Abstract
Purpose
This study aimed to identify potential epidermal growth factor receptor (EGFR) gene mutations in non-small cell lung cancer that went undetected by amplification refractory mutation system-Scorpion real-time PCR (ARMS-PCR).
Materials and Methods
A total of 200 specimens were obtained from the First Affiliated Hospital of Guangzhou Medical University from August 2014 to August 2015. In total, 100 ARMS-negative and 100 ARMS-positive specimens were evaluated for EGFR gene mutations by Sanger sequencing. The methodology and sensitivity of each method and the outcomes of EGFR-tyrosine kinase inhibitor (TKI) therapy were analyzed.
Results
Among the 100 ARMS-PCR-positive samples, 90 were positive by Sanger sequencing, while 10 cases were considered negative, because the mutation abundance was less than 10%. Among the 100 negative cases, three were positive for a rare EGFR mutation by Sanger sequencing. In the curative effect analysis of EGFR-TKIs, the progression-free survival (PFS) analysis based on ARMS and Sanger sequencing results showed no difference. However, the PFS of patients with a high abundance of EGFR mutation was 12.4 months [95% confidence interval (CI), 11.6−12.4 months], which was significantly higher than that of patients with a low abundance of mutations detected by Sanger sequencing (95% CI, 10.7−11.3 months) (p<0.001).
Conclusion
The ARMS method demonstrated higher sensitivity than Sanger sequencing, but was prone to missing mutations due to primer design. Sanger sequencing was able to detect rare EGFR mutations and deemed applicable for confirming EGFR status. A clinical trial evaluating the efficacy of EGFR-TKIs in patients with rare EGFR mutations is needed.
Keywords: Non-small cell lung cancer, EGFR mutation, Sanger sequencing, ARMS
INTRODUCTION
Lung cancer is a leading malignancy in thoracic oncology that causes a majority of deaths both in China and worldwide.1 The prevalence of epidermal growth factor receptor (EGFR) mutations ranges from 5–10% in Caucasians to 60–70% in neversmoking Asian adenocarcinoma patients, indicating that EGFR mutation-positive non-small cell lung cancer (NSCLC) may have a unique disease course.2 In fact, NSCLC patients with sensitive EGFR mutations are highly responsive to EGFR inhibitors, including gefitinib and erlotinib, compared with standard chemotherapy.3,4 Because of inevitable EGFR-tyrosine kinase inhibitor (TKI) resistance, next-generation EGFR-TKIs have been developed, and clinical trials have demonstrated a higher response rate and longer progression-free survival (PFS) and overall survival (OS) among previously treated patients with EGFR-mutant NSCLC.5,6 Therefore, the precise detection of EGFR mutations plays a key role in the clinical management of EGFR mutation-positive NSCLC patients.
Currently, the methods for detecting EGFR mutations include Sanger sequencing,7 amplification refractory mutation system (ARMS),8 pyrosequencing,9 high resolution melting analysis,10 and genome sequencing.11 Sanger sequencing remains the gold standard for EGFR mutation detection in clinical practice and may detect unknown EGFR mutations. The ARMS method, which has also been approved by the China Food and Drug Administration (CFDA), is a highly sensitive and reliable method for detecting EGFR mutations. Due to limitations regarding labor, time, and expertise requirements, as well as low sensitivity, other methods, such as pyrosequencing, high resolution melting analysis, and whole genome sequencing, were excluded from the current clinical EGFR mutation analysis.
In this article, we compared patient outcomes based on EGFR mutation analysis by Sanger sequencing and ARMS in small specimens: both assays have been approved by the CFDA. Upon investigation of the survival data, we found that the curative effect of EGFR-TKIs may be better in lung cancer patients with a high abundance of EGFR mutations than in those with a low mutation abundance. Sanger sequencing could be useful for EGFR mutation detection, and our data support the implementation of secondary genetic testing of EGFR mutation-negative NSCLC patients with a promising response to EGFR-TKI treatment.
MATERIALS AND METHODS
Samples collection
A total of 200 NSCLC patients with an equal number of EGFR ARMS-positive and ARMS-negative cases at The First Affiliated Hospital of Guangzhou Medical University from August 2014 to August 2015 were selected as study participants (IRB number: 2016-29). The two main eligibility criteria were radiologically and pathologically confirmed NSCLC and patient consent. The other inclusion criteria were no previous chemotherapy or radiotherapy and no other severe systemic disease. We also included patients with stage I–III NSCLC who were EGFR ARMS-positive and self-medicated with an EGFR-TKI after refusing adjuvant chemotherapy and radiotherapy. There were 108 male and 92 female patients ranging in age from 48–87 years included in this study. Samples were obtained by CT-guided fine-needle aspiration (n=35) or surgery (n=165). All samples were confirmed to be adenocarcinoma. There were 113 stage I, 52 stage II, 29 stage III, and six stage IV cases.
DNA isolation
DNA was extracted from formalin-fixed, paraffin-embedded tumor tissue using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilsen, Germany) according to the manufacturer's recommendations. Genomic DNA was stored at -20±5℃ after measuring the concentration (ng/mL) thereof and absorbance (A260/280 ratio) using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Cleveland, OH, USA).
Sanger sequencing
Genomic DNA was amplified with four primer pairs targeting exons 18 to 21 and labeled using the EGFR Mutation Detection Kit (Guangzhou Life Technologies Daan Diagnostics Co., Ltd., Guangzhou, China). Sequencing and data collection were performed using an ABI 3100 Genetic Analyzer (Applied Biosystems). All sequence variations were confirmed by multiple independent PCR amplifications and repeat sequencing as previously described.12 The difference between high and low mutation abundance was as previously defined.13
ARMS qPCR
Common EGFR mutations (Del19, L858R and L861Q in exon 21, G719X in exon 18, S768I in exon 20, and three insertions in exon 20) were detected using an ADx-ARMS EGFR 21 Detection Kit (Amoy Diagnostics Co., Ltd., Xiamen, China). qRT-PCR was performed in a StepOne™ PCR System (Thermo Fisher Scientific) according to the manufacturer's instructions.14
Treatment and assessment
Treatment with EGFR-TKIs included oral administration of 250 mg/d gefitinib or 150 mg/d erlotinib, and efficacy was evaluated after treatment by chest CT of the thoracic lesion according to standard clinical practice. Patients with stage I–IIIA disease who self-purchased the targeted drugs after initial disease progression were included in our analysis. According to Response Evaluation Criteria in Solid Tumors, the effects were defined and categorized as complete response, partial response, stable disease, or progressive disease. OS and PFS were defined as the time interval from the beginning of treatment to documented disease progression or death from any cause censored at the last follow-up.15
Statistical analysis
All the analyses were performed using SPSS software, version 22.0 (IBM Corp., Armonk, NY, USA). The Kaplan-Meier method was used to compare median PFS after TKI therapy in the same follow-up group with different detection methods. p-values less than 0.05 were considered statistically significant.
RESULTS
Patient characteristics and samples
From August 2014 to August 2015, 200 patients were screened and met the enrollment criteria. The patient characteristics were as follows: 108 male and 92 female patients ranging in age from 48–87 years were included in this study. Samples were obtained by CT-guided fine-needle aspiration (n=35) or surgery (n=165). All samples were confirmed to be adenocarcinoma. Disease specimens of TNM stage I to IV were included. All patients with an EGFR-sensitive mutation who received a first-generation EGFR-TKI were also included. The patient characteristics are provided in Table 1. Age and TNM stage were well balanced among groups.
Table 1. Clinicopathologic Features of Patients with Lung Adenocarcinoma.
| Number of patients (EGFR positive) | Number of patients (EGFR negative) | Total | p value | |
|---|---|---|---|---|
| Age | 0.67 | |||
| ≥60 | 46 | 49 | 95 | |
| <60 | 54 | 51 | 105 | |
| Gender | 0.00* | |||
| Male | 43 | 65 | 108 | |
| Female | 57 | 35 | 92 | |
| Smoking history | 0.00* | |||
| Non-smoker | 84 | 48 | 132 | |
| Smoker | 16 | 52 | 68 | |
| Stage | 0.131 | |||
| I | 77 | 36 | 113 | |
| II | 17 | 35 | 52 | |
| III | 4 | 25 | 29 | |
| IV | 2 | 4 | 6 | |
| EGFR mutation | 0.00* | |||
| 19-del | 54 | 0 | - | |
| L858R | 46 | 0 | - |
EGFR, epidermal growth factor receptor; 19-del, exon 19 deletion; L858R, arginine for leucine substitution at residue 858.
*p<0.01.
Comparison of mutation detection rates by direct sequencing and ARMS
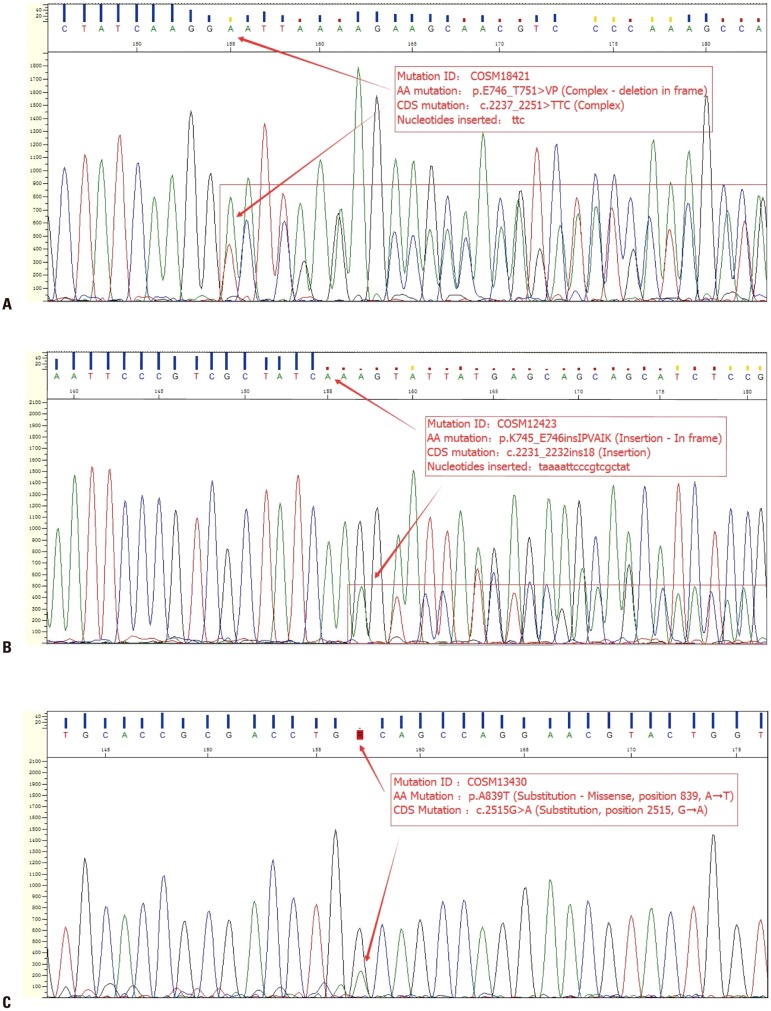
The EGFR mutation statuses of all patients detected by the two methods are summarized in Table 2. Among the 100 ARMS-positive EGFR samples, Sanger sequencing detected mutations in 90 samples; the other 10 were negative. Among the 100 ARMS-negative samples, three were positive for a mutation by the Sanger method, and 97 negative samples were confirmed. Based on the positive likelihood ratio (10.409) and the positive predictive value (96.77%), the ARMS-PCR method can detect EGFR mutations with high efficiency and specificity. Thus, the EGFR mutation rate was higher using ARMS than direct sequencing. Notably, the ARMS method covers only 29 EGFR mutation hotspots in exons 18–21, and Sanger sequencing detected three coding DNA sequence (CDS) mutations in ARMS-negative samples: c.2237_2251>TTC (complex), c.2231_2232ins18 (insertion), and c.2515G>A (substitution, position 2515, G→A) (Fig. 1).
Table 2. Mutation Rate with Different Methods in Our Clinic.
| ARMS | Sanger sequencing | Total | p value | |
|---|---|---|---|---|
| Mutant | Wild-type | |||
| Positive (n=100) | 90 | 10 | 100 | 0.00* |
| Negative (n=100) | 3 | 97 | 100 | |
| Total | 93 | 107 | 200 | |
ARMS, amplification refractory mutation system.
*p<0.05.
Fig. 1. Results of Sanger sequencing of ARMS-negative samples. (A) Patient 1 had a very rare complex inframe deletion: c.2237_2251>TTC (p.E746_T751>VP), which was only reported once in the COSMIC database with mutation Id COSM18421. (B) Patient 2 had another complex inframe insertion: c.2231_2232ins18 (p.K745_E746insIPVAIK, with 18-bp “taaaattcccgtcgctat” inserted), it was reported six times in the COSMIC database with mutation Id COSM12423. (C) Patient 3 had a rare point mutation: c.2515G>A (p.A839T, COSM13430), which was reported four times. ARMS, amplification refractory mutation system. CDS, coding DNA sequence.
EGFR mutation status and clinical outcomes
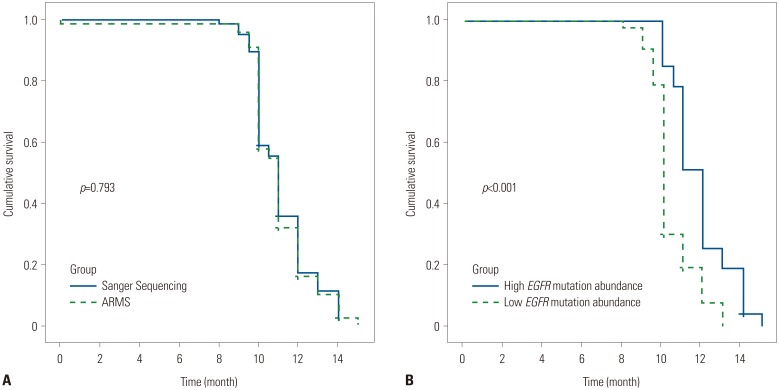
As a higher EGFR mutation abundance may yield better results with EGFR-TKI treatment,16 we compared patient outcomes after EGFR-TKI treatment based on ARMS and Sanger sequencing. In terms of EGFR-TKI treatment, the median PFSs among EGFR-positive patients detected by Sanger sequencing or ARMS were 11.1 months [95% confidence interval (CI), 10.6–11.4 months] and 10.9 months (95% CI, 10.7–11.3 months), respectively; this difference was not significant. The PFS was 12.4 months (95% CI, 11.6–12.4 months) for patients with a high EGFR mutation abundance (n=35), which was longer than that for patients with a low EGFR mutation abundance (95% CI, 10.7–11.3 months) (p<0.001) (Fig. 2). Interestingly, patients with the c.2237_2251>TTC (complex) or c.2231_2232ins18 (insertion) mutation who received EGFR-TKIs had a PFS of 3 months and 6 months, respectively. One patient with a c.2515G>A mutation (substitution, position 2515, G→A) was lost to follow-up after 4 months of EGFR-TKI treatment.
Fig. 2. PFS curves for patients treated with EGFR-TKIs. (A) PFS of patients with EGFR mutation status detected by Sanger sequencing or ARMS (p=0.793). (B) PFS of patients with high or low EGFR mutation abundance detected by Sanger sequencing (p<0.001). PFS, progression-free survival; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; ARMS, amplification refractory mutation system.
DISCUSSION
NSCLC accounts for over 80% of lung cancer cases and includes adenocarcinoma, large cell carcinoma, and squamous cell carcinoma.17 Similar to our results, patients who are female, never smokers, of Asian origin, and present with adenocarcinoma have a higher EGFR mutation frequency,18,19 and this EGFR mutation rate is higher than that in non-adenocarcinoma patients, who have a rate of less than 10%.20 In recent years, NSCLC has been managed according to molecular subtype. In EGFR-mutant NSCLC patients, EGFR-TKI treatment has greatly increased survival compared to those with EGFR wild-type lung cancer.21,22 The predominant EGFR mutations are in exons 18 through 21 and serve as predictors of the efficacy of EGFR-TKIs. Therefore, the identification of an EGFR mutation plays a critical role in NSCLC management.
Although it has been well recognized that EGFR mutations are associated with the therapeutic effect of TKIs in NSCLC patients, current methods do not provide the precision required for clinical practice. Currently, the two main detection methods are ARMS and Sanger sequencing. Although Sanger sequencing remains then gold standard, the ARMS method is considered an alternative because of its high sensitivity in detecting EGFR mutations;23,24 EGFR mutations can be detected in small samples using ARMS. The reason for the high sensitivity with ARMS is its special primer design. One pair of primers amplifies a conserved region, and another primer pair targets the point mutation. ARMS is limited to the detection of known mutations; each reaction system can only detect the pre-specified gene mutation. Therefore, a large number of DNA samples and primer pairs are needed, making this method expensive, if an unknown region must be analyzed. Sanger sequencing can analyze unknown DNA sequences at relatively low cost; the biggest problem is the low sensitivity. Mutations are difficult to detect in specimens with a low content of tumor cells or mutant cells. Moreover, noise within peaks can affect calling EGFR mutations. Therefore, Sanger sequencing is suitable for detecting EGFR mutations in surgical specimens with a high proportion of tumor cells potentially harboring a mutation. The results of this study suggest that Sanger sequencing is recommended for EGFR redetection and for initial detection in surgical specimens.
At least 90% of EGFR mutations occur in exons 19 and 21; the remaining 10% of mutations are in less common sites, and these are called rare EGFR mutations. With the application of EGFR sequencing technology, the discovery of mutations in exons 18–21 is increasing.25 Few treatment strategies have been reported for less common EGFR mutations. For example, first-generation EGFR-TKIs could be used in patients with A763_Y764insFQEA, an exon 20 insertion.26 In our study, we detected 10 EGFR mutation-negative samples by Sanger sequencing among 100 ADx-ARMS-positive samples. Among the 100 ADx-ARMS-negative samples, three were positive for a mutation by Sanger sequencing. Of these, two harbored an exon 19 deletion, and one had an exon 21 c.2515G>A p.A839T mutation (Cosmic ID COSM13430), which might not have been detected by ARMS due to the assay design. The impact of these rare EGFR mutations on EGFR-TKI therapy are far from fully understood. Baek, et al.27 reported that the response to EGFR-TKI treatment and the survival of patients with rare or complex EGFR mutations is worse than those for patients with common mutations. In our study, only two cases with a PFS of 3 months and 6 months are not sufficient to reach a conclusion. Therefore, clinical trials, such as NCT01775943, involving a large number of patients with rare EGFR mutations are warranted to elucidate the efficacy of EGFR-TKIs in these patients.
In this analysis, we also determined that patients with a high EGFR mutation abundance have a better outcome after EGFRTKI treatment. For patients with a high EGFR mutation abundance, the PFS was 12.4 months (95% CI, 11.6–12.4 months), which was higher than that for those with a low EGFR mutation abundance (95% CI, 10.7–11.3 months) (p<0.001). In accordance with a previous report, the EGFR mutation abundance could predict the outcome of EGFR-TKI therapy for advanced NSCLC. Hence, in clinical practice, Sanger sequencing offers additional information for physicians to predict whether the patient may benefit from an EGFR-TKI.
In summary, our results suggest that Sanger sequencing can detect rare EGFR mutations and is applicable for redetermining EGFR status. NSCLC patients with a high mutation burden have a better response to EGFR-TKIs. A clinical trial evaluating the efficacy of EGFR-TKIs in patients with rare EGFR mutations is needed.
ACKNOWLEDGEMENTS
This work was partly supported by the Open Project Program of the State Key Laboratory of Respiratory Disease (SKLRD20160P020, SKLRD20160P020), Clinical application and Translational Research project of the First Affiliated Hospital of Guangzhou Medical University (201515-gyfyy) and Science and Technology Planning Project of Guangdong Province (20140212).
We thank Dr. Xiaoshun Shi from the Affiliated Cancer Hospital & Institute of Guangzhou Medical University for expert medical advice and writing assistance.
Footnotes
The authors have no financial conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Tan DS, Mok TS, Rebbeck TR. Cancer genomics: diversity and disparity across ethnicity and geography. J Clin Oncol. 2016;34:91–101. doi: 10.1200/JCO.2015.62.0096. [DOI] [PubMed] [Google Scholar]
- 3.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive nonsmall-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 5.Lee JY, Sun JM, Lim SH, Kim HS, Yoo KH, Jung KS, et al. A phase Ib/II study of afatinib in combination with nimotuzumab in non-small cell lung cancer patients with acquired resistance to gefitinib or erlotinib. Clin Cancer Res. 2016;22:2139–2145. doi: 10.1158/1078-0432.CCR-15-1653. [DOI] [PubMed] [Google Scholar]
- 6.Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Lu Y, Zhu G, Lei Y, Zheng L, Qin H, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol. 2013;66:1065–1069. doi: 10.1136/jclinpath-2013-201728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Zhao R, Zhang J, Zhang J. ARMS for EGFR mutation analysis of cytologic and corresponding lung adenocarcinoma histologic specimens. J Cancer Res Clin Oncol. 2015;141:221–227. doi: 10.1007/s00432-014-1807-z. [DOI] [PubMed] [Google Scholar]
- 9.Min KW, Kim WS, Jang SJ, Choi YD, Chang S, Jung SH, et al. Comparison of EGFR mutation detection between the tissue and cytology using direct sequencing, pyrosequencing and peptide nucleic acid clamping in lung adenocarcinoma: Korean multicentre study. QJM. 2016;109:167–173. doi: 10.1093/qjmed/hcv103. [DOI] [PubMed] [Google Scholar]
- 10.Do H, Krypuy M, Mitchell PL, Fox SB, Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer. 2008;8:142. doi: 10.1186/1471-2407-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross JS, Cronin M. Whole cancer genome sequencing by nextgeneration methods. Am J Clin Pathol. 2011;136:527–539. doi: 10.1309/AJCPR1SVT1VHUGXW. [DOI] [PubMed] [Google Scholar]
- 12.Hu N, Wang G, Wu YH, Chen SF, Liu GD, Chen C, et al. LDA-SVM-based EGFR mutation model for NSCLC brain metastases: an observational study. Medicine (Baltimore) 2015;94:e375. doi: 10.1097/MD.0000000000000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao ZR, Wang JF, Lin YB, Wang F, Fu S, Zhang SL, et al. Mutation abundance affects the efficacy of EGFR tyrosine kinase inhibitor readministration in non-small-cell lung cancer with acquired resistance. Med Oncol. 2014;31:810. doi: 10.1007/s12032-013-0810-6. [DOI] [PubMed] [Google Scholar]
- 14.Shaozhang Z, Ming Z, Haiyan P, Aiping Z, Qitao Y, Xiangqun S. Comparison of ARMS and direct sequencing for detection of EGFR mutation and prediction of EGFR-TKI efficacy between surgery and biopsy tumor tissues in NSCLC patients. Med Oncol. 2014;31:926. doi: 10.1007/s12032-014-0926-3. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Zhang Q. A Weibull multi-state model for the dependence of progression-free survival and overall survival. Stat Med. 2015;34:2497–2513. doi: 10.1002/sim.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Zhang XC, Chen ZH, Yin XL, Yang JJ, Xu CR, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:3316–3321. doi: 10.1200/JCO.2010.33.3757. [DOI] [PubMed] [Google Scholar]
- 17.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2012;10:1236–1271. doi: 10.6004/jnccn.2012.0130. [DOI] [PubMed] [Google Scholar]
- 18.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 19.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki H, Shimizu S, Endo K, Takada M, Kawahara M, Tanaka H, et al. EGFR and erbB2 mutation status in Japanese lung cancer patients. Int J Cancer. 2006;118:180–184. doi: 10.1002/ijc.21301. [DOI] [PubMed] [Google Scholar]
- 21.Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- 22.Yang X, Yang K, Kuang K. The efficacy and safety of EGFR inhibitor monotherapy in non-small cell lung cancer: a systematic review. Curr Oncol Rep. 2014;16:390. doi: 10.1007/s11912-014-0390-4. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Liu B, Li XY, Li JJ, Qin HF, Tang CH, et al. A comparison of ARMS and direct sequencing for EGFR mutation analysis and tyrosine kinase inhibitors treatment prediction in body fluid samples of non-small-cell lung cancer patients. J Exp Clin Cancer Res. 2011;30:111. doi: 10.1186/1756-9966-30-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lungcancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang JC, Sequist LV, Zhou C, Schuler M, Geater SL, Mok T, et al. Effect of dose adjustment on the safety and efficacy of afatinib for EGFR mutation-positive lung adenocarcinoma: post hoc analyses of the randomized LUX-Lung 3 and 6 trials. Ann Oncol. 2016;27:2103–2110. doi: 10.1093/annonc/mdw322. [DOI] [PubMed] [Google Scholar]
- 26.Voon PJ, Tsui DW, Rosenfeld N, Chin TM. EGFR exon 20 insertion A763-Y764insFQEA and response to erlotinib–Letter. Mol Cancer Ther. 2013;12:2614–2615. doi: 10.1158/1535-7163.MCT-13-0192. [DOI] [PubMed] [Google Scholar]
- 27.Baek JH, Sun JM, Min YJ, Cho EK, Cho BC, Kim JH, et al. Efficacy of EGFR tyrosine kinase inhibitors in patients with EGFR-mutated non-small cell lung cancer except both exon 19 deletion and exon 21 L858R: a retrospective analysis in Korea. Lung Cancer. 2015;87:148–154. doi: 10.1016/j.lungcan.2014.11.013. [DOI] [PubMed] [Google Scholar]