Abstract
Rationale and Objectives
Changes in the composition of body tissues are major aging phenotypes but they have been difficult to study in depth. Here we describe age-related change in abdominal tissues observable in Computed Tomography (CT) scans. We used pattern recognition and machine learning to detect and quantify these changes in a model-agnostic fashion.
Materials and Methods
CT scans of abdominal L4 sections were obtained from Baltimore Longitudinal Study of Aging (BLSA) participants. Age-related change in the constituent tissues were determined by training machine classifiers to differentiate age groups within male and female strata (‘Younger’ at 50-70 y.o., vs. ‘Older’ at 80-99 y.o.). The accuracy achieved by the classifiers in differentiating the age cohorts was used as a surrogate measure of the aging signal in the different tissues.
Results
The highest accuracy for discriminating age differences was 0.76 and 0.72 for males and females, respectively. The classification accuracy was 0.79 and 0.71 for adipose tissue, 0.70 and 0.68 for soft tissue, and 0.65 and 0.64 for bone.
Conclusions
Using image data from a large sample of well characterized pool of participants dispersed over a wide age-range we explored age-related differences in gross morphology and texture of abdominal tissues. This technology is advantageous for tracking effects of biological aging and predicting adverse outcomes when compared to the traditional use of specific molecular biomarkers. Application of pattern recognition and machine learning as a tool for analyzing medical images may provide much needed insight into tissue changes occurring with aging, and further, connect these changes with their metabolic and functional consequences.
Changes in body composition, both in the relative quantities of different tissue and their cellular and matrix content, occur with aging in all living organisms. Humans undergo an overall decline in lean body mass, mostly represented by muscle, and an increase in fat mass that occurs even in individuals who maintain the same weight over long time periods. In addition to global, quantitative changes, there are also qualitative changes that affect the characteristics and regional distribution of fat and muscle. For example, aging men show fat accumulates preferentially within and around the abdomen rather than in the subcutaneous tissue of the limbs and chest. CT and MRI images collected in large epidemiological studies have described tissue-specific changes in density and texture that may be correlated with adverse physiological changes and predict disease outcomes (1, 2). Based on these studies, researchers have suggested that measures of body composition should be included in any attempt to characterize phenotypic aging (1, 2).
A problem with this approach found in CT and MRI images characterizing body composition is the large variability in overall fat content and its regional distribution between individuals. Indeed, no previous studies have used body images to identify characteristics from multiple tissues that in aggregate correlate with chronological age, nor has this approach been validated in independent samples or in subsamples randomly selected from the original study population. Several previous studies used CT and MRI to find age-related differences in brain (3), bone (4), kidney (5) and fat distribution in skeletal muscle (6). All these studies were able to find statistically significant differences in morphology correlated with age, but they involved relatively a small number of participants, and used manual steps or semimanual classification in the analysis pipeline. Thus, until now it remained unclear whether information from body images alone can be used to detect changes in body composition and discern phenotypic age in a fully objective and automated way.
We present an automated analysis framework for abdominal CT scans that uses machine learning to eliminate potential bias from manual image processing steps, uses a much larger data set of over 1200 images, and achieves higher age discrimination accuracy than previous studies. Abdominal CT scans are especially well suited for examining changes in adipose tissue. The characteristic low radiodensity of adipose tissue allows for its easy isolation from other tissues, making it possible to directly compare age-related changes in fat distribution and texture to those in other tissues in the abdomen. Supervised learning is ideal for comparing groups of images with substantial individual differences and a large variety of possible changes between groups (7). We used supervised learning to train classifiers to distinguish between abdominal CT scans of subjects grouped by age, and used classification accuracy as a surrogate measure of the aging signal in abdominal tissues. We compared several different classification techniques used separately and in an ensemble classifier to ensure that our results are independent of the analysis technique used.
We focused our study on age-related changes occurring later in life, thus our “Younger” cohort was 50-70 y.o., and our “Older” cohort was 80-99 y.o, with male and female participants being considered separately. We demonstrated that aging signal could be detected in unmodified scans and that this could be substantially improved with contrast enhancement. We explored differential aging by quantifying the contribution of aging signal from three constituent tissue groups: Adipose Tissue (AT), soft tissues (ST) and bones. We found that AT had the greatest age-related change of the three tissues in both males and females, followed by ST and bones. We also found that males had consistently greater age related change than females in all three abdominal tissues.
Materials and Methods
Our institutional review board approved this retrospective study and waived the requirement for informed consent.
Study population
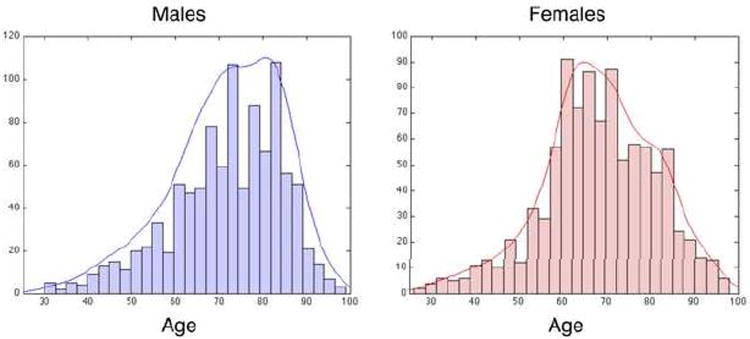
CT scans were from participants in the Baltimore Longitudinal Study of Aging (BLSA) ranging from 30 to 99 years old. Fig. 1 shows the age distribution for male and female participants. Of the 1092 participants with CT scans (544 males and 548 females), we assigned participants into two groups based on their age at the time of exam: “Younger”, age 50-70; and “Older”, age 80-99 (see Table I). We did not further refine or sub-select the BLSA population other than by age, so the demographics inherent in the BLSA (race, income, genetic diversity, BMI, etc.) are reflected in our sub-population. Each of the participants had scans from 1 to 6 visits, such that the total number of scans was 1967.
Figure 1. Age distribution in male and female BLSA subjects.
Table 1. Study population.
| Males | Females | |||||||
|---|---|---|---|---|---|---|---|---|
| Younger | Older | Y±O | Total | Younger | Older | Y±O | Total | |
| # of subjects | 220 | 124 | 344 | 544 | 293 | 75 | 368 | 548 |
| # of samples | 350 | 323 | 673 | 1006 | 465 | 206 | 671 | 961 |
| # of visits per subject | 1.48±0.6 | 2.38±1.5 | 1.81±1.1 | 1.70±1.0 | 1.48±0.6 | 2.26±1.5 | 1.66±0.9 | 1.59±0.9 |
| Mean age | 62.6±5.5 | 84.9±3.9 | 73.3±12 | 71.8±13 | 62.3±5.2 | 85.5±4.5 | 69.4±12 | 68.5±13 |
Number of subjects (row 1) and samples (row 2) are shown for each category (Younger or Older), all used categories (e.g. Younger and Older combined), and all-total as well. Rows 3 and 4 in the table give number of patient visits and average age, respectively.
From raw scans to Hounsfield units
CT images were collected with a Siemens Somatom Sensation 10 CT scanner. The scans were performed with a 500 × 500 mm field of view, 512 × 512 image matrix, and 10-mm slice thickness. Pixel planes used for processing were single-slice CT scans acquired at the L4-L5 vertebrae on the lumbar column. A standard calibration procedure using phantoms in the scanner's bed was performed, so that intensity of the reconstructed pixels was based on the Hounsfield scale. We used shifted Hounsfield units (SHU) where a range in pixel values from 1 to 2000 corresponds to a range of -1000 to 1000 HU.
Automated processing
The data processing was based on conventional pattern recognition as previously described (8). Two basic stages of data flow were image pre-processing and feature computation followed by cross-validation experiments (see Results) using three statistical classifiers (next section).
Image preprocessing (see in Fig. 2) had a biomedical rationale and consisted of several steps: trunk masking (to remove background), rigid-body registration, tissue masking and contrast enhancement. We masked the trunk area by setting a global threshold below the lowest tissue value, choosing the largest connected component (the trunk) and discarding smaller components (including the scanner bed, phantoms, etc.: Fig. 2, panels 1-2, (9)). We implemented a simplified rigid-body registration procedure that includes translation and rotation (Fig. 2, panels 2-3). First, cropping the subject's trunk as above, resulted in the translation transform. The rotational alignment was largely provided by the physical alignment to the scanner's bed, requiring no more than two degrees of additional rotation correction. Due to the relatively small correction required, we used one based on the second-order moments of the binary mask of the trunk (10). Thus, the angle correction was calculated using the binary mask, and then it was applied to the original scan.
Figure 2.
Scheme for generating intensity range data for experiments with tissue-specific scan areas. 1: original scan, 2: background pixels removed, rotation normalizing, 3: clipped, 4: CLAHE adjusted trunk, 5: tissue-specific mask, 6: CLAHE-adjusted and masked image.
Tissue masking was used to define sub-regions of the trunk corresponding to specific tissues. The range of pixel values comprising the mask was defined as a segment in the Hounsfield scale. We used contrast-limited adaptive histogram equalization (CLAHE, (11)) for intensity enhancement. In this approach, the raw scan pixel plane is first split into several tiles (we used 16 tiles), local histogram equalization takes place for each tile, and then interpolation is used at tile boundaries. Importantly, the CLAHE enhancement was performed separately on the whole image, independently of tissue masking (Fig. 2, panels 4-6). We applied the tissue mask to the enhanced scan to select the target tissue. Additionally, the pixels outside the mask were set to the average pixel value of the whole trunk area (Fig. 4B).
Figure 4.
Panel A: analysis of different quantiles over CT scans of all male subjects resulted in three intensity ranges roughly corresponding to adipose tissue, soft tissues and bones. Panel B illustrates segmentation of the whole intensity range according to the selected ranges shown above. Rows 1-3 present Fats, STs, and Bones ranges, respectively. Four male subjects shown correspond to obese 67 y.o. (1), normal 69 y.o. (2), obese 82 y.o. (3), normal 82 y.o. (4).
We used the general image features we previously described (8, 12, 13) to provide a large set of numerical image descriptors for each image. The feature library consists of twelve different algorithms, including features based on histograms from polynomial coefficients (Chebyshev and Chebyshev-Fourier types), coefficients of Zernike polynomials, features derived from Radon transforms, Gabor filters, multi-scale histograms, textures (Tamura and Haralick), edge statistics, and object statistics based on the direct and inverted Otsu threshold. As described before, these image features were also extracted from transforms of the original image, consisting of the fast Fourier transform, Chebyshev transform, and wavelet transform (symlets5, level-1 details), as well as two compound transforms: Fourier of Chebyshev and Fourier of wavelet.
The CHARM descriptors have been a dependable tool for quantitative analysis of diverse biological and biomedical applications (8, 12, 13). The multipurpose nature of the CHARM features allows to avoid biasing the classifier with any pre-conceived notions of the biology responsible for the observed changes. This also allows asking open-ended questions and design assays without a-priori knowledge of underlying mechanism or biology, which is of primary utility when the biology is not well understood.
Statistical classifiers
We employed three statistical classifiers, WND5 (7, 8, 12, 13) (referred to subsequently as WND) and two support vector machine (SVM) classifiers (14-16). When categorizing a given test sample, WND goes over the training data class-by-class and selects the class where the average similarity between all of its training samples and the test sample is largest. In WND, similarity is defined as the inverse-exponential distance in weighted feature space, with the exponent set to 5. Fisher scores were used as weights for the feature space dimensions as before. We have used WND for a variety of pattern recognition applications (8, 12, 13, 17) and we use it here due to its consistent performance. Compared to other classifiers, WND operates in a comparatively high-dimensional space, and it was set to use the top 600 Fisher-weighted features for this study.
SVM converts data into an artificial high-dimensional space and constructs support vectors to build a decision boundary, which is a unique hyperplane maximizing the margin of separation between different classes. The SVM algorithm is based on Cover's theorem stating that data cast in a high-dimensional space non-linearly are more likely to be linearly separable than in the original lower-dimensional space. The design implements very efficient use of this high-dimensional space: during training/testing only pair-wise dot products are used in contrast to explicit use of large vectors in computations. As a specific implementation of the SVM algorithm in our experiments, we used libSVM (14) with the RBF kernel. To be computationally efficient, SVM requires a greater reduction in the dimensionality of the original feature space compared to WND. We used two techniques, both based on the concept of mutual information. The minimal-redundancy maximal relevance (mRMR) (15) algorithm forces inter-class features to be maximally distant (e.g. minimally redundant) while maximizing their relevance to the targets. The conditional mutual information minimization (CMIM, (16)) method operates on similar principles, favoring individually informative and pair-wise weakly dependent features. We set both algorithms to pick the best 30 features from the training sets for use in SVM classifiers.
Classifier evaluation
Classifiers were evaluated using non-exhaustive two-fold cross-validation with 100 randomized splits into training and testing sets. For cross-validation purposes, the presence of multiple scans per participant required that scans from one participant are not used for both training and testing sets. In each split, we randomly selected participants for training and then selected one of their visits at random for the training samples. This resulted in a very close number of training samples for both genders: 420 (e.g. 210 per-class) samples for male participants and 396 (198 per-class) samples for female participants in each randomized split. The remaining scans were then used to test the resulting classifier.
Classifier performance is reported as the overall classification accuracy (CA), which is the fraction of all samples that were called correctly. To avoid overfitting, we used the recursive feature addition, where the features were pre-ranked as described in the previous section. Cross-validation strategy precluded from selection bias. Additionally, we reported per-class accuracies allowing for supplemental analysis of classification errors.
We tested three classification schemes consisting of two classifiers and three feature selection methods (Fisher/WND, mRMR/SVM, and CMIM/SVM). We also defined an ensemble classifier based on the majority vote of the three for each test sample.
Results
Aging signal in raw scans and signal enhancement
Raw scans of whole abdominal areas (labeled Torso) were analyzed first (See Table II). Preprocessing was limited to cropping the raw scans to exclude the area outside of the abdomen. Classification of this raw data resulted in an ensemble CA of 0.688 for male subjects and 0.635 for female subjects.
Table 2. Classification accuracy comparisons.
| Raw | CLAHE | |
|---|---|---|
| Males, WND | 0.655 | 0.718 |
| Males: cmim+svm | 0.677 | 0.762 |
| Males: mrmr+svm | 0.659 | 0.736 |
| Males: ensemble | 0.688 | 0.759 |
| Females: WND | 0. 635 | 0.701 |
| Females: cmim+svm | 0.622 | 0.712 |
| Females: mrmr+svm | 0.595 | 0.698 |
| Females: ensemble | 0.635 | 0.718 |
Comparison of classification accuracy of Torso for two data types: a) original (raw) pixel intensity in the left column, and b) CLAHE-enhanced images in the right column.
Next, we performed image normalization and enhancement (CLAHE pre-processing; see Materials and Methods). We observed that CLAHE enhancement (See Fig. 3) resulted in a significant classification improvement compared to the raw scans, even without rotation correction (not shown). After applying rotation correction, classification improved further but to a much lesser extent than the improvement from contrast enhancement. The CLAHE-enhanced, rotationally corrected images of the whole torso resulted in an ensemble CA of 0.759 for males and 0.718 for females (Table II).
Figure 3.
Representative abdominal CT scans for middle-aged group (age 50-70 y.o., indicated as ‘Y’) and old group (age 80-99 y.o., marked as ‘O’). Each ‘Y’ and ‘O’ group contains 4 subjects. Columns 1,2 show the scans with original (un-manipulated) pixel intensity, while columns 3,4 show CLAHE-enhanced images.
Where is the aging signal?
The Hounsfield scale can be broadly split up into ranges of radiodensity corresponding to bones (1100 - 2000 in shifted Hounsfield units, SHU), adipose tissues (ATs, 850 to 950 SHU), and soft tissues (STs, 1030 to 1060 SHU) (See Figure 4). Bones have a broad range of radiodensities and hence take up a large proportion of the upper range of the Hounsfield scale. Adipose tissues occupy a much smaller portion of the lower range of the Hounsfield scale, while soft-tissues, despite having the largest variety of tissue types, capture a relatively narrow range in the middle of this scale (18). In order to mask these three types of tissue using Hounsfield ranges, we divided the cumulative histogram of Hounsfield units into quartiles, and merged the middle two quartiles to define the three types of tissue (see Fig. 4, panel A). The resulting boundaries (in SHU) were 720-938 for the first segment, 939-1070 for the second segment, and all values above 1071 for the last segment. The three segments largely corresponded to ATs, STs, and Bone tissue ranges.
We compared discrimination between the two age cohorts in all three tissue segments (Bones, STs, ATs) and in the whole torso (labeled Torso), using three different classifiers and an ensemble classifier based on the three classifier votes. The results are shown in Table III, where each cell contains two sub-compartments: overall classification accuracy (CA) in the upper sub-cell and per-class CAs in the lower sub-cell for the Younger and Older classes respectively.
Table 3. Per-tissue classification accuracies in different tissues.
| Torso | ATs | STs | Bones | |
|---|---|---|---|---|
| Males: WND | 0.718 [0.772 0.665] | 0.787 [0.749 0.825] | 0.677 [0.757 0.597] | 0.650 [0.591 0.709] |
| Males: cmim+svm | 0.762 [0.780 0.743] | 0.770 [0.772 0.767] | 0.691 [0.659 0.724] | 0.622 [0.647 0.596] |
| Males: mrmr+svm | 0.736 [0.705 0.767] | 0.753 [0.766 0.741] | 0.670 [0.616 0.723] | 0.609 [0.665 0.553] |
| Males: ensemble | 0.759 [0.770 0.747] | 0.785 [0.769 0.802] | 0.700 [0.704 0.696] | 0.647 [0.646 0.649] |
| Females: WND | 0.701 [0.738 0.644] | 0.709 [0.592 0.824] | 0.672 [0.672 0.671] | 0.639 [0.670 0.608] |
| Females: cmim+svm | 0.712 [0.823 0.601] | 0.689 [0.763 0.614] | 0.655 [0.761 0.549] | 0.632 [0.609 0.654] |
| Females: mrmr+svm | 0.698 [0.769 0.638] | 0.671 [0.724 0.619] | 0.646 [0.722 0.570] | 0.582 [0.601 0.564] |
| Females: ensemble | 0.718 [0.798 0.638] | 0.707[0.705 0.710] | 0.678 [0.738 0.617] | 0.644 [0.654 0.634] |
Upper sub-cells show CA, and lower sub-cells show per-class CAs (for the Younger and Older classes respectively).
The ensemble classifier always reported either the highest overall CA, or was within 0.5% of the highest CA (See Table III). This classifier also had closer and more consistent agreement between the per-class accuracies for Younger and Older subjects. The CAs reported by each of the four classifiers for the different tissues were much more consistent between classifiers than they were between tissues, indicating that different tissues change with age to different extents, and that this is independent of the classification technique used. Thus, the CAs are an indication of the non-uniform aging signal present in different tissues.
In males, the highest aging signal was observed for ATs (0.785, see Table III and Fig. 5), while in females, Torso (0.718) was higher than ATs (0.707). In both males and females, the lowest aging signal was observed in Bones (0.647 and 0.644, respectively). Males had higher aging signals than females in every tissue, with the largest gender difference in ATs (0.078 or 10%) and lowest for Bones (0.003 or 0.5%).
Figure 5.
Per-tissue signal contributions by the ensemble classifier.
Discussion
As indicated in the introduction, the difficulty with characterizing phenotypic aging through direct body composition is in large variability of data within the same age cohort. Large within-class variability requires adequate sampling to capture data variance. In the cross-validation experiments, we trained classifiers on about 400 unique participant-scans to capture the variance and to avoid overfitting to phenotype.
The aging signal in the raw abdominal scans was 0.688 for males and 0.635 for females, which is relatively low compared to a noise level of 0.5. We speculated that the signal strength in raw scans was somewhat compromised by decreased efficiency of feature algorithms on low-contrast images (Fig. 3, columns 1,2). As expected, contrast enhancement resulted in an increased classification signal of 0.759 for males and 0.718 for females. There are several sources of noise in the scan data: variations in subject size, z-section of subjects, and other distortions that are impossible to normalize for. Additionally, only the chronological age of the subjects is known, while it is the unknown physiological age of the subject that affects the observations. In this context, an accuracy of 0.759 should be considered relatively high, and is higher than the age discrimination values reported in previous imaging studies.
Adipose tissue: highest aging signal
Out of the three constituent tissue types (ATs, STs, and Bones) considered, adipose tissue had the largest aging signal in both genders (see Table III and Fig. 5), which is consistent with AT as an aging biomarker. From a pattern recognition perspective this is surprising since AT has a narrow dynamic range as well as (at least by visual inspection) limited variation in texture. This is in contrast to bones, which have the largest dynamic range of the three tissues and a much larger variation in texture, but have little detectable aging signal in either gender. The accuracies we observed are thus reversed from what one may expect from very basic assumptions.
We speculate that the reason for the high aging signal observed in AT may be the age-associated transition of AT from subcutaneous to visceral fat, and probably a change in density and texture that occurs in the visceral fat with aging (19-23). Additionally, the two genders showed substantially different classification accuracies for AT (0.785 in males vs. 0.707 in females), a difference of almost 10%. We speculate that this is due to the difference in the distribution of AT in males vs. females (24). In males, AT typically accumulates in the abdominal area, and abdominal scans are well suited to capture the transition from subcutaneous to visceral fat. In contrast, females have multiple depositions of AT and a more complicated process of fat redistribution during aging that is not necessarily optimally represented in abdominal scans. Thus, the redistribution of AT between subcutaneous and visceral fat may explain the high aging signal in this tissue, while the specific concentration of fat in the abdomen of males vs. a more dispersed distribution in females may explain why the aging signal is substantially higher in males than in females.
Soft tissue
Soft tissue is composed of a variety of tissues largely characterized by their water content (neither bone nor fat). Soft tissue occupies a relatively large percentage of the torso (∼40%), has (by manual observation) a variety of textures and shapes, but also has the narrowest radiodensity range of the three tissues. The observed aging signal (per Table III) was relatively high at 0.700 in males and 0.678 in females. It is possible that muscle is a significant contributor to this aging signal, as myosteatosis is a known marker of aging as well as age-related insulin resistance and hyperinsulinemia (25, 26). However, muscle is not the only component of soft tissue, and is not the main tissue with this range of radiodensities within the abdomen. To our knowledge, ST has not been previously described as an aging marker, and our results indicate that these tissues warrant further study.
Bones: smallest aging signal
As noted above, age discrimination for Bones was the lowest out of the four data sets considered in both genders: 0.647 (males) and 0.644 (females), while the gender difference between the two was in the third digit of their values. Bone tissue is a well-known age marker, and bone loss is a hallmark of the aging process. Bone tissue is also well suited for CT studies because its density spans across half of the Hounsfield scale. Based on these considerations, we would have expected to detect a substantial aging signal in bone. Bone aging is also known to be distinct in the two genders. Bone density with aging decreases in males gradually, while in females bone loss accelerates after mid-life (19, 27, 28). We would expect to detect these differences. Although the aging signal we detected is significantly higher than noise, it is of low magnitude and shows no differences between males and females.
A set of contributing factors may explain our observations. First, in much of the research on bone density loss, the study subject is the femur (29, 30), a weight-bearing bone whose density loss is directly related to functional decline. In this study, the bones consisted of highly variable fragments of lower ribs and the L4 lumbar vertebra. Although the bone in the vertebrae is known to be sensitive to osteoporosis as it consists mostly of trabecular bone, the specific parts of the vertebra and ribs that were captured in the scans was highly variable from sample to sample because they came in and out of view with small changes in body position (See Fig. 4, panel B). These types of abdominal sections are simply not optimal for studying bone structure. Lastly, both the older and younger female cohorts in our dataset consist of post-menopausal women, which may be past the stage of accelerated bone loss, where changes are smaller and more comparable to those in men. We suggest that studies addressing changes in bone would be better performed on scans of the thigh and/or calf.
Where classification errors occur
The per-class accuracy (Table III, values in brackets) allowed for analysis of common misclassification measures, miss rate (e.g. frequency of ‘Y’ misclassified as ‘O’) and fall-out (frequency of ‘O’ misclassified as ‘Y’). These rates can be calculated as complementary to the known per-class accuracy given binary classification problem. For example, the per-class accuracies [0.770 0.747] (Males: ensemble, Torso) give miss rate and fall-out as 0.230 and 0.253, respectively. The per-class misclassifications were rather uniform for male participants (the largest difference is 0.03 for ATs). For female participants, the differences between fall-out and miss values were low (less than 0.02) for ATs and Bones, but were higher for Torso (0.16) and for STs (0.12). Here ‘O’ cohort showed higher misclassification rate, which could be attributed to a greater dispersion of morphology within older participants, given heterogeneous nature of this tissue group (more receptive to age diversion in the constituent tissues).
Conclusions
We present a framework for automated processing of CT scans to evaluate the relative aging signals in tissues present in abdominal sections. The method relies on supervised machine learning and low-level generic image descriptors. Using three different classifiers and ensemble classification we demonstrate that robust aging signals exist in the morphological patterns of contrast-enhanced abdominal CT scans. The analysis showed that aging signal was higher overall in males than in females. In males, the strongest aging signal was observed in adipose tissue, while in females, adipose tissue was a close second to the signal in the whole torso. This was the first aging study based exclusively on image data from the BLSA and is by far the largest aging study based on images thus far conducted in humans.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nikita V. Orlov, National Institute on Aging, National Institute of Health, 251 Bayview Blvd, BRC, Ste 100, Baltimore, MD 21224-6825, U.S. Phone 410-558-8503
Sokratis Makrogiannis, National Institute on Aging, National Institute of Health, 3001 South Hanover St., 5th Fl. Baltimore, MD 21225
Luigi Ferrucci, National Institute on Aging, National Institute of Health, 251 Bayview Blvd, BRC, Office of Scientific Director, Baltimore, MD 21224-6825, U.S. Phone 410-558-8110
Ilya G. Goldberg, National Institute on Aging, National Institute of Health, 251 Bayview Blvd, BRC, Ste 100, Baltimore, MD 21224-6825, U.S
References
- 1.Santanasto AJ, Goodpaster BH, Kritchevsky SB, et al. Body composition remodeling and mortality: the health aging and body composition study. Journals of Gerontology: Medical Sciences. 2016;00(00):1–7. doi: 10.1093/gerona/glw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prado CM, Siervo M, Mire E, et al. A population-based approach to define body-composition phenotypes. The American Journal of Clinical Nutrition. 2014;99(6):1369–77. doi: 10.3945/ajcn.113.078576. [DOI] [PubMed] [Google Scholar]
- 3.Doan NT, Rooden Sv, Versluis MJ, et al. An automated tool for cortical feature analysis: Application to differences on 7 Tesla T2* -weighted images between young and older healthy subjects. Magnetic Resonance in Medicine. 2015;74(1):240–8. doi: 10.1002/mrm.25397. [DOI] [PubMed] [Google Scholar]
- 4.López-Alcaraz M, González P, Aguilera I, López M. Image analysis of pubic bone for age estimation in a computed tomography sample. Int J Legal Med. 2015;129(2):335–46. doi: 10.1007/s00414-014-1034-9. [DOI] [PubMed] [Google Scholar]
- 5.Duan X, Rule AD, Elsherbiny H, et al. Automated assessment of renal cortical surface roughness from computerized tomography images and its association with age. Academic Radiology. 2014;21(11):1441–5. doi: 10.1016/j.acra.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Csapo R, Malis V, Sinha U, Du J, Sinha S. Age-associated differences in triceps surae muscle composition and strength – an MRI-based cross-sectional comparison of contractile, adipose and connective tissue. BMC Musculoskelet Disord. 2014;15(209) doi: 10.1186/1471-2474-15-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shamir L, Delaney J, Orlov N, Eckley DM, Goldberg I. Pattern recognition software and techniques for biological image analysis. PLoS Computational Biology. 2010;6(11):e1000974. doi: 10.1371/journal.pcbi.1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlov N, Shamir L, Macura T, Johnston J, Eckley DM, Goldberg I. WND-CHARM: Multi-purpose image classification using compound image transforms. Pattern Recognition Letters. 2008;29(11):1684–93. doi: 10.1016/j.patrec.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makrogiannis S, Caturegli G, Davatzikos C, Ferrucci L. Computer-aided assessment of regional abdominal fat with food residue removal in CT. Academic Radiology. 2013;20(11):1413–21. doi: 10.1016/j.acra.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teague MR. Image analysis via the general theory of moments. Journal of the Optical Society of America. 1980;70(8):920–30. [Google Scholar]
- 11.Pizer S, Johnston R, Rogers D, Beard D. Effective presentation of medical images on an electronic display station. Radiographics. 1987;7(6):1267–74. doi: 10.1148/radiographics.7.6.3423333. [DOI] [PubMed] [Google Scholar]
- 12.Orlov NV, Weeraratna AT, Hewitt SM, et al. Automatic Detection Of Melanoma Progression By Histological Analysis Of Secondary Sites. Cytometry, Part A. 2012;81A(5):364–73. doi: 10.1002/cyto.a.22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlov N, Chen W, Eckley DM, et al. Automatic classification of lymphoma images with transform-based global features. IEEE Tr on Information Technology in Biomedicine. 2010;14(4):1003–13. doi: 10.1109/TITB.2010.2050695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2(3) [Google Scholar]
- 15.Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2005;27(8):1226–38. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 16.Fleuret F. Fast binary feature selection with conditional mutual information. Journal of Machine Learning Research. 2004;5:1531–55. [Google Scholar]
- 17.Orlov NV, Eckley DM, Shamir L, Goldberg IG. Improving class separability using extended pixel planes: a comparative study. Machine Vision and Applications. 2012;23(5):1047–58. doi: 10.1007/s00138-011-0349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider U, Pedroni E, Lomax A. The calibration of CT Hounsfield units for radiotherapy treatment planning. Physics in Medicine and Biology. 1996;41(1):111–24. doi: 10.1088/0031-9155/41/1/009. [DOI] [PubMed] [Google Scholar]
- 19.Malo M, Rohrbach D, Isaksson H, et al. Longitudinal elastic properties and porosity of cortical bone tissue vary with age in human proximal femur. Bone. 2013;53(2):451–8. doi: 10.1016/j.bone.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Lee P, Brychta R, Collins M, et al. Cold-activated brown adipose tissue is an independent predictor of higher bone mineral density in women. Osteoporosis International. 2013;24(4):1513–8. doi: 10.1007/s00198-012-2110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muruganandan S, Roman A, Sinal C. Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: cross talk with the osteoblastogenic program. Cellular and Molecular Life Sciences. 2009;66(2):236–53. doi: 10.1007/s00018-008-8429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meunier P, Aaron J, Edouard C, Vignon G. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clinical Orthopaedics and Related Research. 1971;80:147–54. doi: 10.1097/00003086-197110000-00021. [DOI] [PubMed] [Google Scholar]
- 23.Bredella M, Gill C, Gerweck A, et al. Ectopic and serum lipid levels are positively assocoated with bone marrow fat in obesity. Radiology. 2013;269(2):534–41. doi: 10.1148/radiol.13130375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enzi G, Gasparo M, Biondetti P, Fiore D, Semisa M, Zurlo F. Subcutaneous and visceral fat distribution according to sex, age, and overweight, evaluated by computed tomography. The American Journal of Clinical Nutrition. 1986;44(6):739–46. doi: 10.1093/ajcn/44.6.739. [DOI] [PubMed] [Google Scholar]
- 25.Boutin RD, Yao L, Canter RJ, Lenchik L. Sarcopenia: current concepts and imaging implications. American Journal of Roentgenology. 2015;205(3):255–66. doi: 10.2214/AJR.15.14635. [DOI] [PubMed] [Google Scholar]
- 26.Miljkovic I, Cauley JA, Wang PY, et al. Abdominal myosteatosis is independently associated to hyperinsulinemia and insulin resistance among older men without diabetes. Obesity. 2013;21(10):2118–25. doi: 10.1002/oby.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieves JW, Formica C, Ruffing J, et al. Males have larger skeletal size and bone mass than females, despite comparable body size. Journal of Bone and Mineral Research. 2005;20(3):529–35. doi: 10.1359/JBMR.041005. [DOI] [PubMed] [Google Scholar]
- 28.Avdagić S, Barić I, Keser I, et al. Differences in peak bone density between male and female students. Archives of Industrial Hygiene and Toxicology. 2009;60(1):79–86. doi: 10.2478/10004-1254-60-2009-1886. [DOI] [PubMed] [Google Scholar]
- 29.Sheth NP, Nelson CL, Paprosky WG. Femoral bone loss in revision total hip arthroplasty: evaluation and management. Journal of the American Academy of Orthopaedic Surgeons. 2013;21(10):601–12. doi: 10.5435/JAAOS-21-10-601. [DOI] [PubMed] [Google Scholar]
- 30.Thomas C, Mayhew P, Power J, et al. Femoral neck trabecular bone: loss with aging and role in preventing fracture. Journal of Bone and Mineral Research. 2009;24(11):1808–18. doi: 10.1359/jbmr.090504. [DOI] [PubMed] [Google Scholar]