Abstract
ALDH16A1 is a novel member of the ALDH superfamily that is enzymatically-inactive and highly expressed in the kidney. Recent studies identified an association between a rare missense single nucleotide variant (SNV) in the ALDH16A1 gene and elevated serum uric acid levels and gout. The present study explores the mechanisms by which ALDH16A1 influences uric acid homeostasis in the kidney. We generated and validated a mouse line with global disruption of the Aldh16a1 gene through gene targeting and performed RNA-seq analyses in the kidney of wild-type (WT) and Aldh16a1 knockout (KO) mice, along with plasma metabolomics. We found that ALDH16A1 is expressed in proximal and distal convoluted tubule cells in the cortex of the kidney and in zone 3 hepatocytes. RNA-seq and gene ontology enrichment analyses showed that cellular lipid and lipid metabolic processes are up-regulated. Three transporters localized in the apical membrane of the proximal convoluted tubule of the kidney known to influence urate/uric acid homeostasis were found to be up-regulated (Abcc4, Slc16a9) or down-regulated (Slc17a3). An initial metabolomics analysis in plasma revealed an altered lipid profile in KO mice that is in agreement with our RNA-seq analysis. This is the first study demonstrating a functional role of ALDH16A1 in the kidney.
Keywords: ALDH16A1, RNA-seq, Metabolomics, Gene enrichment analysis, Renal function, Lipid metabolism
1. Introduction
The aldehyde dehydrogenase (ALDH) superfamily consists of proteins that are mainly enzymes involved in the metabolism of endogenous and exogenous aldehydes [1,2]. Although considered to primarily serve detoxification functions, ALDHs play critical roles in cellular processes by generating important metabolites, such as retinoic acid, betaine and gamma-aminobutyric acid. In addition to aldehyde metabolism, some ALDHs (e.g. ALDH2 and ALDH1A1) exhibit esterase and reductase catalytic activities [3,4]. Finally, ALDHs may also possess non-catalytic functions, such as structural or binding properties [5].
ALDH16A1 is a novel and unique member of the ALDH superfamily for many reasons. The main reason is that it contains two ALDH family domains (Pfam: aldedh) [5], as opposed to the single domain that is typical of other ALDH family members. One of the domains of the ALDH16A1 is full length while the other is truncated, missing most of the region subserving catalytic activity [5]. In addition, the ALDH16A1 gene is not found in fungi and plants, but widely distributed homologues in bacteria, protists, fish, amphibians and mammals can be found [5]. In humans and all vertebrates with the exception of frog, ALDH16A1 is predicted to be a dead enzyme because it is missing Cys-302, the residue critical for catalytic activity [5,6]. In this context, ALDH16A1 has been shown to interact with a number of proteins and these interactions may contribute to its physiological role [7]. A premature truncation of the protein maspardin underlies mast syndrome; while ALDH16A1 co-localizes and interacts with this protein in neuronal cells, the functional implications of the interactions are unknown [8]. There are a number of other proteins that either have been found or are predicted to interact with ALDH16A1. These include SKIP-1 [9], PAAF1 [10], USP1 [11], PRKAG2, ALB, HSP90AB1, SLC2A4, HPRT1, BHMT, and GBAS [5,6] [5,6]. A recent network (BioPlex), that contains interactions among proteins and explores the human interactome, was recently published [12]. According to this BioPlex network (http://wren.hms.harvard.edu/bioplex/index.php, list of interactions through 06/12/2015), the ALDH16A1 interacts directly with adducin 2 (ADD2), endoplasmic reticulum protein 29 (ERP29), family with sequence similarity 117 member B (FAM117B), bacterial lantibiotic synthetase component C-like 1 (LANCL1), protein-beta-aspartate methyltransferase (PCMT1) and toll-like receptor 1 (TLR1). In addition to direct interaction, ALDH16A1 is listed as part of several multi-protein complexes [12].
A recent G-WAS study examined 16 million SNPs and using whole-genome sequencing of 457 Icelanders, identified an association between a rare missense single nucleotide variant (SNV) in the ALDH16A1 gene and elevated serum uric acid levels and gout [13]. Computational analyses suggest that this is likely due to altered interactions between the mutant ALDH16A1 and hypoxanthine phosphoribosyltransferase 1 (HPRT1) [5].
The aim of the present study was to explore the physiological role of ALDH6A1 protein by conducting RNA-sequencing analysis in the kidney of Aldh16a1 knockout (KO) mice. In addition, a preliminary plasma metabolomic analysis was performed to identify possible pathways affected by the loss of ALDH16A1. Our results support a functional role of ALDH16A1 in the kidney.
2. Material and methods
2.1. Reagents
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) with the exception of acetonitrile (Baker Analyzed Ultra LC/MS™, VWR, Radnor, PA), formic acid (Optima™ LC/MS grade, Fisher Scientific, Waltham, MA, USA) and water (Milli-Q - Ultrapure Water, EMD, Millipore, Billerica, MA).
2.2. Development of Aldh16a1 “knockout-first” mouse model
A “knockout-first” conditional allele for Aldh16a1 has been developed by Skarnes et al. in highly germline-competent C57BL/6N embryonic stem (ES) cells [14]. Frozen mouse sperm harboring the Aldh16a1 “knockout-first” allele were purchased from the MRC Mary Lyon Centre and Helmholtz Zentrum München (stock C57BL/6NTac-Aldh16a1<tm1a(EUCOMM)Wtsi>/WtsiH) and used to generate the global Aldh16a1 knockout (KO) mouse colony through in vitro fertilization by the University of Colorado Gene Targeting Core. Heterozygotes were crossed to generate littermates of wild-type (WT) and KO animals. Genotyping was performed by standard PCR using tail DNA. Mice were group-housed, maintained in a temperature-controlled room (21–22 °C) on a 12 h light/dark cycle and supplied with food and water ad libitum. Male mice (3 mo age) were euthanized by CO2 asphyxiation and tissues (kidney, liver and brain) were quickly removed en bloc. One kidney and one piece of liver from the central lobe were fixed in 4% paraformaldehyde for histology. All other tissues were immediately flash frozen in liquid nitrogen for subsequent gene and protein expression analyses. All animal use was conducted in compliance with Institutional Animal Care and Use Committee of the Yale University and was performed according to the published National Institutes of Health guidelines.
2.2.1. Western blot analysis
Frozen tissues were homogenized in RIPA buffer (150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 0.1% SDS, 50 mM Tris, 1 mM EDTA, 1 mM PMSF, protease inhibitor cocktail, pH 7.4) on ice with a tissue disruptor (BioSpec Products, Bartlesville). Tissue homogenates were centrifuged at 12,000 rpm at 4 °C for 20 min and the supernatant was collected. Supernatant proteins (20 μg) were resolved by 10% SDS-PAGE and immunoblotted using a rabbit polyclonal anti-human ALDH16A1 antibody (Cell Signaling Technology, Danvers, MA) at a 1:5000 dilution. HRP-conjugated secondary antibody (Abcam, Cambridge, UK). Protein bands were visualized using a chemiluminescence imaging system (PerkinElmer Life and Analytical Sciences, Waltham, MA).
2.2.2. Histological and immunohistochemical (IHC) analysis
Tissues were formalin-fixed and paraffin-embedded by the Department of Pathology at the Yale University (New Haven, CT) using standard procedures for histochemical and immunohistochemical (IHC) staining. Paraffin tissue sections (4 μm) were deparaffinized, rehydrated and processed for hematoxylin and eosin (H&E) or IHC staining. Tissue sections were examined by an experimental pathologist (D.O.) blinded to the mouse phenotype. For IHC staining, antibodies were used at dilutions of 1:50 for the rabbit polyclonal anti-human ALDH16A1 (see above) and 1:500 for goat polyclonal anti-rabbit IgG (Sigma, St. Louis, MO).
2.3. RNA-seq and gene enrichment analyses
Total RNA was extracted from mouse tissues (N = 3, WT and KO) using RNeasy Plus Mini Kit (Qiagen, Valencia, CA) and purified using the RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA). Total RNA quality was determined by estimating the A260/A280 and A260/A230 ratios using a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., USA). RNA integrity was verified by the relative abundance of 18S and 28S subunits of ribosomal RNA (Agilent 2100 Bioanalyzer). Six sequencing libraries were produced from the purified total RNA samples by the Illumina TruSeq stranded protocol. The libraries underwent 76bp single-end sequencing using Illumina HiSeq 2500. The raw RNA-sequencing reads were first assessed for quality using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). FASTX-toolkit [15] was then used to trim off Illumina adaptor sequences and low quality bases. Reads passing the quality control were aligned to the Ensembl annotation of mouse genome reference sequence GRCm38 by TopHat 2 [16] and Bowtie [17] alignment engines. Reads mapped to multiple locations were discarded from further analysis. To quantify the abundance of each transcript, FPKM (analogous fragments per kilobase of transcript per million mapped reads) was used to normalize the read count of a transcript by both its length and total mapped reads using Cufflinks [18]. Differential expression analysis was performed using Cuffdiff [19] with default options. Genes with fold change > log2 1 and false discovery rate (FDR) controlled p-value ≤ 0.05 were considered differentially expressed in KO mice (relative to WT mice). Genes with fold change < − log2 1 and false discovery rate (FDR) controlled p-value ≤ 0.05 were considered to be down-regulated. MetaCore™ Bioinformatics software from Thomson Reuters (https://portal.genego.com/) was used for the gene function categories and pathway analysis of the differentially expressed genes. Pathways with FDR adjusted p-value ≤ 0.05 were considered significant.
2.4. Metabolomics
2.4.1. Animals and sample preparation
Female mice (3 mo age, N = 4), were euthanized by CO2 asphyxiation. Immediately thereafter, blood was collected by cardiac puncture into K3-EDTA tubes (MiniCollect, Greiner Bio-One, Gmbh, Austria). The blood was then transferred to a 1 mL tube (Protein LoBind, Eppendorf), centrifuged at 4 °C for 15 min at 13,000 rpm, and the plasma was collected. Plasma samples were then precipitated with 100% acetonitrile (1:1 ratio), vortexed for 30 s, centrifuged at 4 °C for 10 min at 13,000 rpm. The supernatant was collected, dried in a Speed Vac (SPD111V P1, Thermo Scientific), reconstituted in 50 μL acetonitrile:water (1:1 %v/v), and subsequently transferred into HPLC vials for analysis.
2.4.2. UPLC-MS analysis
Chromatography was performed on a UPLC (Acquity UPLC® I-Class System, Waters, Milford, MA) coupled to a quadrupole time-of-flight (QTOF) mass spectrometry (MS), (Xevo G2-XS, Waters, Milford, MA). Three μL of extracted plasma was injected onto the column (Acquity BEH, C18 130 Å, 1.7 μm, 2.1 mm × 100 mm, Waters, Milford, MA) which was operating at 50 °C. The sample manager temperature was maintained at 4 °C. In brief, the following mobile-phase linear gradient consisting of 0.1% (v/v) formic acid in water (A) and acetonitrile containing 0.1% formic acid (B) was used at a flow rate of 0.28 mL/min: 1% B for 1 min to 10% B at 3.0 min, to 40% B at 18 min to 1% B at 21 min and continuing at 1% B for an additional 7 min. The total run was 28 min in duration. The mass spectrometer was operating in positive ionization mode and under the following conditions: desolvation temperature at 500 °C, desolvation gas flow at 1000 L/h, source temperature was maintained at 150 °C and source voltage of 2.5 kV.
2.4.3. Data analysis
The software used to obtain the data was Waters MassLynx™ Software (Waters Corporation, Milford, MA). UPLC-MS acquired data were analyzed using Progenesis QI software (Waters Corporation, Milford, MA) for peak alignment, peak picking and data normalization. Peak picking thresholds were set between 0.5 and 25 min. An output table was subsequently generated to include paired m/z retention times and raw and normalized peak intensities for pooled and individual samples. These metabolic features were exported to EZ info software (Waters Corporation, Milford, MA) for principal component analysis (PCA). Discriminatory metabolites were investigated by using exact mass when searching HMDB (http://www.hmdb.ca/) and the METLIN (https://metlin.scripps.edu/) online databases. Metabolites with a p-value ≤0.05 were used for subsequent orthogonal partial least-squares discriminate analyses (O-PLS-DA). The ions identified were distributed in the S-plot for visual signal identification.
3. Results and discussion
3.1. Aldh16a1 “knockout-first” mouse model: development and validation
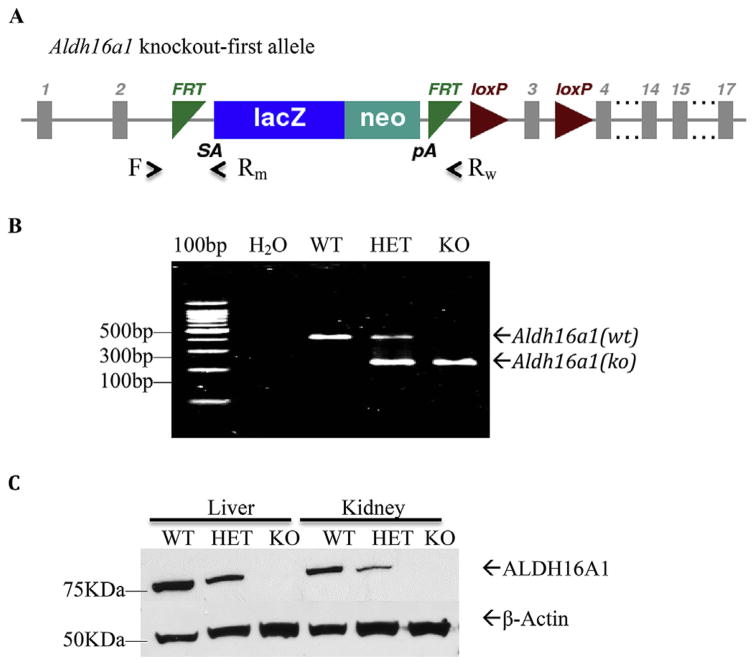
To begin to explore the pathophysiological roles of ALDH16A1 in vivo, we generated a mouse line harboring a “knockout-first” conditional allele for Aldh16a1 (Fig. 1). The main features of the gene-trapping conditional Aldh16a1 allele include the insertion of a LacZ-Neo trapping cassette flanked by Frt sites in intron 2 and two LoxP sites flanking the exon 3 (Fig. 1A). The presence of LacZ-Neo trapping cassette leads to the expression of reporter gene LacZ and termination of Aldh16a1 expression at the Aldh16a1 locus. Genotyping strategy was designed to differentiate Aldh16a1 alleles in a single multiplex PCR reaction where primer sets F/Rw and F/Rm amplify a 350-bp and 230-bp DNA fragments from WT and KO alleles, respectively (Fig. 1B). Western blot analysis confirmed that ALDH16A1 protein was undetectable in the examined tissues of KO mice (Fig. 1C).
Fig. 1. Global Aldh16al knockout (KO) mouse model.
(A) Scheme of the Aldh16a1 knockout-first allele containing an insertion of a LacZ-Neo trapping cassette flanked by Frt sites in intron 2 and two LoxP sites flanking the exon 3. The presence of the trapping cassette leads to the expression of reporter gene LacZ and termination of Aldh16a1 expression. A single multiplex PCR reaction was designed to genotype Aldh16a1 alleles, in which primer sets F/Rw and F/Rm amplify 350-bp and 230-bp DNA fragments (respectively) from WT and KO alleles. (B) Representative image of PCR genotyping using tail DNA. (C) Western blot analysis of ALDH16A1 protein in whole cell lysates from kidney, liver and brain tissues confirming that this protein was not expressed in KO mice. WT, wild-type; HET, Aldh16a1 heterozygote; KO: Aldh16a1 knockout.
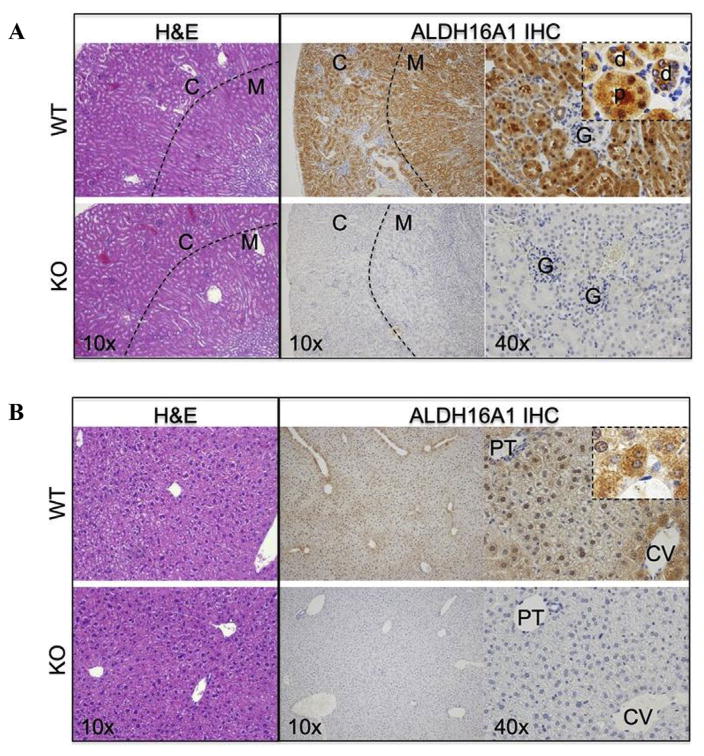
ALDH16A1 protein was expressed in kidney and liver tissues of WT mice (Fig. 1C). The gross morphology of the kidney and liver of KO mice was similar to that of WT mice (Fig. 2A and B). In the WT mouse kidney, immunohistochemical staining revealed ALDH16A1 to be expressed by proximal and distal convoluted tubule cells in the cortex, and by the S3 proximal convoluted tubule cells, in the thick ascending tubule cells and collecting tubules in the medulla (Fig. 2A, upper panel). By contrast, ALDH16A1 was absent from Bowman’s capsule parietal and visceral epithelial cells, the thin descending loops of Henle, and all vasculature, including endothelial cells, smooth muscle cells, pericytes of interlobular, arcate and intralobular arteries. In the WT mouse liver, ALDH16A1 was expressed by zone 3 hepatocytes (centrilobular pattern). Stellate cells, Kupffer cells, periportal fibroblasts, cholangiocytes and vasculature did not express ALDH16A1 (Fig. 2B, upper panel). As expected, no ALDH16A1 immunopositive signal was detected in the tissues of KO mice (Fig. 2A and B, lower panels). The proximal renal tubule cells make up a significant portion of the kidney and carry out diverse regulatory and endocrine functions.
Fig. 2. General histology and immunohistochemical (IHC) staining for ALDH16A1.
Tissue sections from wild-type (WT) and Aldh16a1 knockout (KO) mice were stained with hematoxylin and eosin (H&E) or for immunohistochemical (IHC) localization of ALDH16A1. Representative microscopic images of tissue stained with H&E (10x, left panel) and for ALDH16A1 (10x, middle panel; 40x, right panel) are shown for kidney (A) and liver (B) from WT (upper panel) and KO (lower panel) mice. In kidney tissue (A), the dashed line in 10x images represents the border between the cortex (C) and medulla (M) of the kidney. In the 40x images, examples of the proximal (p) and distal (d) convoluted tubule cells and glomerulus (G) of the cortex are identified. In liver tissue 40x IHC images (B), the central vein (CV) and portal triad (PT) are identified. A higher magnification (100×) of ALDH16A1 IHC in zone 3 hepatocytes of WT liver is shown in the right panel inset.
The ALDHs are expressed in many tissues, including liver, kidney, lung, stomach, duodenum, jejunum, ileum, large intestine, heart, brain, gonads (testes and ovaries), placenta, and uterus [1]. A systematic study of 15 ALDH mRNA expression in the kidneys of mice has shown ALDH4A1, ALDH6A1, ALDH7A1, ALDH8A1 and ALDH9A1 mRNAs to be highly expressed, ALDH1A2, ALDH1A3, ALDH1A7 ALDH1B1 AND ALDH3B1 mRNA to have low expression and ALDH1A1 and ALD18A1 mRNA to not be expressed [20]. All of the ALDHs expressed in the kidney are localized in the proximal tubule cells [21].
Molecular characteristics of transporters in the proximal tubules play an important role in progressive kidney diseases, such as Fanconi’s syndrome, renal tubular acidosis, cystinuria and other amino acid transport disorders [22]. A recent in silico study showed that the ALDH16 protein contains two ALDH domains, four transmembrane and one coiled-coil domains [5]. Taking into account the structure of the transmembrane domains of ALDH16A1, the location of its expression (renal tubule cells), and recent studies showing its interaction with other proteins, it may be speculated that ALDH16A1 plays a role as a transporter or possibly contributes to transporting molecules in the tubule region of the kidney.
3.2. Differential gene expression in kidney
Kidney tissue from KO mice was used to characterize the transcriptional consequences of the loss of the Aldh16a1 gene. To determine changes in expressed genes relative to WT mice, experiments focused on the read-covered transcripts derived from genes annotated in ENSEMBL; this resulted in identification of 548 genes. The number of differentially-expressed genes induced in KO mice was 286 (Table 1) [23]. Two of the ten most up-regulated genes belong to the solute carrier (SLC) family Slc7a13 and Slc22a30 (28.2- and 24.11-fold change in log2 scale, respectively). Slc7a13 codes a protein that is a member of the heterodimeric amino acid transporters [24]. It mediates sodium-independent aspartate/glutamate and is part of the antiporter system x(c)(−), which imports the oxidized form of cysteine into the cells [24]. Cysteine is the rate-limiting substrate for the important antioxidant glutathione (GSH). Cyp2j13 was up-regulated 28.83-fold in the KO mouse kidney (Table 1). It codes for an enzyme active in the metabolism of arachidonic acid, linoleic acid, as well as other lipids and xenobiotics [25,26]. Interestingly, Acsm3 (Acyl-CoA synthetase medium-chain family member 3) was also up-regulated (27.45-fold change, Table 1). It codes for an enzyme that catalyzes the initial reaction in fatty acid metabolism, which allows for the participation of fatty acids in anabolic and catabolic pathways [27]. There is very limited information for Gm12839 gene, which showed a 25.21-fold change (Table 1). Its official name is Cyp4b1-ps2 (cytochrome P450) and the gene is located in chromosome 4. Pzp (Pregnancy-Zone Protein, alpha-2-macroglobulin like), also up-regulated, is a protein coding gene that normally is strongly up-regulated during pregnancy and in numerous inflammatory states. It is expressed in both genders and is a panprotease inhibitor capable of binding, capturing, and inhibiting all classes of proteases [28,29]. Despite its original identification many decades ago, the precise biological importance still remains unclear. However, recent studies have found a strong association between elevated concentrations of Pzp and Alzheimer’s disease [30,31]. Col6a6 (Collagen Type VI Alpha 6), a non-fibrillar collagen has recently been identified as a component of the extracellular matrix of the human skeletal muscle [32]. Tbx10 is a member of the mouse T-box gene family and plays an important role in early vertebrate development [33].
Table 1.
Up-regulated, differentially-expressed genes in the kidney of Aldh16a1 KO mice. Gene transcription profiling was obtained by RNA-sequencing of renal RNA from wild-type (WT) and Aldh16a1 knockout (KO) mice. Genes with fold change > log2 1and false discovery rate (FDR) controlled p-value ≤ 0.05 in KO samples relative to WT samples were arbitrarily defined as being differentially-expressed. The transcripts in KO mice that differed most from WT mice is ranked in fold-change, with the ten genes that were most up-regulated being listed.
| Gene symbol | Gene ID (Mus musculus) | Log2 FC | q-valuea |
|---|---|---|---|
| Cyp2j13 | 230459 | 8.83 | 2.78 × 10−3 |
| Slc7a13 | 74087 | 8.20 | 2.78 × 10−3 |
| Acsm3 | 20216 | 7.45 | 2.78 × 10−3 |
| Gm12839 | 631037 | 5.21 | 2.78 × 10−3 |
| Pzp | 11287 | 4.80 | 2.78 × 10−3 |
| Ugt2b5 | 22238 | 4.65 | 2.78 × 10−3 |
| Col6a6 | 245026 | 4.56 | 2.78 × 10−3 |
| Tbx10 | 109575 | 4.36 | 8.96 × 10−3 |
| Slc22a30 | 319800 | 4.11 | 2.78 × 10−3 |
| Timd2 | 171284 | 4.10 | 2.78 × 10−3 |
FDR controlled p-value.
Expression of Gal, Hand1 and Phox2b were suppressed in the KO mice (Table 2). Galanin (GAL), the neuropeptide product of Gal, exhibits diverse physiological functions, including energy homeostasis. High levels of endogenous GAL in transgenic mice has been associated with obesity, as well as lipid metabolism alterations [34]. In addition, decreased GAL expression is associated with decreased appetite and increased levels of leptin, in agreement with the lipid metabolism alterations that have been shown in other studies [35,36]. There is limited information regarding Hand1. Its function and status in the Reference Sequence (RefSeq) collection is provisional. Cyp21a2, which was also down-regulated (2−9.14-fold change, Table 2), encodes steroid 21-hydroxylase (21-OH) which is necessary for glucocorticoid and mineralocorticoid synthesis. Its deficiency can lead to adrenal hyperplasia which is characterized by salt-wasting and overproduction of androgens [37]. A member of the SLC family (Slc7a12) was down-regulated (2−7.14-fold change) as was Chga (2−6.7-fold change). The protein encoded by Chga, chromogranin A, is a member of the chromogranin/secretogranin family of neuroendocrine secretory proteins and a precursor to three biologically-active peptides, vasostatin, pancreastatin and parastatin. The first inhibits angiogenesis and suppresses tumor growth, the second strongly inhibits glucose-induced insulin release from the pancreas, and the third inhibits parathyroid cell secretion [38–40]. The extent of down-regulation of the other genes shown on Table 2 were greater than 2−4.37-fold change. An interesting finding was the down-regulation of Abcc3, a gene of the ABC-family of transporters. Increased expression of the ABC-family is predominantly implicated in cancer drug resistance, which leads to chemotherapy failure [41].
Table 2.
Down-regulated, differentially-expressed genes in the kidney of Aldh16a1KO mice. Gene transcription profiling was obtained by RNA-sequencing of renal RNA from wild-type (WT) and Aldh16a1 knockout (KO) mice. Genes with fold change > log2 1and false discovery rate (FDR) controlled p-value ≤ 0.05 in KO samples relative to WT samples were arbitrarily defined as being differentially-expressed. The transcripts in KO mice that differed most from WT mice are ranked in fold change, with the ten genes that were most down-regulated being listed.
| Gene symbol | Gene ID (Mus musculus) | Log2 FC | q-valuea |
|---|---|---|---|
| Gal | 14419 | ∞ | 2.78 × 10−3 |
| Hand1 | 15110 | ∞ | 2.78 × 10−3 |
| Phox2b | 18935 | ∞ | 2.78 × 10−3 |
| Cyp21a1 | 13079 | −9.14 | 7.22 × 10−3 |
| Slc7a12 | 140918 | −7.14 | 2.78 × 10−3 |
| Chga | 12652 | −6.70 | 2.78 × 10−3 |
| Dlk1 | 13386 | −6.02 | 2.78 × 10−3 |
| Scg2 | 20254 | −6.00 | 2.78 × 10−3 |
| Mc2r | 17200 | −4.75 | 1.4 × 10−2 |
| Abcc3 | 76408 | −4.37 | 2.78 × 10−3 |
FDR controlled p-value.
3.3. Gene ontology analysis
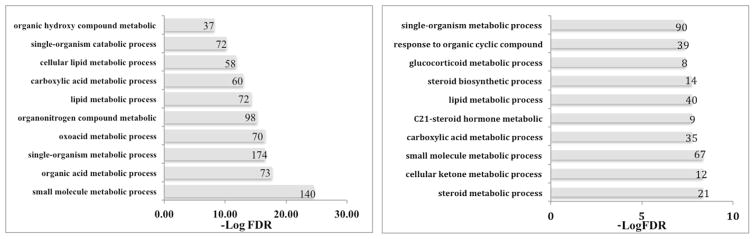
Gene ontology enrichment analysis was performed for kidney on 1189 genes that were up- (n = 335) or down-regulated (n = 854). The ten most significant pathways are shown in Fig. 3 along with their FDR adjusted p-values and the number of regulated genes. In the kidney, nine of the ten most significantly up-regulated gene ontology processes were metabolic, such as small molecule, organic acid and lipid metabolism; only one was a single-organism catabolic process, i.e., involves pathways resulting in the breakdown of substances which involves a single organism (Fig. 3). The up-regulation of the cellular lipid and the lipid metabolic processes could possibly explain the up-regulation of the Cyp2j13 and the Acsm3 mRNA (Table 1), which are both involved in lipid metabolism. The down-regulated gene ontology enrichment analysis followed the same pattern as for the up-regulated, with eight of the ten being metabolic processes. Three of these ten are related to steroids (i.e., steroid, C-21 steroid hormone and glucocorticoid metabolic processes) and steroid biosynthesis was also down-regulated.
Fig. 3.
Gene ontology enrichment analysis in the kidney. Gene ontology processes that are up-regulated and down-regulated in the kidney of Aldh16a1 knockout mice relative to wild-type mice are shown. False Discovery Rate (FDR) is the adjusted p-value. Numbers next to each bar indicates the number of genes altered in each process.
RNA-seq revealed changes in three renal transporters (Abcc4, Slc16a9 and Slc17a3) (Table 3) that are localized to the apical membrane of the proximal convoluted tubule cells and influence uric acid homeostasis. In KO mice, changes in two of the transporters (Abcc4 1.14, and Slc16a9 −1.11-fold change in log2 scale) would favor development of hyperuricemia, while the change in the third transporter (Slc17a3, −1.44-fold change in log2 scale) would favor enhanced urate/uric acid excretion [42]. Extending the present results in the KO mice to the study in humans demonstrating an association between an ALDH16A1 SNV and hyperuricemia, it may be rationalized that reduced ALDH16A1 leads to an alteration in proximal tubule uric acid transport that favors a change from net secretion to net reabsorption. The observed high expression of ALDH16A1 in the renal proximal tubule in WT mice is consistent with a role in modulating the above solute transporters. Molecular modeling predicts that both the short and long forms of human ALDH16A1 protein may interact with the hypoxanthine-guanine phosphoribosyltransferase (HPRT1) protein, a key enzyme involved in uric acid metabolism and gout [5]. Whether ALDH16A1 influences these proximal tubular transporters by a similar mechanism remains to be established.
Table 3.
Transporter genes affected in the kidney of Aldh16a1 knockout mice.
| Gene | Transport Direction | Log2 Fold Change (KO vs. WT) | q-valuea | Predicted Effect |
|---|---|---|---|---|
| Abcc4 | Secretion | 1.14 | 0.028 | Hyperuricemia |
| Slc16a9 | Reabsorption | −1.11 | 0.03 | Hyperuricemia |
| Slc17a3 | Secretion | −1.44 | 0.025 | Hypouricemia |
FDR controlled p-value.
3.4. Metabolomics: multivariate analysis and ion profile changes
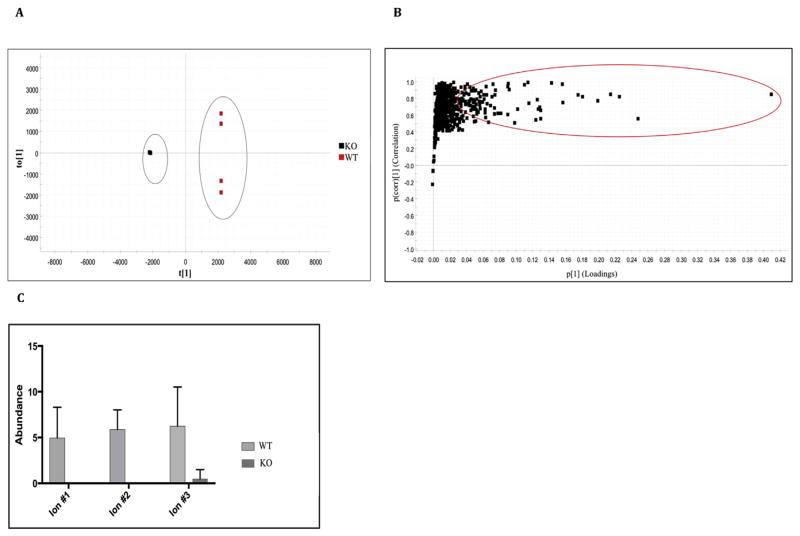
The orthogonal partial least squares discriminant analysis (OPLS-DA) plot was used to build predictive models capable of discriminating between KO and WT mice (Fig. 4A) and demonstrated separation between the two groups; the R2 value reflecting the goodness-of-fit of the model was 0.8815. An S-Plot was generated (Fig. 4B) to identify the ions contributing to the differences observed between KO and WT mice. Of the original 42,608 metabolic features, 3427 were assigned for comparison of the KO mice with the WT mice; 339 were found to be higher in the WT mice. The S-plot generated, revealed that there were ions significantly down-regulated in or completely absent from KO mice (maximum fold change > 2, p ≤ 0.05). This is shown by the absence of ions on the lower left side of the plot (Fig. 4B). The statistically significant ions (correlation coefficient (pcorr value) greater than 0.8 and p ≤ 0.05) are highlighted by the red oval on the S-plot (upper right). The identities of some of these putative ions that are absent from the KO mice are shown in Table 4. This initial pilot study revealed an altered lipid profile in the KO mice and, interestingly, the ion abundance was lower in KO mice than in WT mice (Fig. 4C). These findings are intriguing given that membrane lipids can act as precursors for second messengers. For example, diacylglycerol (DAG) is rapidly phosphorylated to phosphatidic acid (GPA), which is an important intermediate in phospholipid biosynthesis and a potent regulator of enzyme function and the bilayer structure [43,44]. This is an ongoing project and we are currently investigating all of the ions and lipids in KO mice that differ from WT mice.
Fig. 4.
A) OPLS-DA scores plot displaying the separation for Aldh16a1 knockout (KO, black squares) and wild-type (WT, red squares) mice. They grey ovals show the separation of the two groups. B) S-Plot displaying ions of interest in KO (−1, lower left side) and WT mice (+1, upper right side). Ions of interest were selected from 42,608 metabolic features for KO (−1, left side) and WT (+1, right side) mice in total. Statistically significant ions (correlation coefficient (pcorr value) > 0.8 and p < 0.05) are highlighted by the red circle. C) Ion abundance of three selected ions in KO and WT mice. Ions 1 and 2 are completely absent from KO mice, while ion 3 has very low ion abundance in KO mice. Data represent mean and associated SD (N = 4). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 4. Putative ions.
increased in WT mice and absent from or of very low abundance in knockout mice. Neutral mass, mass to charge ratio (m/z), retention time, max fold change and significance (p) are shown.
| Compound# | Possible identification | Formula | Neutral mass | Mass to charge ratio (m/z) | Retention time (min) | Anova (p) (WT vs. KO) | Maximum fold change |
|---|---|---|---|---|---|---|---|
| 1 |
a PS (18:4(6Z,9Z,12Z,15Z)/13:0) PS (13:0/18:4(6Z,9Z,12Z,15Z)) |
C37H64NO10P | 793.4007 | 758.3868 | 14.66 | 0.00395 | ∞ |
| 2 |
b DG (23:0/26:2/0:0) DG (26:2/23:0/0:0) |
C52H98O5 | 802.734 | 841.6971 | 16.19 | 0.000505 | 4.06 × 106 |
| 3 |
c GPA (24:4/26:1) GPA(26:1/24:4) |
C53H95O8P | 890.6632 | 873.6599 | 14.77 | 0.008 | 2750 |
Diacylglycerophosphoserines.
1,2-diacylglycerol (Glycerolipid).
Phosphatidic acid (Glycerophospholipid).
3.5. Conclusions
We have successfully generated a mouse line harboring a “knockout-first” conditional allele for the Aldh16a1 gene and validated the model by general histological examination. ALDH16A1 was shown to be expressed in the kidney by proximal and distal convoluted tubule cells in the cortex and by zone 3 hepatocytes in the liver. RNA-seq data was used for the first time to provide comprehensive gene information on the Aldh16a1 KO mouse model at the transcriptional level. The differential gene expression revealed 548 genes in kidney that were altered. The gene ontology enrichment analysis of the RNA-seq data showed cellular lipid and lipid metabolic processes were up-regulated. RNA-Seq data also revealed three urate/uric acid transporters localized to the apical membrane of the proximal convoluted tubule of the kidney to be altered in Aldh16a1 KO mice. More specifically, Abcc4 was down-regulated, while both Slc16a9 and Slc17a3 were up-regulated. An initial metabolomics analysis revealed an altered lipid profile in Aldh16a1 KO mice, which supports the RNA-seq data and the gene ontology enrichment analysis. This ongoing project is shedding light on the physiological role of ALDH16A1 protein. From the current data, it seems that this protein affects many processes and more research is needed to delineate its function in the kidney and in lipid metabolism.
Acknowledgments
We would like to thank Dr. Caroline Johnson for valuable discussions relating to metabolomic analyses. This work was supported by NIH/NIAMS AR064137, NIH/NIAAA 5R24 AA02205-06 and China Scholarship Council (CSC, No.201508140059).
References
- 1.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin drug metabolism. Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum genomics. 2011;5:283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasiliou V, Thompson DC, Smith C, Fujita M, Chen Y. Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells. Chemicobiological Interact. 2013;202:2–10. doi: 10.1016/j.cbi.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, Thompson DC, Vasiliou V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56:89–101. doi: 10.1016/j.freeradbiomed.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasiliou V, Sandoval M, Backos DS, Jackson BC, Chen Y, Reigan P, Lanaspa MA, Johnson RJ, Koppaka V, Thompson DC. ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein-protein interactions with HPRT1. Chem Biol Interact. 2013;202:22–31. doi: 10.1016/j.cbi.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011;5:283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2:138–143. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanna MC, Blackstone C. Interaction of the SPG21 protein ACP33/maspardin with the aldehyde dehydrogenase ALDH16A1. Neurogenetics. 2009;10:217–228. doi: 10.1007/s10048-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wistow J, Piatigorsky J. Recruitment of Enzymes as Lens Structural Proteins. [DOI] [PubMed] [Google Scholar]
- 10.Yang QO, Kottgen A, Dehghan A, Smith AV, Glazer NL, Chen MH, Chasman DI, Aspelund T, Eiriksdottir G, Harris TB, Launer L, Nalls M, Hernandez D, Arking DE, Boerwinkle E, Grove ML, Li M, Kao WHL, Chonchol M, Haritunians T, Li G, Lumley T, Psaty BM, Shlipak M, Hwang SJ, Larson MG, O’Donnell CJ, Upadhyay A, van Duijn CM, Hofman A, Rivadeneira F, Stricker B, Uitterlinden AG, Pare G, Parker AN, Ridker PM, Siscovick DS, Gudnason V, Witteman JC, Fox CS, Coresh J. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circulation-Cardiovascular Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zinovieva RD, Tomarev SI, Piatigorsky J. Aldehyde dehydrogenase-derived omega-crystallins of squid and octopus. Specialization for lens expression. J Biol Chem. 1993;268:11449–11455. [PubMed] [Google Scholar]
- 12.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wuhr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulem P, Gudbjartsson DF, Walters GB, Helgadottir HT, Helgason A, Gudjonsson SA, Zanon C, Besenbacher S, Bjornsdottir G, Magnusson OT, Magnusson G, Hjartarson E, Saemundsdottir J, Gylfason A, Jonasdottir A, Holm H, Karason A, Rafnar T, Stefansson H, Andreassen OA, Pedersen JH, Pack AI, de Visser MC, Kiemeney LA, Geirsson AJ, Eyjolfsson GI, Olafsson I, Kong A, Masson G, Jonsson H, Thorsteinsdottir U, Jonsdottir I, Stefansson K. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat Genet. 2011;43:1127–1130. doi: 10.1038/ng.972. [DOI] [PubMed] [Google Scholar]
- 14.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- 16.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotech. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci. 2008;101:51–64. doi: 10.1093/toxsci/kfm280. [DOI] [PubMed] [Google Scholar]
- 21.Lindgren D, Bostrom AK, Nilsson K, Hansson J, Sjolund J, Moller C, Jirstrom K, Nilsson E, Landberg G, Axelson H, Johansson ME. Isolation and characterization of progenitor-like cells from human renal proximal tubules. Am J Pathol. 2011;178:828–837. doi: 10.1016/j.ajpath.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakhoul N, Batuman V. Role of Proximal Tubules in the Pathogenesis of Kidney Disease. doi: 10.1159/000313944. [DOI] [PubMed] [Google Scholar]
- 23.Elraghy O, Baldwin WS. Repression of multiple CYP2D genes in mouse primary hepatocytes with a single siRNA construct. Vitro Cell Dev Biol Anim. 2015;51:9–14. doi: 10.1007/s11626-014-9803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande M, Smith SB, Ganapathy V, Maher P. The cystine/glutamate antiporter system x(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal. 2013;18:522–555. doi: 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graves JP, Gruzdev A, Bradbury JA, DeGraff LM, Li H, House JS, Hoopes SL, Edin ML, Zeldin DC. Quantitative polymerase chain reaction analysis of the mouse Cyp2j subfamily: tissue distribution and regulation. Drug Metab Dispos. 2015;43:1169–1180. doi: 10.1124/dmd.115.064139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graves JP, Edin ML, Bradbury JA, Gruzdev A, Cheng J, Lih FB, Masinde TA, Qu W, Clayton NP, Morrison JP, Tomer KB, Zeldin DC. Characterization of four new mouse cytochrome P450 enzymes of the CYP2J subfamily. Drug Metab Dispos. 2013;41:763–773. doi: 10.1124/dmd.112.049429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acylcoenzyme A synthetase genes in the human genome. J Lipid Res. 2007;48:2736–2750. doi: 10.1194/jlr.M700378-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs DM. alpha2-macroglobulin in late-onset Alzheimer’s disease. Exp Gerontol. 2000;35:473–479. doi: 10.1016/s0531-5565(00)00113-3. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt AR, Cater JH, Ranson M. PZP and PAI-2: structurally-diverse. functionally similar pregnancy proteins? Int J Biochem Cell Biol. 2016;79:113–117. doi: 10.1016/j.biocel.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Nijholt DA, Ijsselstijn L, van der Weiden MM, Zheng PP, Sillevis Smitt PA, Koudstaal PJ, Luider TM, Kros JM. Pregnancy zone protein is increased in the Alzheimer’s disease brain and associates with senile plaques. J Alzheimer’s Dis JAD. 2015;46:227–238. doi: 10.3233/JAD-131628. [DOI] [PubMed] [Google Scholar]
- 31.Ijsselstijn L, Dekker LJ, Stingl C, van der Weiden MM, Hofman A, Kros JM, Koudstaal PJ, Sillevis Smitt PA, Ikram MA, Breteler MM, Luider TM. Serum levels of pregnancy zone protein are elevated in presymptomatic Alzheimer’s disease. J Proteome Res. 2011;10:4902–4910. doi: 10.1021/pr200270z. [DOI] [PubMed] [Google Scholar]
- 32.Chu ML, Conway D, Pan TC, Baldwin C, Mann K, Deutzmann R, Timpl R. Amino acid sequence of the triple-helical domain of human collagen type VI. J Biol Chem. 1988;263:18601–18606. [PubMed] [Google Scholar]
- 33.Xue XD, Kimura W, Wang B, Hikosaka K, Itakura T, Uezato T, Matsuda M, Koseki H, Miura N. A unique expression pattern of Tbx10 in the hindbrain as revealed by Tbx10(LacZ) allele. Genes (New York, N Y) 2010;2000(48):295–302. doi: 10.1002/dvg.20615. [DOI] [PubMed] [Google Scholar]
- 34.Sandoval-Alzate HF, Agudelo-Zapata Y, Gonzalez-Clavijo AM, Poveda NE, Espinel-Pachon CF, Escamilla-Castro JA, Marquez-Julio HL, Alvarado-Quintero H, Rojas-Rodriguez FG, Arteaga-Diaz JM, Eslava-Schmalbach JH, Garces-Gutierrez MF, Vrontakis M, Castano JP, Luque RM, Dieguez C, Nogueiras R, Caminos JE. Serum galanin levels in young healthy lean and obese non-diabetic men during an oral glucose tolerance test. Sci Rep. 2016;6:31661. doi: 10.1038/srep31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahu A. Intracellular leptin-signaling pathways in hypothalamic neurons: the emerging role of phosphatidylinositol-3 kinase-phosphodiesterase-3B-cAMP pathway. Neuroendocrinology. 2011;93:201–210. doi: 10.1159/000326785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahu A. Minireview: a hypothalamic role in energy balance with special emphasis on leptin. Endocrinology. 2004;145:2613–2620. doi: 10.1210/en.2004-0032. [DOI] [PubMed] [Google Scholar]
- 37.Naiki Y, Miyado M, Horikawa R, Katsumata N, Onodera M, Pang S, Ogata T, Fukami M. Extra-adrenal induction of Cyp21a1 ameliorates systemic steroid metabolism in a mouse model of congenital adrenal hyperplasia. Endocr J. 2016;63:897–904. doi: 10.1507/endocrj.EJ16-0112. [DOI] [PubMed] [Google Scholar]
- 38.Nikoopour E, Krougly O, Lee-Chan E, Mansour Haeryfar SM, Singh B. Detection of Vasostatin-1-specific CD8(+) T Cells in Non-obese Diabetic Mice that Contribute to Diabetes Pathogenesis. doi: 10.1111/cei.12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volante M, Tota D, Giorcelli J, Bollito E, Napoli F, Vatrano S, Buttigliero C, Molinaro L, Gontero P, Porpiglia F, Tucci M, Papotti M, Berruti A, Rapa I. Androgen deprivation modulates gene expression profile along prostate cancer progression. Hum Pathol. 2016;56:81–88. doi: 10.1016/j.humpath.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Fasciotto BH, Trauss CA, Greeley GH, Cohn DV. Parastatin (porcine chromogranin A347-419), a novel chromogranin A-derived peptide, inhibits parathyroid cell secretion. Endocrinology. 1993;133:461–466. doi: 10.1210/endo.133.2.8344192. [DOI] [PubMed] [Google Scholar]
- 41.Balaji SA, Udupa N, Chamallamudi MR, Gupta V, Rangarajan A. Role of the drug transporter ABCC3 in breast cancer chemoresistance. PLoS One. 2016;11:e0155013. doi: 10.1371/journal.pone.0155013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reginato AM, Mount DB, Yang I, Choi HK. The genetics of hyperuricaemia and gout, Nature reviews. Rheumatology. 2012;8:610–621. doi: 10.1038/nrrheum.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo B, Regier DS, Prescott SM, Topham MK. Diacylglycerol kinases. Cell Signal. 2004;16:983–989. doi: 10.1016/j.cellsig.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 44.Athenstaedt K, Daum G. Phosphatidic acid, a key intermediate in lipid metabolism. Eur J Biochem. 1999;266:1–16. doi: 10.1046/j.1432-1327.1999.00822.x. [DOI] [PubMed] [Google Scholar]