Abstract
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder. Mutations in the DJ-1, including L166P, are responsible for recessive early-onset PD. Many lines of evidence have shown that L166P is not only a loss-of-function mutant, but also a pro-apoptotic-like protein that results in mitochondrial dysfunction. L166P has been reported to be unstable and to mislocalize to mitochondria. However, the mechanisms underlying the instability of L166P compared to wild-type DJ-1 remain largely unknown. Here, we showed that Omi/HtrA2, a mitochondrial serine protease that has also been linked to the pathogenesis of PD, contributed to L166P instability. Omi directly interacted with and cleaved L166P in mitochondria to decrease the L166P level. However, Omi did not bind and cleave wild-type DJ-1. Moreover, Omi cleaved L166P at both serine residues 3 and 121, while L166P-induced cell death under H2O2 treatment was alleviated by over-expression of Omi. Our data reveal a bridge between DJ-1 and Omi, two PD-associated genetic factors, which contributes to our understanding of the pathogenesis of PD.
Keywords: Parkinson’s disease, DJ-1, L166P, Instability, Omi/HtrA2, Cleavage
Introduction
Parkinson’s disease (PD) is one of the most common neurodegenerative diseases, affecting 1%–2% of the general population more than 65 years old [1, 2]. It is due to the loss of dopaminergic neurons in the pars compacta of the substantia nigra and decreased dopamine levels in the striatum. Although most cases of PD are sporadic forms, our knowledge of its causes has come largely from the dysfunctions of genes that are responsible for familial PD, such as DJ-1, SNCA/α-Synuclein, Parkin, PINK1, and ATP12A2 [3, 4]. At the cellular level, mitochondrial dysfunction is thought to be a common and crucial factor associated with the pathogenesis of PD [5, 6].
DJ-1, a conserved protein composed of 189 amino-acids, was initially identified as an oncoprotein that functionally collaborates with the activated small G-protein ras in NIH3T3 cells [7]. It has been reported that DJ-1 is able to protect cells from death by multiple cellular pathways including transcriptional regulation [8–11], anti-apoptotic activity [12–15], anti-oxidative activity [16–18], RNA binding [19], and chaperone activity [20, 21]. Deletions or a point mutation in the human DJ-1 gene that results in a substitution of proline for leucine at residue 166 (L166P) has been reported to be responsible for recessive early-onset PD [22]. Moreover, other DJ-1 mutations have also been associated with sporadic early-onset PD [23]. Interestingly, the most commonly studied PD-associated L166P mutant is unstable in cells [24–26]. The crystal structures of wild-type DJ-1 and L166P demonstrate that mutation of L166P prevents the normal folding of the C-terminal region [27–29]. DJ-1 forms soluble dimers, whereas the L166P mutant exists as a monomer in cells [25]. The structural instability of the L166P mutant leads to rapid degradation by the ubiquitin-proteasome system or proteasomal endoproteolytic cleavage [26, 30]. In addition, wild-type DJ-1 mainly occurs in the cytoplasm and the nucleus, as well as partly on mitochondria [7, 30–32]. However, the L166P mutant is predominantly localized in mitochondria [22, 33, 34]. Furthermore, many studies have shown that L166P not only loses functions in comparison to wild-type DJ-1, but also has a pro-apoptotic property [14, 16, 34–36]. Decrease of protein levels and mislocation to mitochondria are responsible for the pathogenic effects of L166P in PD. However, the mechanisms by which the L166P mutant is more unstable than wild-type DJ-1 are largely unknown.
Omi/HtrA2 is a serine protease belonging to the high-temperature requirement factor A (HtrA) family [37]. Although Omi is mainly localized in mitochondria, endoplasmic reticulum and Golgi localization have also been reported [38, 39]. Although Omi is pro-apoptotic in some somatic cells due to its release from mitochondria to the cytosol in response to apoptotic stimuli [39–43], mitochondrial Omi plays a physiological cytoprotective role in neurons [44–46]. Loss of Omi function leads to neurodegeneration in mouse models and has been linked to the pathogenesis of PD [45, 47, 48].
In this study, we demonstrated that the PD-associated L166P mutant, but not wild-type DJ-1, directly binds to and is cleaved by the mitochondrial serine protease Omi at both serine residues 3 and 121. In addition, L166P-induced cellular toxicity under oxidative stress was alleviated by Omi. Our findings reveal a relationship between two PD-associated genetic factors, DJ-1 and Omi.
Materials and Methods
Cell Cultures, Transfection, and Reagents
Mouse neuroblastoma Neuro 2a (N2a), human embryonic kidney 293 (HEK293), and human cervical carcinoma HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY) containing 10% fetal bovine serum (Hyclone Laboratories, Logan, UT) with streptomycin (100 µg/mL) and penicillin (100 U/mL) (Gibco). H1299 cells were maintained in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum with streptomycin (100 µg/mL) and penicillin (100 U/mL). The cells were transfected with expression plasmids using the Lipofectamine2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The Omi protease inhibitor UCF101 was from Calbiochem (Darmstadt, Germany).
Small Interfering RNAs (siRNAs) and Plasmids
The double-stranded oligonucleotides used in this study were designed against human Omi mRNA or negative control (si-NC) and synthesized by Shanghai GenePharma (Shanghai, China) as described previously [49]. The transfection was performed using Oligofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. pGEX-5x-1-DJ-1, p3×Flag-myc-cmv-24-DJ-1, and p3×Flag-myc-cmv-24-L166P have been described previously [9, 50]. For HA-tagged DJ-1 or L166P construct, DJ-1 or L166P was generated by sub-cloning the PCR products amplified from p3×Flag-myc-cmv-24-DJ-1 or p3×Flag-myc-cmv-24-L166P with the primers 5′-CGGGATCCATGGCTTCCAAAAGAG-3′ and 5′-CGGAATTCTCAGTCTTTAAGAACAAGTG-3′ into pKH3-HA at the BamHI/EcoRI sites. For mitochondria-targeted HA-DJ-1 or HA-L166P, the mitochondrial targeting sequence from the cytochrome c oxidase subunit 8A was amplified by PCR with the primers 5′-CCCAAGCTTACCATGTCCGTCCTGACGCCG-3′ and 5′-AACTGCAGCAACGAATGGATCTTGG-3’ from the pDsRed-Mito vector and then inserted into pKH3-HA-DJ-1 or pKH3-HA-L166P at the PstI/HindIII sites. All constructs expressing Omi have been described previously [49, 51]. The fidelity of all constructs was confirmed by sequencing.
Immunoblot Analysis and Antibodies
Immunoblot analysis was performed as described previously [52]. The following primary antibodies were used: monoclonal antibodies anti-DJ-1 (Santa Cruz Biotechnology), anti-Flag (Sigma, St. Louis, MO), anti-GST (Santa Cruz Biotechnology), anti-HA (Santa Cruz Biotechnology), anti-His (Abmart, Shanghai, China), anti-Tom20 (Santa Cruz Biotechnology), anti-PARP (Cell Signaling Tech, Danvers, MA), and anti-α-tubulin (Merck Calbiochem, San Diego, CA), as well as polyclonal antibody against Omi raised by immunizing New Zealand White rabbits as described previously [49]. The secondary antibodies, sheep anti-mouse IgG-HRP and anti-rabbit IgG-HRP were from Amersham Pharmacia Biotech (Arlington Heights, IL). The proteins were visualized using an ECL detection kit (Amersham Pharmacia Biotech).
Immunoprecipitation
Immunoprecipitation was as described previously [12]. Briefly, cells were lysed in RIPA lysis buffer at 4 °C. After centrifugation at 12,000 g for 15 min, the supernatant was used for immunoprecipitation with appropriate antibodies coupled to protein G Sepharose beads (Roche, Basel, Switzerland). The immunoprecipitates were then washed 5 times and subjected to immunoblot analysis. The input represents 10% of the supernatant used in the co-immunoprecipitation experiments.
GST Pulldown Assays
Twenty micrograms of GST (glutathione S-transferase), GST-DJ-1, or GST-L166P protein expressed by E. coli strain DH5α was incubated with 25 μL of glutathione agarose beads (GE Healthcare Life Sciences, Piscataway, NJ) for 30 min on ice. After washing three times with PBS, the beads were incubated with 50 μg His-Omi expressed by E. coli strain BL21 for 3 h on ice. Then the beads were washed five times with ice-cold HNTG buffer (20 mmol/L Hepes-KOH, 100 mmol/L NaCl, 0.1% Triton X-100, 10% glycerol, pH 7.5). Then the samples were subjected to immunoblot analysis with anti-His antibody. The input represents 10% of the protein that was incubated with GST or GST-fused proteins. The purified GST, GST-DJ-1, and GST-L166P were stained with anti-GST antibody.
In vitro Proteolytic Cleavage Assays
Bacterially-expressed GST, GST-DJ-1, and GST-L166P were purified using glutathione agarose beads. His-Omi and His-S276C were purified with nickel-nitrilotriacetic acid affinity resin according to the manufacturer’s instructions. GST, GST-DJ-1, or GST-L166P was incubated with His-Omi or His-S276C in protease cleavage buffer (20 mmol/L Na2HPO4 (pH 8.0), 10% glycerol, 200 mmol/L NaCl). Four hours after incubation at 37 °C, the reaction was terminated with SDS sample buffer. Reaction products were then subjected to immunoblot analysis.
Subcellular Fractionation Assay
Mitochondrial and cytosolic fractions were isolated using the Cell Mitochondria Isolation Kit (Beyotime, Nantong, China) according to the manufacturer’s instructions. The total cell lysates, mitochondrial and cytosolic fractions were subjected to immunoblot analysis. Tom20 served as a mitochondrial marker and α-tubulin as a cytosolic marker.
Immunofluorescence
HEK 293 cells were washed with PBS and fixed in 4% paraformaldehyde for 10 min at room temperature followed by permeabilization in 0.25% Triton X-100 PBS for 10 min. After pre-blocking with 5% fetal bovine serum for 1 h, the cells were incubated with anti-HA antibody or anti-Omi antibodies overnight at 4 °C followed by incubation with Rho-conjugated donkey anti-rabbit secondary antibody (Santa Cruz Biotechnology) or Alexa Fluor-350-labeled goat anti-mouse IgG (Invitrogen). Finally, the cells were observed under a fluorescence microscope (Olympus, IX71, Tokyo, Japan).
MTT Cell Viability Assay
Cells were washed with PBS and incubated with 0.5 mg/mL MTT (3-(4,5)-dimethylthiahiazo(-z-y1)-3,5-di-phenytetrazoliumromide, Sigma) dissolved in DMEM without phenol red. After 3 h, the medium was removed and the formazan crystals were dissolved in DMSO (Dimethyl sulfoxide) by incubation at 37 °C for 30 min. The absorbance was measured using a photometer at 570 nm and the background at 630 nm was subtracted. The data were normalized to the control and the ratios were presented as the mean ± SEM from three independent experiments.
Statistical Analysis
Densitometric analysis of immunoblots from three independent experiments was calculated using Photoshop 7.0 software (Adobe, San Jose, CA). The data were analyzed using Origin 6.0 software (Originlab, Northampton, MA). Statistical analysis was performed using one-way analysis of variance (ANOVA). Student’s t-test was used for comparisons between two groups. A P value of < 0.05 was considered statistically significant. All data were presented as mean ± SEM.
Results
Omi is Involved in the Instability of L166P
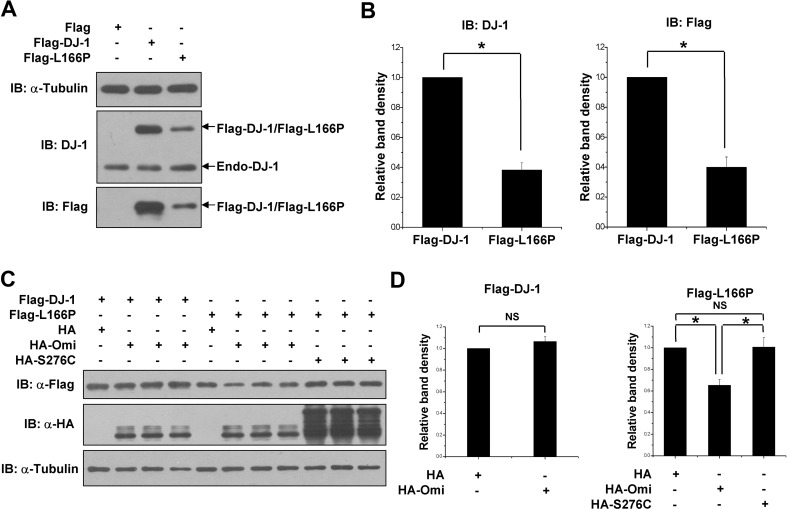
It has been reported that the steady-state levels of the PD-linked L166P mutant were lower than wild-type DJ-1 [24, 25, 30]. We performed transfection experiments using the N-terminal Flag-tagged DJ-1 or L166P to verify this effect. N2a cells were transfected with equal amounts of DJ-1 or L166P plasmid. Immunoblot analysis showed that the protein levels of L166P were dramatically lower than DJ-1 (Fig. 1A, B). Similar results were obtained using either anti-DJ-1 antibody or anti-Flag antibody that were used to assess the levels of Flag-DJ-1 and Flag-L166P (Fig. 1A, B).
Fig. 1.
Omi is involved in the instability of L166P. A Immunoblots of lysates of N2a cells transfected with equal amounts of Flag-DJ-1 or Flag-L166P construct for 36 h and using the indicated antibodies. B Ratios of Flag-DJ-1 or Flag-L166P relative to α-tubulin from (A) from densitometric analysis Data are from three independent experiments (mean ± SEM; *P < 0.05; ns, no statistical significance, one-way ANOVA). C Immunoblots of lysates of H1299 cells co-transfected with Flag-DJ-1 or Flag-L166P and HA, HA-Omi or HA-S276C for 36 h as indicated. D Ratios of Flag-DJ-1 or Flag-L166P relative to α-tubulin from (C) from densitometric analysis. Data from three independent experiments (mean ± SEM, *P < 0.05; ns, no statistical significance, one-way ANOVA)
The L166P DJ-1 mutant that is unstable and mislocalizes to mitochondria is involved in the mitochondrial dysfunction of PD [14, 16, 34, 35]. Interestingly, the mitochondrial Omi has cytoprotective effects and plays important roles in mitochondrial quality control [44–47, 53]. We wondered whether Omi is involved in the instability of L166P and therefore measured the protein levels of DJ-1 or L166P in H1299 cells co-transfected with Flag-DJ-1 or Flag-L166P along with HA, HA-Omi, or HA-S276C (a protease-deficient mutant) [45]. To attain expression levels comparable to wild-type DJ-1, we transfected the cells with amounts of L166P plasmids triple that of wild-type DJ-1 as described previously [12, 13]. Overexpression of Omi, but not S276C Omi, led to a remarkable decrease of the L166P level (Fig. 1C, D). In addition, over-expression of Omi had no effect on the protein levels of wild-type DJ-1 (Fig. 1C, D).
Omi Inhibition Increases the Protein Level of L166P
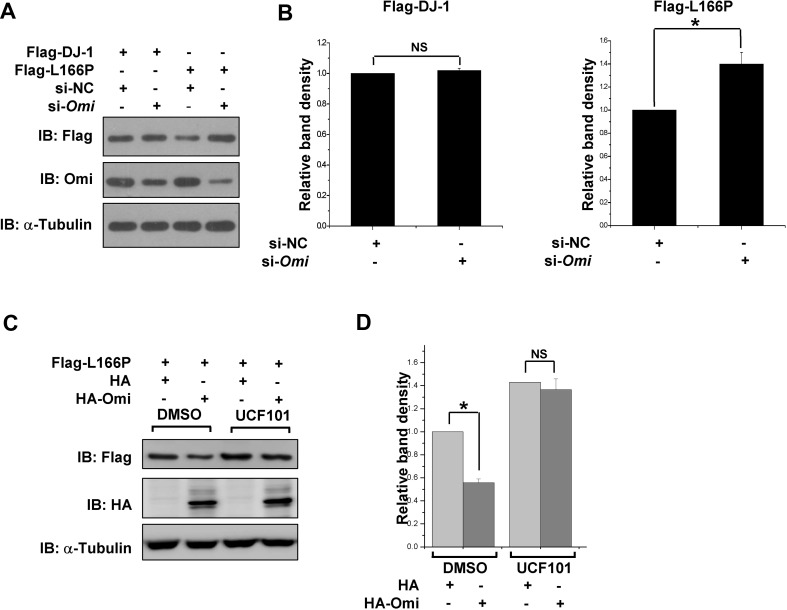
We further assessed the effects of Omi on the protein level of L166P using a small interfering RNA (siRNA) that targets Omi, as described previously [49]. Omi levels were greatly decreased after transfection of si-Omi, while the wild-type DJ-1 levels did not change (Fig. 2A, B). However, knockdown of Omi led to a significant increase of L166P protein levels (Fig. 2A, B). Interestingly, the reduction of L166P protein levels by overexpressing Omi was reversed by UCF101, an Omi protease inhibitor that shows high selectivity against the serine protease activity of Omi (Fig. 2C, D) [54], further suggesting that Omi reduces L166P protein levels through its protease activity.
Fig. 2.
Omi inhibition increases L166P protein levels. A Immunoblots with the indicated antibodies of lysates 48 h after H1299 cells were first transfected with si-NC or si-Omi and 24 h later, Flag-DJ-1 and Flag-L166P were transfected into the cells. B Ratios of Flag-DJ-1 or Flag-L166P relative to α-tubulin from (A) from densitometric analysis. Data are from three independent experiments (mean ± SEM, *P < 0.05; ns, no statistical significance, one-way ANOVA). C Immunoblots of lysates of H1299 cells harboring Flag-L166P transfected with HA or HA-Omi. After transfection, the cells were treated with DMSO or UCF101 (20 μmol/L) for 24 h. D Ratios of Flag-L166P relative to α-tubulin from (C) from densitometric analysis. Data are from three independent experiments (mean ± SEM, *P < 0.05; ns, no statistical significance, one-way ANOVA)
Omi Interacts with and Cleaves L166P DJ-1, But not Wild-Type DJ-1
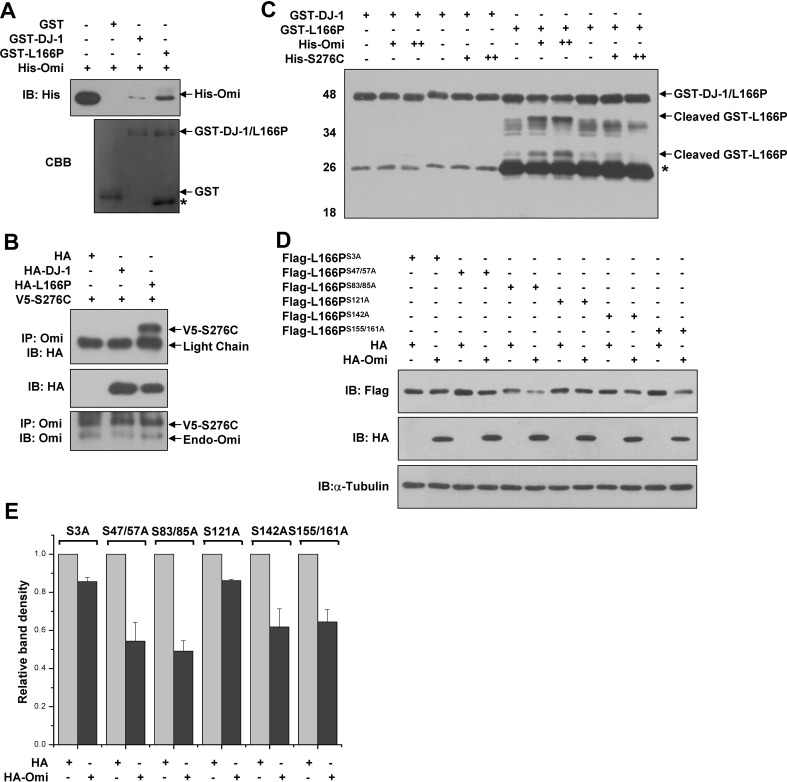
To investigate whether Omi decreases the protein levels of L166P through their direct interaction, we first performed in vitro pull-down assays. GST-L166P, but not GST alone, interacted with His-Omi expressed by E. coli. Notably, wild-type DJ-1 hardly interacted with Omi compared with L166P (Fig. 3A). To further confirm the interaction between L166P and Omi, we performed co-immunoprecipitation experiments in HEK293 cells. To avoid cleavage of L166P by Omi in co-immunoprecipitation experiments, the protease-defective mutant S276C was used to detect the interaction. We found that HA-L166P was co-immunoprecipitated when S276C Omi was precipitated using Omi antibodies, whereas wild-type DJ-1 was not detected in the immunoprecipitates (Fig. 3B).
Fig. 3.
Omi decreases the protein levels of L166P, but not wild-type DJ-1 through direct interaction. A Omi interacts with L166P in vitro. Pull-down assays used purified GST, GST-DJ-1, or GST-L166P along with His-Omi expressed in E. coli. Immunoblot analysis used anti-His antibody. B HEK 293 cells co-transfected with HA, DJ-1-HA, or L166P-HA along with V5-S276C were lysed and immunoprecipitated with anti-Omi antibodies. C Omi cleaves L166P in vitro. Cleavage assays were performed using equal amounts of purified GST, GST-DJ-1, or GST-L166P and increasing amounts of purified His-Omi or His-S276C. The proteins were incubated in the protease buffer at 37 °C for 4 h. Immunoblot analysis was performed using anti-GST antibody. D H1299 cells were co-transfected with HA or HA-Omi along with serine-to-alanine mutants of L166P. After 36 h, the cell lysates were subjected to immunoblot analysis with anti-Flag antibody. E Ratios of serine-to-alanine mutants of L166P relative to α-tubulin from (E) from densitometric analysis
Next, we performed in vitro cleavage assays to determine whether L166P is directly cleaved by Omi. Equal amounts of GST-DJ-1 or GST-L166P (N-terminal GST tag) were incubated with increasing amounts of His-Omi expressed in E. coli. GST-L166P but not GST-DJ-1 was cleaved by His-Omi protease (Fig. 3C). Immunoblot analysis showed that the two bands of cleaved GST-L166P products increased with increasing amounts of His-Omi but not His-S276C (Fig. 3C). Together, these data suggested that Omi directly cleaves L166P, and this is dependent on its protease activity.
As Omi is a serine protease, we constructed different mutants in which each serine was converted to alanine to identify Omi cleavage site(s) in L166P. Flag-L166PS3A and Flag-L166PS121A, but not Flag-L166P and other mutants, were resistant to Omi proteolytic activity (Fig. 3D and E). Thus, our data suggested that Omi cleaves L166P at both serine residues 3 and 121.
Mitochondrial Localization and Misfolded Structure Contribute to L166P Instability
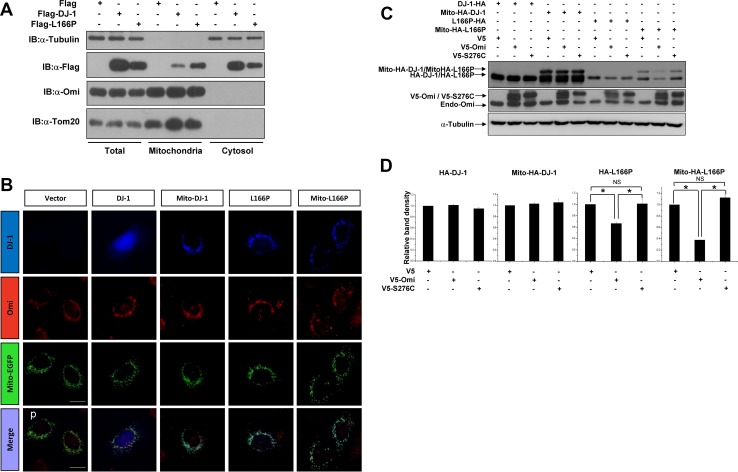
To investigate the influence of subcellular distribution of DJ-1 and L166P on their interaction with and cleavage by Omi, we first performed subcellular fractionation assays. HEK293 cells transfected with Flag, Flag-DJ-1, or Flag-L166P were subjected to fractionation assays to examine their distribution in mitochondria and the cytosol. Consistent with previous studies [22, 34], wild-type DJ-1 was mainly localized in the cytosol, and partly in mitochondria. In contrast to wild-type DJ-1, L166P was mainly localized in mitochondria, with much less in the cytosol (Fig. 4A). In addition, Omi was mostly localized in mitochondria (Fig. 4A). These results indicated that L166P but not wild-type DJ-1 is prone to be present in mitochondria and binds to mitochondrial serine protease Omi.
Fig. 4.
Mitochondrial targeting L166P facilitates its cleavage by Omi. A Subcellular fractionation assays of H1299 cells after transfection with Flag, Flag-DJ-1, or Flag-L166P for 36 h. B HEK 293 cells were co-transfected with plasmids expressing HA, HA-DJ-1, Mito-HA-DJ-1, HA-L166P, or Mito-HA-L166P along with Mito-EGFP as indicated. After 36 h, the cells were subjected to immunocytochemical analysis with anti-HA antibody (blue) and anti-Omi antibodies (red). Mito-EGFP was used as a mitochondrial marker (green). Scale bar, 10 μm. C Immunoblots of lysates of H1299 cells co-transfected with HA-DJ-1, Mito-HA-DJ-1, HA-L166P, or Mito-HA-L166P along with V5, V5-Omi, or V5-S276C for 36 h. D Ratios of A-DJ-1, Mito-HA-DJ-1, HA-L166P, or Mito-HA-L166P relative to α-tubulin from densitometric analysis of (B). Data from three independent experiments (mean ± SEM, *P < 0.05; ns, no statistical significance, one-way ANOVA)
To further determine whether L166P cleavage by Omi is due to its mislocalization to mitochondria, HA-DJ-1 and HA-L166P with an N-terminal mitochondria-targeting sequence (Mito-HA-DJ-1 and Mito-HA-L166P) were created. Immunofluorescent staining showed that HA-DJ-1 was distributed diffusely throughout cells, and that HA-L166P was present mainly in mitochondria and to a certain extent in the cytosol (Fig. 4B). Meanwhile, Mito-HA-DJ-1 and Mito-HA-L166P exhibited typical mitochondrial localization, showing co-localization with Mito-EGFP (Fig. 4B). Although Mito-HA-DJ-1 was distributed in mitochondria, overexpression of Omi had no effect on the protein levels of Mito-HA-DJ-1, similar to that on wild-type HA-DJ-1 (Fig. 4C, D); however, overexpression of Omi, but not S276C, resulted in a greater decrease of Mito-HA-L166P than HA-L166P. These data indicated that not only mislocalization to mitochondria but also its misfolded structure influences L166P recognition and cleavage by Omi.
L166P-Induced Cell Death Under H2O2 Treatment is Inhibited by Omi
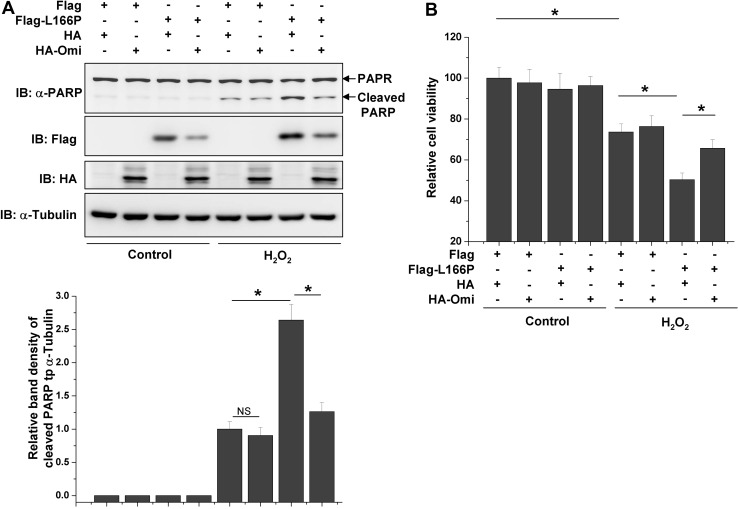
L166P promotes cell death under various stress conditions such as oxidative stress and apoptotic stimuli [14, 16, 34–36]. We next examined whether Omi protects cells against L166P-induced cell death. Under normal conditions, overexpression of L166P did not induce significant cell death, as indicated by poly ADP-ribose polymerase (PARP) cleavage and cell viability (Fig. 5A, B). However, L166P dramatically promoted PARP cleavage and reduced cell viability in response to H2O2 treatment (Fig. 5A, B). Moreover, Omi reduced L166P-induced PARP cleavage and cell death under H2O2 treatment, although overexpression of Omi alone did not significantly influence cell survival (Fig. 5A, B).
Fig. 5.
L166P-induced cell death under H2O2 treatment is inhibited by Omi. A, B N2a cells were transfected with Flag, Flag-L166P along with HA or HA-Omi for 24 h as indicated. The cells were then treated with or without 200 μmol/L H2O2 for 24 h. After treatment the cell lysates were subjected to immunoblot analysis (A) or the cells were subjected to MTT assay (B)
Discussion
In the present study, we demonstrated that the PD-linked mutant L166P, but not wild-type DJ-1, was cleaved by Omi in mitochondria through direct interaction, resulting in a reduction of L166P protein levels. In addition, Omi cleaved L166P at both serine residues at positions 3 and 121. Moreover, L166P-induced cell death in response to oxidative stress was repressed by Omi overexpression. Thus, our results indicated that Omi plays a role in the regulation of L166P in mammalian cells.
Omi dysfunction is tightly associated with neurodegeneration, especially in the pathogenesis of PD [48, 53, 55]. Mutations in the Omi gene have been associated with PD [47, 56, 57]. Omi is a serine protease important for mitochondrial quality control [53]. Serine 276 is located in the protease domain of Omi and the S276C mutation results in a loss of Omi protease activity [45]. Omi knockout mice and mnd2 mice that harbor the S276C mutation both exhibit progressive neurodegeneration and motor deficits similar to PD [45, 51]. Omi but not S276C decreases L166P in mitochondria, indicating that Omi regulates the steady-state level of L166P through its serine protease activity. Since there is no report showing that DJ-1 and Omi mutations occur simultaneously in PD patients, it would be interesting to investigate whether the Omi activity is changed in patients (or animal models) harboring the L166P mutant, which may further validate the linkage between these two PD factors.
DJ-1 forms homodimers in cells [29], which is essential for the pro-survival functions of DJ-1 [9, 12, 13, 15, 16]. Mutations in the DJ-1 gene, including deletions and point mutations, have been linked to recessive familial forms of PD [22]. The most commonly studied PD-associated mutant L166P was reported to be unstable and exhibited improper subcellular localization [22]. The L166P mutant has a spontaneously unfolded structure that is incapable of forming a stable homodimer as DJ-1 [29, 33, 58]. Using GST pull-down assays, we demonstrated that Omi tightly interacted with L166P; whereas wild-type DJ-1 hardly interacted with Omi (Fig. 3A, B). It is possible that the unfolded structure of L166P leads to exposure of its binding site(s) to Omi, which is responsible for the enhanced association of L166P with Omi.
Wild-type DJ-1 is diffusely distributed throughout cells, but L166P is mainly localized to mitochondria [7, 22, 30–32, 34]. The unfolded structure-mediated monomeric state may be responsible for its mitochondrial localization [59]. Omi is a mitochondrial serine protease that cleaves its substrate [38, 39]. Our data provided evidence that mitochondrial L166P, but not wild-type DJ-1, interacted with and was cleaved by Omi. Although the mitochondrial L166P is more susceptible to proteolysis by Omi, the mitochondrial wild-type DJ-1, in which a mitochondrial targeting signal is introduced, is also resistant to the proteolytic activity of Omi. These data suggest that the conformation of L116P is the essential factor for its interaction with and proteolysis by Omi, rather than the mitochondrial localization. L166P is unstable and subject to the UPS degradation or proteasomal endoproteolytic cleavage [24–26, 30]. However, the factors involved in L166P instability have not been identified. From our results, inhibition of Omi protease activity by its inhibitor UCF101 or conversion of serine 3 or 121 to alanine increased the steady-state level of L166P, suggesting that the proteolysis of L166P by Omi is also a means of regulating L166P stability. It is also likely that the cleavage by Omi leads to more structural instability of L166P, and recognition by its E3 ligase(s), therefore promoting L166P degradation by the ubiquitin proteasome system.
L166P has pro-apoptotic actions in mitochondria, promoting cell death in response to diverse stimuli [14, 16, 34–36]. In our results, overexpression of L166P accelerated H2O2-induced cell death. However, overexpression of Omi significantly blocked this accelerated cell death, suggesting that the regulation of mitochondrial L166P by Omi has a protective effect on mitochondria.
In summary, our study demonstrates that L166P, but not wild-type DJ-1, is cleaved by the mitochondrial serine protease Omi through direct interaction. In addition, the unfolded structure and mislocalization to mitochondria may be responsible for the cleavage of L166P by Omi. Our results provide a mechanistic explanation for L166P instability and bridges two PD-associated genetic factors in mammalian cells, which may be helpful for our understanding of the pathogenesis of PD.
Acknowledgements
This work was supported by the National Key Scientific R&D Program of China (2016YFC1306000), the National Natural Sciences Foundation of China (31471012, 81761148024, 31330030, and 81371393), the Suzhou Clinical Research Center of Neurological Disease (Szzx201503), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Kai Fu and Yanfei Wang have contributed equally to this work.
Contributor Information
Guanghui Wang, Email: wanggh@suda.edu.cn.
Haigang Ren, Email: rhg@suda.edu.cn.
References
- 1.Lansbury PT, Jr, Brice A. Genetics of Parkinson’s disease and biochemical studies of implicated gene products. Curr Opin Genet Dev. 2002;12:299–306. doi: 10.1016/S0959-437X(02)00302-7. [DOI] [PubMed] [Google Scholar]
- 2.Tao K, Wang B, Feng D, Zhang W, Lu F, Lai J, et al. Salidroside protects against 6-hydroxydopamine-induced cytotoxicity by attenuating ER stress. Neurosci Bull. 2016;32:61–69. doi: 10.1007/s12264-015-0001-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delamarre A, Meissner WG. Epidemiology, environmental risk factors and genetics of Parkinson’s disease. Presse Med. 2017;46:175–181. doi: 10.1016/j.lpm.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Button RW, Luo S, Rubinsztein DC. Autophagic activity in neuronal cell death. Neurosci Bull. 2015;31:382–394. doi: 10.1007/s12264-015-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose A, Beal MF. Mitochondrial dysfunction in Parkinson’s disease. J Neurochem. 2016;139(Suppl 1):216–231. doi: 10.1111/jnc.13731. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Q, Yang X, Cai D, Ye L, Hou Y, Zhang L, et al. echinacoside protects against MPP(+)-induced neuronal apoptosis via ROS/ATF3/CHOP pathway regulation. Neurosci Bull. 2016;32:349–362. doi: 10.1007/s12264-016-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SM, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 8.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer-and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan J, Ren H, Jia N, Fei E, Zhou T, Jiang P, et al. DJ-1 decreases Bax expression through repressing p53 transcriptional activity. J Biol Chem. 2008;283:4022–4030. doi: 10.1074/jbc.M707176200. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Taira T, Niki T, Seino C, Iguchi-Ariga SM, Ariga H. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J Biol Chem. 2001;276:37556–37563. doi: 10.1074/jbc.M101730200. [DOI] [PubMed] [Google Scholar]
- 11.Giaime E, Sunyach C, Druon C, Scarzello S, Robert G, Grosso S, et al. Loss of function of DJ-1 triggered by Parkinson’s disease-associated mutation is due to proteolytic resistance to caspase-6. Cell Death Differ. 2010;17:158–169. doi: 10.1038/cdd.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu K, Ren H, Wang Y, Fei E, Wang H. DJ-1 inhibits TRAIL-induced apoptosis by blocking pro-caspase-8 recruitment to FADD. Oncogene. 2010;31:1311–1322. doi: 10.1038/onc.2011.315. [DOI] [PubMed] [Google Scholar]
- 13.Junn E, Taniguchi H, Jeong BS, Zhao X, Ichijo H, Mouradian MM. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc Natl Acad Sci U S A. 2005;102:9691–9696. doi: 10.1073/pnas.0409635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo JS, Kim MY, Ann EJ, Hong JA, Park HS. DJ-1 modulates UV-induced oxidative stress signaling through the suppression of MEKK1 and cell death. Cell Death Differ. 2008;15:1030–1041. doi: 10.1038/cdd.2008.26. [DOI] [PubMed] [Google Scholar]
- 15.Kim RH, Peters M, Jang Y, Shi W, Pintilie M, Fletcher GC, et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, Mizusawa H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312:1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 18.Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, Thomas B, et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc Natl Acad Sci U S A. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Brug MP, Blackinton J, Chandran J, Hao LY, Lal A, Mazan-Mamczarz K, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci U S A. 2008;105:10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SJ, Kim SJ, Kim IK, Ko J, Jeong CS, Kim GH, et al. Crystal structures of human DJ–1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J Biol Chem. 2003;278:44552–44559. doi: 10.1074/jbc.M304517200. [DOI] [PubMed] [Google Scholar]
- 21.Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2:e362. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 23.da Costa CA. DJ-1: a newcomer in Parkinson’s disease pathology. Curr Mol Med. 2007;7:650–657. doi: 10.2174/156652407782564426. [DOI] [PubMed] [Google Scholar]
- 24.Macedo MG, Anar B, Bronner IF, Cannella M, Squitieri F, Bonifati V, et al. The DJ-1L166P mutant protein associated with early onset Parkinson’s disease is unstable and forms higher-order protein complexes. Hum Mol Genet. 2003;12:2807–2816. doi: 10.1093/hmg/ddg304. [DOI] [PubMed] [Google Scholar]
- 25.Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson’s disease, confers reduced protein stability and impairs homo-oligomerization. J Neurochem. 2003;87:1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez-Castelao B, Munoz C, Sanchez I, Goethals M, Vandekerckhove J, Castano JG. Reduced protein stability of human DJ-1/PARK7 L166P, linked to autosomal recessive Parkinson disease, is due to direct endoproteolytic cleavage by the proteasome. Biochim Biophys Acta. 2012;1823:524–533. doi: 10.1016/j.bbamcr.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc Natl Acad Sci U S A. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honbou K, Suzuki NN, Horiuchi M, Niki T, Taira T, Ariga H, et al. The crystal structure of DJ-1, a protein related to male fertility and Parkinson’s disease. J Biol Chem. 2003;278:31380–31384. doi: 10.1074/jbc.M305878200. [DOI] [PubMed] [Google Scholar]
- 29.Anderson PC, Daggett V. Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson’s disease. Biochemistry. 2008;47:9380–9393. doi: 10.1021/bi800677k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller DW, Ahmad R, Hague S, Baptista MJ, Canet-Aviles R, McLendon C, et al. L166P mutant DJ-1, causative for recessive Parkinson’s disease, is degraded through the ubiquitin-proteasome system. J Biol Chem. 2003;278:36588–36595. doi: 10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Shimoji M, Thomas B, Moore DJ, Yu SW, Marupudi NI, et al. Mitochondrial localization of the Parkinson’s disease related protein DJ-1: implications for pathogenesis. Hum Mol Genet. 2005;14:2063–2073. doi: 10.1093/hmg/ddi211. [DOI] [PubMed] [Google Scholar]
- 32.Ren H, Fu K, Wang D, Mu C, Wang G. Oxidized DJ-1 interacts with the mitochondrial protein BCL-XL. J Biol Chem. 2011;286:35308–35317. doi: 10.1074/jbc.M110.207134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorner K, Holtorf E, Waak J, Pham TT, Vogt-Weisenhorn DM, Wurst W, et al. Structural determinants of the C-terminal helix-kink-helix motif essential for protein stability and survival promoting activity of DJ-1. J Biol Chem. 2007;282:13680–13691. doi: 10.1074/jbc.M609821200. [DOI] [PubMed] [Google Scholar]
- 34.Ren H, Fu K, Mu C, Zhen X, Wang G. L166P mutant DJ-1 promotes cell death by dissociating Bax from mitochondrial Bcl-XL. Mol Neurodegener. 2012;7:40. doi: 10.1186/1750-1326-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deeg S, Gralle M, Sroka K, Bahr M, Wouters FS, Kermer P. BAG1 restores formation of functional DJ-1 L166P dimers and DJ-1 chaperone activity. J Cell Biol. 2010;188:505–513. doi: 10.1083/jcb.200904103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Geng J, Liu J. Adiponectin offers protection against L166P mutant DJ-1-induced neuronal cytotoxicity mediated by APPL1-dependent AMPK activation. Int J Neurosci. 2014;124:350–361. doi: 10.3109/00207454.2013.846340. [DOI] [PubMed] [Google Scholar]
- 37.Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell. 2002;10:443–455. doi: 10.1016/S1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 38.Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, et al. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem. 2000;267:5699–5710. doi: 10.1046/j.1432-1327.2000.01589.x. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki Y, Takahashi-Niki K, Akagi T, Hashikawa T, Takahashi R. Mitochondrial protease Omi/HtrA2 enhances caspase activation through multiple pathways. Cell Death Differ. 2004;11:208–216. doi: 10.1038/sj.cdd.4401343. [DOI] [PubMed] [Google Scholar]
- 40.Martins LM, Turk BE, Cowling V, Borg A, Jarrell ET, Cantley LC, et al. Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J Biol Chem. 2003;278:49417–49427. doi: 10.1074/jbc.M308659200. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/S1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 42.van Loo G, van Gurp M, Depuydt B, Srinivasula SM, Rodriguez I, Alnemri ES, et al. The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity. Cell Death Differ. 2002;9:20–26. doi: 10.1038/sj.cdd.4400970. [DOI] [PubMed] [Google Scholar]
- 43.Hartkamp J, Carpenter B, Roberts SG. The Wilms’ tumor suppressor protein WT1 is processed by the serine protease HtrA2/Omi. Mol Cell. 2010;37:159–171. doi: 10.1016/j.molcel.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, et al. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, et al. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425:721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- 46.Liu MJ, Liu ML, Shen YF, Kim JM, Lee BH, Lee YS, et al. Transgenic mice with neuron-specific overexpression of HtrA2/Omi suggest a neuroprotective role for HtrA2/Omi. Biochem Biophys Res Commun. 2007;362:295–300. doi: 10.1016/j.bbrc.2007.07.118. [DOI] [PubMed] [Google Scholar]
- 47.Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum Mol Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 48.Skorko-Glonek J, Zurawa-Janicka D, Koper T, Jarzab M, Figaj D, Glaza P, et al. HtrA protease family as therapeutic targets. Curr Pharm Des. 2013;19:977–1009. doi: 10.2174/1381612811319060003. [DOI] [PubMed] [Google Scholar]
- 49.Li B, Hu Q, Wang H, Man N, Ren H, Wen L, et al. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ. 2010;17:1773–1784. doi: 10.1038/cdd.2010.55. [DOI] [PubMed] [Google Scholar]
- 50.Ren H, Fu K, Mu C, Li B, Wang D, Wang G. DJ-1, a cancer and Parkinson’s disease associated protein, regulates autophagy through JNK pathway in cancer cells. Cancer Lett. 2010;297:101–108. doi: 10.1016/j.canlet.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Xu R, Hu Q, Ma Q, Liu C, Wang G. The protease Omi regulates mitochondrial biogenesis through the GSK3beta/PGC-1alpha pathway. Cell Death Dis. 2014;5:e1373. doi: 10.1038/cddis.2014.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo D, Ying Z, Wang H, Chen D, Gao F, Ren H, et al. Regulation of autophagic flux by CHIP. Neurosci Bull. 2015;31:469–479. doi: 10.1007/s12264-015-1543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dagda RK, Chu CT. Mitochondrial quality control: insights on how Parkinson’s disease related genes PINK1, parkin, and Omi/HtrA2 interact to maintain mitochondrial homeostasis. J Bioenerg Biomembr. 2009;41:473–479. doi: 10.1007/s10863-009-9255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cilenti L, Lee Y, Hess S, Srinivasula S, Park KM, Junqueira D, et al. Characterization of a novel and specific inhibitor for the pro-apoptotic protease Omi/HtrA2. J Biol Chem. 2003;278:11489–11494. doi: 10.1074/jbc.M212819200. [DOI] [PubMed] [Google Scholar]
- 55.Yuan Y, Zhang X, Zheng Y, Chen Z. Regulation of mitophagy in ischemic brain injury. Neurosci Bull. 2015;31:395–406. doi: 10.1007/s12264-015-1544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unal Gulsuner H, Gulsuner S, Mercan FN, Onat OE, Walsh T, Shahin H, et al. Mitochondrial serine protease HTRA2 p. G399S in a kindred with essential tremor and Parkinson disease. Proc Natl Acad Sci U S A. 2014;111:18285–18290. doi: 10.1073/pnas.1419581111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian JY, Guo JF, Wang L, Sun QY, Yao LY, Luo LZ, et al. Mutation analysis of LRRK2, SCNA, UCHL1, HtrA2 and GIGYF2 genes in Chinese patients with autosomal dorminant Parkinson’s disease. Neurosci Lett. 2012;516:207–211. doi: 10.1016/j.neulet.2012.03.086. [DOI] [PubMed] [Google Scholar]
- 58.Olzmann JA, Brown K, Wilkinson KD, Rees HD, Huai Q, Ke H, et al. Familial Parkinson’s disease–associated L166P mutation disrupts DJ-1 protein folding and function. J Biol Chem. 2004;279:8506–8515. doi: 10.1074/jbc.M311017200. [DOI] [PubMed] [Google Scholar]
- 59.Maita C, Maita H, Iguchi-Ariga SM, Ariga H. Monomer DJ-1 and its N-terminal sequence are necessary for mitochondrial localization of DJ-1 mutants. PLoS One. 2013;8:e54087. doi: 10.1371/journal.pone.0054087. [DOI] [PMC free article] [PubMed] [Google Scholar]