Abstract
Out of 103 microsatellite markers used for studying the genetic diversity among local landraces of Luffa species, 56 were found polymorphic, including 38 gSSR and 18 eSSR, respectively. A total of 197 amplification products were obtained. The mean number of alleles per locus was 3.52. The PIC ranged from 0.037 to 0.986, while size of amplified product ranged from 105 to 500 bp. Cucumber-derived SSRs were amplified within L. acutangula (68%), L. aegyptiaca (61.16%), and L. hermaphrodita (60.2%), with an average of 63.12% cross-transferability. The Jaccard’s coefficient ranged from 0.66 to 0.97, with an average of 0.81. High genetic variability was observed for node of 1st hermaphrodite flower (6.4–17), days to 1st hermaphrodite flower (38–52.1), days to 1st fruit harvest (43–65), number of fruit per cluster (1–5.9), fruit length (3.9–25 cm), fruit weight (18.4–175 g), number of fruit per plant (20–147.5), and yield per plant (2.2–4.7 kg). Two sub-populations were identified including 21 genotypes (sub-population I) and 06 genotypes (sub-population II), these two sub-populations showed 0.608–0.395% of the ancestral relationship to each other. This study provides information for future exploration, collection, and utilization of Luffa genotypes, as well as the polymorphic markers identified could be available for the study of landmarks in linkages, genomic structures, evolutionary ecology, and marker-assisted selection (MAS) in Luffa species.
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1030-0) contains supplementary material, which is available to authorized users.
Keywords: Dendrogram, Molecular markers, Polymorphic, Satputia, Quantitative data
Introduction
The genus Luffa (Cucurbitaceous) is distributed mainly in the tropical regions of the world (Chakravarty 1982) and consisted of nine species worldwide, of which seven species are native to India. Out of seven species, four are wild (L. echinata Roxb, L. graveolens Roxb, L. tuberosa Roxb., and L. umbilata M. Roem), two are major cultivated species in the plains and low hills of the India (Ridge gourd; L. acutangula L. Roxb. and Sponge gourd; L. aegyptiaca Mill.) (Chandra 1995), whereas hermaphrodite Luffa comes under ridged gourd group has smaller size fruits in cluster (cf. L. hermaphrodita; ‘Satputia’ in Hindi-meaning seven children) is commercially cultivated in Eastern Uttar Pradesh and Bihar (Misra et al. 2016). The fruits of Luffa species are very nutritious and a good source of vitamin-A, calcium, phosphorus, ascorbic acid, and iron. Its medicinal properties are used as toothache, disinfectant, antihelmintic, anti-diarrhoea, anti-syphilitic, purgative, cardiotonic, laxative, and also potentially cure to diabetes and hypertension (Choudhary et al. 2011). Smooth gourd dried fruits are popular and wide-reaching as bath sponges, largely used as substitute biomaterial for packaging, ethanol production, and as an adsorbent of heavy metal present in industrial water (Papanicolaou et al. 2015). The collection and conservation of hermaphrodite Luffa in India has not been attempted properly.
Morphological variability is evident in the cultivated species of Luffa (Prakash et al. 2013). However, diversity analysis based on morphology, presence of flavonoids and breeding system classified L. acutangula and L. aegyptiaca into a single clade suggesting a different species status, apart from other five Luffa species (Schilling and Heiser 1981). Despite its economic importance and presence of considerable variability, no systemic information is accessible on the genetic amelioration of L. hermaphrodita. Thus, the genetic structure and diversity of the hermaphrodite Luffa is very much credential for the present day exploratory research in the crop improvement program.
Furthermore, assessment of genetic diversity based on phenotypic traits has limitations, since these traits are greatly influenced by environmental factors and developmental stages of plant. Molecular marker has been used for the assessment of genetic diversity in ridge gourd, i.e., random amplified polymorphic DNA (RAPD; Hoque and Rabbani 2009), inter-simple sequence repeat (ISSR; Sikdar et al. 2010; Prakash et al. 2014), and ISSR and DAMD (Direct Amplification of Minisatellite DNA) markers (Misra et al. 2016), so far there is no report on the assessment of genetic diversity in hermaphrodite Luffa, using genomic SSRs (gSSRs) and EST-derived SSRs (eSSRs). Among the PCR-based molecular markers, microsatellite markers in general have high level of transferability and neutral, and therefore, these markers are significantly valuable (Varshney et al. 2005). The eSSRs offers advantages over gSSRs that can be applied for analysis of functional diversity in gene rich region of genome (Zhang et al. 2005).
The present study explores the genetic diversity among local landraces of hermaphrodite Luffa spp. using morphological and molecular markers and to discuss the implications of present finding for conservation efforts as well as improvement of this species in India.
Materials and methods
Plant materials
A set of 31 accessions including, 27 accessions of hermaphrodite Luffa (L. hermaphrodita), two lines of L. acutangula (Ridge gourd), and two lines of L. aegyptiaca (Sponge gourd), were used in this study (Table 1; Fig. S1). The seed samples of these accessions were maintained at Indian Institute of Vegetable Research (IIVR), Varanasi, India.
Table 1.
Description of Luffa accessions and the mean of the eight quantitative characters averaged over 2 years (2012 and 2013)
| S. no. | Genotype name | Character | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NFHF | DFHF | Days to first fruit harvest | No. of fruits/cluster | Fruit length (cm) | Fruit weight (g) | No. of fruits/plant | Yield/plant (kg) | |||
| Uttar Pradesh population | L. hermaphrodita genotypes | District | ||||||||
| 1 | VRS-1 | Kushinagar | 6.40 | 41.13 | 45.07 | 5.93 | 16.42 | 31.78 | 147.53 | 4.69 |
| 2 | VRS-2 | Deoria | 7.33 | 50.87 | 51.40 | 5.00 | 12.42 | 28.81 | 134.13 | 3.87 |
| 3 | VRS-3 | Basti | 8.47 | 51.07 | 56.67 | 4.07 | 10.28 | 31.99 | 122.93 | 3.92 |
| 5 | VRS-6 | Varanasi | 7.27 | 48.47 | 54.73 | 4.40 | 11.65 | 26.82 | 138.27 | 3.71 |
| 6 | VRS-7 | Deoria | 6.47 | 43.00 | 48.33 | 5.27 | 15.60 | 33.64 | 129.80 | 4.36 |
| 7 | VRS-12 | Kushinagar | 7.27 | 47.93 | 53.13 | 3.93 | 13.35 | 29.92 | 126.13 | 3.77 |
| 8 | VRS-11 | Gorakhpur | 7.20 | 44.33 | 50.60 | 5.20 | 14.69 | 38.64 | 122.94 | 4.67 |
| 9 | VRS-14 | Deoria | 7.47 | 48.73 | 53.27 | 4.40 | 14.28 | 24.53 | 141.53 | 3.47 |
| 11 | VRS-21 | Basti | 6.47 | 46.60 | 51.40 | 4.27 | 11.50 | 28.48 | 142.67 | 4.06 |
| 12 | VRS-24 | Deoria | 7.53 | 50.00 | 54.13 | 4.40 | 3.90 | 18.95 | 145.20 | 2.75 |
| 13 | VRS-23 | Kushinagar | 8.67 | 51.13 | 56.87 | 4.53 | 13.81 | 25.49 | 141.27 | 3.60 |
| 14 | VRS-26 | Varanasi | 6.47 | 48.87 | 52.13 | 4.20 | 4.88 | 18.42 | 128.67 | 2.37 |
| 15 | VRS-25 | Kushinagar | 8.20 | 51.07 | 55.20 | 4.13 | 4.84 | 19.29 | 143.53 | 2.77 |
| 17 | VRS-29 | Deoria | 6.47 | 46.53 | 51.40 | 4.33 | 12.24 | 27.30 | 138.93 | 3.79 |
| 18 | VRS-32 | Kushinagar | 8.27 | 45.93 | 50.47 | 4.47 | 13.60 | 29.53 | 139.87 | 4.12 |
| 20 | VRS-35 | Kushinagar | 7.47 | 49.20 | 53.60 | 4.87 | 13.47 | 29.12 | 139.33 | 4.06 |
| 26 | VRS-22 | Kushinagar | 6.90 | 46.33 | 51.27 | 4.60 | 11.22 | 23.92 | 140.80 | 3.37 |
| 27 | VRS-9 | Varanasi | 7.40 | 50.33 | 55.87 | 4.00 | 12.66 | 27.87 | 136.87 | 3.81 |
| 29 | VRS-30 | Balia | 6.53 | 46.07 | 51.60 | 4.60 | 12.47 | 28.69 | 143.80 | 4.12 |
| 30 | VRS-34 | Varanasi | 8.60 | 51.07 | 56.13 | 4.53 | 11.80 | 27.35 | 143.20 | 3.92 |
| 24 | VRS-19 | Gorakhpur | 7.10 | 48.13 | 53.40 | 4.20 | 12.27 | 25.86 | 145.27 | 3.76 |
| Bihar population | L. hermaphrodita genotypes | |||||||||
| 4 | VRS-5 | Gopalganj | 8.20 | 52.13 | 57.27 | 4.13 | 14.48 | 31.18 | 133.33 | 4.16 |
| 10 | VRS-16 | West Champaran | 9.33 | 48.60 | 53.47 | 5.07 | 12.27 | 23.68 | 138.27 | 3.27 |
| 16 | VRS-27 | West Champaran | 7.33 | 47.27 | 51.67 | 4.00 | 4.81 | 20.15 | 146.93 | 2.96 |
| 19 | VRS-33 | Gopalganj | 9.40 | 50.13 | 55.60 | 3.87 | 12.72 | 28.59 | 142.27 | 4.07 |
| 28 | VRS-8 | Gopalganj | 7.20 | 47.07 | 52.47 | 4.07 | 11.73 | 26.13 | 140.13 | 3.66 |
| 31 | VRS-13 | West Champaran | 6.40 | 49.87 | 53.93 | 4.20 | 12.89 | 25.33 | 136.67 | 3.46 |
| Other sp. | L. acutangula genotypes | |||||||||
| 21 | Swarna Manjari | HARP (Jharkhand) | 14.00 | 52.00 | 65.00 | 1.00 | 24.00 | 175.00 | 20.00 | 2.50 |
| 22 | Pusa Nasdar | IARI (New Delhi) | 15.00 | 44.00 | 50.00 | 1.00 | 25.00 | 114.00 | 24.00 | 2.63 |
| Other sp. | L. aegyptiaca genotypes | |||||||||
| 23 | PusaChickni | IARI (New Delhi) | 17.00 | 38.00 | 43.00 | 1.00 | 19.00 | 108.00 | 28.00 | 3.14 |
| 25 | Kalyanpur Hari Chikni | Kanpur | 16.00 | 45.00 | 55.00 | 1.00 | 24.00 | 110.00 | 25.00 | 2.21 |
| Mean | 8.51 | 47.77 | 53.03 | 4.02 | 13.17 | 39.95 | 123.46 | 3.58 | ||
| Standard error | 0.52 | 0.59 | 0.70 | 0.23 | 0.90 | 6.42 | 7.07 | 0.12 | ||
DFHF days to 1st hermaphrodite/female flower, NFHF node at which 1st hermaphrodite/female flower appeared
Field evaluation
The field evaluations of Luffa accessions were conducted at Research Farm of IIVR, Varanasi, India. The experiment was laid out in a complete randomized block design (CRBD) with three replications, each with 15 individual plants. Five individual plants were randomly chosen and data were recorded on eight quantitative traits namely, node at which 1st hermaphrodite/female flower appeared, days taken to 1st hermaphrodite/female flower, days to first fruit harvest, number of fruit/cluster, fruit length (cm), fruit weight (g), and number of fruit/plant and yield/plant (kg). Fruits were harvested at edible maturity for recording the fruit traits. The 2-year pooled data on quantitative traits were analyzed using SPSS, Version 16.0 (SPSS Inc, Chicago) and estimates of correlation coefficients were obtained.
DNA extraction and SSRs genotyping
The genomic DNA was extracted from 10-day young leaf tissues using DNeasy Plant Mini Kit (Qiagen, Germany) following the manufacturer’s instructions. A total of 103 SSRs (82 gSSRs and 21 eSSRs) were used in the present study (82 gSSRs from Ren et al. 2009 and 21 eSSRs from Hu et al. 2010). These SSRs were used to be amplified following the protocol of Pandey et al. (2013) among 31 genotypes of Luffa and the amplification fragments were observed in 3% metaphore agarose (Lonza Rockland, Inc., USA) gel, submerged in chilled 1× TBE buffer and electrophoresed for about 3–4 h at 6 V/cm using horizontal electrophoresis system. After electrophoresis, the finely resolved SSR products were visualized under UV transilluminator and photographed using the Stratagene Eagle Eye still video system or Kodak (1D 3.5) imaging system. After gel electrophoresis and imaging, good-quality gel photographs were used to score all the visible and unambiguously scorable fragments amplified by SSR primers. The 100 bp DNA ladder (MBI Fermentas) was used as molecular size marker for calculating the allelic size.
SSRs data analysis
Clear and unambiguous SSR bands were scored according to their sizes inform of a binary data matrix. The data matrix was further subjected to analysis using NTSYS-PC version 2.11W (Rohlf 1998). To calculate the pair wise Jaccard’s similarities coefficient (Jaccard 1908), SIMQUAL function was used. Then, this similarity matrix was utilized to form UPGMA tree to estimate similarity indices and genetic relatedness among the collected genotypes. A dendrogram was constructed using the unweighted pair group method with arithmetic mean (UPGMA) cluster algorithm. Polymorphic information content (PIC) of molecular markers was calculated as per the formula: , where P ij is the frequency of the jth pattern for marker j and the summation extends over n patterns.
Hierarchical analysis of molecular variance (AMOVA) based on the pair wise squared Euclidean distances among different alleles was conducted using ARLEQUIN version 3.01 (Excoffier et al. 1992; Schneider et al. 2000) to further quantify the amount of genetic variation residing at two levels (i.e., among and within populations). The same program was used to generate the matrix of pairwise F ST values, which indicate the genetic differentiation between populations.
Population structure
To estimate the number of natural populations (K) and the proportion of assignment of individuals from each of the assumed populations to each of the inferred natural genetic clusters, population structure analysis was conducted using Bayesian model-based clustering method implemented in the program STRUCTURE version 2.2 (Pritchard et al. 2000) with burning length of 30,000 followed by 100,000 iterations in the admixture model. Based on the previous knowledge of the hermaphrodite Luffa collection, the population structure analysis was done assuming the number of sub-populations (K) from 1 to 5. To estimate the real number of ‘K’ and the ancestry membership of each individual in the inferred cluster, the program was run 10 times for each ‘K’. We further used delta K, an ad hoc quantity related to the second-order rate of change of the log probability of data with respect to the number of clusters to predict the real number of clusters (Evanno et al. 2005).
Results
Performance of genotypes based on quantitative traits
The range of all the quantitative traits revealed that there is a wide variation among the Luffa genotypes studied (Table 1). The first hermaphrodite flower was appeared at early node for VRS-1 and VRS-13 (6.4), whereas Pusa Chickni had female flower at 17th node. However, days taken to first female flower were found to be a minimum of 38 days in Pusa Chickni. The genotype, Swarna Manjari was found to be most delayed (65 days), while Pusa Chickni (43 days) has earliest harvesting. The highest number of fruits per cluster was found in VRS-1 (5.93), but Pusa Nasdar showed maximum length and weight (25 cm and 114 g) of fruit. Highest number of fruits/plant was recorded in VRS-1 (147.53), while lowest in Swarna Manjari (20). The maximum yield per plant was recorded in VRS-1 (4.69 kg) followed by VRS-11 (4.67 kg) (Table 1). Estimates of correlation study indicated that yield per plant had a highly significant positive association with the number of fruits per plant (r = 0.551), and number of fruits per cluster (r = 0.671). Whereas, fruit length, days to first fruit harvest and days to first hermaphrodite female flower showed positive but nonsignificant association with yield (Supplementary Table S1).
SSRs’ allele amplification and their polymorphism
A total of 103 SSR markers were deployed to evaluate genetic diversity in a collection of 27 accessions of L. hermaphrodita along with two lines each of L. acutangula (Ridge gourd) and L. aegyptiaca (Sponge gourd). Out of these, 70 SSRs (~ 68%) showed successful PCR amplification/cross-species transferability, while rest failed to amplify. These 70 SSRs were further analyzed for polymorphism among 31 genotypes and 56 SSRs (38 gSSR and 18 eSSR) were found polymorphic, while rest 14 SSRs (13 gSSRs and 01 eSSR) were monomorphic (Table 2). A total of 197 alleles were amplified by 56 polymorphic markers in 31 genotypes of Luffa species, which revealed ~ 3.52 alleles per polymorphic SSR locus. The PIC value of SSR varied with the least (0.037) for CK758649 to the highest (0.994) for SSR14649, with an average of 0.55, across all polymorphic SSRs combinations. The size of amplified alleles ranged between 105 bp (SSR11909) and 500 bp (SSR14649). The five most informative SSRs with high PIC values were SSR14649 (0.994), SSR16301 (0.986), SSR18530 (0.932), AF202378 (0.887), and DN910437 (0.834) (Table 2).
Table 2.
Details of SSRs used, their gene bank accession number, number of amplified alleles and PIC values
| Sl. no. | SSR and their gene bank accession number | No. of alleles amplified | PIC |
|---|---|---|---|
| 1 | SSR05492 | 3 | 0.251 |
| 2 | SSR18362 | 2 | 0.336 |
| 3 | SSR11909 | 2 | 0.492 |
| 4 | SSR13131 | 3 | 0.687 |
| 5 | SSR30665 | 2 | 0.603 |
| 6 | SSR22273 | 3 | 0.687 |
| 7 | SSR10783 | 3 | 0.720 |
| 8 | SSR16301 | 4 | 0.986 |
| 9 | SSR00670 | 3 | 0.128 |
| 10 | SSR19511 | 3 | 0.503 |
| 11 | SSR19493 | 5 | 0.893 |
| 12 | SSR19430 | 2 | 0.927 |
| 13 | SSR22172 | 3 | 0.657 |
| 14 | SSR06660 | 3 | 0.888 |
| 15 | SSR16842 | 6 | 0.864 |
| 16 | SSR13251 | 5 | 0.075 |
| 17 | SSR13189 | 3 | 0.535 |
| 18 | SSR33278 | 2 | 0.898 |
| 19 | SSR11742 | 3 | 0.113 |
| 20 | SSR06609 | 3 | 0.128 |
| 21 | SSR01179 | 6 | 0.123 |
| 22 | SSR02384 | 3 | 0.898 |
| 23 | SSR17823 | 8 | 0.084 |
| 24 | SSR13105 | 4 | 0.728 |
| 25 | SSR04255 | 2 | 0.906 |
| 26 | SSR05195 | 3 | 0.614 |
| 27 | SSR01148 | 7 | 0.716 |
| 28 | SSR02460 | 2 | 0.393 |
| 29 | SSR19755 | 5 | 0.555 |
| 30 | SSR07284 | 4 | 0.094 |
| 31 | SSR14247 | 3 | 0.629 |
| 32 | SSR11439 | 3 | 0.139 |
| 33 | SSR01498 | 5 | 0.122 |
| 34 | SSR18530 | 3 | 0.932 |
| 35 | SSR24696 | 4 | 0.687 |
| 36 | SSR14649 | 5 | 0.994 |
| 37 | SSR17219 | 2 | 0.931 |
| 38 | SSR02895 | 3 | 0.691 |
| 39 | DN910157 | 2 | 0.593 |
| 40 | DN909514 | 5 | 0.786 |
| 41 | DN909840 | 2 | 0.252 |
| 42 | DN910067 | 2 | 0.198 |
| 43 | CK700725 | 3 | 0.238 |
| 44 | DN910665 | 4 | 0.414 |
| 45 | DN910469 | 3 | 0.301 |
| 46 | DN910437 | 3 | 0.834 |
| 47 | CK758579 | 2 | 0.268 |
| 48 | AF202378 | 2 | 0.887 |
| 49 | CV000928 | 3 | 0.300 |
| 50 | BI740103 | 8 | 0.650 |
| 51 | EW968287 | 4 | 0.691 |
| 52 | AY942801 | 3 | 0.641 |
| 53 | CK758649 | 4 | 0.825 |
| 54 | CK758649 | 8 | 0.037 |
| 55 | CO998209 | 2 | 0.499 |
| 56 | AB112672 | 2 | 0.503 |
| Average | 3.52 | 0.550 |
Marker transferability and genetic clustering among Luffa species
To delineate cross-species/genera transferability, 103 cucumber-derived SSRs were tested on a panel of three species of Luffa, i.e., L. hermaphrodita, L. aegyptiaca, and L acutangula.. Among three Luffa species, maximum cross-transferability of SSR markers was found in L. acutangula (68%).
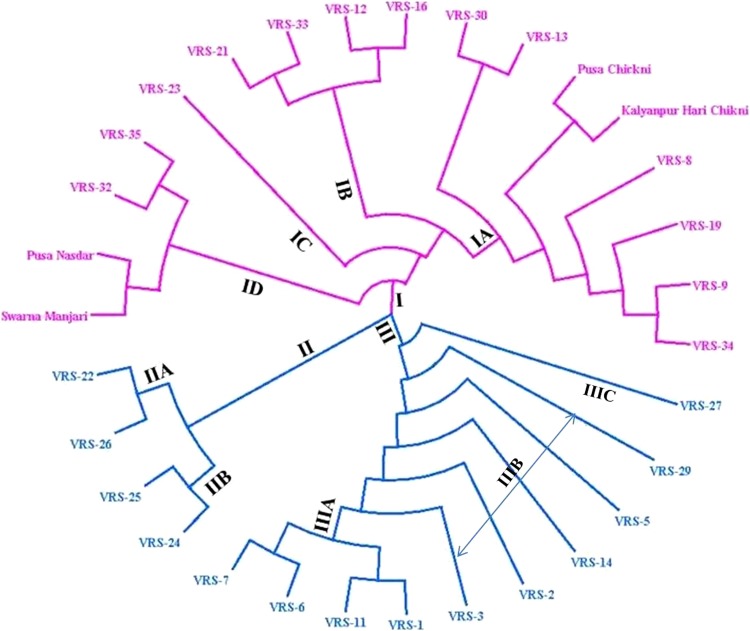
The Jaccard’s coefficient of similarity ranged from 0.66 (VRS-5 vs VRS-6) to 0.97 (VRS-33 vs VRS-26) with an average of 0.81 (Supplementary Table S2). Based on Jaccard’s coefficient of similarity, a dendrogram of 31 Luffa genotypes was constructed (Fig. 1). Thirty-one Luffa genotypes were grouped into three major clusters. Cluster I consisted of 17 genotypes, including 13 genotypes of L. hermaphrodita and two genotypes each of L. aegyptiaca and L. acutangula. Cluster II (04) and III (10) solely consisted of L. hermaphrodita genotypes (Fig. 1).
Fig. 1.
UPGMA dendrogram of 31 genotypes of Luffa based on the 56 polymorphic SSR markers
The major cluster I was further sub divided into four subclusters: hermaphrodite Luffa genotypes VRS-34, VRS-9, VRS-19, VRS-8, VRS-13, VRS-30, and two genotypes of L. aegyptiaca formed cluster IA, while VRS-16, VRS12, VRS-33, VRS-21 formed Cluster IB, one genotypes VRS-23 formed Cluster IC and VRS-35, VRS-32, and two genotypes of L. acutangula made Cluster ID. Cluster II was further sub divided into cluster IIA with VRS-22 and VRS-26 and cluster IIB with VRS-25 and VRS-24.Cluster III was further subdivided into three subclusters, i.e., IIIA (04 genotypes; VRS-7, VRS-6, VRS-11 and VRS-1), IIIB (05 genotypes; VRS-3, VRS-2, VRS-14, VRS-5, and VRS-29) and IIIC (01 genotype; VRS-27), (Fig. 1).
Analysis of population structure
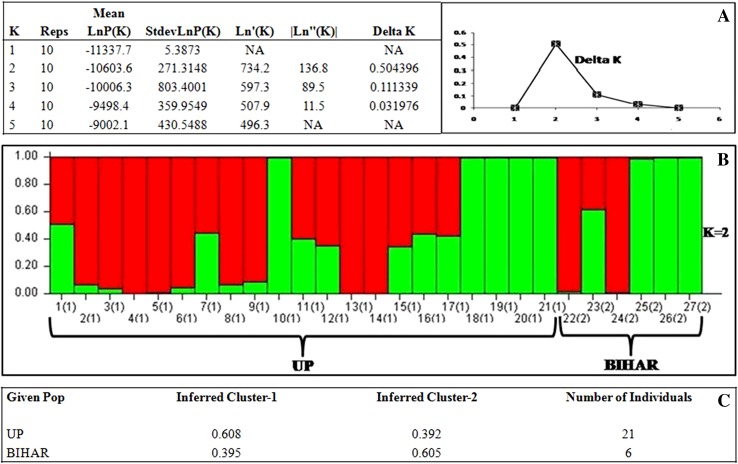
A genetic structural analysis using Bayesian model was done by software STRUCTURE v. 2.2. For each genotype, the proportion of its genome derived from different clusters was estimated and genotypes were assigned to a cluster when 60% or more of their inferred genome belongs to the cluster. Genotypes with lower percentage were considered to be admixed. STRUCTURE software overestimates the number of sub-populations for inbred lines of self-pollinated species (Pritchard and Wen 2003), it is difficult to choose the “correct” K from LnP(K). However, the second-order likelihood delta K was also calculated following (Evanno et al. 2005). The average LnP(K) value increased with increasing value of K and it decreased after K = 2 (Fig. 2a). Thus, the Evanno test of 27 genotypes of hermaphrodite Luffa resulted into two sub-populations and these two sub-populations identified by STRUCTURE mainly reflected their ancestral origins (Fig. 2b). Sub-population I (UP-Uttar Pradesh) included 21 genotypes collected from six districts of UP (Fig. 2b), while sub-population II (Bihar) included 06 genotypes collected from three districts of Bihar. However, using a threshold of > 60% for the designation of group representative, 13 genotypes out of 21 genotypes were assigned to UP population and the remaining 08 genotypes (~ 39%) were assigned as admixed genotypes. Similarly, 04 genotypes out of 06 were assigned to Bihar population and remaining two genotypes were considered as admixed (Fig. 2c).
Fig. 2.
Graphical representation of genetic structure of Indian Luffa spp. and the out group inferred from a combined analysis of SSR markers using the Bayesian clustering software STRUCTURE ver. 2.3. a Evanno test results showing estimated delta K. b Bar plots showing representation of ancestry membership coefficients of all individuals in the two inferred genetic clusters. c The average membership fractions of the individuals from the taxa of Luffa spp. in the two inferred genetic clusters
Eventually, the proportion of assignment of each assumed population to each of the inferred genetic clusters showed a weak structuring pattern. All the two assumed sub-populations had 0.608–0.395% of the ancestry membership to one of the two genetic clusters (Fig. 2c). In summary, the hermaphrodite Luffa genotypes clustered into two populations which generally reflect the ancestral origin of the accessions including the use of smaller genotypes by growers, as supported by the co ancestry values and the influence of geographic distribution.
Results of AMOVA revealed negative variance components and association fixation in between two sub-populations, thus indicated the absence of genetic structure (Table 3). However, the same was positive within population, indicating that there was a weak genetic structure in whole population. A negative or nearly zero (− 0.04) value of F ST obtained in the present study indicates less or no heterozygous genotypes present in these two populations (Table 3).
Table 3.
Summary of AMOVA, fixation index and specific fixation index for Luffa hermaphrodita populations estimated from SSR markers using ARLEQUIN 3.5
| Source of variation | df | Sum of squares | Variance components | Percentage of variation | F ST | Population specific F ST |
|---|---|---|---|---|---|---|
| Among populations | 1 | 40.021 | − 1.98304 Va | − 3.51 | − 0.04 | − 0.04 (UP) − 0.01 (BIHAR) |
| Within populations | 25 | 1463.238 | 58.52952 Vb | 103.51 | ||
| Total | 26 | 1503.259 | 56.54649 |
Discussion
The expression of genetic diversity in hermaphrodite Luffa genotypes makes it possible to develop breeding strategies for high yielding cultivar of hermaphrodite Luffa (Choudhary et al. 2011). Efforts have been made to speed up genetic improvement by studying morphological diversity in hermaphrodite Luffa (Choudhary et al. 2011) and through genetic diversity based on RAPD and ISSR markers in Luffa acutangula (Hoque and Rabbani 2009). However, there is no report of genetic diversity study in hermaphrodite Luffa using SSR markers. Detailed knowledge of genetic relationship among the genotypes underlying variation based on SSR markers is required to partition the genetic diversity and to study the population structure for an efficient breeding efforts in hermaphrodite Luffa.
Germplasm characterization is an important component of breeding programs for an effective management and utilization of plant genetic resources. The germplasm used in the present study showed a wide variation in terms of quantitative traits. Two genotypes, VRS-1 and VRS-11, showed maximum potential for yield due to high fruit weight and numbers. Genotype VRS-1 had been recommended by the Institute Technology Identification Committee for commercial cultivation in UP, Jharkhand and Bihar states of India. The genotype, VRS-1 could be utilized in future breeding programs for development of cultivars in hermaphrodite Luffa having more number of fruit, sequential and cluster bearing fruiting, and early harvest.
The present study successfully amplified a large number of SSR markers (54.36%) out of the total used to be amplified across the Luffa spp. Percentage of transferability of SSRs is very much similar to transferability in watermelon (57.1%), pumpkin (53.6%), and gourds, (60.7%), but much lower as compared to melons (92.9%) (Hu et al. 2010). In general, successful cross-species amplification of SSRs occurs in relative species within the genus, but in the present study, a high percentage of a cross-genus amplification (> 50%) demonstrated that used SSR markers have a high transferability across genus.
In the present study, 56 polymorphic SSR loci were used to quantify the genetic diversity of Luffa genotypes. A high percentage of fixed alleles were observed in the sub-populations identified by the Bayesian and similarity methods. A total of 197 SSR alleles were found with a mean of 3.52 alleles per polymorphic SSR marker with an amplification product size ranged from 105 to 500 bp. Similar SSR product size range of 101–609 bp has been reported in gherkin (Matsumoto et al. 2012). In the present study, the mean polymorphic content (PIC) was 0.55, which is smaller as obtained in cucumber (0.664) with different set of gSSR (Hu et al. 2011). The Jaccord’s similarity coefficient ranged from 0.64 to 0.91 with a mean of 0.77. The value of genetic similarity ranged between 0.542 and 0.941 with a mean of 0.792 as calculated from eSSR by Hu et al. (2010) in cucumber. Out of 27 accessions of Luffa, hermaphrodita 24 accessions grouped in major cluster I, which indicated a low level of genetic diversity in L. hermaphrodita collection based on molecular markers. Morphologically, the two cultivated species L. aegyptiaca and L. acutangula closely resembled each other, except for ridges on the fruit. However, it is worth to mention that, these two species grouped separately in cluster I. The present study also indicated that accessions collected from different districts of UP are genetically diverse. Genetic diversity analysis using molecular markers is very useful for an effective and efficient management of plant genetic resources and conservation priority in cucurbits (Pandey et al. 2013).
In situ population studies of crops are essential for the conservation of plant genetic resources, especially in regions with high genetic diversity (Adugna et al. 2013). The level of population structure in a species has implications for the design and analysis of association mapping studies (Newell et al. 2010). The sub-population identified by STRUCTURE analysis was very close to UPGMA based clustering. The combination of these two methods revealed such information that could be a much better option as compared to similarity-based grouping methodologies alone. Though, three major groups found through UPGMA as compared to two groups in STRUCTURE analysis was due to intermediate type of genotypes generated through out crossing.
Divergent selection, which could play a major role in the production of locally adapted populations, can be attained by measures of population genetic differentiation (McAssey et al. 2016). The lack of a clear geographic signal among Luffa genotypes of UP and Bihar suggests a history of extensive gene flow across the native range of satputia landraces. This may be due to unusual distribution of this species via human and other means (Asch 1993). Our results also revealed an evidence of genetic substructure within the hermaphrodite Luffa collection. Despite the occurrence of significant population genetic structure consisted of two groups, the UPGMA neighbour-joining tree showed no discernible phylogenetic structure amongst accession, suggesting that there is no clear cut division within the Luffa gene pool. Similar report has been earlier published by Mandel et al. (2011). AMOVA analysis revealed negative variance component and similar result in the case of F ST. This signifies the much inbreeding in L. hermaphrodita for which there is rare heterozygous genotypes in these two populations.
This study is the first such effort that described the genetic relationship among Luffa species using SSRs and transferability of cucumber SSR markers to Luffa species. Genetic structure revealed that different Luffa species are differentiated in due course of evolution, but genetic diversity is narrow. A more detailed study with larger representation of wild and cultivated taxa/landraces required for exact relationship among different Luffa species. The present finding evidences the applicability of SSR markers for diversity analysis and characterization of the Luffa germplasm.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1 Distribution of Luffa species genotypes as per geographic origin. Each collection site is indicated by a dot. (JPEG 42 kb)
Acknowledgements
Authors are thankful to the Director, ICAR-Indian Institute of Vegetable Research for providing necessary facilities in the field and laboratory to carry out this work.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1030-0) contains supplementary material, which is available to authorized users.
References
- Adugna A, Snow AA, Sweeney PM, Bekele E, Mutegi E. Population genetic structure of in situ wild Sorghum bicolor in its Ethiopian center of origin based on SSR markers. Genet Resour Crop Evol. 2013;60:1313–1328. doi: 10.1007/s10722-012-9921-8. [DOI] [Google Scholar]
- Asch DL (1993) Common sunflower (Helianthus annuus L.): the pathway toward domestication. In: Proceedings of the 58th annual meeting of the society for American archaeology, St. Louis, pp 1–15
- Chakravarty HL (1982) Fascicles of flora of India: fascicle II. Cucurbitaceae, Botanical Survey of India, West Bengal, India, pp 85–116
- Chandra U. Distribution, domestication and genetic diversity of Luffa gourd in Indian sub-continent. Indian J Plant Genet Resour. 1995;8:189–196. [Google Scholar]
- Choudhary BR, Pandey S, Singh PK, Singh R. Genetic divergence in hermaphrodite ridge gourd (Luffa acutangula) Veg Sci. 2011;38:68–72. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque S, Rabbani MG. Assessment of genetic relationship among landraces of Bangladeshi ridge gourd (Luffa acutangula Roxb.) using RAPD markers. J Sci Res. 2009;1:615–623. doi: 10.3329/jsr.v1i3.1968. [DOI] [Google Scholar]
- Hu J, Li J, Liang F, Liu L, Si S. Genetic relationship of a cucumber germplasm collection revealed by newly developed EST-SSR markers. J Genet. 2010;89:e28–e32. [PubMed] [Google Scholar]
- Hu J, Wang L, Li J. Comparison of genomic SSR and EST-SSR markers for estimating genetic diversity in cucumber. Biol Plant. 2011;55:577–580. doi: 10.1007/s10535-011-0129-0. [DOI] [Google Scholar]
- Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vard Sci Nat. 1908;44:223–270. [Google Scholar]
- Mandel JR, Dechaine JM, Marek LF, Burke JM. Genetic diversity and population structure in cultivated sunflower and a comparison to its wild progenitor, (Helianthus annuus L.) Theor Appl Genet. 2011;123:693–704. doi: 10.1007/s00122-011-1619-3. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Watanabe N, Kuboyama T. Cross-species transferability of 86 cucumber (Cucumis sativus L.) microsatellite markers to gherkin (C. anguria L.) Sci Hortic. 2012;136:110–114. doi: 10.1016/j.scienta.2012.01.009. [DOI] [Google Scholar]
- McAssey EV, Corbi J, Burke JM. Range-wide phenotypic and genetic differentiation in wild sunflower. BMC Plant Biol. 2016;16:1–11. doi: 10.1186/s12870-016-0937-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Srivastava AK, Verma S, Pandey S, Bargali SS, Rana TS, Nair KN. Phenetic and genetic diversity in Indian Luffa (Cucurbitaceae) inferred from morphometric, ISSR and DAMD markers. Genet Resour Crop Evol. 2016;123:1–16. [Google Scholar]
- Newell MA, Cook D, Tinker NA, Jannink JL. Population structure and linkage disequilibrium in oat (Avena sativa L.): implications for genome-wide association studies. Theor Appl Genet. 2010;122:623–632. doi: 10.1007/s00122-010-1474-7. [DOI] [PubMed] [Google Scholar]
- Pandey S, Ansari WA, Mishra VK, Singh AK, Singh M. Genetic diversity in Indian cucumber based on microsatellite and morphological markers. Biochem Syst Ecol. 2013;51:19–27. doi: 10.1016/j.bse.2013.08.002. [DOI] [Google Scholar]
- Papanicolaou GC, Psarra E, Anastasiou D. Manufacturing and mechanical response optimization of epoxy resin/Luffa cylindrica composite. J Appl Polym Sci. 2015;132:41992. [Google Scholar]
- Prakash K, Pandey A, Radhamani J, Bisht IS. Morphological variability in cultivated and wild species of Luffa (Cucurbitaceae) from India. Genet Resour Crop Evol. 2013;60:2319–2329. doi: 10.1007/s10722-013-9999-7. [DOI] [Google Scholar]
- Prakash K, Pati K, Arya L, Pandey A, Verma M. Population structure and diversity in cultivated and wild Luffa species. Biochem Syst Ecol. 2014;56:165–170. doi: 10.1016/j.bse.2014.05.012. [DOI] [Google Scholar]
- Pritchard JK, Wen W (2003) Documentation for structure software : version 2. Available from http://pritch.bsd.uchicago.edu. Accessed 19 July 2002
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Zhang ZH, Liu JH, Staub JE, Han YH, et al. An integrated genetic and cytogenetic map of the cucumber genome. PLoS One. 2009;4:57–95. doi: 10.1371/annotation/bbfdac40-32cc-4c3a-a049-436796875bf4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf FJ (1998) NTSYS-pc. Numerical taxonomy and multivariate analysis system, version 2.02 Exeter software, Setauket, New York, USA
- Schilling EE, Heiser CB. Flavonoides and systematics of Luffa. Biochem Syst Ecol. 1981;9:263–285. doi: 10.1016/0305-1978(81)90006-5. [DOI] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. ARLEQUIN Ver. 2000. A software population genetics data analysis. Geneva: Genetics and Biometry Laboratory, University of Geneva; 2000. [Google Scholar]
- Sikdar B, Bhattacharya M, Mukherjee A, Banerjee A, Ghosh E, Ghosh B, Roy SC. Genetic diversity in important members of Cucurbitaceae using isozyme, RAPD and ISSR markers. Biol Plant. 2010;54:135–140. doi: 10.1007/s10535-010-0021-3. [DOI] [Google Scholar]
- Varshney RK, Graner A, Sorrells MW. Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005;23:48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bernard M, Leroy P, Feuillet C, Sourdille P. High transferability of bread wheat EST-derived SSRs to other cereals. Theor Appl Genet. 2005;111:677–687. doi: 10.1007/s00122-005-2041-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Distribution of Luffa species genotypes as per geographic origin. Each collection site is indicated by a dot. (JPEG 42 kb)