Abstract
Heat-stroke is a serious form of hyperthermia with high mortality, and can induce severe central nervous system disorders. The neurovascular unit (NVU), which consists of vascular cells, glial cells, and neurons, controls blood-brain barrier (BBB) permeability and cerebral blood flow, and maintains the proper functioning of neuronal circuits. However, the detailed function of each BBB component in heat-stroke remains unknown. In order to interpret alterations caused by heat stress, we performed transcriptome comparison of neuron and astrocyte primary cultures after heat treatment. Differentially-expressed genes were then selected and underwent Gene Ontology annotation and Kyoto Encyclopedia of Genes and Genomes pathway analysis. Gene-act networks were also constructed, and the expression of pivotal genes was validated by quantitative PCR, as well as single-cell qPCR in heat-stroke rats. Our work provides valuable information on the transcriptional changes in NVU cells after heat stress, reveals the diverse regulatory mechanisms of two of these cellular components, and shows that a cell-type-specific approach may be a promising therapeutic strategy for heat-stroke treatments.
Electronic supplementary material
The online version of this article (doi:10.1007/s12264-017-0156-8) contains supplementary material, which is available to authorized users.
Keywords: Heat stroke, RNA-seq, Neuron, Astrocyte, Primary culture, Single-cell qPCR
Introduction
Heat-stroke is a life-threatening illness characterized by an elevated core body temperature that rises above 40 °C and central nervous system (CNS) dysfunction that results in delirium, convulsions, or coma [1]. Multiple organ dysfunction syndromes can occur in patients with severe heat-stroke, and its mortality is particularly high. During the 2003 heat-wave event in Europe, the high temperature caused an estimated 30,000 deaths [2]. Heat-stroke can induce severe CNS injury which is characterized by cognitive and motor deficits, and permanent brain damage and coma [3]. The CNS, which is particularly vulnerable to direct thermal effects, can also be injured by disruption of the blood-brain barrier (BBB) and brain edema [4].
Heat-stroke results from exposure to a high environmental temperature (in which case it is called classic, or nonexertional, heat-stroke) or from strenuous exercise (in which case it is called exertional heat-stroke). Global warming is already causing heat-waves in temperate climates, and the threat of heat-stroke is increasing [1]. Greater knowledge of the cellular and molecular responses to heat stress will help point to novel preventive measures and a new paradigm of immunomodulation.
The neurovascular unit (NVU) comprises vascular cells (endothelium, pericytes, and vascular smooth muscle cells), glial cells (astrocytes, microglia, and oligodendroglia), and neurons [5]. The NVU controls BBB permeability and cerebral blood flow, and maintains the chemical composition of the neuronal milieu, which is required for the proper functioning of neuronal circuits. Heat stress induces cerebral ischemia and BBB disruption that result in extravasation of substances into the brain extracellular compartment causing vasogenic edema and cell injury [6]. As the most abundant cell type within the CNS, astrocytes play essential roles in maintaining normal brain function. They are a critical structural and functional part of the tripartite synapses and the NVU, and communicate with neurons, oligodendrocytes, and endothelial cells [7]. It is known that after an ischemic stroke, astrocytes perform multiple functions, both detrimental and beneficial for neuronal survival, during the acute phase. However, the role of astrocytes in heat stress-induced BBB and cerebral damage remains unknown. And the cellular and molecular changes of each BBB component are complex, and insufficiently understood. Thus it is urgent to obtain a global perspective on the changes in each BBB component following heat stress.
In this study, we investigated the transcriptional profiles of neuron and astrocyte cultures after heat stress.
Materials and Methods
Neuron and Astrocyte Cultures
Cortical neurons were prepared from Sprague-Dawley rats on embryonic day (E) 18. After treatment with 0.1 mg/mL trypsin (Thermo Fisher Scientific, Waltham, MA) and 0.6 μg/mL DNase (Sigma-Aldrich, St. Louis, MO) for 10 min at 37 °C and mechanical dissociation, cortical cells were plated at 5×105 cells/mL on culture dishes coated with 0.1 mg/mL poly-DL-lysine. The cells were cultured in neurobasal medium containing 2% B27 supplement (Thermo Fisher Scientific, Waltham, MA), 0.5 mmol/L glutamine, and 1% penicillin-streptomycin, under 5% CO2/10% O2 at 37 °C. Forty-eight hours later, 20 μg/mL 5-fluorodeoxyuridine was added to the medium to inhibit the growth of glial cells.
Rat astrocytes were purified from E18 Sprague-Dawley rat forebrains and cultured. Briefly, cortices were enzymatically then mechanically dissociated to generate a single-cell suspension. Cortical cells were incubated in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. When the cells covered 50%–60% of the dishes, oligodendrocytes were removed by shaking at 100 rpm for 3 h at 22 °C.
All animal experiments complied with the Animal Research Reporting of In Vivo Experiments guidelines and were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978). Approval was obtained from the Animal Care Committee of General Hospital of Jinan Military Region.
High Temperature Exposure
The heat stressed neuron and astrocyte cultures were placed in a 5% CO2 incubator adjusted to 38 °C, 39 °C, 40 °C, or 42 °C for 30 min on days 7–10. After the heat stress exposure, the cultures were moved back to a 37 °C incubator to recover for 2 h. Control neuron and astrocyte cultures remained in the 37 °C incubator throughout. After recovery, the cultures were collected in TRIzol reagent (Thermo Fisher Scientific, Waltham, MA).
Heat-Stroke Rats and Single-Cell Transcriptional Profiling
Adult male Sprague-Dawley rats (8 weeks old) were housed in individual Plexiglas cages in an animal room maintained under a constant light-dark cycle (lights on from 07:00 to 19:00), temperature (22 ± 1 °C), and humidity (45%–55%). Food and water were provided ad libitum. To obtain heat-stroke rats, conscious, unrestrained rats were exposed to an ambient temperature of 40 °C and 60% humidity in a floor-standing incubator, in the absence of food and water, until a core body temperature of 40 °C was reached (~100 min). Control rats were placed in the same incubator at an ambient temperature of 22 °C and 60% humidity for 100 min.
Then the rats were deeply anesthetized with 10% chloral hydrate. Upon sacrifice, the hypothalamus was freshly dissected and dissociated with a Neural Tissue Dissociation Kit (Miltenyi Biotec, Shanghai, China). The isolated cells of choice were collected under a microscope. Then samples underwent reverse transcription and target-specific amplification, followed by quantitative PCR (qPCR) as previously reported [8].
RNA Extraction, Reverse Transcription, and Quantitative PCR
Total cellular RNA was isolated from cultures with the Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA). Reverse transcription was performed with the ABM 5× All-In-One RT MasterMix kit (Applied Biological Materials, Waltham, MA). The qPCR was performed with the KAPA SYBR Fast qPCR Master Mix kit (Kapa Biosystems, Wilmington, MA). The primers used for qPCR are listed in Supplemental Table S1.
RNA-seq and Bioinformatics Analysis
RNA extracted from neuron and astrocyte cultures was used to prepare mRNA libraries using the KAPA Stranded mRNASeq Kit (Illumina platform, Kapa Biosystems, Wilmington, MA). The RNA-seq libraries were sequenced on the Hiseq2500 platform. Raw sequencing data were first evaluated by FastQC [9], and then mapped to the Rattus norvegicus genome (rn6) using Tophat 2.0 [10] based on Bowtie2 [11]. The read count for each gene was obtained from the mapping results using Cufflinks [12] and normalized to reads per kilobase of exon model per million mapped reads (RPKM). The read counts were normalized with limma [13] and analyzed with edgeR [14]. Differentially expressed genes (DEGs) between the heat-treated samples and controls were determined with FDR (false discovery rate) <0.1 and fold change >1.5 or <1/1.5. Heatmap and Hierarchical Clustering analyses were performed with MultiExperiment Viewer (MeV) 4.9 [15]. GO analyses were performed with DAVID Bioinformatics Resources 6.8 [16, 17] and WEGO [18]. Pathway enrichment analyses were performed with Enrichr [19, 20]. Gene-act network analyses were performed with STRING [21] and Cytoscape [22]. All raw sequence data have been submitted to the Sequence Read Archive of the National Institutes of Health (USA) (accession number: SRP093962).
Statistical Analysis
All qPCR data are presented as mean ± SEM. Student’s t-test, the Mann-Whitney rank sum test, or one-way analysis of variance (ANOVA) was used for the statistical analysis of qPCR data. Student-Newman-Keuls’s post hoc analysis was performed after one-way ANOVA.
Results
40 °C and 42 °C Stress Increase HSP70 Transcription
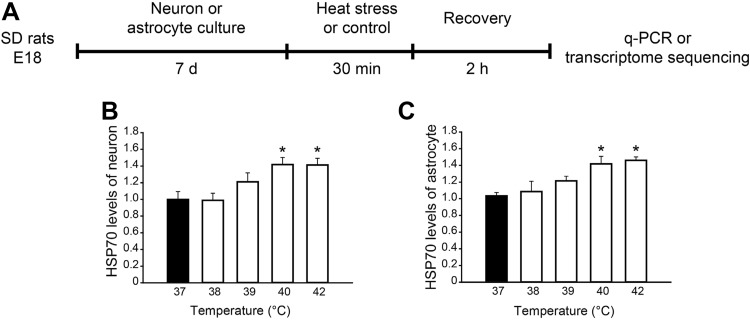
To explore the responses of neurons and astrocytes to high temperature, cultures were exposed to heat stress at 38 °C, 39 °C, 40 °C, and 42 °C, and then the transcription levels of heat shock protein HSP70, a marker of the heat stress response [23], were assessed with qPCR (Fig. 1). At 40 °C and 42 °C, the transcription of HSP70 in neurons and astrocytes increased significantly compared with 37 °C controls (Fig. 1B and C), indicating that they undergo remarkable responses to heat stress at 40 °C and above.
Fig. 1.
Experimental design of high temperature exposure. A Experimental protocol. B, C Fold changes of HSP70 transcription in neuron and astrocyte cultures exposed to different temperatures (n = 3/group). For neurons, F(4, 10) = 5.312, *P = 0.015 (one-way ANOVA); 40 °C vs 37 °C *P = 0.038, 42 °C vs 37 °C *P = 0.024 (Student’s-Newman-Keuls post hoc analysis). For astrocytes, F(4, 10) = 5.700, *P = 0.012 (one-way ANOVA); 40 °C vs 37 °C *P = 0.028, 42 °C vs 37 °C *P = 0.038 (Student’s-Newman-Keuls post hoc analysis).
Heat Stress Induces Transcriptional Regulation in Neurons and Astrocytes
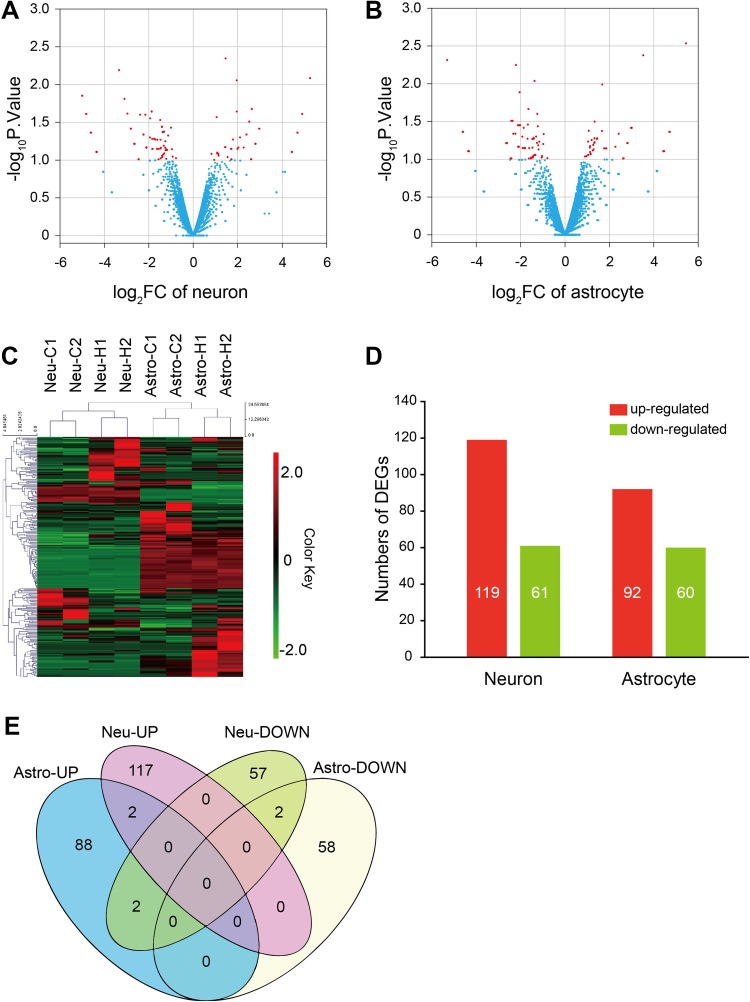
To investigate the transcriptional events in neurons and astrocytes exposed to high temperature, the cultures were stimulated at 40 °C and harvested to extract total RNA samples. Transcriptional profiles were detected by next-generation sequencing, and the DEGs between heat-treated samples and controls were determined (Fig. 2A–C). After heat stress, 119 genes were up-regulated in neurons, while 61 were down-regulated. As for astrocytes, 92 genes were up-regulated, while 60 were down-regulated (Fig. 2D). The transcriptional regulation of neurons and astrocytes differed greatly (Fig. 2C and E). Most DEGs were different in neurons and astrocytes, except for 6 genes, among which Apobec2 and ENSRNOG00000001066 were up-regulated in both, C6 and Ccl3 were down-regulated in neurons but up-regulated in astrocytes, and LOC689081 and ENSRNOG00000046059 were down-regulated in both. These results suggested that high-temperature stress activates different regulatory mechanisms in neurons and astrocytes, which may play different roles in the damage induced by heat exposure.
Fig. 2.
Gene transcription changes after high temperature exposure. A, B Volcano plots of DEGs of cultured neurons and astrocytes. Red dots indicate significant DEGs. C Heatmap showing hierarchical clustering of DEGs. D Summary of the significant DEGs in cultured neurons and astrocytes. E Venn diagram of cell-type-specific DEGs in cultured neurons and astrocytes.
Heat Stress Alters Diverse Pathways in Neurons and Astrocytes
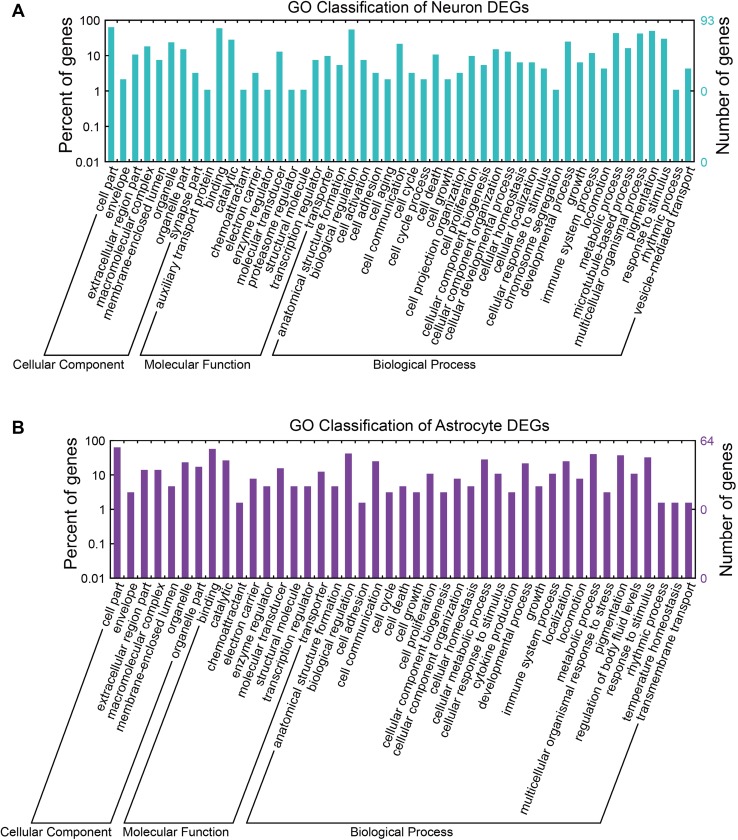
To investigate the regulatory mechanisms in neurons and astrocytes during heat stress, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Genes regulated in neurons and astrocytes were associated with important cellular components and processes (Fig. 3A and B). To gain further insight into the biological functions of the DEGs, functional annotation clustering was performed using DAVID 6.8. The neuronal DEGs were divided into 12 clusters (Table S2). The main enriched clusters were hemostasis and blood coagulation (PROCR, FGA, and F9), vision and sensory transduction (ARR3, PDC, RCVRN, RHO, and OLR1029), and oxidation-reduction process (NOX4, FMO1, AASS, DCXR, CYBB, RASSF9, and AOC3). On the other hand, the astrocytic DEGs were divided into 9 clusters (Table S3), and the main enriched clusters were iron ion binding and oxidoreductase (CYP2AC1, FAR2, CYP2J16, CYP2D5, DOXL1, etc.), leukocyte chemotaxis and cytokine-cytokine signaling (CCL12, CCL3, PTGIR, CCR1, CRHBP, CXCL2, CCL4, etc.), and transmembrane related process (MILL1, CYP2D5, SYNDIG1, RGD1304770, NPY2R, etc.).
Fig. 3.
Gene Ontology (GO) for genes regulated following heat stress. The GO terms cellular component, molecular function, and biological process for neurons (A) and astrocytes (B).
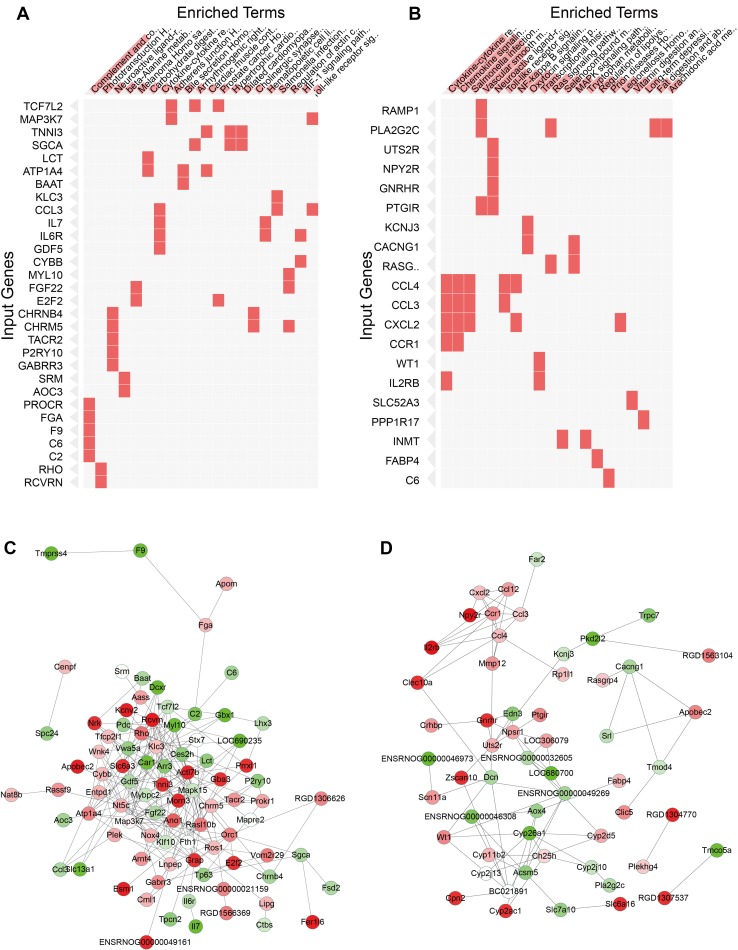
Meanwhile, the most enriched pathways in neurons in the Enrichr KEGG pathway analysis were complement and coagulation cascades (FGA, PROCR, F9, C6, and C2), phototransduction (RCVRN and RHO), and neuroactive ligand-receptor interaction (GABRR3, P2RY10, CHRNB4, CHRM5, and TACR2) (Fig. 4A, Table 1), indicating that signal transduction associated with complement and coagulation cascades is activated after heat stress in neurons. In contrast, the most enriched pathways in astrocytes were cytokine-cytokine receptor interaction and chemokine signaling pathways (CCR1, IL2RB, CCL4, CCL3, and CXCL2). The Toll-like receptor signaling pathway and NF-κB signaling pathway were also included (CCL4, CCL3, and CXCL2; Fig. 4B, Table 2), which suggested that astrocytes might play a role in the inflammatory reaction and NF-κB signaling-induced cellular responses.
Fig. 4.
KEGG pathway enrichment analyses and protein-protein interaction networks of DEGs. A,B Pathway terms with high Enrichr enrichment scores of DEGs between control and heat-treated cultures of neurons (A) and astrocytes (B) are shown on the horizontal coordinates. Input genes were permuted based on KEGG clustering. C, D Protein-protein interaction network analyses of DEGs of neurons (C) and astrocytes (D) after heat stress. Green, down-regulated genes; red, up-regulated genes. The densities of node colors indicate levels of down-regulation or up-regulation.
Table 1.
KEGG pathway analysis of neuronal DEGs.
| ID | Term | KEGG ID | Overlap | P-value | Adjusted P-value | Z-score | Combined Score | Genes |
|---|---|---|---|---|---|---|---|---|
| 1 | Complement and coagulation cascades | hsa04610 | 5/79 | 0.0002 | 0.0274 | −1.824 | 6.559 | Fga, Procr, F9, C6, C2 |
| 2 | Phototransduction | hsa04744 | 2/27 | 0.0157 | 0.6136 | −1.96 | 0.957 | Rcvrn, Rho |
| 3 | Neuroactive ligand-receptor interaction | hsa04080 | 5/277 | 0.0384 | 0.6136 | −1.821 | 0.89 | Gabrr3, P2ry10, Chrnb4, Chrm5, Tacr2 |
| 4 | Beta-alanine metabolism | hsa00410 | 2/31 | 0.0201 | 0.6136 | −1.785 | 0.872 | Aoc3, Srm |
| 5 | Melanoma | hsa05218 | 2/71 | 0.0847 | 0.6136 | −1.686 | 0.823 | E2f2, Fgf22 |
| 6 | Carbohydrate digestion and absorption | hsa04973 | 2/45 | 0.0388 | 0.6136 | −1.674 | 0.818 | Atp1a4, Lct |
| 7 | Cytokine-cytokine receptor interaction | hsa04060 | 4/265 | 0.1029 | 0.6136 | −1.588 | 0.776 | Il7, Ccl3, Gdf5, Il6r |
| 8 | Adherens junction | hsa04520 | 2/74 | 0.0907 | 0.6136 | −1.568 | 0.766 | Tcf7l2, Map3k7 |
| 9 | Bile secretion | hsa04976 | 2/71 | 0.0847 | 0.6136 | −1.531 | 0.748 | Atp1a4, Baat |
| 10 | Arrhythmogenic right ventricular cardiomyopathy | hsa05412 | 2/74 | 0.0907 | 0.6136 | −1.475 | 0.721 | Tcf7l2, Sgca |
| 11 | Cardiac muscle contraction | hsa04260 | 2/78 | 0.099 | 0.6136 | −1.443 | 0.705 | Atp1a4, Tnni3 |
| 12 | Prostate cancer | hsa05215 | 2/89 | 0.1226 | 0.6136 | −1.438 | 0.703 | Tcf7l2, E2f2 |
| 13 | Hypertrophic cardiomyopathy | hsa05410 | 2/83 | 0.1095 | 0.6136 | −1.425 | 0.696 | SgcA, Tnni3 |
| 14 | Dilated cardiomyopathy | hsa05414 | 2/90 | 0.1248 | 0.6136 | −1.322 | 0.646 | Sgca, Tnni3 |
| 15 | Cholinergic synapse | hsa04725 | 2/111 | 0.1732 | 0.6136 | −1.29 | 0.63 | Chrnb4, Chrm5 |
| 16 | Hematopoietic cell lineage | hsa04640 | 2/88 | 0.1204 | 0.6136 | −1.29 | 0.63 | Il7, Il6r |
| 17 | Salmonella infection | hsa05132 | 2/86 | 0.116 | 0.6136 | −1.249 | 0.61 | Klc3, Ccl3 |
| 18 | Regulation of actin cytoskeleton | hsa04810 | 3/214 | 0.1749 | 0.6136 | −1.228 | 0.6 | Chrm5, Myl10, Fgf22 |
| 19 | HIF-1 signaling pathway | hsa04066 | 2/103 | 0.1544 | 0.6136 | −1.198 | 0.585 | Cybb, Il6r |
| 20 | Toll-like receptor signaling pathway | hsa04620 | 2/106 | 0.1614 | 0.6136 | −1.196 | 0.584 | Ccl3, Map3k7 |
Table 2.
KEGG pathway analysis of astrocytic DEGs.
| ID | Term | KEGG ID | Overlap | P-value | Adjusted P-value | Z-score | Combined Score | Genes |
|---|---|---|---|---|---|---|---|---|
| 1 | Cytokine-cytokine receptor interaction | hsa04060 | 5/265 | 0.0045 | 0.1614 | −1.877 | 3.424 | Ccr1, Il2rb, Ccl4, Ccl3, Cxcl2 |
| 2 | Chemokine signaling pathway | hsa04062 | 4/187 | 0.0073 | 0.1614 | −1.876 | 3.423 | Ccr1, Ccl4, Ccl3, Cxcl2 |
| 3 | Salmonella infection | hsa05132 | 3/86 | 0.0056 | 0.1614 | −1.695 | 3.092 | Ccl4, Ccl3, Cxcl2 |
| 4 | Vascular smooth muscle contraction | hsa04270 | 3/120 | 0.0137 | 0.2254 | −1.789 | 2.666 | Ptgir, Pla2g2c, Ramp1 |
| 5 | Neuroactive ligand-receptor interaction | hsa04080 | 4/277 | 0.0269 | 0.355 | −1.797 | 1.861 | Uts2r, Ptgir, Npy2r, Gnrhr |
| 6 | Toll-like receptor signaling pathway | hsa04620 | 2/106 | 0.0724 | 0.4737 | −1.685 | 1.259 | Ccl4, Ccl3 |
| 7 | NF-kappa B signaling pathway | hsa04064 | 2/93 | 0.0578 | 0.4737 | −1.625 | 1.214 | Ccl4, Cxcl2 |
| 8 | Oxytocin signaling pathway | hsa04921 | 2/158 | 0.1401 | 0.4737 | −1.579 | 1.18 | Cacng1, Kcnj3 |
| 9 | Transcriptional misregulation in cancer | hsa05202 | 2/180 | 0.1719 | 0.4737 | −1.302 | 0.973 | Wt1, Il2rb |
| 10 | Ras signaling pathway | hsa04014 | 2/227 | 0.243 | 0.4737 | −1.258 | 0.94 | Pla2g2c, Rasgrp4 |
| 11 | MAPK signaling pathway | hsa04010 | 2/255 | 0.2862 | 0.4737 | −1.132 | 0.846 | Cacng1, Rasgrp4 |
| 12 | Tryptophan metabolism | hsa00380 | 1/40 | 0.1561 | 0.4737 | −1.088 | 0.813 | Inmt |
| 13 | Regulation of lipolysis in adipocytes | hsa04923 | 1/56 | 0.2104 | 0.4737 | −1.086 | 0.812 | Fabp4 |
| 14 | Prion diseases | hsa05020 | 1/35 | 0.1384 | 0.4737 | −1.043 | 0.779 | C6 |
| 15 | Legionellosis | hsa05134 | 1/55 | 0.2071 | 0.4737 | −1.036 | 0.774 | Cxcl2 |
| 16 | Selenocompound metabolism | hsa00450 | 1/17 | 0.0717 | 0.4737 | −1.029 | 0.769 | Inmt |
| 17 | Vitamin digestion and absorption | hsa04977 | 1/24 | 0.0982 | 0.4737 | −1.016 | 0.759 | Slc52a3 |
| 18 | Long-term depression | hsa04730 | 1/60 | 0.2235 | 0.4737 | −0.994 | 0.743 | Ppp1r17 |
| 19 | Fat digestion and absorption | hsa04975 | 1/41 | 0.1596 | 0.4737 | −0.934 | 0.698 | Pla2g2c |
| 20 | Arachidonic acid metabolism | hsa00590 | 1/62 | 0.2299 | 0.4737 | −0.912 | 0.682 | Pla2g2c |
Protein-Protein Interaction Network Analysis and qPCR Validation
Protein-protein interaction networks were built with the genes affected by heat exposure in order to investigate the regulatory relationships between them in neurons and astrocytes (Fig. 4C and D). The networks were composed of up-regulated and down-regulated genes. We found that genes involved in important signaling pathways were in the center of the neuronal protein-protein interaction network (Fig. 4C), including MAPK15, WNK4, RHO, ARR3, and ROS1. The most enriched genes in the complement and coagulation cascades (FGA, PROCR, F9, C6, and C2) formed a branch of the network, suggesting this pathway might be an independent regulatory mechanism in neurons after heat stress.
On the other hand, the genes regulated in astrocytes comprised several sub-networks, one of which consisted of genes enriched in cytokine-cytokine receptor interaction, chemokine signaling pathway, Toll-like receptor signaling pathway, and NF-κB signaling pathway (CCR1, IL2RB, CCL4, CCL3, and CXCL2). This supported the point that astrocytes are associated with the inflammatory reaction and NF-κB signaling-induced cellular responses to heat stress.
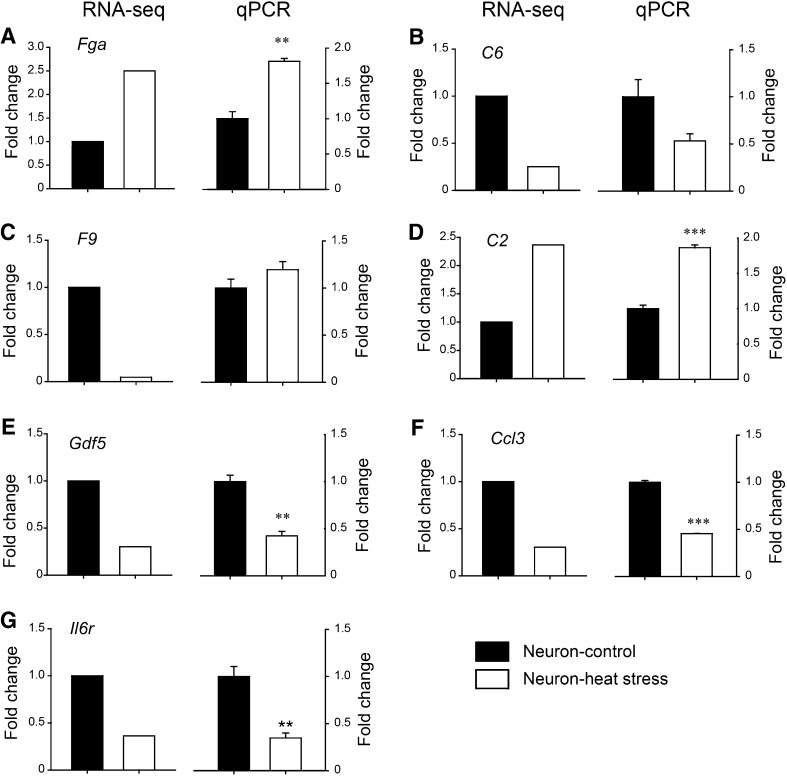
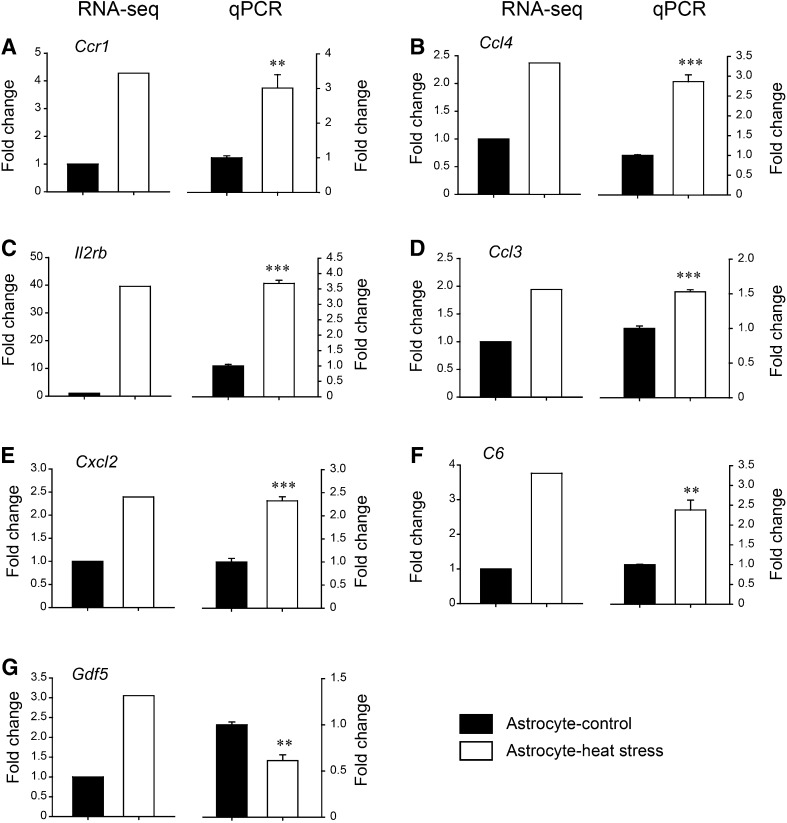
Eleven of the most enriched DEGs were chosen to validate the transcript sequencing results, in which the expression changes were detected using qPCR. The results demonstrated that the transcription levels of Fga and C2 were up-regulated, while C6, Ccl3, Gdf5, and Il6r were down-regulated in neuron cultures, consistent with the RNA-seq data, except for F9 (Fig. 5; P < 0.05 for Fga, C2, Gdf5, Ccl3, and Il6r; P = 0.076 for C6, P = 0.20 for F9 (t-test)). On the other hand, C6 and the inflammation-related genes Ccr1, Ccl4, Il2rb, and Ccl3 were up-regulated in astrocyte cultures, consistent with the RNA-seq data, except for Gdf5 (Fig. 6; P < 0.05 for Ccr1, Ccl4, Il2rb, Ccl3, Cxcl2, C6, and Gdf5 (t-test)). Quantitative PCR analysis was in accordance with the transcriptome sequencing results.
Fig. 5.
Validation of DEGs in heat-stressed neurons. Selected DEGs in neurons were detected by qPCR (n = 3/group) to validate the results of RNA-seq. Left panels: fold changes of RNA-seq; right panels: fold changes in qPCR.: A **P = 0.0017 for Fga, B P = 0.076 for C6, C P = 0.200 for F9, D ***P < 0.001 for C2, E **P = 0.0022 for Gdf5, F ***P < 0.001 for Ccl3, and G **P = 0.0054 for Il6r (Student’s t-test).
Fig. 6.
Validation of DEGs in heat-stressed astrocytes. Selected DEGs in astrocytes were detected by qPCR (n = 3/group) to validate the results of RNA-seq. Left panels: fold changes of RNA-seq; right panels: fold changes in qPCR. A **P = 0.0067 for Ccr1, B ***P < 0.001 for Ccl4, C ***P < 0.001 for Il2rb, D ***P < 0.001 for Ccl3, E ***P < 0.001 for Cxcl2, F **P = 0.0053 for C6, and G **P = 0.0051 for Gdf5 (Student’s t-test).
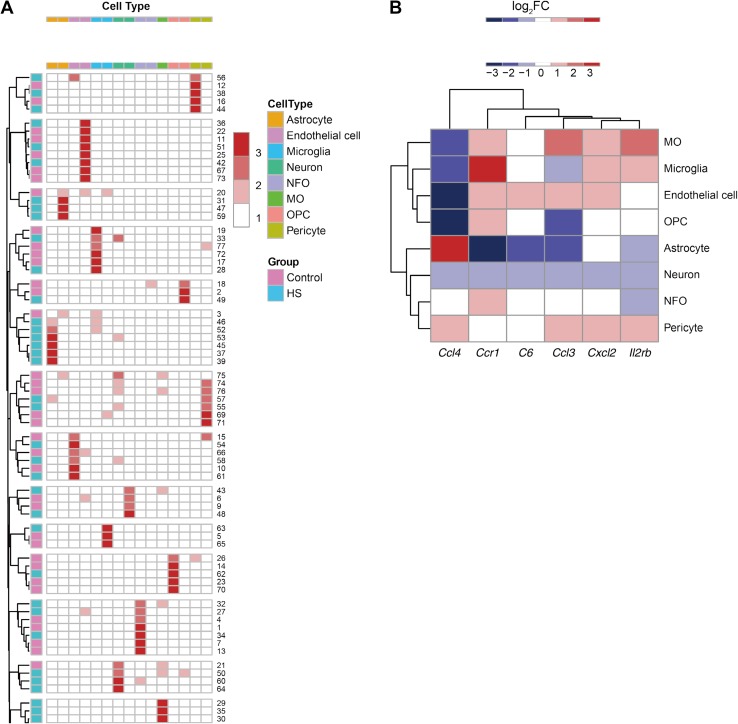
Single-Cell qPCR Reveals Different Roles of Astrocytes In Vitro and In Vivo
Besides astrocytes, the NVU contains other cell types, like microglia, pericytes, and endothelial cells, which may also play roles in brain inflammation. And fetal primary cultures may not be mature and healthy enough to mirror corresponding adult cells in vivo. To investigate the roles of different NVU cells in the adult CNS during heat-stroke, we determined the single-cell transcriptional profiles in the hypothalamus of adult male rats with heat-stroke. Fifteen cell-type-specific markers were chosen to identify the cells that underwent single-cell qPCR, based on a previous report [24] (primers listed in Supplemental Table S1). The transcriptional levels of the markers were assessed in all the cells, and identification was performed (Fig. 7A). Samples with no evident transcription of any marker were removed. Then the pivotal genes were assessed via qPCR. We found that the transcription of C6 and Ccr1 were down-regulated in neurons, as well as C6, Ccl3, and Ccr1 in astrocytes (Fig. 7B and Table 3). The down-regulation of these genes in astrocytes was not in accordance with the situation in primary culture reveled by RNA-seq and qPCR, indicating a complex regulatory mechanism in astrocytes during heat-stroke.
Fig. 7.
Single-cell profiling of heat-stroke rats. The transcriptional levels in the hypothalamus of heat-stroke rats were assessed by single-cell qPCR. A The types of hypothalamic cells were determined by the transcription patterns of cell-type-specific markers. B Heatmap of the fold changes of pivotal genes in hypothalamic cells.
Table 3.
Single-cell transcriptional profiling.
| Genes | Astrocytes | Endothelial cells | Microglia | Neurons | NFO | MO | OPC | Pericytes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| log2FC | P | log2FC | P | log2FC | P | log2FC | P | log2FC | P | log2FC | P | log2FC | P | log2FC | P | |
| C6 | −1.92 | 0.045 * | 0.54 | 0.32 | −0.16 | 0.88 | −1.48 | 0.002 ** | −0.34 | 0.417 | −0.59 | – | −0.37 | 0.631 | −0.51 | 0.66 |
| Ccl3 | −1.86 | 0.048 * | 0.31 | 0.711 | −0.92 | 0.514 | −1.6 | 0.099 | −0.17 | 0.83 | 1.79 | – | −1.8 | 0.384 | 0.27 | 0.704 |
| Ccl4 | 2.14 | 0.409 | −3.73 | 0.36 | −2.21 | 0.35 | −1.57 | 0.065 | 0.07 | 0.914 | −1.99 | – | −3.1 | 0.506 | 0.36 | 0.679 |
| Ccr1 | −2.98 | 0.045 * | 0.29 | 0.616 | 3.05 | 0.196 | −1.44 | 0.044 * | 0.2 | 0.708 | 1.05 | – | 0.74 | 0.226 | −0.13 | 0.877 |
| Cxcl2 | −0.58 | 0.195 | 0.43 | 0.455 | 0.39 | 0.73 | −0.99 | 0.149 | −0.49 | 0.499 | 0.42 | – | −0.74 | 0.541 | 0.95 | 0.421 |
| Il2rb | −1.26 | 0.126 | −0.75 | 0.468 | 0.14 | 0.839 | −1.06 | 0.174 | −1.34 | 0.245 | 1.7 | – | −0.55 | 0.5 | 0.48 | 0.534 |
NFO, newly-formed oligodendrocytes; MO, myelinating oligodendrocytes; OPC, oligodendrocyte precursor cells; FC, fold change; * P < 0.05; ** P < 0.01.
Discussion
The NVU and the BBB are considered to be essential units participating in maintaining the functions of the CNS. A better understanding of the cellular and molecular regulation of the NVU and BBB components after heat exposure is one of the critical steps for new therapeutic strategies in heat-stroke.
In this work, we investigated the transcriptional profiles of neuron and astrocyte cultures after heat stress, and found that they underwent different transcriptional changes. Transcriptome sequencing showed that heat stress caused a number of transcriptional regulations in both neurons and astrocytes. Most of the significant DEGs of the two cell cultures were not the same, suggesting that different response mechanisms are activated in neurons and astrocytes by heat stress.
Previous work has demonstrated increased immune responses during heat-stroke [25]. Studies on rodent models found increased expression of cytokines, COX enzymes, chemokines, and chemokine receptors in the brain, liver, and heart, including IL-1β, IL-6, TNF-α, IL-10, and iNOS [25–27]. Attenuating the systemic inflammatory response can alleviate neuronal damage and reduce mortality in rodents. As these studies were carried out on whole tissues, the detailed molecular processes and regulatory mechanisms of different cell types remained unclear. Ours is the first study to identify the detailed changes in gene expression of different BBB components and the function of astrocytes after heat stress.
Functional annotation revealed enriched clusters of leukocyte chemotaxis and cytokine-cytokine signaling in astrocytic but not neuronal DEGs. Further KEGG pathway analysis demonstrated enriched cytokine/chemokine, Toll-like receptor, and NF-κB signaling pathways in astrocytes. It is worth noting that the chemokine-related genes were found in astrocytes but not neurons, and these genes combined into a regulatory sub-network in the protein-protein interaction network of astrocytic DEGs. These DEGs were composed of the C-X-C motif chemokine ligand (CXCL2), C-C motif chemokine ligand (CCL3, CCL4, and CCL12), C-C chemokine receptor (CCR1), interleukin receptor (IL2RB), and neuropeptide Y receptor (NPY2R), which are important in the immune system, and also play roles in the CNS.
Chemokines are small proteins classified into four families on the basis of the number and spacing of the conserved cysteine residues in the N-terminal position. These families are named CXC, CC, CX3C, and C, in accordance with systematic nomenclature. Chemokines exert their biological effects through cell surface receptors from the G protein-coupled receptor superfamily [28]. It is known that chemokines and their receptors are constitutively expressed in the developing and adult nervous systems. Recent studies have suggested that chemokines and their receptors may contribute to homeostasis in the mature brain, through the modulation of neurotransmitter release, thereby regulating cell survival and synaptic transmission [29, 30]. They also contribute to disruption of the integrity of the BBB, neuroprotection, development and migration of adult neuronal progenitor stem cells, and axonal sprouting and elongation. In terms of pathology, chemokines and chemokine receptors are involved in many CNS disorders, including Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, and amyotrophic lateral sclerosis [28]. A previous study revealed significant changes in the expression of 25 cytokines and chemokines in the hypothalamus of heat-stroke rats [3]. Our results showed that these changes may occur in astrocytes, rather than neurons, so the former may thereby contribute to neuroinflammation and aggravate neural injury.
It is known that the situation differs between cells in vivo and in vitro, thus the fetal cells may not mirror corresponding adult cells. And the NVU contains other cell types that could also be involved in brain inflammation, like microglia, pericytes, and endothelial cells. To address this, we performed single-cell profiling of the hypothalamus in adult heat-stroke rats. The hypothalamus plays a pivotal role in thermoregulation, and ischemic and oxidative damage to the hypothalamus may be responsible for heat-stroke [25]. Hence, we focused on measuring heat-stroke-induced transcriptional changes in different hypothalamic cells from adult rats. Single-cell qPCR revealed significant changes of pivotal genes in astrocytes. Significant changes of cytokine- or chemokine-related genes were not found in the other cell types, except for Ccr1 in neurons. It was surprising that C6, Ccl3, and Ccr1 were all up-regulated in RNA-seq and qPCR in primary astrocyte culture after heat stress, but down-regulated in heat-stroke rats. According to previous work on whole hypothalamic tissue, the transcriptional levels of CCL3 and CCR1 are down-regulated in heat-stroke rats [3]. In addition, down-regulation of C5, CCL12, CCL4, and CCR3 was found, while CCL6, CCR8, and CXCL13 were up-regulated. Maybe the primary cells separated from E18 brain were not mature and healthy enough to mimic the adult properties. On the other hand, considering that the inflammation-related pathways are composed of large gene networks, and the number of pivotal genes detected in the qPCR was limited, perhaps the results of qPCR did not reveal the overall perspective of inflammatory regulation in astrocytes. There may be a correlation between astrocytes and inflammation with complex mechanisms in vivo, such as interactions between different cells. In addition, microglia are known to be resident macrophage-like immune cells in the CNS and play vital roles in both physiological and pathological conditions, regulating multiple aspects of inflammation, such as repair, cytotoxicity, regeneration, and immunosuppression, due to their different kinds of activation states or phenotypes [31]. It was surprising to find no significant changes of pivotal genes in the single-cell transcriptional profiling of microglia. This may be due to the fact that the well-established markers of microglia at present, such as TNF-α, CCL3, and CD11b [24], are all inflammation-related genes, whose expression levels might be regulated by heat stress. To investigate the role of microglia in heat stroke, more precise investigations are needed.
Astrocytes have traditionally been considered to be a homogenous cellular population, and it was assumed that astrocytes from different CNS regions are functionally interchangeable. However, glial biologists now have an appreciation of the important ways in which astrocytes are functionally diverse [32]. Recent evidence has shown that the heterogeneity of astrocytes exists not only in morphology and function but also many other aspects, including development, genomes, astrogliosis, and cell-cell interactions [33]. Several subtypes of astrocytes are important in the neuroinflammation of CNS trauma and multiple sclerosis, such as white matter astrocytes with high expression of GFAP, CD44, vimentin, and nestin, and low expression of NDRG2. The subventricular zone astrocytes with high expression of GFAP, GLAST, and nestin can migrate towards a lesion site and give rise to new neurons following injury [33]. Perhaps different subtypes of astrocytes from different CNS regions or with different expression of subtype-specific markers play diverse roles in heat-stroke. Detection of inflammatory genes of different subtypes of astrocytes may be helpful in understanding the detailed roles of astrocytes in heat-stroke.
Another well-established fact is that mRNA expression does not always reflect the actual synthesis of the respective protein. As our work was performed with transcriptome sequencing and qPCR, it is possible that the protein expression of pivotal genes might not precisely mirror the gene expression data reported.
In conclusion, astrocytes may be an anti-inflammatory target for heat-stroke. We believe that this work provides information for a cell-type-specific approach to heat-stroke treatment, while further exploration of the detailed functions and regulatory mechanisms of each BBB component is warranted.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Qiumin Le (Fudan University, China), Dr. Biao Yan, and Professors Guowei Le and Yonghui Shi (Jiangnan University, China) for technical assistance. This work was supported by the 12th Five-Year Plan of the PLA (BWS11J062), the China Postdoctoral Science Foundation (2015M572806), and the Director’s Fund of the General Hospital of Jinan Military Region, China (2014ZX03).
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12264-017-0156-8) contains supplementary material, which is available to authorized users.
Contributor Information
Huaiqiang Hu, Email: huhuaiqiang@126.com.
Bingzhen Cao, Email: cbzxia2011@163.com.
References
- 1.Bouchama A, Knochel JP. Heat stroke. N Engl J Med. 2002;346:1978–1988. doi: 10.1056/NEJMra011089. [DOI] [PubMed] [Google Scholar]
- 2.Kovats RS, Kristie LE. Heatwaves and public health in Europe. Eur J Public Health. 2006;16:592–599. doi: 10.1093/eurpub/ckl049. [DOI] [PubMed] [Google Scholar]
- 3.Audet GN, Dineen SM, Quinn CM, Leon LR. Altered hypothalamic inflammatory gene expression correlates with heat stroke severity in a conscious rodent model. Brain Res. 2016;1637:81–90. doi: 10.1016/j.brainres.2016.01.048. [DOI] [PubMed] [Google Scholar]
- 4.Argaud L, Ferry T, Le QH, Marfisi A, Ciorba D, Achache P, et al. Short-and long-term outcomes of heatstroke following the 2003 heat wave in Lyon. France. Arch Intern Med. 2007;167:2177–2183. doi: 10.1001/archinte.167.20.ioi70147. [DOI] [PubMed] [Google Scholar]
- 5.Zlokovic BV. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat Rev Neurosci. 2011;12:723–738. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein Y, Roberts WO. The pathopysiology of heat stroke: an integrative view of the final common pathway. Scand J Med Sci Sports. 2011;21:742–748. doi: 10.1111/j.1600-0838.2011.01333.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Chopp M. Astrocytes, therapeutic targets for neuroprotection and neurorestoration in ischemic stroke. Prog Neurobiol. 2016;144:103–120. doi: 10.1016/j.pneurobio.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citri A, Pang ZP, Sudhof TC, Wernig M, Malenka RC. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat Protoc. 2011;7:118–127. doi: 10.1038/nprot.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrew S. FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 15 July 2016.
- 10.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA–Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open–source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 16.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016. [DOI] [PMC free article] [PubMed]
- 21.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horowitz M, Robinson SD. Heat shock proteins and the heat shock response during hyperthermia and its modulation by altered physiological conditions. Prog Brain Res. 2007;162:433–446. doi: 10.1016/S0079-6123(06)62021-9. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA–sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CY, Chen JY, Chen SH, Cheng TJ, Lin MT, Hu ML. Therapeutic treatment with ascorbate rescues mice from heat stroke-induced death by attenuating systemic inflammatory response and hypothalamic neuronal damage. Free Radic Biol Med. 2016;93:84–93. doi: 10.1016/j.freeradbiomed.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Biedenkapp JC, Leon LR. Increased cytokine and chemokine gene expression in the CNS of mice during heat stroke recovery. Am J Physiol Regul Integr Comp Physiol. 2013;305:R978–R986. doi: 10.1152/ajpregu.00011.2013. [DOI] [PubMed] [Google Scholar]
- 27.Quinn CM, Audet GN, Charkoudian N, Leon LR. Cardiovascular and thermoregulatory dysregulation over 24 h following acute heat stress in rats. Am J Physiol Heart Circ Physiol. 2015;309:H557–H564. doi: 10.1152/ajpheart.00918.2014. [DOI] [PubMed] [Google Scholar]
- 28.Reaux-Le Goazigo A, Van Steenwinckel J, Rostene W, Melik Parsadaniantz S. Current status of chemokines in the adult CNS. Prog Neurobiol. 2013;104:67–92. doi: 10.1016/j.pneurobio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Miller RJ, Rostene W, Apartis E, Banisadr G, Biber K, Milligan ED, et al. Chemokine action in the nervous system. J Neurosci. 2008;28:11792–11795. doi: 10.1523/JNEUROSCI.3588-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rostene W, Kitabgi P, Parsadaniantz SM. Chemokines: a new class of neuromodulator? Nat Rev Neurosci. 2007;8:895–903. doi: 10.1038/nrn2255. [DOI] [PubMed] [Google Scholar]
- 31.Du L, Zhang Y, Chen Y, Zhu J, Yang Y, Zhang HL. Role of microglia in neurological disorders and their potentials as a therapeutic target. Mol Neurobiol 2016. doi:10.1007/s12035-016-0245-0. [DOI] [PubMed]
- 32.Ben Haim L, Rowitch DH. Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci. 2017;18:31–41. doi: 10.1038/nrn.2016.159. [DOI] [PubMed] [Google Scholar]
- 33.Hu X, Yuan Y, Wang D, Su Z. Heterogeneous astrocytes: Active players in CNS. Brain Res Bull. 2016;125:1–18. doi: 10.1016/j.brainresbull.2016.03.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.