately one-third) at pH 6.0 than at either pH 7.0 or 8.0 (Fig. 4C). This suggests that polysulfide production from H2S and SOD at pH 6.0 was likely underestimated. These experiments suggest that SOD preferentially reacts with dissolved H2S, not the hydrosulfide anion. It also appeared that as pH increased, which also increased hydrosulfide anion concentration, peak SSP4 fluorescence was progressively shifted to lower H2S concentrations and at the highest H2S concentrations H2S oxidation became more inhibited. This suggests that the hydrosulfide anion directly inhibits SOD oxidation of H2S.
3.7. Effect of H2O2 on SOD oxidation of H2S (Fig. 5)
Fig. 5.
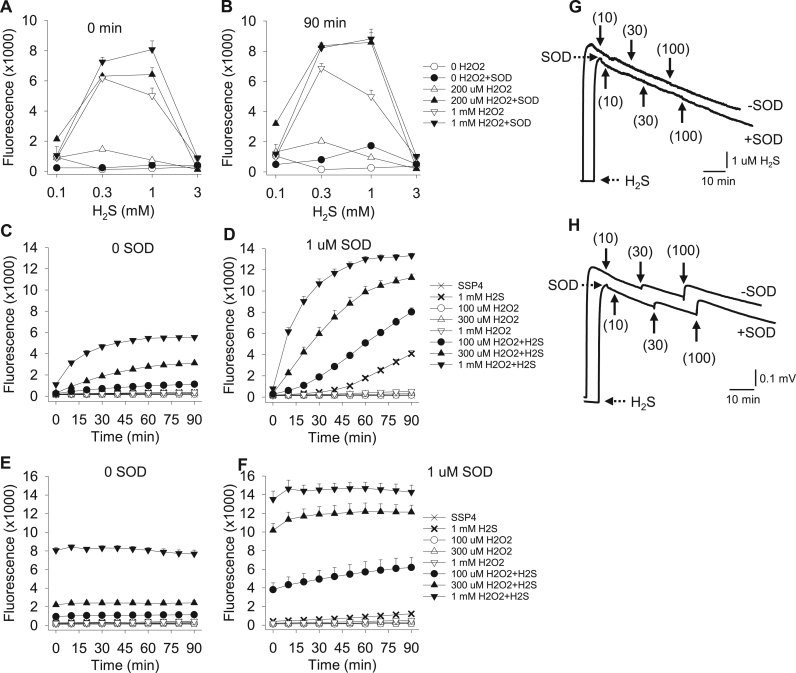
Efficacy of H2O2 as a substrate for SOD metabolism of H2S and polysulfide production (SSP4 fluorescence). (A, B) Comparison of fluorescence produced after 90 min incubation in hypoxia and immediately transferred to plate reader (A; 0 min, i.e., immediately after transfer) versus an additional 90 min in air. Fluorescence did not significantly increase after air exposure indicating that oxidation was completed in hypoxia. C-F) Time-dependent change in fluorescence in normoxia (C, D) or after 90 min in hypoxia and subsequent transfer to normoxia (E, F) without (C, E) or with (D, F) SOD. H2S is effectively oxidized by H2O2 independent of O2 and this is enhanced by SOD. All points are mean +SE, n = 4 replicates. (G-H) Amperometric traces of H2S metabolism. (G) H2S sensor trace showing addition of H2O2 (μM concentration added in parentheses) to 10 μM H2S does not affect the decrease in H2S concentration either with or without 3 μM SOD (+SOD or -SOD, respectively). (H) H2O2 sensor trace showing that addition of H2O2 (concentrations in parentheses) to 10 μM H2S produces a concentration-dependent increase in H2O2 independent of the presence of 3 μM SOD. These traces were recorded simultaneously with those of panel A. Also note the H2O2 sensor is considerably more sensitive to H2S than to H2O2.
Because the canonical product of superoxide dismutation by SOD is H2O2, we first examined if SOD could catalyze the reaction between H2S and H2O2 either with or without O2, and this clearly appeared to occur (Fig. 5). Substantial SSP4 fluorescence was already evident from the combination of 200 μM H2O2 and 300 μM H2S immediately after removing the plate from the hypoxia chamber (Fig. 5A). This concentration of H2O2 (200 μM) is roughly equivalent to the concentration of dissolved O2 in normoxia at room temperature (~190 μM assuming an O2 solubility coefficient of 1.72 μM l−1 mmHg−1 [42]). Increasing the H2O2 concentration to 1 mM and/or increasing the H2S concentration to 1 mM did not significantly affect fluorescence, indicating that the maximum yield of polysulfide was produced with 200 μM H2O2 and 300 μM H2S. Although 1 mM H2O2 plus either 0.3 or 1 mM H2S also oxidized SSP4 in the absence of SOD, lower H2O2 concentrations did not (H2O2 alone did not affect SSP4 fluorescence). This indicates that the lower concentrations of H2O2 in the reaction between H2O2 and H2S involve interaction with SOD. SSP4 fluorescence after the plate had been in room air for 90 min (Fig. 5B) did not increase from the initial t = 0 min immediately after removing the plate from hypoxia confirming that the reaction had gone to completion while in the hypoxia chamber and that H2O2 was the reactant.
We next compared the time-dependent SOD-mediated catalysis of 1 mM H2S and variable H2O2 in the presence and absence of O2 to further characterize this reaction (Fig. 5C-F). Under normoxic conditions, H2O2 concentration-dependently increased SSP4 fluorescence (Fig. 5C) but maximal fluorescence was less than half of that in the presence of 1 μM SOD (Fig. 5D). SSP4 fluorescence increased at a faster rate in the presence of H2O2, H2S and SOD in normoxia (Fig. 5D) than it did with similar concentrations of H2S and SOD in normoxia but without H2O2 (Fig. 5D, thick X; also compare to Fig. 1). For example, with 1 mM H2O2 fluorescence rapidly increased and was near maximal within 45 min. By 60 min fluorescence had plateaued and at this time it was over seven-fold greater than in the absence of H2O2. SOD catalyzed polysulfide production with H2O2 was inhibited at high (3 mM) H2S (Fig. 5A,B) consistent with that observed with 3 mM H2S in normoxia without H2O2 (cf. Fig. 1, Fig. 2).
When samples were incubated in hypoxia for 90 min the initial (t = 0 min) fluorescence was already maximal and it only slightly increased for the subsequent 90 min in normoxia (Fig. 5E,F). This shows that the reaction between H2S and H2O2 is independent of O2. Although polysulfides were produced in the absence of SOD (Fig. 5E), five times as much production was observed with 300 μM H2S and twice as much with 1 mM H2S in the presence of 1 μM SOD (Fig. 5F) showing that SOD readily catalyzes polysulfide formation from H2S and H2O2 without O2.
To determine if SOD consumed H2O2 during metabolism of H2S we monitored this reaction in real time with the H2S and H2O2 amperometric sensors (Fig. 5G,H). We hypothesized that when H2O2 was added to the H2S sensor, in the presence of H2S, there would be a noticeable drop in H2S concentration with, but not without SOD, and SOD would prevent the increase in H2O2 response when monitored with the H2O2 sensor. Consecutive additions of 10, 30 and 100 μM H2O2 did not affect the rate of 10 μM H2S metabolism by 3 μM SOD (Fig. 5G) indicating that there was no immediate reaction between H2S and H2O2. Using the H2O2 sensor (Fig. 5H) it was also evident that addition of 30 μM H2O2 in the absence or presence of 3 μM SOD produced a similar increase in the recorded voltage (0.059 ± 0.008, n = 4 and 0.051 ± 0.004, n = 5 mV, respectively) as did 100 μM H2O2 (0.17 ± 0.016, n = 4 and 0.17 ± 0.012, n = 5 mV, respectively). Ten μM H2O2 did not produce a noticeable signal at these settings. These results also indicate that H2O2 does not directly participate in SOD metabolism of H2S, confirming the earlier findings of Searcy et al. [43]. Clearly, the contribution of H2O2 needs further investigation. In addition, Fig. 5H clearly shows that the H2O2 sensor is more sensitive to H2S than to H2O2 (32 fold in these experiments), confirming our previous observations [28].
3.8. Effects of possible electron donors and acceptors on SOD oxidation of H2S (Fig. 6)
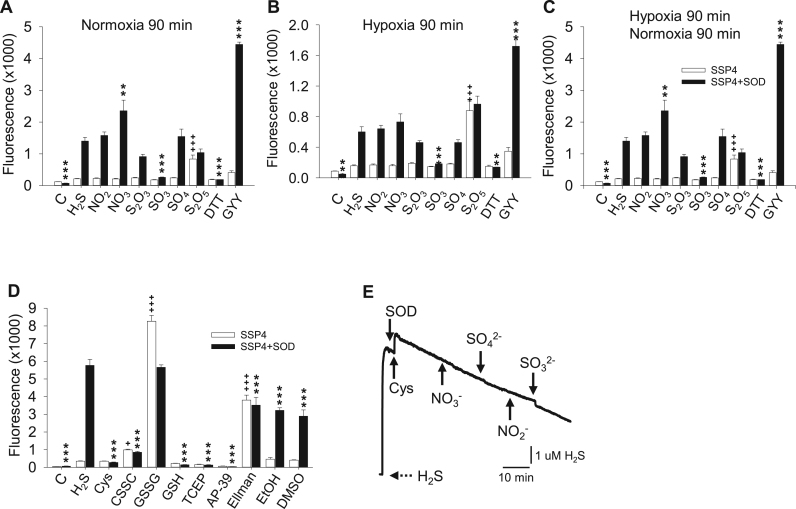
Fig. 6.
Effects of possible electron doors and acceptors on polysulfide production (SSP4 fluorescence) from oxidation of H2S with (solid bars) or without (open bars) SOD in normoxia and hypoxia (A-D). SOD (1 μM), H2S (1 mM) and 1 mM sodium salts of nitrite (NO2), nitrate (NO3), thiosulfate (S2O3), sulfite (SO3), sulfate (SO4), metabisulfate (S2O5) and 1 mM dithiothreitol (DTT) and GYY 4137 (GYY), (C control, no H2S A-C) were incubated for 90 min in normoxia (A), 90 in in hypoxia and examined immediately (B) or 90 min hypoxia followed by 90 min normoxia in a covered plate (C). D) 0.75 mM cysteine (Cys), cystine (CSSC), oxidized glutathione (GSSG), glutathione (GSH), the non-sulfur reductant, tris(2-carboxyethyl)phosphine, (TCEP), a mitochondria-targeted H2S donor (AP-39), Ellman's reagent (Ellman), ethanol (EtOH) and dimethyl sulfoxide (DMSO) were incubated in normoxia with 0.75 mM H2S. All points are mean +SE, n = 4 replicates; *, **, and ***, significantly different from 1 mM H2S plus SOD at p < 0.5, 0.01, or 0.001, respectively; +++, significantly different from 1 mM H2S without SOD at p < 0.001). (E) Amperometric trace showing the lack of effect of cysteine (1 mM), nitrite (NO2-; 1 mM), nitrate (NO3-1 mM), sulfite (SO32-; 1 mM) and sulfate (SO42-; 1 mM) on 3 μM SOD metabolism of 10 μM H2S.
Polysulfide production (SSP4 fluorescence) and amperometric measurement of H2S was used to determine if possible electron donors/acceptors other than O2 or H2O2 affected SOD oxidation of H2S. These compounds were selected because they either could have contributed to RSS metabolism early in evolution, or because of their use in present-day experiments. In the first series of experiments H2S (0.75 or 1 mM) was incubated with a variety of equal molar concentrations of possible electron donors/acceptors in the presence or absence of 1 μM SOD and under normoxic and hypoxic conditions to determine if they affected polysulfide production (Fig. 6A-D). The slow releasing H2S donor, GYY4137 (GYY) consistently increased polysulfide formation in all conditions, possibly because it provided a continual source of H2S [44] or sulfane sulfur. Nitrate (NO3-) increased SSP4 fluorescence when O2 was present, whereas fluorescence was decreased with sulfite (SO32-), DTT, cysteine (Cys), cystine (CSSC), glutathione (GSH), the sulfur-free reductant tris(2-carboxyethyl)phosphine (TCEP), the mitochondrial H2S donor AP-39, and to a lesser extent with Ellman's reagent, ethanol and DMSO. Fluorescence was not affected by nitrite (NO2-), thiosulfate (S2O32-) or sulfate (SO42-). Metabisulfate (S2O52-) increased SSP4 fluorescence with or without SOD. The inhibition of SOD oxidation of H2S by sulfite and metabisulfite in normoxia and hypoxia, suggests that their action is not just due to scavenging O2. Oxidized glutathione (GSSG) greatly increased fluorescence in the absence of SOD. Since SSP4 reacts with persulfides as well as polysulfides this observation confirms that the reaction of H2S with GSSG produces a persulfide (GSSH) and a free thiol (GSH; [45]). With SOD there was no difference in fluorescence between H2S and H2S plus GSSG. However, there was a significant (p < 0.001) decrease between GSSG without and with SOD suggesting that GSSG also inhibits SOD-mediated polysulfide production or that there was some GSSG contamination that could also react with SSP4. Alternatively, SOD might compete with GSSG for H2S, and when it does the polysulfides produced may affect fluorescence as much as the persulfides made by GSSG alone. In order to determine if 1 mM cysteine, nitrate, nitrite, sulfite or sulfate directly reacted with 10 μM H2S in the presence of 3 μM SOD we monitored H2S in real-time with the amperometric sensor. As shown in Fig. 6E, none of these compounds affected the H2S response suggesting there is no direct reaction within the 10–30 min time frame of the experiment.
3.9. H2S and polysulfide metabolism by MnSOD (Fig. 7)
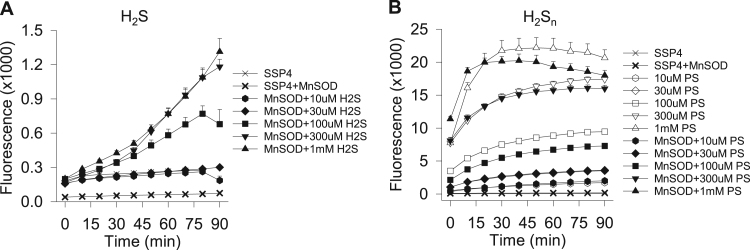
Fig. 7.
H2S and polysulfide (H2Sn) metabolism by 1 μM MnSOD. MnSOD produces polysulfides (SSP4 fluorescence) from H2S (A) but does not metabolize polysulfides (B). Mean +SE, n = 4 replicates.
In order to determine if H2S metabolism was a general property of SODs we examined the effects of manganese SOD (MnSOD) on H2S metabolism. Our results show that polysulfides (SSP4 fluorescence) were concentration-dependently produced from H2S by 1 μM MnSOD (Fig. 7A), whereas MnSOD did not appear to metabolize K2Sn-type polysulfides (Fig. 7B). These results suggest that sulfide/sulfur metabolism by MnSOD is similar to that by Cu/ZnSOD.
4. Discussion
Superoxide dismutases (SODs) are well-known antioxidant enzymes that catalyze the dismutation of superoxide (O2•-) to H2O2 and O2 (Eq. (2)). Three SODs are present in vertebrates, the mammalian cytosolic and soluble forms use a Cu/Zn center whereas Mn is in the mitochondrial SOD [34]. The present experiments suggest that SOD also metabolizes sulfide and the initial product of this reaction is persulfide (H2S2). This reaction requires an oxidant such as oxygen, peroxide or sulfane sulfur and appears to utilize H2S rather than hydrosulfide anion. The persulfides generated may eventually react with additional sulfane sulfur to form longer-chain polysulfides (H2Sn where n>2). These studies support the hypothesis that one, if not the original, function of SOD was in sulfur metabolism in the pre-oxic Earth using an as yet unidentified electron acceptor and that this legacy persists to the present day in utilizing O2 or H2O2 as the electron acceptor.
It is not clear how SOD catalyzes this reaction. One possibility is the general reaction;
| H2S + O2 → HS• + O2•- + H+, | (5) |
This reaction (Eq. (5)) is reminiscent of superoxide dismutation (Eq. (2)) in reverse and it may proceed through the following steps;
| H2S + Cu++(SOD) → HS• Cu+(SOD) + H+, | (6) |
| HS• Cu+(SOD) → HS• + Cu+(SOD), | (7) |
| 2HS• → H2S2· | (8) |
| Cu+(SOD) + O2 → Cu++(SOD) + O2•- | (9) |
| 2Cu++(SOD) + 2O2•- + 2H+ → Cu++(SOD) + O2 + H2O2 | (10) |
In addition, hydrogen peroxide could be consumed by excess hydrogen sulfide to produce sulfane sulfur, which would react with hydrogen sulfide or polysulfides to extend the polysulfide chain length. This would explain why we did not detect hydrogen peroxide produced by reaction 10 (Fig. 5).
We did not try to determine if SOD also catalyzed reaction 5 in reverse, although the formation of persulfide, which can readily occur by the combination of 2 thiyl radicals, would decrease the probability of this occurring. Liochev and Fridovich [46] measured the reduction of tetranitromethane by O2•- and observed that the superoxide dismutase reaction could also be reversed under these conditions. However, the rate constant they obtained was so slow that the authors concluded that there would be little if any significant amount of O2•- production.
In an earlier study Searcy [39] showed that Cu,Zn SOD utilized O2 to oxidize H2S with a 1:1 O2:H2S ratio. In “dilute” solutions (0.01 μM SOD, 280 μM O2, 500 µm H2S), the reaction, which was monitored by O2 consumption, appeared to accelerate (i.e., an “augmented” reaction) such that after 1 h the rate of H2S oxidation was 600 times faster than the initial rate. With 10 μM SOD, however, the reaction progressively decreased. When this reaction was monitored by H2S absorbance, the SOD threshold was 0.01 μM but the rate did not increase in proportion to SOD concentration until SOD exceeded 1 μM (3 μM). Sulfane sulfur (S0) and H2O2 were produced by this reaction. The “augmented” reaction was not affected by azide (10 mM), benzoate (25 mM), formate (50 mM) or catalase (50 nM) but it was completely inhibited by the radical spin-trap N-tert-butyl-α-phenylnitrone (PBN, 50 mM) suggesting a radical was involved in the “augmented” reaction. However the authors could not detect any paramagnetic resonance after 1 h, and according to the authors the PBN results were “still open to interpretation”. Cyanide (10 mM) completely inhibited the initial reaction but only partially inhibited the “augmented” reaction suggesting that it was to some extent non-enzymatic. The augmented reaction could be sustained if stored for 23 h in anoxia but not in normoxia. H2S (500 μM) and H2O2 were equally consumed (approximately by half) with or without SOD indicating that SOD is not a peroxidase. In a related study Searcy et al. [43] showed that hydrosulfide anion (HS-), not dissolved H2S, was responsible for the SOD catalyzed reaction between H2S and superoxide and proposed that the reaction with O2 was;
| HS- + H+ + O2 → S0 + H2O2, | (11) |
but that the initial reaction produced a sulfhydryl radical (HS•) and reduced SOD Cu(II) which then combined with another HS- to produce a H2S2• radical that then reduced a second Cu(II). They also suggested that the reaction with H2S was saturable.
Our studies confirm those of Searcy [39] and Searcy et al. [43] in that SOD catalyzes the oxidation of H2S and forms polysulfides, but our results and conclusions also differ in a number of respects. First, we observed that the reaction appeared to have two components, an initial fast reaction followed by a slower one. We did not find any evidence for an augmented reaction or evidence of the ability of the spin-trap DMPO to affect the rate of persulfide production (Fig. 3E, F). Second, our study suggests that dissolved H2S, not HS-, reacts with SOD, whereas HS- inhibits the reaction (Fig. 4). Third, we found that H2S2, but not H2S3 or H2S4 was the initial product suggesting that this is derived from the reaction of two thiyl radicals (Fig. 1, Fig. 2), although other reaction mechanisms are also possible (see below). If the reaction generates S0 as proposed by Searcy [39] and Searcy et al. [43], this either proceeds slowly or occurs concomitant with H2S2 production. Fourth, we did not see an increase in H2O2 production with the H2O2 electrode during the reaction (Fig. 5), although it is possible that we could have overlooked this due to the electrode's greater sensitivity for H2S than H2O2.
The ability to change the ratio of dissolved H2S to hydrosulfide anion (HS-) one hundred-fold, from 10:1 to 1:10 between pH 6 and 8, afforded us the opportunity to examine reaction of SOD with these two forms of sulfide. Our results indicate that dissolved H2S appears to be the preferred the substrate. Although HS- would seem to be the more likely candidate for this reaction (and may well be involved in Eq. (6), following initial metal complexation of H2S and subsequent internal electron transfer reaction to produce a thiyl radical formally bound to Cu+) it paradoxically appears to inhibit SOD, at least at higher concentrations. The reason(s) for this is (are) not evident. Clearly, the pH sensitivity of fluorescent SSP4 cannot account for this discrepancy as SSP4 fluorescence is decreased at pH 6, not increased. HS- could inhibit SOD if it forms a relatively stable bond with the SOD Cu. The apparent preference for H2S at pH 6 would then be the indirect result of less SOD inhibition due to lower relative concentration of HS-. HS- binding to zinc may also be inhibitory. Regardless, further studies are necessary to resolve this issue.
It is also possible that sulfide inhibition of SOD or even polysulfide formation occurs, not at the catalytic Cu site but through interaction with SOD cysteines. Cu/Zn SOD monomers have four cysteines, Cys6, Cys57, Cys111 and Cys146. Cys6 is buried within the protein and relatively unreactive, Cys57 and Cys 146 form an intramolecular disulfide bridge important for the high structural stability of the protein, and Cys111 is a free “reactive” cysteine with a low pKa that renders it susceptible to oxidation. Cys111 from adjacent monomers forms a heptasulfane bridge that can be cleaved by thiols and reformed by elemental sulfur [47]. It is possible that the polysulfane bridge that forms involving Cys111 contributes to persulfide formation via sulfhydryl-disulfide exchange reactions. This may require elevated H2S concentrations and could account for our observed 0.3 μM H2S threshold. If this does indeed occur it appears to be a specific property of SOD because a free thiol, such as that found in albumin [40] is not sufficient to generate polysulfides on its own (Fig. S1).
H2S has been shown to inhibit O2•- dismutation by bovine erythrocyte SOD with an estimated IC50 of approximately 10 mM [48], and this inhibition could be partially (20–50%) reversed by dialysis in water for 8–12 h at 4 °C. SOD was also inhibited by SO32-(IC50 ~18 mM) and SO42- (27% inhibition with 20 mM). Our study shows that under normoxic conditions polysulfide production from H2S oxidation by SOD is also inhibited by H2S (Fig. 2) and that the enzyme appears to be nearly tenfold more sensitive to H2S (IC50 = 1.25 mM) and SO32-(essentially complete inhibition at 1 mM), but not affected by SO42-(Fig. 2, Fig. 6). We also observed considerable inhibition by cysteine, cystine, glutathione, the mitochondrial targeted H2S donor, AP-39, and by the thiol-dependent reductant DTT and the sulfhydryl-free reductant TCEP. The reaction was unaffected by S2O32- or S2O52- Collectively, these results suggest that polysulfide production by SOD oxidation of H2S is regulated by a number of H2S metabolites including H2S itself and that this is more in tune to the sulfur moiety than it is to their redox properties which suggests their target is SOD cysteines, not the reactive Cu/Zn motif. This is supported by our observation that neither SO32-, SO42-, NO2- nor NO3- directly affect H2S concentration in the presence of SOD when H2S is measured amperometrically.
5. Biological relevance
As described in the introduction, the efficacy of polysulfides as signaling molecules is now well established. In a series of seminal studies Kimura's group identified specific polysulfides produced enzymatically from 3-mercaptopyruvate (3-MP) and by direct reaction with nitric oxide (NO). In COS cells or murine neurons 3-mercaptopyruvate sulfurtransferase (3-MST) catalyzed formation of H2S3 and H2S from 3-MP in a 10:1 ratio, with little production of either H2S2 or H2S5 [49]. They also observed that H2S3 could be produced from H2S by either 3-MST or rhodanese, whereas the latter did not metabolize 3-MP. The 3-MST catalyzed production of H2S3 increased linearly with 3-MP concentration between 400 μM and 1.5 mM, but it did not increase further with 1.75 mM and it was completely inhibited by 2.0 mM. This abrupt inhibition at 2.0 mM 3-MP (as well as the 3-MP concentration) is strikingly similar to our observation of H2S inhibition of polysulfide production by SOD (Fig. 2) but differs in that Cu/ZnSOD catalyzes the formation of H2S2 not H2S3. We did not identify the polysulfide(s) produced by MnSOD but it seems resonable to assume that H2S2 was also the primary product. As 3-MST is found in both the cytosol and mitochondria and Cu/ZnSOD is cytosolic and MnSOD is mitochondrial, this would allow formation of H2S2 and H2S3 in both compartments. H2S2 and H2S3, are produced, apparently in nearly equal amounts, when H2S is added to NO and this has been suggested as another mechanism for polysulfide signaling [50], confirming earlier reports of polysulfide formation by us [51]. This would also increase H2S2 and H2S3 independently of either 3-MST or SOD. Although polysulfide-specific fluorophores were used in both studies by Kimura's group to demonstrate an increase in cellular polysulfides [49], [50], the resolution was not sufficient to distinguish between a mitochondrial or cytosolic origin. Reactive organic hydroper- and hydropolysulfides may also be generated from cystine by cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE) in the cytoplasm [52], or possibly in mitochondria after stress-induced increases in CBS and CSE in the latter [53], [54]. This requires additional study.
The biological relevance of H2S metabolism by SOD remains to be clarified. We show in buffer solutions that this reaction is dependent on the concentrations of both SOD and H2S with a SOD threshold of ~0.3 μM and a H2S threshold of ~100 μM. With 1 μM SOD the EC50 for H2S is 380 μM and maximum polysulfide formation is achieved at ~750 μM H2S. Cu/ZnSOD concentration in rat hepatocyte cytosol is 1.36 mg/ml [55] and in human red blood cells it is 7.0 μg/mg protein [56], which is equivalent to 42 and 75 μM, respectively. This is considerably higher than the 1 μM SOD we used and 140–250 times greater than the threshold for H2S oxidation in the present experiments. We [38] previously showed that RBC from mammals and sub-mammalian vertebrates rapidly removes H2S from plasma or buffer and the present results indicate that RBC SOD could be an effective mediator of this process. Furthermore, H2S oxidation by MnSOD in the mitochondrion may be especially significant in dealing with mitochondrial H2S. Collectively, our results suggest that SOD is effectively positioned to oxidize H2S produced within cells or entering from an external source.
A number of questions remain including the considerable differences between the concentrations of H2S we used in this study and those anticipated to exist within the cell which are < 1 μM [57], [58] and the amount of H2S2 produced which is relatively low given the initial 1 mM H2S concentration. Several factors could account for these discrepancies. First, intracellular H2S concentrations are expected, and predicted [59] to be considerably greater in the vicinity of their production, but this information is currently unavailable. Second, SSP4 is relatively insensitive and slow to react. Third, H2S is quite volatile [60] and the actual H2S concentration during an experiment is likely to be considerably lower than assumed (which may also contribute to the fall in H2S concentration in the absence of SOD in Fig. 1). Fourth, it is possible (probable) that sulfur moieties, in addition to polysulfides are produced but remain undetected. Clearly, these issues need to be resolved to understand the physiological relevance of H2S metabolism by SOD.
We also observed that H2S metabolism by SOD is inhibited by H2S in excess of 1 mM with an IC50 of 1.25 mM H2S and complete inhibition at 1.75 mM. This may be attributable to the high [HS-] at pH 7.4 (cf. Fig. 4). Although is seems likely that SOD retains functionality, under normal physiological conditions, it may lose its effectiveness when inhibited by exogenous H2S; a point to be considered in designing experiments.
6. Perspectives
The physiological relevance of SOD in sulfide metabolism may have its foundations in evolution. Life began in an anoxic and reducing environment around 3.8 bya and these conditions generally persisted until the advent of oxidative photosynthesis in cyanobacteria which increased atmospheric oxygen to 0.5–1% and heralded in the “great oxidation event” (GOE) around 2.3 bya [31], [32], [33]. It is thought that in the absence of O2, and, therefore, O2•-, prior to this period, there was either no need for SOD enzymes to evolve [34] or they acquired importance when O2 became prevalent [35]. However, the fact that SODs appear to have evolved well before the GOE [34], [35] suggests that they performed important biochemical functions on substrates other than ROS. The identity of one or more of the putative electron acceptors in these ancient organisms remains to be determined as our studies failed to find a likely candidate from several of the more obvious oxidants. This feature could have been lost over the past 600 million years evolution in oxic environments, however, the observation that H2O2 can substitute for O2 suggests there is some latitude in specificity.
It is also possible that one of these primordial electron acceptors may be more applicable to catalysis by an ancient SOD, such as FeSOD [34] and this remains to be explored. Other variations in SOD catalysts are also known. NiSOD uses electron rich thiolate ligation between Cys2 and Cys6 to adjust the reduction potential instead of an aqua ligand [34]. Although NiSOD appears to be a case of convergent evolution [34], it is an interesting precedent for a more extensive role of sulfur in the ancient world.
The evolutionary pedigree of SOD appears to be shared with another “antioxidant” enzyme, catalase. We recently showed that catalase, which, like SOD, also appeared long before the GOE, is sulfide/sulfur oxido/reductase that can utilize either O2 or H2O2 to oxidize H2S, or in the absence of O2 regenerate H2S from other endogenous sources such as thioredoxin, or even from dithiothreitol [29]. Thus while SOD appears to unidirectionally oxidize H2S, catalase can both oxidize H2S and recover it depending on environmental circumstances. This could provide an interesting regulatory loop in which under certain circumstances, such as normoxia, H2S is oxidized to a polysulfide by both SOD and catalase, whereas in other situations, such as hypoxia, SOD metabolism is inhibited and catalase recovers H2S from these or other sources.
Collectively, our findings provide additional support for the hypothesis that the primordial substrates of ancient redox enzymes were sulfur-based, and most likely H2S. Our work also suggest that both SOD and catalase may remain biochemical effectors of sulfide/sulfur metabolism in modern-day cells.
Acknowledgments
This research was supported by National Science Foundation Grant No. IOS 1446310 (KRO) and by the German Research Council (DFG CO 1305/2-1, and CO 1305/3-1 / GO 1367/3-1, and DFG SFB1116 TP06 (MMC-K).
Acknowledgments
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
Author contributions
K.R.O. and M.F. designed the studies, M.M.C.-K. provided intellectual and conceptual input, K.R.O. wrote the manuscript, K.R.O., M.M. and M.M.C.-K. criticallt revised the manuscript, F.A., K.A., S.P., Y.G., E.D. and T.R.S. performed the studies, all authors interpreted the data and commented on the manuscript.
Footnotes
Note: unless stated otherwise for convenience H2S, H2S2 and H2Sn are used throughout to represent various dissociated forms, i.e., HS-, HS2-, HSn- and S2-, S22-, Sn2-.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.11.009.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Brandes N., Tienson H., Lindemann A., Vitvitsky V., Reichmann D., Banerjee R., Jakob U. Time line of redox events in aging postmitotic cells. Elife. 2013;2:e00306. doi: 10.7554/eLife.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown D.I., Griendling K.K. Regulation of signal transduction by reactive oxygen species in the cardiovascular system. Circ. Res. 2015;116:531–549. doi: 10.1161/CIRCRESAHA.116.303584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y.R., Zweier J.L. Cardiac mitochondria and reactive oxygen species generation. Circ. Res. 2014;114:524–537. doi: 10.1161/CIRCRESAHA.114.300559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chouchani E.T., Pell V.R., James A.M., Work L.M., Saeb-Parsy K., Frezza C., Krieg T., Murphy M.P. A unifying mechanism for mitochondrial superoxide production during ischemia-reperfusion injury. Cell Metab. 2016;23:254–263. doi: 10.1016/j.cmet.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Circu M.L., Aw T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Circu M.L., Aw T.Y. Intestinal redox biology and oxidative stress. Semin. Cell Dev. Biol. 2012;23:729–737. doi: 10.1016/j.semcdb.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson S.B. Investigating the role of reactive oxygen species in regulating autophagy. Methods Enzymol. 2013;528:217–235. doi: 10.1016/B978-0-12-405881-1.00013-6. [DOI] [PubMed] [Google Scholar]
- 8.Goncalves R.L., Quinlan C.L., Perevoshchikova I.V., Hey-Mogensen M., Brand M.D. Sites of superoxide and hydrogen peroxide production by muscle mitochondria assessed ex vivo under conditions mimicking rest and exercise. J. Biol. Chem. 2015;290:209–227. doi: 10.1074/jbc.M114.619072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambeth J.D., Neish A.S. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu. Rev. Pathol. 2014;9:119–145. doi: 10.1146/annurev-pathol-012513-104651. [DOI] [PubMed] [Google Scholar]
- 10.Li X., Fang P., Mai J., Choi E.T., Wang H., Yang X.F. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013;6:19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 12.Olschewski A., Weir E.K. Redox regulation of ion channels in the pulmonary circulation. Antioxid. Redox Signal. 2014 doi: 10.1089/ars.2014.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radi E., Formichi P., Battisti C., Federico A. Apoptosis and oxidative stress in neurodegenerative diseases. J. Alzheimer's Dis.: JAD. 2014;42:S125–S152. doi: 10.3233/JAD-132738. [DOI] [PubMed] [Google Scholar]
- 14.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz E., Wenzel P., Munzel T., Daiber A. Mitochondrial redox signaling: interaction of mitochondrial reactive oxygen species with other sources of oxidative stress. Antioxid. Redox Signal. 2014;20:308–324. doi: 10.1089/ars.2012.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma K. Obesity and diabetic kidney disease: role of oxidant stress and redox balance. Antioxid. Redox Signal. 2016;25:208–216. doi: 10.1089/ars.2016.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winterbourn C.C. Are free radicals involved in thiol-based redox signaling? Free Radic. Biol. Med. 2014;80:164–170. doi: 10.1016/j.freeradbiomed.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Olson K.R., Straub K.D. The role of hydrogen sulfide in evolution and the evolution of hydrogen sulfide in metabolism and signaling. Physiology. 2016;31:60–72. doi: 10.1152/physiol.00024.2015. [DOI] [PubMed] [Google Scholar]
- 19.Lo C.M., Carroll K.S. The redox biochemistry of protein sulfenylation and sulfinylation. J. Biol. Chem. 2013;288:26480–26488. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura H. Signaling molecules: hydrogen sulfide and polysulfide. Antioxid. Redox Signal. 2015;22:362–376. doi: 10.1089/ars.2014.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy P. Mechanistic chemical perspective of hydrogen sulfide signaling. Methods Enzymol. 2015;554:3–29. doi: 10.1016/bs.mie.2014.11.036. [DOI] [PubMed] [Google Scholar]
- 22.Paul B.D., Snyder S.H. Protein sulfhydration. Methods Enzymol. 2015;555:79–90. doi: 10.1016/bs.mie.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Cortese-Krott M.M., Koning A., Kuhnle G.G., Nagy P., Bianco C., Pasch A., Wink D.A., Fukuto J., Jackson A.A., van Goor H., Olson K.R., Feelisch M. The reactive species interactome: evolutionary emergence, biological significance, and opportunities for redox metabolomics and personalized medicine. Antioxid. Redox Signal. 2017 doi: 10.1089/ars.2017.7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filipovic M.R. Persulfidation (S-sulfhydration) and H2S. Handb. Exp. Pharmacol. 2015;230:29–59. doi: 10.1007/978-3-319-18144-8_2. [DOI] [PubMed] [Google Scholar]
- 25.Ono K., Akaike T., Sawa T., Kumagai Y., Wink D.A., Tantillo D.J., Hobbs A.J., Nagy P., Xian M., Lin J., Fukuto J.M. Redox chemistry and chemical biology of HS, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic. Biol. Med. 2014;17:82–94. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toohey J.I., Cooper A.J. Thiosulfoxide (sulfane) sulfur: new chemistry and new regulatory roles in biology. Molecules. 2014;19:12789–12813. doi: 10.3390/molecules190812789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palinkas Z., Furtmuller P.G., Nagy A., Jakopitsch C., Pirker K.F., Magierowski M., Jasnos K., Wallace J.L., Obinger C., Nagy P. Interactions of hydrogen sulfide with myeloperoxidase. Br. J. Pharmacol. 2015;172:1516–1532. doi: 10.1111/bph.12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLeon E.R., Gao Y., Huang E., Arif M., Arora N., Divietro A., Patel S., Olson K.R. A case of mistaken identity: are reactive oxygen species actually reactive sulfide species? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016;310:R549–R560. doi: 10.1152/ajpregu.00455.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson K.R., Gao Y., DeLeon E.R., Arif M., Arif F., Arora N., Straub K.D. Catalase as a sulfide-sulfur oxido-reductase: an ancient (and modern?) regulator of reactive sulfur species (RSS) Redox Biol. 2017;12:325–339. doi: 10.1016/j.redox.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wachtershauser G. Groundworks for an evolutionary biochemistry: the iron-sulphur world. Prog. Biophys. Mol. Biol. 1992;58:85–201. doi: 10.1016/0079-6107(92)90022-x. [DOI] [PubMed] [Google Scholar]
- 31.Canfield D.E., Ngombi-Pemba L., Hammarlund E.U., Bengtson S., Chaussidon M., Gauthier-Lafaye F., Meunier A., Riboulleau A., Rollion-Bard C., Rouxel O., Asael D., Pierson-Wickmann A.C., El A.A. Oxygen dynamics in the aftermath of the Great Oxidation of Earth's atmosphere. Proc. Natl. Acad. Sci. USA. 2013;110:16736–16741. doi: 10.1073/pnas.1315570110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hohmann-Marriott M.F., Blankenship R.E. Evolution of photosynthesis. Annu. Rev. Plant Biol. 2011;62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- 33.Planavsky N.J., Reinhard C.T., Wang X., Thomson D., McGoldrick P., Rainbird R.H., Johnson T., Fischer W.W., Lyons T.W. Earth history. low mid-proterozoic atmospheric oxygen levels and the delayed rise of animals. Science. 2014;346:635–638. doi: 10.1126/science.1258410. [DOI] [PubMed] [Google Scholar]
- 34.Sheng Y., Abreu I.A., Cabelli D.E., Maroney M.J., Miller A.F., Teixeira M., Valentine J.S. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014;114:3854–3918. doi: 10.1021/cr4005296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller A.F. Superoxide dismutases: ancient enzymes and new insights. FEBS Lett. 2012;586:585–595. doi: 10.1016/j.febslet.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoops B., Loumaye E., Van D.E., V Evolution of the peroxiredoxins. Subcell. Biochem. 2007;44:27–40. doi: 10.1007/978-1-4020-6051-9_2. [DOI] [PubMed] [Google Scholar]
- 37.Lu J., Holmgren A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 38.Whitfield N.L., Kreimier E.L., Verdial F.C., Skovgaard N., Olson K.R. Reappraisal of H2S/sulfide concentration in vertebrate blood and its potential significance in ischemic preconditioning and vascular signaling. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;294:R1930–R1937. doi: 10.1152/ajpregu.00025.2008. [DOI] [PubMed] [Google Scholar]
- 39.Searcy D.G. HS-:O2 oxidoreductase activity of Cu,Zn superoxide dismutase. Arch. Biochem. Biophys. 1996;334:50–58. doi: 10.1006/abbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 40.Turell L., Radi R., Alvarez B. The thiol pool in human plasma: the central contribution of albumin to redox processes. Free Radic. Biol. Med. 2013;65:244–253. doi: 10.1016/j.freeradbiomed.2013.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson K.R. A practical look at the chemistry and biology of hydrogen sulfide. Antioxid. Redox Signal. 2012;17:32–44. doi: 10.1089/ars.2011.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boutilier R.G., Heming T.A., Iwama G.K. Physiochemical parameters for use in fish respiratory physiology. In: Hoar W.S., Randall D.J., editors. Fish Physiology Vol X, Gills Part A. Academic Press; Orlando: 1984. pp. 401–430. [Google Scholar]
- 43.Searcy D.G., Whitehead J.P., Maroney M.J. Interaction of Cu,Zn superoxide dismutase with hydrogen sulfide. Arch. Biochem. Biophys. 1995;318:251–263. doi: 10.1006/abbi.1995.1228. [DOI] [PubMed] [Google Scholar]
- 44.Li L., Whiteman M., Guan Y.Y., Neo K.L., Cheng Y., Lee S.W., Zhao Y., Baskar R., Tan C.H., Moore P.K. Characterization of a novel, water-soluble hydrogen sulfide-releasing molecule (GYY4137): new insights into the biology of hydrogen sulfide. Circulation. 2008;117:2351–2360. doi: 10.1161/CIRCULATIONAHA.107.753467. [DOI] [PubMed] [Google Scholar]
- 45.Francoleon N.E., Carrington S.J., Fukuto J.M. The reaction of H(2)S with oxidized thiols: generation of persulfides and implications to H(2)S biology. Arch. Biochem. Biophys. 2011;516:146–153. doi: 10.1016/j.abb.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Liochev S.I., Fridovich I. Reversal of the superoxide dismutase reaction revisited. Free Radic. Biol. Med. 2003;34:908–910. doi: 10.1016/s0891-5849(03)00024-8. [DOI] [PubMed] [Google Scholar]
- 47.You Z., Cao X., Taylor A.B., Hart P.J., Levine R.L. Characterization of a covalent polysulfane bridge in copper-zinc superoxide dismutase. Biochemistry. 2010;49:1191–1198. doi: 10.1021/bi901844d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan A.A., Schuler M.M., Coppock R.W. Inhibitory effects of various sulfur compounds on the activity of bovine erythrocyte enzymes. J. Toxicol. Environ. Health. 1987;22:481–490. doi: 10.1080/15287398709531087. [DOI] [PubMed] [Google Scholar]
- 49.Kimura Y., Toyofuku Y., Koike S., Shibuya N., Nagahara N., Lefer D., Ogasawara Y., Kimura H. Identification of H2S3 and H2S produced by 3-mercaptopyruvate sulfurtransferase in the brain. Sci. Rep. 2015;5:14774. doi: 10.1038/srep14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto R., Koike S., Takano Y., Shibuya N., Kimura Y., Hanaoka K., Urano Y., Ogasawara Y., Kimura H. Polysulfides (H2Sn) produced from the interaction of hydrogen sulfide (H2S) and nitric oxide (NO) activate TRPA1 channels. Sci. Rep. 2017;7:45995. doi: 10.1038/srep45995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortese-Krott M.M., Kuhnle G.G., Dyson A., Fernandez B.O., Grman M., DuMond J.F., Barrow M.P., McLeod G., Nakagawa H., Ondrias K., Nagy P., King S.B., Saavedra J.E., Keefer L.K., Singer M., Kelm M., Butler A.R., Feelisch M. Key bioactive reaction products of the NO/H2S interaction are S/N-hybrid species, polysulfides, and nitroxyl. Proc. Natl. Acad. Sci. USA. 2015 doi: 10.1073/pnas.1509277112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ida T., Sawa T., Ihara H., Tsuchiya Y., Watanabe Y., Kumagai Y., Suematsu M., Motohashi H., Fujii S., Matsunaga T., Yamamoto M., Ono K., varie-Baez N.O., Xian M., Fukuto J.M., Akaike T. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA. 2014 doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu M., Zhang W., Wu L., Yang G., Li H., Wang R. Hydrogen sulfide (H2S) metabolism in mitochondria and its regulatory role in energy production. Proc. Natl. Acad. Sci. USA. 2012;109:2943–2948. doi: 10.1073/pnas.1115634109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teng H., Wu B., Zhao K., Yang G., Wu L., Wang R. Oxygen-sensitive mitochondrial accumulation of cystathionine beta-synthase mediated by Lon protease. Proc. Natl. Acad. Sci. USA. 2013;110:12679–12684. doi: 10.1073/pnas.1308487110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang L.Y., Slot J.W., Geuze H.J., Crapo J.D. Molecular immunocytochemistry of the CuZn superoxide dismutase in rat hepatocytes. J. Cell Biol. 1988;107:2169–2179. doi: 10.1083/jcb.107.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowling A.C., Barkowski E.E., McKenna-Yasek D., Sapp P., Horvitz H.R., Beal M.F., Brown R.H., Jr. Superoxide dismutase concentration and activity in familial amyotrophic lateral sclerosis. J. Neurochem. 1995;64:2366–2369. doi: 10.1046/j.1471-4159.1995.64052366.x. [DOI] [PubMed] [Google Scholar]
- 57.Olson K.R., DeLeon E.R., Liu F. Controversies and conundrums in hydrogen sulfide biology. Nitric Oxide. 2014;41:11–26. doi: 10.1016/j.niox.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Wedmann R., Bertlein S., Macinkovic I., Boltz S., Miljkovic J.L., Munoz L.E., Herrmann M., Filipovic M.R. Working with "HS": facts and apparent artifacts. Nitric Oxide. 2014 doi: 10.1016/j.niox.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Olson K.R. A theoretical examination of hydrogen sulfide metabolism and its potential in autocrine/paracrine oxygen sensing. Respir. Physiol. Neurobiol. 2013;186:173–179. doi: 10.1016/j.resp.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 60.DeLeon E.R., Stoy G.F., Olson K.R. Passive loss of hydrogen sulfide in biological experiments. Anal. Biochem. 2012;421:203–207. doi: 10.1016/j.ab.2011.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material