Abstract
Communication, the effective delivery of information, is fundamental to life across all scales and species. Nervous systems (by necessity) may be most specifically adapted among biological tissues for high rate and complexity of information transmitted, and thus, the properties of neural tissue and principles of its organization into circuits may illuminate capabilities and limitations of biological communication. Here, we consider recent developments in tools for studying neural circuits with particular attention to defining neuronal cell types by input and output information streams—i.e., by how they communicate. Complementing approaches that define cell types by virtue of genetic promoter/enhancer properties, this communication-based approach to defining cell types operationally by input/output (I/O) relationships links structure and function, resolves difficulties associated with single-genetic-feature definitions, leverages technology for observing and testing significance of precisely these I/O relationships in intact brains, and maps onto processes through which behavior may be adapted during development, experience, and evolution.
Introduction
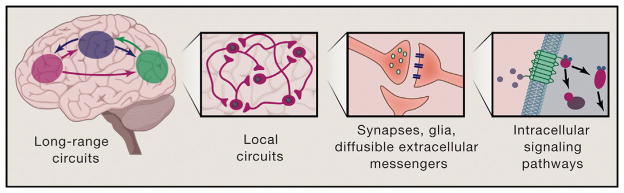
Nervous systems are designed for communication over many scales (Figure 1), beginning at the most fundamental level shared by all cellular systems in biology, in which communication occurs via protein-protein interactions, movement of second messengers within cells, and local release and detection of diffusible transmitters between cells. Nervous systems become clearly unique in their communication properties only at the tissue and organ level, in which billions of cells may work together as an intricately organized, interconnected circuit. It is through the organization of cells into these neural circuits that the brain supports the vast diversity of animal behavior, up to and including human consciousness, cognition, and emotion.
Figure 1. Nervous Systems Are Designed for Communication over Many Scales.
Nervous systems communicate at the brainwide level, the circuit level, the intercellular (synaptic) level, and the intracellular level (shown left to right). While the latter levels are fundamental to all biological systems, the more complex brainwide and circuit levels of communication distinguish the nervous system and support the unique function of this highly specialized tissue. Opportunities for new discovery in neural communication are abundant across these scales of analysis.
Neural circuits are both extremely complex and exquisitely specific, and the connectivity motifs used to build these circuits vary widely even within a single organism. Contrast the mammalian cerebellar granule neuron, which may receive only five mossy fiber inputs (Llinas et al., 2004) with the mammalian cortical pyramidal neuron, which receives thousands of inputs from a broad array of cortical and subcortical brain regions (Ballesteros-Yáñez et al., 2006). As with inputs, output structuring of neuronal types is also highly diverse, with a broad range of numbers and distributions of both local downstream neurons and distant postsynaptic partners across the nervous system. Indeed, each neuron type might be viewed as a distinct elemental device, definable in part by how it communicates via receiving, processing, and disseminating information. Understanding communication in the nervous system will require analyzing the input/output organization of these elements within larger neural circuits, observing the actual operation of these elements during behavior, and testing hypotheses built on this knowledge with model-guided perturbations targeted to these elements to determine the behaviorally relevant dynamics of information flow and processing.
Given the fundamental necessity of cell-cell communication for brain function, neuroscientists have long devoted substantial effort to developing and deploying technologies for exploring the structure and function of brain communication networks. Although many decades of neuroanatomical research have provided foundational principles underlying neural circuit organization, much remains to be discovered, and opportunities for discovery are particularly abundant at the borders between communication scales (Figure 1). Recent technological developments are indeed beginning to allow neuroscientists to connect neuronal circuit architecture and activity information across different scales and modalities. These methods are advancing the understanding of circuits in behaviorally relevant contexts, while at the same time heightening the need for cell typology that is more tightly linked to function, in order to define the cellular properties that are most relevant for nervous system operation. In this primer, we focus on currently available and rapidly evolving technologies for such structural and functional circuit-level analysis—with attention to both opportunities and limitations—and highlight the concept of the input/output (I/O)-defined circuit element (IODE) as a basic and recent experimentally tractable building block for the study and understanding of nervous system communication across scales.
Structural Definition of Communicating Circuit Elements: Molecules and Wiring
Which neuron types communicate with which other neuron types, and how is this relevant to behavior? For more than 100 years, dating back to the first elegant and prescient hand-drawn arrows depicting putative information flow between specific kinds of neurons (defined by shape and location) from Santiago Ramon y Cajal and his students, neuroscientists have presumed that the study of brain function will depend in part on the identification of cellular connections that mediate information transfer. Ramon y Cajal was able to combine a simple and robust neuronal visualization technique (the Golgi stain) with his keen observer’s eye and a systematic workflow to infer a great deal about neuronal communication despite the many limitations of the methodology.
Since Cajal’s era, many other anatomical tracing methods, which to various extents address some of the limitations of Golgi staining, have become widely utilized in the field (Table S1). Established techniques include dyes such as FluoroGold and other injectable markers taken up by cells that can give rise to fluorescent, pigmented, or electron-dense signals suitable for examination of long-range projections across the brain (Honig and Hume, 1989; Katz and Iarovici, 1990; Katz et al., 1984; Naumann et al., 2000; Reiner et al., 2000). Several proteins have also been adapted for neuronal tracing (Table S1; Conte et al., 2009; Gerfen and Sawchenko, 1984; Kissa et al. 2002; LaVail and LaVail, 1972; Schwab et al., 1978). Although these protein tracers are not solely retrograde or anterograde in all systems, they can be effective when used in the context of separately validated circuit anatomy (e.g., Gradinaru et al., 2010; Gunaydin et al., 2014; Xu et al., 2012). Some of these tracers (e.g., wheat germ agglutinin [WGA]) provide the additional leverage of trans-synaptic labeling, in which the cell bodies of neurons synaptically connected to a “starter-cell” population (those cells which initially contained the marker) are labeled, although restriction of the label only to monosynaptic connections cannot in principle be guaranteed by these proteins alone without further engineering and genetic targeting (e.g., using the GAL4-UAS system in Drosophila or zebrafish or recombinase-driver lines and engineered viral vectors in mammals as discussed below).
The relatively recent availability of engineered viral vectors for circuit tracing has driven rapid and substantial progress in the investigation of neural circuits, particularly in mammals. One of the most commonly used vector types is derived from adeno-associated viruses (AAVs). AAVs are now a workhorse tool for circuit mapping since they can be engineered to safely deliver genes encoding protein markers to neurons within a practical size limit set by the viral capsid capacity (~5 kB). Moreover, they are relatively cheap, can be concentrated to high titers (~1013 viral genomes/mL), and are safe (BSL-1). A simple but powerful example of the use of AAVs for circuit mapping comes from the Allen Brain Institute’s Mouse Connectivity Database (http://connectivity.brain-map.org), a growing collection of projection-mapping experiments (Oh et al., 2014). These experiments are carried out by injecting AAVs expressing yellow fluorescent protein (YFP) as a cell-filling marker, under the control of a partially cell-type-specific promoter or in a recombinase-dependent manner when injected into recombinase-driver mouse lines. Projections of the YFP-expressing population can then be visualized in individual serial sections or with 3D rendering across many sections.
The power of this resource derives from its remarkable breadth; experiments in the Mouse Connectivity Database are not hypothesis-driven, but rather serve as an openly accessible resource for hypothesis generation and testing by other labs. A limitation of the YFP cell-filling approach is that the traced cells, though specified by cell-body location and, in some cases, also by a genetic feature, are not specified by critically important properties of neurons: input and output. For example, it is unclear if the traced cells (which have axonal projections observed in a particular region) actually give rise to axonal terminations in the slice corresponding to that region. To address this issue, other circuit-tracing strategies have been developed that take advantage of the ability of certain viruses (e.g., rabies, herpes simplex virus [HSV], and canine adenovirus [CAV]) to efficiently transduce axon terminals, thus specifying neurons by their outputs. Modern anatomical methods can thus be used to deepen our understanding of communication pathways in the brain by resolving such output-defined elements (ODEs; an abbreviation useful in this context) of the circuit (Figure 2).
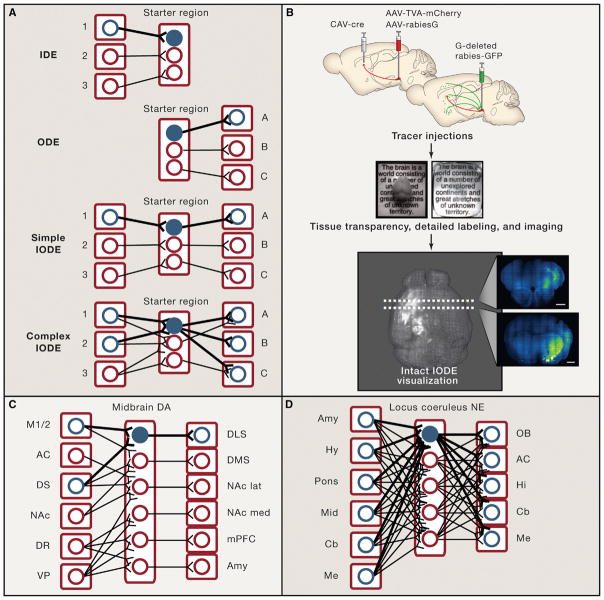
Figure 2. Input/Output-Defined Elements in the Nervous System.
(A) Input and output defined elements (highlighted in blue) as nervous system building blocks are schematized. An input-defined element (IDE) is a cell-type defined by location, cell origin, and/or activity of its afferent structures. An output-defined element (ODE) is a cell-type defined by location, cell target, and/or activity of its efferent structures. An I/O-defined element (IODE) is specified by both input and output anatomy and activity as defined above. In a simple case, a cell might serve the purpose of processing and relaying information from input site 1 to output site A. In a more complex case, a cell might integrate and process information from input sites 1 and 2, then relay its output to multiple brain regions (output sites A, B, and C).
(B) Intact-system methods for visualizing IODEs. After IODE tracer injections (e.g., those involved in implementing TRIO; Schwarz et al., 2015), whole brains can be clarified (e.g., using CLARITY; adapted with permission from Chung et al., 2013) and intact IODEs can be visualized in the fully intact organ (Lerner et al., 2015, Menegas et al., 2015). Scale bars on optical coronal sections are 1 mm. The IODE visualized here shows inputs from motor cortex and striatum to DLS-projecting midbrain dopamine neurons, as schematized in C (adapted with permission from Lerner et al., 2015).
(C and D) IODEs observed in recent studies of the midbrain dopamine (DA) system (B) and the locus coeruleus norepinephrine (NE) system (C). Midbrain dopamine neurons form distinct, though sometimes complex, IODEs (Beier et al., 2015; Lerner et al., 2015), whereas locus coeruleus norepinephrine neurons are not readily distinguishable by either input or output (Schwarz et al., 2015). M1/2, primary and secondary motor cortices; AC, anterior cingulate; DS, dorsal striatum; DLS, dorsolateral striatum; DMS, dorsomedial striatum; NAc, nucleus accumbens; NAc lat, NAc lateral shell; NAc med, NAc medial shell; DR, dorsal raphe; VP, ventral pallidum; mPFC, medial prefrontal cortex; Amy, amygdala; Hy, hypothalamus; Mid, midbrain; Cb, cerebellum; Me, medulla; OB, olfactory bulb; Hi, hippocampus.
Viral definition of ODEs can be further refined by layering onto output definition an additional cell-type characteristic, such as neurotransmitter production defined by a genetic marker. One such early approach to circuit mapping based on both an axonal target and a genetic feature (Fenno et al., 2014) involved delivering an HSV (expressing Flp recombinase in a Cre-recombinase-dependent manner) to the axon target region of interest (in this case, nucleus accumbens [NAc] of tyrosine hydroxylase [TH]-Cre recombinase-driver mice). Only the dopaminergic (TH+) cells arising from the ventral tegmental area (VTA), into which a separate Flp-dependent construct carried by an AAV had been introduced, were able to express the payload from the AAV, since only these cells had been able to produce Flp from the Cre-dependent construct delivered via HSV. In this way, cells were triply defined and targeted based on cell body location (VTA), a genetic feature (TH+), and an axon termination target (NAc). This example illustrates how multiple features are required to identify cell types, since NAc-projecting VTA cells may be dopaminergic, GABAergic, or even glutamatergic, and VTA dopamine cells may project to the prefrontal cortex, amygdala, and dorsal striatum in addition to the NAc. It is only when the axonal target and genetic criteria are combined that one may specifically isolate NAc-projecting dopamine cells in the VTA for study.
CAV (Soudais et al., 2001) has also been used to direct recombinases (e.g., Cre or Flp) to neurons projecting to a particular output region of interest and can be combined with transgenic mouse driver lines (Beier et al., 2015; Lerner et al., 2015; Schwarz et al., 2015). For example, CAV has been used to examine central monoaminergic cells defined by projection target in order to ask whether noradrenergic or dopaminergic neurons that project to one region of the brain also send collateral projections to other regions (Beier et al., 2015; Lerner et al., 2015; Schwarz et al., 2015). These results revealed disparate circuit properties of different neuromodulator cell populations. While all noradrenergic neurons in the locus coeruleus collateralize broadly, distinct dopaminergic neuron types (especially those intermixed in the substantia nigra pars compacta) form separable output pathways to distinct downstream targets. These examples demonstrate the productive investigation of ODEs using axon-transducing viruses such as HSV and CAV.
Viral tracing is especially versatile because the targeting properties of the virus itself (e.g., axon transduction) can be easily multiplexed with targeting capabilities afforded by the proteins it is engineered to express. For example, AAVs can be used to deliver not just single-marker proteins such as YFP, but also trans-synaptic tracer proteins like WGA or the components of even more refined neuronal tracing systems. One important example of the latter is GRASP (GFP-reconstitution across synaptic partners), an elegant split-GFP technique that allows fluorescent marking of close (likely synaptic) contacts between membranes of two cells. Though originally developed in C. elegans (Feinberg et al., 2008), GRASP has been adapted for use in mammals (mGRASP; Kim et al., 2012). This system works by tethering split-GFP fragments to synaptic-targeting proteins in two cell populations suspected of forming connections onto one another. In mGRASP, one GFP fragment is fused with the intracellular targeting domain of neurexin-1β to target presynaptic sites, while the other GFP fragment is fused with a sequence from neuroligin-1 to target postsynaptic sites. The pre- and postsynaptic components of the mGRASP system can be directed specifically to two cell populations of interest using AAVs. Connections between these populations are then detected when the GFP fragments come into close contact at synapses and fluoresce as reconstituted GFP. A similar approach, SynView, operates by labeling only those connections where neurexin-1β and neuroligin-1 or neuroligin-2 first bind each other, giving specific information about synapses that naturally use these synaptic adhesion molecules (Tsetsenis et al., 2014). Both mGRASP and SynView can offer information about synaptic locations for enriching information gained by visualizing processes and thus can be used to define with some precision pre- and postsynaptic partners in potential communicative events.
Another neuronal labeling technique that enriches anatomical information beyond single-color labeling is Brainbow (Livet et al., 2007). Brainbow mice have been engineered to express randomized combinations of fluorophores in each cell, a property that allows researchers to trace processes of individual neurons even among other densely packed processes. Brainbow technology is not only available for tracing in mice, but also has been widely adapted for use in Drosophila and zebrafish (Hadjieconomou et al., 2011; Hampel et al., 2011; Pan et al., 2011). Recent improvements (e.g., Brainbow 3.0, 3.1, and 3.2; Cai et al., 2013) have enhanced expression and detection; not only can individual fluorophores be stained using specific antibodies to further enhance signals, but also the “default” fluorophore present in nontargeted [i.e., Cre recombinase-negative] cells has been eliminated in favor of a non-fluorescent marker protein (Phi-YFP), which can be visualized by immunostaining if desired. Thus, by crossing Brainbow 3.2 transgenic mice with a Cre-driver line or by injecting a Cre-expressing AAV, one can visualize only a subset of neurons identified by recombinase expression and further distinguish individual cells within that group. The detailed single-axon analyses permitted by Brain-bow—and complementary technologies such as MAGIC Marker or CLoNe (García-Moreno et al., 2014; Loulier et al., 2014)—may open the door to applications resolving axon distribution diversity across development and hence accessing different ODEs within circuitry (Figure 2).
The techniques discussed so far primarily involve tracing the outputs of single cells or, in the case of GRASP/mGRASP, identifying the connections of two predefined partner types. In contrast, newer trans-synaptic tracing techniques allow the broad labeling of cells across the brain with axons forming connections onto a postsynaptic starter-cell population—the former (labeled afferent) cells are defined by a feature of their output and hence are also ODEs. To the extent that trans-synaptic tracing experiments with different starter-cell populations reveal different afferent patterns, the latter (postsynaptic starter-cell) populations can in turn be contrasted and thus considered input-defined elements (IDEs).
Certain of the tracers mentioned above (e.g., WGA and PHA-L) are able to cross synapses; however, with time, these can cross multiple synapses in series and thus the identification of direct connections using these reagents alone is not assured. To limit input tracing to monosynaptic connections, Callaway and colleagues (Wickersham et al., 2007) developed a system in which a modified rabies virus lacks an essential glycoprotein needed for trans-synaptic transport. The glycoprotein can then be provided only to the population of starter cells from which input tracing will occur, ensuring rigorous single-synapse definition of the afferent ODEs. Rabies viruses can also be further engineered to refine the starter-cell population (and hence the ODEs synapsing onto the starter-cell population) by pseudotyping with the coat protein EnvA, which causes infection to depend on TVA (avian tumor virus receptor), an avian receptor not found in the mammalian brain. Exogenous TVA in turn can be selectively expressed in the desired starter-cell subpopulation, such as midbrain dopamine neurons (Ogawa et al., 2014; Watabe-Uchida et al., 2012), serotonin neurons (Ogawa et al., 2014; Pollak Dorocic et al., 2014; Weissbourd et al., 2014), director indirect-pathway striatal projection neurons (Wall et al., 2013), and striatal cholinergic interneurons (Guo et al., 2015).
Recently, several groups have extended this circuit-building toolbox with different approaches to make starter-cell populations not only potentially input defined, but also output defined by integrating the concept of projection-targeted recombinase delivery (Fenno et al., 2014). In one such approach (Beier et al., 2015; Lerner et al., 2015; Schwarz et al., 2015) termed TRIO (tracing the relation between input and output; Schwarz et al., 2015), it is possible to examine monosynaptic inputs to starter-cell neural populations under conditions in which the latter are also defined by their output targets (e.g., using retrograde transport of Cre or Flp recombinase packaged in a CAV). Differing-input starter cells resolved in this way can then be considered IODEs (Figure 2). Using this approach Lerner et al. (2015) showed that the inputs to midbrain dopamine neuron starter cells are biased depending on the projection target of these starter cells, thus resolving IODEs (this result contrasted with noradrenergic neurons in the locus coeruleus, which receive relatively homogeneous inputs regardless of output; Schwarz et al., 2015). In combination with studies of neural collateralization (in the same papers and described above), it was possible to conclude that dopamine neurons are equipped to communicate specific input signals tailored to distinct output brain structures, whereas noradrenergic neurons may broadcast communications more generally across the brain. As in Fenno et al. (2014), TRIO also can be engineered for yet further refinement of the starter cell population by a genetic feature in addition to axonal target (conditional TRIO or cTRIO; e.g., in DAT::cre driver mice; Beier et al., 2015).
Another form of IODE tracing has also recently been published (Menegas et al., 2015). Here, instead of using retrograde transport of a recombinase to specify starter cells, the authors used retrograde transport of TVA, packaged into an AAV (some AAVs can transduce CNS axons, though more variably and weakly than CAV or HSV). It is important to note when using this strategy that the rabies glycoprotein should ideally also be delivered according to a retrograde strategy to prevent tracing from cells not belonging to the correct ODE; otherwise, disynaptic tracing can occur due to connectivity properties among glycoprotein-expressing cells within the local microcircuitry. Many other strategies for IODE definition are now possible (see Table S1 for a summary of available IODE building blocks), which should all involve careful consideration of the scientific question at hand and the relevant circuit anatomy.
Though the above brainwide methods are heavily dependent on fluorescent markers and thus may be generally constrained by the limits of light microscopy, where indicated, these may be followed up with higher-resolution local studies (e.g., leveraging super-resolution light microscopy, electron microscopy, and/or array tomography for detailed synaptic analysis; Atasoy et al., 2014; Kim et al., 2014b; Maglione and Sigrist, 2013; Micheva and Smith, 2007; Ragan et al., 2012) to further resolve the fine structure of IODEs. While engineered-virus, tracer protein, and dye-based methods are generally limited to animal models, the results arising may be compared as needed with corresponding (though less precisely defined) structural features in human brains inferred from diffusion-weighted imaging (DWI), fMRI, or histology (Table S1). Such comparisons may be useful for translating basic findings on circuit structure in animal models into clinical diagnostic tools or interventions.
High-resolution anatomical studies enabled by the technologies described above are substantially advancing the understanding of neural circuit connectivity, yet the throughput of such experiments becomes an issue when dealing with brain-wide investigations and in light of the need to be confident that (for example) the source or origin of a population of axons has not been missed or misidentified. Here, intact-brain analyses (as enabled by CLARITY and other whole-organ tissue transparency methods; Chung et al., 2013; Ertürk et al., 2012; Hama et al., 2011; Renier et al., 2014; Susaki et al., 2014; Richardson and Lichtman, 2015), especially when compatible with rich molecular information as may be obtained with multiplexed protein and RNA analysis, have provided key leverage, along with high-speed and high-resolution light-sheet microscopy methods such as COLM (CLARITY-optimized light-sheet microscopy; Tomer et al., 2014; Lerner et al., 2015; Tomer et al., 2015) and emerging methods for automated counting and quantitative analysis of circuit components (Kim et al., 2015b; Menegas et al., 2015). Integration of circuit-labeling techniques (Table S1) with analyses enabled by whole-brain transparency, labeling, imaging, and analysis methods (Table S2) promises to accelerate progress in dissecting IODEs, setting the stage for functional circuit investigation.
Functional Definition of Circuit Elements: Wiring-Dependent Optogenetic Control
The anatomical methods outlined above have greatly enhanced investigation of communication pathways in the brain by allowing highly refined definition of potential sources and targets. However, these methods alone lack access to the actual content and functional significance of the communication. Thus, neuroscientists increasingly seek to combine anatomical observations with both activity perturbations and activity readouts to form a complete picture of circuit information processing, which may include computations performed at the cellular level on incoming information, as well as modulation of the global dynamics of information flow among neural circuit components in vivo. Integration of anatomical maps of communication pathways, with complementary functional approaches to observe and control activity events themselves, has proceeded along several dimensions, all enabled by recent technology development.
The method of optogenetics (Yizhar et al., 2011; Deisseroth 2014) has allowed functional characterization of connectivity motifs by enabling temporally precise manipulation of defined neural circuit elements in living systems, both in slice preparations and in vivo. Optogenetics involves the expression of single microbial proteins, which permit light-activated regulation of ion flow in genetically targeted neurons, resulting in cell-type-specific neuronal control during behavior. Optogenetics thus dovetails well with the structural methods described above that involve expression of single fluorescent marker proteins in cell-type-specific neuronal populations; other capabilities, limitations, and technical considerations of optogenetics have been recently reviewed (Grosenick et al., 2015; Deisseroth, 2015). Crucial to the success of optogenetics for functional circuit mapping, microbial opsins such as channelrhodopsins and halorhodopsins are trafficked into axons (a process typically enhanced with molecular engineering) and can also be delivered by recombinase-activated labeling strategies traveling retrograde from synapses; hence, the functional communication of ODEs can be modulated using the leverage of their defined anatomical outputs (Figure 3).
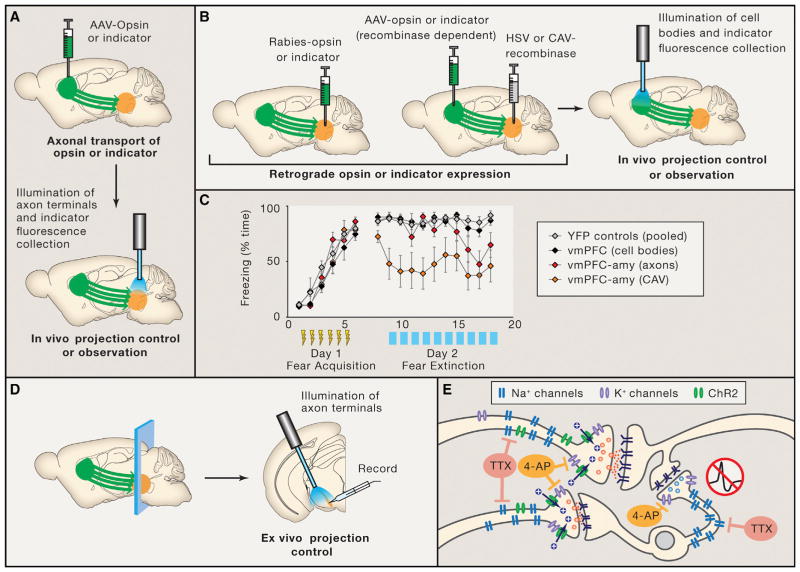
Figure 3. Functional Methods for Circuit Element Mapping In Vivo and Ex Vivo.
(A) The expression of opsins or activity indicators in axon terminals allows for functional and anatomical circuit element definition in vivo, achieved by specific fiberoptic-based illumination of axon terminals.
(B) Specific stimulation or observation of a projection can also be achieved by using a retrograde virus such as rabies, HSV, or CAV to express an opsin or activity indicator only in cells that have a specific efferent target. In this case, cell bodies may be illuminated directly.
(C) An example of free mouse behavior during optical control of output-defined elements (ODEs). In this case, the ventromedial prefrontal cortex (vmPFC) to amygdala (amy) projection was manipulated during fear conditioning and extinction using three different strategies. First, non-specific stimulation of vmPFC cell bodies (black symbols); second, stimulation of vmPFC axon terminals in the amygdala (red symbols); third, stimulation of vmPFC cell bodies which send projections to the amygdala (orange symbols; identified by CAV-cre injections in the basomedial amygdala; Adhikari et al., 2015). Increasingly specific output definition of the stimulation circuit element elicits increasingly potent effects on the cued freezing (fear memory) behavior. Yellow bolts indicate six shock-tone pairings given on the training day. Blue bars indicate the time of blue light stimulation of the target circuit element on the extinction day (when tones are played, but no shocks delivered). Gray symbols, YFP (no-opsin) control cohort. Adapted with permission.
(D) Expression of opsins in axons’ terminals also allows for optical control of defined circuit elements in the ex vivo slice electrophysiology preparation.
(E) Acute slice preparations allow for fine circuit dissection in controlled conditions, for example using TTX/4-AP to definitively isolate monosynaptic connections (Petreanu et al., 2009; Lerner et al., 2015; Adhikari et al., 2015). First, TTX blocks action-potential-dependent release, preventing disynaptic stimulation through non-opsin-expressing neurons. Second, 4-AP increases terminal excitability by blocking K+ channels. Channelrhodopsin (ChR) optical drive then induces action-potential-independent depolarization at ChR-expressing terminals only, while patch clamping of different target cells thus allows definition of afferent fibers as output-defined elements (ODEs).
Not only can the axons of genetically specified groups of neurons then be controlled (stimulated or inhibited) by light delivered directly to projections during behavior, but subsequent living acute slice preparations can be taken from the target regions and used to study the functional connectivity of these projections in isolation with single-cell resolution (Figure 3D). For example, in cases where it is of interest to define the monosynaptic (direct) target of ODEs, drugs may be applied to the extracellular recording solution in slice preparations to prevent the occurrence of polysynaptic events (Petreanu et al., 2009). In one version of this method, sodium channels are blocked using tetrodotoxin (TTX), which prevents action-potential-driven release events, while at the same time, voltage-dependent potassium channels are blocked using 4-aminopyridine (4-AP) to facilitate direct depolarization of only channelrhodopsin-expressing axon terminals by blue light, detected by whole-cell patch clamp in putative, directly postsynaptic cells (Figure 3E). This approach is useful for isolating monosynaptic connections between cell types and brain regions as well as for subcellular mapping of functional synapses (Little and Carter, 2012; 2013; MacAskill et al., 2012; Petreanu et al., 2009; Sun et al., 2014), but inasmuch as this approach involves direct actuation in nerve terminals of channelrhodopsin (which fluxes Ca2+ ions as well as Na+, K+, and H+), it is important to not draw detailed conclusions about natural synaptic release dynamics (which are highly Ca2+-sensitive) from this sort of work. An as yet unidentified Ca2+-impermeable channelrhodopsin, which would still depolarize the presynaptic terminal strongly enough to drive natural Ca2+ influx via voltage-gated Ca2+ channels, might be of value in some cases where the focus is not on simple presence or absence of direct synaptic connections.
Nevertheless, this approach to defining direct communication partners has been useful, for example, in facilitating direct IODE definition in the study discussed above (Lerner et al., 2015) that isolated direct striatal inputs to midbrain dopamine neurons and functionally contrasted the inputs arriving from distinct subregions of the striatum to output-defined subpopulations of dopamine cells. Adhikari et al. (2015) also employed this optogenetic monosynaptic connectivity method together with CLARITY and viral tracing to discover a direct connection between ventromedial prefrontal cortical (mPFC) and basomedial amygdala, which turned out to be behaviorally important for top-down regulation of fear and anxiety responses. Also studying communication between the mPFC and amygdala, Little and Carter (2013) used two-photon optogenetic methods to determine the density and distribution of amygdalar inputs onto mPFC cells, demonstrating how optogenetics can be employed for detailed functional mapping of synaptic locations. Demonstrating the potential health relevance of these functional approaches to circuit mapping, optogenetic recruitment of cells and synapses defined by a specific connectivity feature (distinct long-range afferent projections to the NAc) has been applied to probe detailed hypotheses on the synaptic basis of cocaine addiction (Britt et al., 2012; Creed et al., 2015; Pascoli et al., 2014).
Most of the above patch-clamp studies focused on defining direct monosynaptic neuronal connections, but an integrative view of circuit-element output might further consider effects of diverse interacting downstream cell populations, since both monosynaptic and polysynaptic connectivity associated with the output brain region will be important in sculpting elicited activity. Reduced slice preparations may be selected if a specific hypothesis is to be tested regarding local circuit modulation of activity (e.g., the role of sparse interneuron populations recruited by feedback inhibition to modulate local dynamics in slice preparations; Sohal et al., 2009). Conversely, more exploratory brainwide analyses of functional connectivity may be conducted, in which global outputs (resulting from activity in an optogenetically defined cell population) can be measured in a regionally unbiased fashion throughout the brain. The latter approach has been taken using electrophysiological postsynaptic readouts (Chuhma et al., 2011; Mingote et al., 2015) or fMRI (Ferenczi et al., 2016; Lee et al., 2010); in the fMRI case, focal regions in mPFC were found to exert specific and behaviorally relevant influence over the manner in which distant brain regions communicated with each other (Ferenczi et al., 2016). A brainwide database of these second-order interactions was provided (Ferenczi et al., 2016), illustrating how precise modulation of intended direct targets in a brain region (as will occur in natural or experimental settings) exerts influence by accessing the intact brain as a dynamical system, with relevance in this case to top-down control of physiological behavioral state transitions.
Optogenetic analysis of communication in brain circuitry has been employed not only with the physiology readouts noted above, but also in freely moving animals to provide information on the causal relationships between neural circuit activity patterns and behavior (reviewed in Deisseroth, 2014 and Steinberg et al., 2015). Using (for example) fiberoptic neural interfaces, optogenetics can be used in the behavioral setting to stimulate, inhibit, or modulate a population of cell bodies within a brain region or to address a specific ODE in vivo (using either selective illumination of opsin-expressing projection terminals or the retrograde-opsin-expression strategy to recruit cells by a feature of their connectivity; Figures 3A and 3B; Deisseroth, 2014; Steinberg et al., 2015). Behavioral optogenetics experiments have demonstrated the utility of these approaches for resolving the effects of defined circuit elements; indeed, more specific behavioral effects are often observed when resolving cells by projection target instead of generally illuminating cell bodies without regard to projection target (Adhikari et al., 2015; Kim et al., 2013; Warden et al., 2012). For example, Warden et al. (2012) found that optogenetic activation of prefrontal cortical projections to the dorsal raphe nucleus selectively modulated behavioral state (favoring active coping defined by motivated escape behavior in the forced swim test, while not generally increasing locomotor activity) in a manner that depended on specific activation of that pathway (in contrast, nonspecific stimulation of the prefrontal cortex or dorsal raphe, or stimulation of other projections from prefrontal cortex, did not cause the same specific effect profile). In a separate study, Kim et al. (2013) examined several projections arising from a single brain area (the bed nucleus of the stria terminalis [BNST]) that each selectively recruited distinct features (risk avoidance, respiratory rate, or conditioning value) of anxiety-related behavioral state transitions. Upstream of the BNST, Adhikari et al. (2015) studied fear and anxiety modulated by recruitment of distinct cortico-amygdalar projections, observing that fear extinction was enhanced by specific top-down pathway recruitment, but not by non-specific stimulation of cortical cell bodies (Figure 3C).
These studies, and many others like them, indicate that defining circuit elements by structural I/O features (alone or in combination with other features such as genetic markers) is a tractable experimental approach that maps onto nervous system structure-function relationships more precisely than simple regional stimulation. Notably, optical stimuli (as delivered to these structural elements of interest) can be readily mapped in parametric fashion by varying light intensity and timing. Of course, without incorporation of pre-existing knowledge of native activity patterns, experimenter-defined activity traffic along a given neural communication pathway is unlikely to precisely match the natural dynamics of the pathway. Nevertheless, specific modulation of relevant complex behaviors is still typically observed, revealing that defining these elements by their detailed I/O structure alone may describe meaningful communication in the circuit. When data are available on endogenous activity patterns relating to encoding or transformation of information, optogenetic methods can additionally take into account these data, as discussed next.
Activity Readouts for Delineating Input- and Output-Defined Circuit Elements
A complete picture of I/O properties for specific circuit elements would include not just anatomy, but also activity in the form of naturally occurring neuronal signals along the anatomical I/O pathways of interest. A diverse array of compatible tools for reading out activity and examining the information processing as executed by the circuit (Table S3) indeed now allows layering of this crucial dimension onto anatomical circuit maps. Such an integrated approach allows certain questions across a range of scales to be addressed that would be difficult to answer from anatomy or from optogenetics alone.
Traditional electrophysiological approaches to examining activity in vivo bring the highest temporal resolution but are fundamentally limited in terms of accessibility of cell type and wiring information; moreover, these are not readily able to monitor activity in axons, which would be important for providing pathway specificity just as modulation of axons has provided pathway specificity in optogenetic studies. Crucially, then, it is difficult to cast electrophysiological data in the same framework as the structural (physical and molecular) and optogenetic-control datastreams discussed above. Although temporal resolution with fluorescence Ca2+ signals (for example, as recorded with genetically encoded Ca2+ indicators) is not as high as with electrophysiology, the cell-type and pathway specificity provided is invaluable when interfacing activity data with anatomical information. This field is rapidly advancing (e.g., Chen et al., 2013) and now extends to faster genetically encoded voltage sensors as well (e.g., Gong et al., 2015); for the purpose of the primer, we focus here on developments that are most immediately and directly linked to the anatomical and optogenetic methods described above for delineating brainwide I/O defined elements.
A fluorescence recording approach, termed fiber photometry (Gunaydin et al., 2014), has been developed for genetically encoded activity sensors and is particularly well suited for monitoring specific ODEs in cell bodies (Cui et al., 2013) and even in deep-brain genetically defined fiber tracts during behavior (Gunaydin et al., 2014; Kim et al., 2016). Fiber photometry is a photon-counting (photometric) strategy built on a fiberoptic interface targeted to the brain area or axonal tract of interest, with optics designed to detect even small-activity fluorescence signals arising from genetically encoded Ca2+-indicator proteins in axons deep in the brain of behaving mammals (Gunaydin et al., 2014). Fiber photometry permits real-time observations of the activity along specific axonal projections defined by origin and target and complements methods for imaging superficially exposed axons with conventional objectives in behaving animals (De Paola et al., 2006; Grutzendler et al., 2002; Lovett-Barron et al., 2014). In the initial demonstration of fiber photometry in deep projections (Gunaydin et al., 2014), Ca2+-indicator expression was targeted to mouse VTA dopamine neurons and activity in the projections of these neurons to the NAc was monitored; thus, the circuit element was defined by outgoing projection anatomy as well as by neurotransmitter phenotype. It was observed that endogenous activity of this output stream was robustly modulated during social interaction but much less so during novel-object interaction in the same mice; optogenetic control over the same projection then revealed that this was a causally significant signal in the behavior. A next-generation version of this method, frame-projected independent-fiber photometry (FIP), has now been developed for recording fluorescence activity signals from many brain regions or deep-brain fiber tracts simultaneously in behaving mice and for tuning optogenetic perturbation to elicit dynamics matching patterns occurring naturally in behavior (Kim et al., 2016). Together, these fiber photometry examples illustrate the utility of optical readouts for resolving activity magnitude and timing in projection-defined elements during free behavior.
In vivo photometry has been applied to many target elements throughout the brain (including cell bodies as well; Chen et al., 2015; Cui et al., 2013; Lerner et al., 2015; Zalocusky et al., 2016; Lütcke et al., 2010; Schulz et al., 2012) and has also recently been used to delineate IDEs, in the sense of differing activity observed in defined circuit elements during behavior. For example, if the somata of two genetically similar cell populations exhibit opposite-direction activity changes in response to the same behavioral stimulus, these two populations are likely to at some level receive different input stimuli and therefore represent IDEs. A recent study (Lerner et al., 2015) demonstrated such IDEs alongside anatomical analysis of inputs and outputs, which also differed for the same populations; using fiber photometry in mice, substantia nigra pars compacta neurons that project to dorsolateral striatum were observed to exhibit activity elevations in response to both appetitive and aversive stimuli, while those projecting to dorsomedial striatum exhibited activity elevations in response to appetitive stimuli but decreased activity in response to aversive stimuli. These two cell populations, though neither genetically nor spatially separable, were in fact thus shown to be communicating separable streams of information (Lerner et al., 2015). This finding opens up avenues for further exploration and illustrates the informative directions that can be taken as IODE characterization reframes models of circuit organization.
Although fiber photometry was designed for ease of use in freely moving behavior as well as direct compatibility with typical anatomical tracing and optogenetic control datastreams, cellular-resolution imaging can be applied as desired for more detailed and complementary information (just as anatomy and optogenetics readily also allow cellular-resolution work in more restricted fields of view). In an example linking behavior, anatomy, optogenetics, and cellular-resolution imaging, Rajasethupathy et al. (2015) found that an ODE from anterior cingulate cortex selectively influences a sparse population of “hub” neurons in the hippocampus that are highly correlated with other cells in the local network during memory retrieval. The discovery of this rare cell type and the observation of local circuit dynamics required the use of single-cell-resolution two-photon imaging to effectively link the anatomical and functional lines of evidence pursued.
Such high-resolution and highly local optical readout of activity elicited by control of defined circuit elements is currently complemented by more global (even brainwide) readouts that are also well suited to reporting on effects of ODE activity. Brainwide activity readout (sacrificing spatial and temporal resolution) can be achieved via optogenetic fMRI (ofMRI) BOLD ( blood oxygen level dependent) signals (Lee et al., 2010) or (achieving single-cell resolution while still further sacrificing temporal resolution) via immediate early gene (IEG)-based readouts (e.g., IEG-immunohistochemical labeling or IEG-promoter-driven expression of cell-filling fluorescent proteins, as in the TRAP (targeted recombination in active populations) method; Guenthner et al., 2013). Together, these examples illustrate how activity readouts can complement I/O mapping of neural circuit elements and set the stage for diverse research directions combining activity readouts with functional manipulations (e.g., optogenetics) alongside structural anatomical studies (including via whole-brain tissue clearing, pathway tracing, and molecular labeling).
Mesoscale Elements of Communication: the Input/Output-Defined Cell Type
Certain anatomical and functional approaches to circuit mapping rely to some extent on a simplifying assumption: that the group of cells being labeled and traced belongs to a discrete “type.” How these cell types are defined can profoundly influence the interpretation of experiments, yet our definitions of cell types are rapidly evolving. A survey of cell types in the brain is among the early goals of the US BRAIN Initiative (Jorgenson et al., 2015), yet consensus is still lacking as to how to best define and organize such categories. Many of the viral/genetic strategies for circuit mapping described above rely, as a matter of practicality, on single-feature recombinase-driver lines to define cell type. Other studies have focused on careful quantifications of morphology to create categories. Nevertheless, each neuron, like each snowflake, is unique. Were fully detailed criteria to be applied, each cell would form its own new class, but such excessively detailed categorization would not provide useful overarching principles describing circuit function (for example, each cell in each mammal’s hippocampus is slightly different, but in general, hippocampi appear to solve the problem of spatial navigation similarly). Therefore, neuroscientists must make educated judgments about which elements of cell-type definition are likely to be most meaningful for developing workable theories of brain communication.
We suggest here that the genetic and morphology markers used most commonly thus far are proxies for the neural elements that really matter for circuit function: inputs and outputs defined by wiring and activity. Other markers are not unsuitable by any means, but the field should move where possible toward defining cell types directly in terms of their circuit function. Among other useful aspects, thinking about cell types in IODE terms will create organizing links between molecular/cellular neuroscientists and systems neuroscientists and draw attention to molecular and cellular elements that give neurons particular input- and output-defined circuit properties. Working from such shared concepts may facilitate synthesis of findings and productive interchanges and even promote engagement of computational and theoretical neuroscientists since the resulting datasets will be well-suited to closed-loop and system-identification approaches (Grosenick et al., 2015; Figure 4).
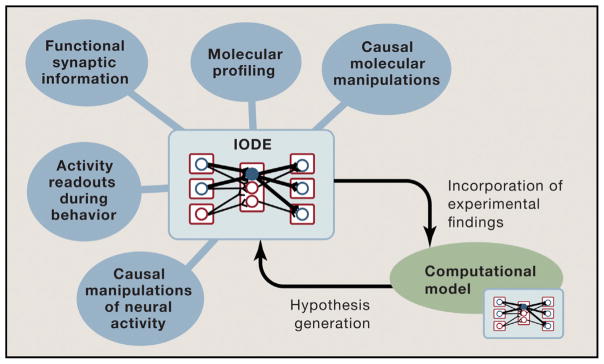
Figure 4. Organizing Principles for Cross-Modal Investigation of Neural Circuits.
IODEs may help provide a behaviorally relevant and experimentally tractable framework for guiding and integrating information about neural circuits across many levels of investigation. Once the basic physical structure of an IODE is understood from the anatomical tools described here (including the most relevant convergence and divergence of information through collaterals when considering a specific behavior), activity information during behavior from physiology, imaging, and molecular datastreams can be collected using targeting tools aligned with the IODE structure and then layered onto the diagram to form a more complete understanding of I/O relationships. Computational analyses may facilitate registration and joint interpretation of information gathered by these disparate techniques, as well as generation of higher-order hypotheses to guide further data collection (e.g., through system-identification strategies; Grosenick et al., 2015). Iterations of this data-collection and hypothesis-generation cycle, and crucially the linking of distinct IODEs into loops and more complex topological structures, may continue until experimental and theoretical concepts converge.
To bring these ideas to a concrete example, we note that dopamine neurons are currently undergoing a revolution in their classification that illustrates how and why shifts in cell-type definition take place. Until recently, dopamine neurons (as suggested by the name) had been defined primarily by their production and release of the neurotransmitter dopamine; TH, the rate-limiting enzyme in catecholamine production, is often used as one of several molecular markers. Yet, to be useful as a conceptual building block toward understanding brain function, this definition should imply that all dopamine neurons have at least somewhat similar roles in their brain circuits, an assumption that is widely understood in neuroscience to be false, for at least three critical reasons.
First and most simply, dopamine neurons can be subdivided based on their outputs to distinct brain regions, which include the prefrontal cortex, amygdala, NAc core, NAc medial shell, NAc lateral shell, dorsomedial striatum, and dorsolateral striatum, the pituitary gland, the chemoreceptor trigger zone, and many other targets. The dopaminergic projections to these different output regions are largely parallel, meaning that information communicated by a dopamine neuron will be received largely by just a single target output region. It is only when dopamine neurons are viewed from this structural-output-defined perspective that observations of opposite-valence responses to stimuli by different subsets of dopamine neurons become interpretable (Kim et al., 2014a; Lerner et al., 2015; Matsumoto and Hikosaka, 2009).
Second, though the neurotransmitter released might be considered (and is) important, different dopamine neurons also release diverse other neurotransmitters, including GABA and glutamate, which profoundly influence how these neurons participate in a circuit. For example, cholinergic interneurons in the striatum inhibit striatal projection neurons by stimulating release of GABA from dopamine neuron terminals (Nelson et al., 2014). As above, connectivity matters: it appears that dopamine neurons projecting to the NAc, but not to the dorsal striatum, co-release glutamate (Chuhma et al., 2014; Mingote et al., 2015; Stuber et al., 2010). Furthermore, this glutamate release causes burst firing behavior in ventral, but not dorsal, striatal cholinergic interneurons in response to dopamine neuron stimulation (Chuhma et al., 2014), with profound significance for circuit function. This example illustrates the importance of the structural-output- and activity-output-defined perspective for discriminating cell types.
Third and finally, input definitions also turn out to be critical for understanding dopamine neurons. Projection-defined populations of dopamine neurons receive a different balance of inputs from other brain regions, in terms of the numbers of afferents, functional strength, and functional consequences of representing completely different appetitive or aversive (rewarding or punishing) environmental stimuli (Beier et al., 2015; Lerner et al., 2015; Watabe-Uchida et al., 2012). The example of dopamine neurons clearly delineates the shortcomings of defining a cell type by a molecular feature only, as with transgenic mouse lines. However, it is not meant to suggest that other aspects of a cell’s phenotype are irrelevant, and I/O must be considered functionally as well as anatomically. For example, VTA GABA neurons may have similar input and output anatomy at some level of inspection compared to VTA dopamine neurons (Beier et al., 2015) but are clearly a different cell type as would be readily distinguished by the effect of their output on downstream structures.
The true power of the IODE-centered definition of different neuronal cell types derives in large part from its ability to interface with other modes of cell-type investigation and will not replace but rather build upon molecular labels. Again turning to the midbrain dopamine system as a model for use of this framework, it has been found that defining dopamine neurons by projection target immediately leads to appreciation of diversity in subtype-specific expression of the dopamine transporter (DAT), dopamine D2 autoreceptors, GIRK channels, and HCN channels (Lammel et al., 2011; Lerner et al., 2015; Margolis et al., 2008). As a result of this diversity, subtypes of dopamine neurons may also differ in their pacemaking mechanisms, an observation that may help explain the progressive pattern of degeneration in Parkinson’s disease (Chan et al., 2007; Khaliq and Bean, 2010; Puopolo et al., 2007). Further molecular investigations of dopaminergic IODEs, e.g., via molecular-profiling techniques (Ekstrand et al., 2014; Namburi et al., 2015), are likely to yield even more detailed insights into the overall organization of the system, which is not possible when dopamine neuron subpopulations are grouped as one. Though some unbiased automated discovery of cell types may be possible from single-cell sequencing data (Grün and van Oudenaarden, 2015), it may also be fruitful, perhaps even more so, to pursue unbiased cell-type discovery from connectivity datasets (Jonas and Kording, 2015). At the very least, the use of single genetic or anatomical features in isolation may cause neuroscientists to ignore meaningful sources of variability in data and thus hamper progress toward deeper understanding of the fundamental and versatile building blocks of communication and computation in the brain.
Summary: Confronting Realities of Communication Complexity and Scale in the Brain
Transitioning to I/O definition of cell types is no longer fully technologically limited, but formidable barriers remain. Among these barriers is that conceptual and analytical models have lagged behind experimental and technological advances. The resulting limitation manifests at many levels, ranging from data handling to guiding and interpreting experiments.
Regarding data handling, fast progress in circuit analysis is already dramatically accelerating the rate of dataset acquisition, as well as the size of individual discrete and compressed datasets. Even from a single laboratory, modern structural and activity datasets can reach the petabyte scale each year, creating challenges for both storage and processing. Clearly, utility will be limited by the ability of individual labs as well as the broader community to access and work with these datasets. How can neuroscientists best coordinate data handling to allow for the emergence of transformative new theories of brain function? Central open-access organization may be crucial, and some larger efforts at brain mapping, such as the Allen Institute’s efforts, have developed platforms for scientists to search, view, and manipulate the data generated. The Human Connectome Project (http://www.humanconnectomeproject.org/) also maintains an accessible database of human imaging data for download. Nevertheless, there is no single database where reliable data from all levels of analysis (e.g., anatomy, control, and activity) is incorporated together in a common language, nor is there an interface that allows easy back-and-forth communication between experimental and theoretical approaches to circuit function, as new information becomes available. We suggest that a unifying database employing the IDE/ODE/IODE framework, in which data from these different modalities can be expressed in the same terms, may help advance the type of rapid-cycle communication between experimentalists and theorists that will soon become indispensible as the complexity of our circuit diagrams increases.
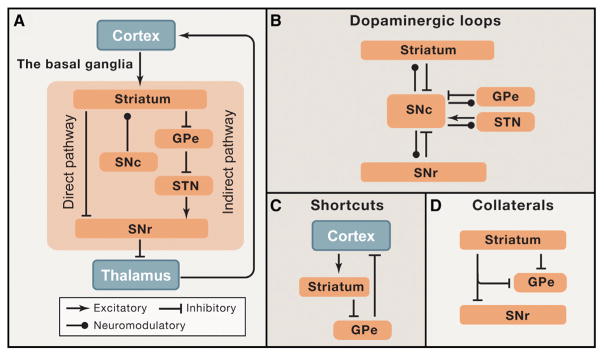
As a current pressing example of circuit complexity, we turn to the basal ganglia (BG), a highly interconnected group of subcortical nuclei that may play a role in (among other behaviors) action selection and motor learning. What circuit-level organizing concepts currently exist for BG? Many discussions of the function of the BG to date have relied on the simple concept of a “direct” and an “indirect” pathway offering two alternative, opposing streams of information flow from the input nucleus of the BG (striatum) to the output nucleus (SNr; Figure 5A). Yet reality is well known to be substantially more complex, involving multiple feedback loops and spirals among nuclei (Figures 5B–D; Alexander et al., 1986; Cazorla et al., 2014; Haber, 2003; Kupchik et al., 2015; Mallet et al., 2012; Nambu et al., 2002; Saunders et al., 2015). The richness of this structure is both enticing and intimidating; there are clearly many more discoveries to be made about BG circuit function, yet already the complexity of our simplified diagram makes forming an intuitive understanding of circuit dynamics (and hence experimental design and data interpretation) very difficult. Of course, due to rich collateralization, individual IODES in the BG and beyond may play separable roles in different brainwide structures, with topologies that become increasingly challenging to visualize and to represent.
Figure 5. Example: Different Levels of Inspection for Basal Ganglia Circuitry.
(A) Simplified diagram of basal ganglia (BG) circuitry depicts the “direct” and “indirect” pathways, which have opposing influences on BG output. While the concepts of the direct and indirect pathways have yielded important insights, the reality of BG circuitry is much more complex. (B–D) Examples of additional circuit complexity in the BG.
(B) Dopamine neurons in the substantia nigra pars compacta (SNc) project not only to the striatum but to other BG nuclei. These dopamine neurons also receive direct projections back from these nuclei. (C) Information need not loop all the way through the cortico-BG-thalamic circuitry. Several shortcuts are available, including the one pictured in which the globus pallidus external segment (GPe) sends projections back to the cortex (Saunders et al., 2015). (D) The “direct” and “indirect” pathways are not absolute. For example, some “direct pathway” striatal neurons also send collaterals to the GPe (Cazorla et al., 2014). A more sophisticated understanding of BG circuit dynamics may emerge as we build testable hypotheses based on a more realistic picture of the circuitry as shown in (B–D). Such an approach will be facilitated by defining, controlling, and observing cells based on their input and output properties in the intact functioning system (see Figure 4).
To break the standoff between accessibility and realism, computational approaches are a key part of the solution (Figure 4). Such approaches can be used to generate models of neural circuit communication that can be tested further by experimentalists. Results generated by experimentalists can then support further refinement of the circuit model, iterating until the experimental and theoretical concepts converge (see Phillips, 2015 for discussion relevant to biology more generally). As this iteration proceeds, it will motivate and incorporate new technological innovations as well, such as the advent of single-cell control with two-photon and spatial-light modulator-based play-in of optogenetic control over cell ensembles in vivo (Prakash et al., 2012; Packer et al., 2012; Rickgauer et al., 2014; Packer et al., 2015; Reutsky-Gefen et al., 2013; Szabo et al., 2014) or controlling ensembles based on their past involvement in behavior in the same animal (e.g., Liu et al., 2012). Such progress may also eventually bring insight into the development and plasticity of neural circuitry, as IDEs, ODEs, and IODEs may be well suited to serve as primitive building blocks that self-assembling circuitry can employ and adapt, to evolve loops and more intricate (perhaps as yet undiscovered) topologies as well as complex behaviors from individual cells and cell pairs.
In summary, although the challenges involved in understanding intact brain function remain formidable, there are considerable opportunities on the horizon for breakthroughs that may have a substantial impact on basic research as well as on the understanding of disease. Technological developments across many modalities, including progress in anatomical tracing and molecular-profiling techniques, innovations in optogenetic control, and advances in diverse activity readouts, are driving fundamental changes in the way that neuroscientists work. Organized thinking about communication in neural circuits may in itself help in organizing ties among researchers operating within these different modalities and from other biology and engineering disciplines.
Supplementary Material
Acknowledgments
T.N.L. is supported by an NRSA Postdoctoral Fellowship (1F32MH105053-01). K.D. is supported by the National Institute of Mental Health, National Institute on Drug Abuse, the Wiegers Family Fund, the Nancy and James Grosfeld Foundation, the H.L. Snyder Medical Foundation, the Samuel and Betsy Reeves Fund, and the U.S. Army Research Laboratory and Defense Advanced Research Projects Agency (Cooperative Agreement Number W911NF-14-2-0013); nothing in this material represents official views or policies of our funders. We acknowledge R. Tomer and A. Adhikari for their work on the data cited and shown in Figures 2 and 3, respectively, and we thank R. Poldrack, B. Knutson, and the entire Deisseroth lab for advice and support. All tools and methods are distributed and supported freely (www.optogenetics.org, clarityresourcecenter.org).
Footnotes
Supplemental Information includes three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.02.027.
AUTHOR CONTRIBUTIONS
T.N.L. and K.D. wrote the paper. L.Y. contributed to the summary of tissue transparency methods.
References
- Adhikari A, Lerner TN, Finkelstein J, Pak S, Jennings JH, Davidson TJ, Ferenczi E, Gunaydin LA, Mirzabekov JJ, Ye L, et al. Basomedial amygdala mediates top-down control of anxiety and fear. Nature. 2015;527:179–185. doi: 10.1038/nature15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderón NC, Esposti F, Borghuis BG, Sun XR, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander GE, DeLong MR, Strick PL. Parallel organization of functionally segregated circuits linking basal ganglia and cortex. Annu Rev Neurosci. 1986;9:357–381. doi: 10.1146/annurev.ne.09.030186.002041. [DOI] [PubMed] [Google Scholar]
- Atasoy D, Betley JN, Li WP, Su HH, Sertel SM, Scheffer LK, Simpson JH, Fetter RD, Sternson SM. A genetically specified connectomics approach applied to long-range feeding regulatory circuits. Nat Neurosci. 2014;17:1830–1839. doi: 10.1038/nn.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Yáñez I, Benavides-Piccione R, Elston GN, Yuste R, DeFelipe J. Density and morphology of dendritic spines in mouse neocortex. Neuroscience. 2006;138:403–409. doi: 10.1016/j.neuroscience.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, Luo L. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell. 2015;162:622–634. doi: 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt JP, Benaliouad F, McDevitt RA, Stuber GD, Wise RA, Bonci A. Synaptic and behavioral profile of multiple glutamatergic inputs to the nucleus accumbens. Neuron. 2012;76:790–803. doi: 10.1016/j.neuron.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10:540–547. [PubMed] [Google Scholar]
- Cazorla M, de Carvalho FD, Chohan MO, Shegda M, Chuhma N, Rayport S, Ahmari SE, Moore H, Kellendonk C. Dopamine D2 receptors regulate the anatomical and functional balance of basal ganglia circuitry. Neuron. 2014;81:153–164. doi: 10.1016/j.neuron.2013.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Guzman JN, Ilijic E, Mercer JN, Rick C, Tkatch T, Meredith GE, Surmeier DJ. ‘Rejuvenation’ protects neurons in mouse models of Parkinson’s disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin YC, Kuo TW, Knight ZA. Sensory detection of food rapidly modulates arcuate feeding circuits. Cell. 2015;160:829–841. doi: 10.1016/j.cell.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuhma N, Tanaka KF, Hen R, Rayport S. Functional connectome of the striatal medium spiny neuron. J Neurosci. 2011;31:1183–1192. doi: 10.1523/JNEUROSCI.3833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuhma N, Mingote S, Moore H, Rayport S. Dopamine neurons control striatal cholinergic neurons via regionally heterogeneous dopamine and glutamate signaling. Neuron. 2014;81:901–912. doi: 10.1016/j.neuron.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte WL, Kamishina H, Reep RL. Multiple neuroanatomical tract-tracing using fluorescent Alexa Fluor conjugates of cholera toxin subunit B in rats. Nat Protoc. 2009;4:1157–1166. doi: 10.1038/nprot.2009.93. [DOI] [PubMed] [Google Scholar]
- Creed M, Pascoli VJ, Lüscher C. Addiction therapy Refining deep brain stimulation to emulate optogenetic treatment of synaptic pathology. Science. 2015;347:659–664. doi: 10.1126/science.1260776. [DOI] [PubMed] [Google Scholar]
- Cui G, Jun SB, Jin X, Pham MD, Vogel SS, Lovinger DM, Costa RM. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–242. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paola V, Holtmaat A, Knott G, Song S, Wilbrecht L, Caroni P, Svoboda K. Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 2006;49:861–875. doi: 10.1016/j.neuron.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Deisseroth K. Circuit dynamics of adaptive and maladaptive behaviour. Nature. 2014;505:309–317. doi: 10.1038/nature12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–1225. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Nectow AR, Knight ZA, Latcha KN, Pomeranz LE, Friedman JM. Molecular profiling of neurons based on connectivity. Cell. 2014;157:1230–1242. doi: 10.1016/j.cell.2014.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt HU. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczi EA, Zalocusky KA, Liston C, Grosenick L, Warden MR, Amatya D, Katovich K, Mehta H, Patenaude B, Ramakrishnan C, et al. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 2016;351:aac9698. doi: 10.1126/science.aac9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Moreno F, Vasistha NA, Begbie J, Molnár Z. CLoNe is a new method to target single progenitors and study their progeny in mouse and chick. Development. 2014;141:1589–1598. doi: 10.1242/dev.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Sawchenko PE. An anterograde neuroanatomical tracing method that shows the detailed morphology of neurons, their axons and terminals: immunohistochemical localization of an axonally transported plant lectin, Phaseolus vulgaris leucoagglutinin (PHA-L) Brain Res. 1984;290:219–238. doi: 10.1016/0006-8993(84)90940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, Schnitzer MJ. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 2015;350:1361–1366. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosenick L, Marshel JH, Deisseroth K. Closed-loop and activity-guided optogenetic control. Neuron. 2015;86:106–139. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, van Oudenaarden A. Design and Analysis of Single-Cell Sequencing Experiments. Cell. 2015;163:799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Wang D, He X, Feng Q, Lin R, Xu F, Fu L, Luo M. Whole-brain mapping of inputs to projection neurons and cholinergic interneurons in the dorsal striatum. PLoS ONE. 2015;10:e0123381. doi: 10.1371/journal.pone.0123381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber SN. The primate basal ganglia: parallel and integrative networks. J Chem Neuroanat. 2003;26:317–330. doi: 10.1016/j.jchemneu.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat Methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hampel S, Chung P, McKellar CE, Hall D, Looger LL, Simpson JH. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nat Methods. 2011;8:253–259. doi: 10.1038/nmeth.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig MG, Hume RI. Dil and diO: versatile fluorescent dyes for neuronal labelling and pathway tracing. Trends Neurosci. 1989;12:333–335. 340–341. [PubMed] [Google Scholar]
- Jonas E, Kording K. Automatic discovery of cell types and microcircuitry from neural connectomics. eLife. 2015;4:e04250. doi: 10.7554/eLife.04250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson LA, Newsome WT, Anderson DJ, Bargmann CI, Brown EN, Deisseroth K, Donoghue JP, Hudson KL, Ling GSF, MacLeish PR, et al. The BRAIN Initiative: developing technology to catalyse neuroscience discovery. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0164. pii: 20140164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, Iarovici DM. Green fluorescent latex microspheres: a new retrograde tracer. Neuroscience. 1990;34:511–520. doi: 10.1016/0306-4522(90)90159-2. [DOI] [PubMed] [Google Scholar]
- Katz LC, Burkhalter A, Dreyer WJ. Fluorescent latex micro-spheres as a retrograde neuronal marker for in vivo and in vitro studies of visual cortex. Nature. 1984;310:498–500. doi: 10.1038/310498a0. [DOI] [PubMed] [Google Scholar]
- Khaliq ZM, Bean BP. Pacemaking in dopaminergic ventral tegmental area neurons: depolarizing drive from background and voltage-dependent sodium conductances. J Neurosci. 2010;30:7401–7413. doi: 10.1523/JNEUROSCI.0143-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Zhao T, Petralia RS, Yu Y, Peng H, Myers E, Magee JC. mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat Methods. 2012;9:96–102. doi: 10.1038/nmeth.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Adhikari A, Lee SY, Marshel JH, Kim CK, Mallory CS, Lo M, Pak S, Mattis J, Lim BK, et al. Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature. 2013;496:219–223. doi: 10.1038/nature12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HF, Ghazizadeh A, Hikosaka O. Separate groups of dopamine neurons innervate caudate head and tail encoding flexible and stable value memories. Front Neuroanat. 2014a;8:120. doi: 10.3389/fnana.2014.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Greene MJ, Zlateski A, Lee K, Richardson M, Turaga SC, Purcaro M, Balkam M, Robinson A, Behabadi BF, et al. EyeWirers. Space-time wiring specificity supports direction selectivity in the retina. Nature. 2014b;509:331–336. doi: 10.1038/nature13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 2015b;10:292–305. doi: 10.1016/j.celrep.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CK, Yang SJ, Pichamoorthy N, Young NP, Kauvar I, Jennings JH, Lerner TN, Berndt A, Lee SY, Ramakrishnan C, et al. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat Methods. 2016 doi: 10.1038/nmeth.3770. Published online February 15, 2016 http://dx.doi.org/10.1038/nmeth.3770. [DOI] [PMC free article] [PubMed]
- Kissa K, Mordelet E, Soudais C, Kremer EJ, Demeneix BA, Brûlet P, Coen L. In vivo neuronal tracing with GFP-TTC gene delivery. Mol Cell Neurosci. 2002;20:627–637. doi: 10.1006/mcne.2002.1141. [DOI] [PubMed] [Google Scholar]
- Kupchik YM, Brown RM, Heinsbroek JA, Lobo MK, Schwartz DJ, Kalivas PW. Coding the direct/indirect pathways by D1 and D2 receptors is not valid for accumbens projections. Nat Neurosci. 2015;18:1230–1232. doi: 10.1038/nn.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Ion DI, Roeper J, Malenka RC. Projection-specific modulation of dopamine neuron synapses by aversive and rewarding stimuli. Neuron. 2011;70:855–862. doi: 10.1016/j.neuron.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail JH, LaVail MM. Retrograde axonal transport in the central nervous system. Science. 1972;176:1416–1417. doi: 10.1126/science.176.4042.1416. [DOI] [PubMed] [Google Scholar]
- Lee JH, Durand R, Gradinaru V, Zhang F, Goshen I, Kim DS, Fenno LE, Ramakrishnan C, Deisseroth K. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465:788–792. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, Deisseroth K. Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell. 2015;162:635–647. doi: 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JP, Carter AG. Subcellular synaptic connectivity of layer 2 pyramidal neurons in the medial prefrontal cortex. J Neurosci. 2012;32:12808–12819. doi: 10.1523/JNEUROSCI.1616-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JP, Carter AG. Synaptic mechanisms underlying strong reciprocal connectivity between the medial prefrontal cortex and basolateral amygdala. J Neurosci. 2013;33:15333–15342. doi: 10.1523/JNEUROSCI.2385-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Llinas RR, Walton KD, Lang EJ. Cerebellum. Synaptic Organization of the Brain. 2004;Chapter 7 [Google Scholar]; Synaptic Organization of the Brain. Chapter 7. Springer; US: [Google Scholar]
- Loulier K, Barry R, Mahou P, Le Franc Y, Supatto W, Matho KS, Ieng S, Fouquet S, Dupin E, Benosman R, et al. Multiplex cell and lineage tracking with combinatorial labels. Neuron. 2014;81:505–520. doi: 10.1016/j.neuron.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Lovett-Barron M, Kaifosh P, Kheirbek MA, Danielson N, Zaremba JD, Reardon TR, Turi GF, Hen R, Zemelman BV, Losonczy A. Dendritic inhibition in the hippocampus supports fear learning. Science. 2014;343:857–863. doi: 10.1126/science.1247485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H, Murayama M, Hahn T, Margolis DJ, Astori S, Zum Alten Borgloh SM, Göbel W, Yang Y, Tang W, Kügler S, et al. Optical recording of neuronal activity with a genetically-encoded calcium indicator in anesthetized and freely moving mice. Front Neural Circuits. 2010;4:9. doi: 10.3389/fncir.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAskill AF, Little JP, Cassel JM, Carter AG. Subcellular connectivity underlies pathway-specific signaling in the nucleus accumbens. Nat Neurosci. 2012;15:1624–1626. doi: 10.1038/nn.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglione M, Sigrist SJ. Seeing the forest tree by tree: super-resolution light microscopy meets the neurosciences. Nat Neurosci. 2013;16:790–797. doi: 10.1038/nn.3403. [DOI] [PubMed] [Google Scholar]
- Mallet N, Micklem BR, Henny P, Brown MT, Williams C, Bolam JP, Nakamura KC, Magill PJ. Dichotomous organization of the external globus pallidus. Neuron. 2012;74:1075–1086. doi: 10.1016/j.neuron.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis EB, Mitchell JM, Ishikawa J, Hjelmstad GO, Fields HL. Midbrain dopamine neurons: projection target determines action potential duration and dopamine D(2) receptor inhibition. J Neurosci. 2008;28:8908–8913. doi: 10.1523/JNEUROSCI.1526-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Hikosaka O. Two types of dopamine neuron distinctly convey positive and negative motivational signals. Nature. 2009;459:837–841. doi: 10.1038/nature08028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Bergan JF, Ogawa SK, Isogai Y, Umadevi Venkataraju K, Osten P, Uchida N, Watabe-Uchida M. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife. 2015;4:e10032. doi: 10.7554/eLife.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingote S, Chuhma N, Kusnoor SV, Field B, Deutch AY, Rayport S. Functional Connectome Analysis of Dopamine Neuron Glutamatergic Connections in Forebrain Regions. J Neurosci. 2015;35:16259–16271. doi: 10.1523/JNEUROSCI.1674-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu A, Tokuno H, Takada M. Functional significance of the cortico-subthalamo-pallidal ‘hyperdirect’ pathway. Neurosci Res. 2002;43:111–117. doi: 10.1016/s0168-0102(02)00027-5. [DOI] [PubMed] [Google Scholar]
- Namburi P, Beyeler A, Yorozu S, Calhoon GG, Halbert SA, Wichmann R, Holden SS, Mertens KL, Anahtar M, Felix-Ortiz AC, et al. A circuit mechanism for differentiating positive and negative associations. Nature. 2015;520:675–678. doi: 10.1038/nature14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann T, Härtig W, Frotscher M. Retrograde tracing with Fluoro-Gold: different methods of tracer detection at the ultrastructural level and neurodegenerative changes of back-filled neurons in long-term studies. J Neurosci Methods. 2000;103:11–21. doi: 10.1016/s0165-0270(00)00292-2. [DOI] [PubMed] [Google Scholar]
- Nelson AB, Hammack N, Yang CF, Shah NM, Seal RP, Kreitzer AC. Striatal cholinergic interneurons Drive GABA release from dopamine terminals. Neuron. 2014;82:63–70. doi: 10.1016/j.neuron.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa SK, Cohen JY, Hwang D, Uchida N, Watabe-Uchida M. Organization of monosynaptic inputs to the serotonin and dopamine neuromodulatory systems. Cell Rep. 2014;8:1105–1118. doi: 10.1016/j.celrep.2014.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Peterka DS, Hirtz JJ, Prakash R, Deisseroth K, Yuste R. Two-photon optogenetics of dendritic spines and neural circuits. Nat Methods. 2012;9:1202–1205. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HW, Häusser M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Methods. 2015;12:140–146. doi: 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YA, Livet J, Sanes JR, Lichtman JW, Schier AF. Multi-color Brainbow imaging in zebrafish. Cold Spring Harb Protoc. 2011;2011:t5546. doi: 10.1101/pdb.prot5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoli V, Terrier J, Espallergues J, Valjent E, O’Connor EC, Lüscher C. Contrasting forms of cocaine-evoked plasticity control components of relapse. Nature. 2014;509:459–464. doi: 10.1038/nature13257. [DOI] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R. Theory in Biology: Figure 1 or Figure 7? Trends Cell Biol. 2015;25:723–729. doi: 10.1016/j.tcb.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Farah MJ. Progress and challenges in probing the human brain. Nature. 2015;526:371–379. doi: 10.1038/nature15692. [DOI] [PubMed] [Google Scholar]
- Pollak Dorocic I, Fürth D, Xuan Y, Johansson Y, Pozzi L, Silberberg G, Carlén M, Meletis K. A whole-brain atlas of inputs to serotonergic neurons of the dorsal and median raphe nuclei. Neuron. 2014;83:663–678. doi: 10.1016/j.neuron.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, Goshen I, Packer AM, Peterka DS, Yuste R, Schnitzer MJ, Deisseroth K. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puopolo M, Raviola E, Bean BP. Roles of subthreshold calcium current and sodium current in spontaneous firing of mouse midbrain dopamine neurons. J Neurosci. 2007;27:645–656. doi: 10.1523/JNEUROSCI.4341-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Sankaran S, Marshel JH, Kim CK, Ferenczi E, Lee SY, Berndt A, Ramakrishnan C, Jaffe A, Lo M, et al. Projections from neocortex mediate top-down control of memory retrieval. Nature. 2015;526:653–659. doi: 10.1038/nature15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Veenman CL, Medina L, Jiao Y, Del Mar N, Honig MG. Pathway tracing using biotinylated dextran amines. J Neurosci Methods. 2000;103:23–37. doi: 10.1016/s0165-0270(00)00293-4. [DOI] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Reutsky-Gefen I, Golan L, Farah N, Schejter A, Tsur L, Brosh I, Shoham S. Holographic optogenetic stimulation of patterned neuronal activity for vision restoration. Nat Commun. 2013;4:1509. doi: 10.1038/ncomms2500. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Deisseroth K, Tank DW. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat Neurosci. 2014;17:1816–1824. doi: 10.1038/nn.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Oldenburg IA, Berezovskii VK, Johnson CA, Kingery ND, Elliott HL, Xie T, Gerfen CR, Sabatini BL. A direct GABAergic output from the basal ganglia to frontal cortex. Nature. 2015;521:85–89. doi: 10.1038/nature14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz K, Sydekum E, Krueppel R, Engelbrecht CJ, Schlegel F, Schröter A, Rudin M, Helmchen F. Simultaneous BOLD fMRI and fiber-optic calcium recording in rat neocortex. Nat Methods. 2012;9:597–602. doi: 10.1038/nmeth.2013. [DOI] [PubMed] [Google Scholar]
- Schwab ME, Javoy-Agid F, Agid Y. Labeled wheat germ agglutinin (WGA) as a new, highly sensitive retrograde tracer in the rat brain hippocampal system. Brain Res. 1978;152:145–150. doi: 10.1016/0006-8993(78)90140-3. [DOI] [PubMed] [Google Scholar]
- Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, Luo L. Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature. 2015;524:88–92. doi: 10.1038/nature14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J. 2001;15:2283–2285. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- Steinberg EE, Christoffel DJ, Deisseroth K, Malenka RC. Illuminating circuitry relevant to psychiatric disorders with optogenetics. Curr Opin Neurobiol. 2015;30:9–16. doi: 10.1016/j.conb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber GD, Hnasko TS, Britt JP, Edwards RH, Bonci A. Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. J Neurosci. 2010;30:8229–8233. doi: 10.1523/JNEUROSCI.1754-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun QQ, Wang X, Yang W. Laserspritzer: a simple method for optogenetic investigation with subcellular resolutions. PLoS ONE. 2014;9:e101600–e101608. doi: 10.1371/journal.pone.0101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Szabo V, Ventalon C, De Sars V, Bradley J, Emiliani V. Spatially selective holographic photoactivation and functional fluorescence imaging in freely behaving mice with a fiberscope. Neuron. 2014;84:1157–1169. doi: 10.1016/j.neuron.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R, Lovett-Barron M, Kauvar I, Andalman A, Burns VM, Sankaran S, Grosenick L, Broxton M, Yang S, Deisseroth K. SPED Light Sheet Microscopy: Fast Mapping of Biological System Structure and Function. Cell. 2015;163:1796–1806. doi: 10.1016/j.cell.2015.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsenis T, Boucard AA, Araç D, Brunger AT, Südhof TC. Direct visualization of trans-synaptic neurexin-neuroligin interactions during synapse formation. J Neurosci. 2014;34:15083–15096. doi: 10.1523/JNEUROSCI.0348-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, De La Parra M, Callaway EM, Kreitzer AC. Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron. 2013;79:347–360. doi: 10.1016/j.neuron.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden MR, Selimbeyoglu A, Mirzabekov JJ, Lo M, Thompson KR, Kim SY, Adhikari A, Tye KM, Frank LM, Deisseroth K. A prefrontal cortex-brainstem neuronal projection that controls response to behavioural challenge. Nature. 2012;492:428–432. doi: 10.1038/nature11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Weissbourd B, Ren J, DeLoach KE, Guenthner CJ, Miyamichi K, Luo L. Presynaptic partners of dorsal raphe serotonergic and GABAergic neurons. Neuron. 2014;83:645–662. doi: 10.1016/j.neuron.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann KK, Young JAT, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Morishita W, Buckmaster PS, Pang ZP, Malenka RC, Südhof TC. Distinct neuronal coding schemes in memory revealed by selective erasure of fast synchronous synaptic transmission. Neuron. 2012;73:990–1001. doi: 10.1016/j.neuron.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Zalocusky KA, Ramakrishnan C, Lerner TN, Davidson TJ, Knutson B, Deisseroth K. Nucleus accumbens D2R neurons signal past unfavorable outcomes in decision-making. Nature. 2016 doi: 10.1038/nature17400. Published online March 23, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.