Abstract
Microbes produce a variety of secondary metabolites to be explored for herbicidal activities. We investigated an endophyte Pseudomonas viridiflava CDRTc14, which impacted growth of its host Lepidium draba L., to better understand the possible genetic determinants for herbicidal and host-interaction traits. Inoculation tests with a variety of target plants revealed that CDRTc14 shows plant-specific effects ranging from beneficial to negative. Its herbicidal effect appeared to be dose-dependent and resembled phenotypically the germination arrest factor of Pseudomonas fluorescens WH6. CDRTc14 shares 183 genes with the herbicidal strain WH6 but the formylaminooxyvinylglycine (FVG) biosynthetic genes responsible for germination arrest of WH6 was not detected. CDRTc14 showed phosphate solubilizing ability, indole acetic acid and siderophores production in vitro and harbors genes for these functions. Moreover, genes for quorum sensing, hydrogen cyanide and ACC deaminase production were also found in this strain. Although, CDRTc14 is related to plant pathogens, we neither found a complete pathogenicity island in the genome, nor pathogenicity symptoms on susceptible plant species upon CDRTc14 inoculation. Comparison with other related genomes showed several unique genes involved in abiotic stress tolerance in CDRTc14 like genes responsible for heavy metal and herbicide resistance indicating recent adaptation to plant protection measures applied in vineyards.
Introduction
Worldwide, weeds compromise crop yield and increase costs of crop production1. The weed problem on global food production has become severe due to increased use of synthetic herbicides, the rapidly evolving weed resistance against herbicides and negative impacts on the environment2,3. The employment of bioherbicides within the framework of an integrated weed management strategy has the potential to offer a number of benefits such as higher target specificity, faster degradation and lower (or no) toxicity than chemical herbicides. Despite the efforts of many researchers to find new bioherbicidal candidates in vitro and to test them under field conditions, only thirteen authorized bioherbicides derived from microorganisms or natural molecules are currently available on the market. Among them, only three are based on bacteria, ten on fungi, one is based on a virus and one contains natural plant extracts4,5.
Bacterial herbicides have many benefits over fungal herbicides, particularly due to quick growth of bacteria, relatively simple propagation and storage conditions, and greater amenability to genetic manipulation6,7. Many bacterial species/strains have been investigated as natural weed control agents, especially Pseudomonas fluorescens and Xanthomonas campestris, but only P. fluorescens strain WH6 has been investigated to understand the genetic basis of its herbicidal activity8. A culture filtrate of strain WH6 arrests the germination of a number of weedy grass species, including the annual bluegrass (Poa annua L.)9,10. The herbicidal activity of strain WH6 has been attributed to the production of formylaminooxyvinylglycine (FVG), originally referred to as the germination arrest factor, and the gvg gene cluster9,11 was reported to be responsible for the production of FVG11.
Pseudomonas viridiflava is a pectinolytic bacterium in the P. syringae species complex. Strains belonging to this species generally show a broad host range and can live either as pathogen and saprophyte12. Pathogenic strains of P. viridiflava generally have two pathogenicity islands [single (S-PAI) and tripartite (T-PAI)] and/or the avrE effector of the type III secretion system13. Despite of being the disease causative agent of multiple plant species, some P. viridiflava strains exhibit biocontrol activities against various human and plant pathogenic fungi14,15. Additionally, some strains have been shown to degrade various pesticides (Fenvalerate, an organophosphate) and polycyclic aromatic hydrocarbons (PAHs)16–18. Although P. viridiflava belongs to the well-known P. syringae complex, the biology and functioning of this species is not thoroughly investigated and only nine draft P. viridiflava genomes are available in the NCBI database.
In this study we particularly addressed strain CDRTc14, which we previously isolated from the weed Lepidium draba L. and which showed inhibiting effects towards its host. Based on whole genome average nucleotide identity (ANI), CDRTc14 was assigned to P. viridiflava 19. As this strain is related to plant pathogens and inhibited growth of its plant host we aimed to better understand the activities of this strain, its interaction with plants and underlying mechanisms by employing in-depth genome analysis, comparative genomics and functional tests.
Results and Discussion
Herbicidal activity
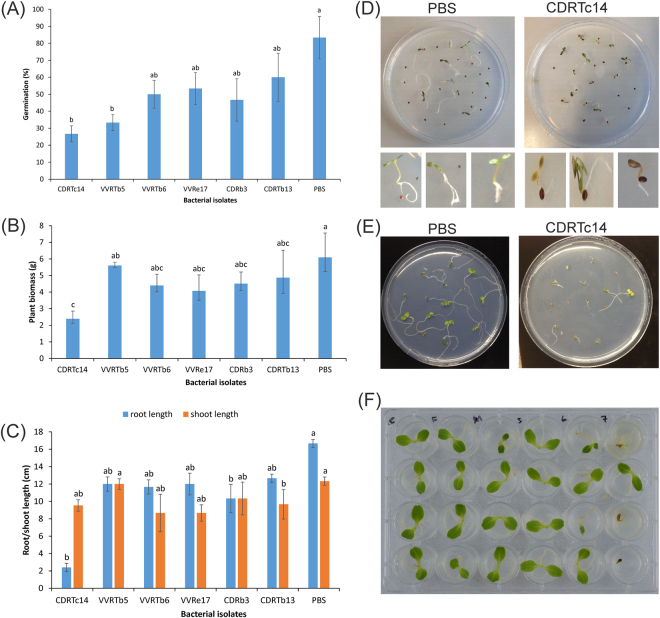
Generally, bacterial herbicides are known to exhibit a narrow host range, and they impact mostly closely related plant species8. Here, we evaluated the herbicidal effect of bacteria isolated from L. draba on its host and closely related plant species. Overall 98 isolates were tested for their phytotoxic effect on L. draba but only six isolates (P. viridiflava CDRTc14, P. fluorescens CDRTb13, Agromyces humatus CDRb3, Arthrobacter sp. VVRe17, Rhizobium sp. VVRTb6, Bosea sp. VVRTb5, Table S6) showed growth inhibition on L. draba in vitro and under greenhouse conditions (Fig. 1). Among them, CDRTc14 showed the most pronounced effect on L. draba (both in vitro and greenhouse). Further investigation revealed that CDRTc14 has a negative effect on lettuce (Lactuca sativa L.) growth but not on Arabidopsis thaliana (Fig. S1). To obtain more information on the host range of CDRTc14, we tested its effect on nine different plant species, which are close relatives of L. draba and belong to the Brassicaceae family. Interestingly, we observed differential effects (including positive, neutral or negative) on these plant species due to CDRTc14 inoculation (Table 1). For example, CDRTc14 significantly inhibited (−56%) the germination of the weed Sisymbrium officinale (belonging to the Brassicaceae linage I (Fig. S3)) as compared to the mock treatment. However, this strain significantly enhanced root growth (69%) and plant biomass (52%) of the Brassica napus variety Cleopatra also belonging to the Brassicaceae linage I. In contrast, CDRTc14 significantly improved root length (76%) and seedling biomass (148%), respectively, of the phylogenetically closest relatives (linage III) Lepidium meyenii and Erysimum odoratum. CDRTc14 did not cause a significant effect on other Brassicaceae species (B. napus variety Pharao (linage I), Hesperis matronalis and Lepidium sativum (linage II). Our results are not in accordance with the centrifugal phylogenetic method (CPM)20, postulating that near-neighbor species to the target are at greater risk of attack than distant species. However, similar results were reported for the herbicidal strain of Burkholderia andropogonis, which showed differential effects on species of the Caryophyllales and Fabaceae family21. Also the biocontrol agent P. fluorescens strain D7 specifically inhibits downy brome growth while it stimulates the growth of rapeseed (Brassica napus L.)22. However, the CPM has been considered as the standard for host-specificity testing of biological control agents since 1974 but it has been suggested to revise this hypothesis23 and also our results contradict this hypothesis.
Figure 1.
Bioherbicidal activity of CDRTc14 and other strains. (A–C) Results of greenhouse experiment showing germination percentage, plant biomass, root and shoot length. (D) Herbicidal effect of CDRTc14 on L. draba weed in vitro (E) Herbicidal effect of CDRTc14 on lettuce in vitro. (F) Dose dependent effect of CDRTc14 on lettuce, showing phytotoxic effect at 107 CFU ml−1. PBS (Phosphate-buffered saline) was used as control.
Table 1.
Percent change in plant growth from different Brassicaceae with P. viridiflava CDRTc14 inoculation compared to uninoculated control.
| Plant Species | Germination percentage | Root Length | Shoot Length | Seedling weight |
|---|---|---|---|---|
| Lepidium meyenii | −11.1 | 76.9** | 0.7 | 44.0 |
| Hesperis matronalis | −3.0 | −19.2 | −19.0 | −34.5 |
| Isatis tinctoria | −4.2 | −26.7 | 5.9 | 1.0 |
| Erysimum odoratum | 35.7 | −10.1 | 27.6 | 148** |
| Sisymbrium officinale | −56.3** | 5.5 | −1.3 | −25.4 |
| Lepidium sativum | 8.3 | 17.1 | 5.2 | 3.2 |
| B. napus var. Cleopatra | 30.4 | 69.0** | −3.3 | 52.1* |
| B. napus. var. Pharao | −3.3 | −2.8 | −5.3 | 4.6 |
Note: Values in the table indicates percent change compare to untreated control. The Student’s T−tests was applied to compare treatment means with control (PBS) and significant values are indicated as *p < 0.05, **p < 0.01, ***p < 0.001.
Genomic features and phylogenetic analysis
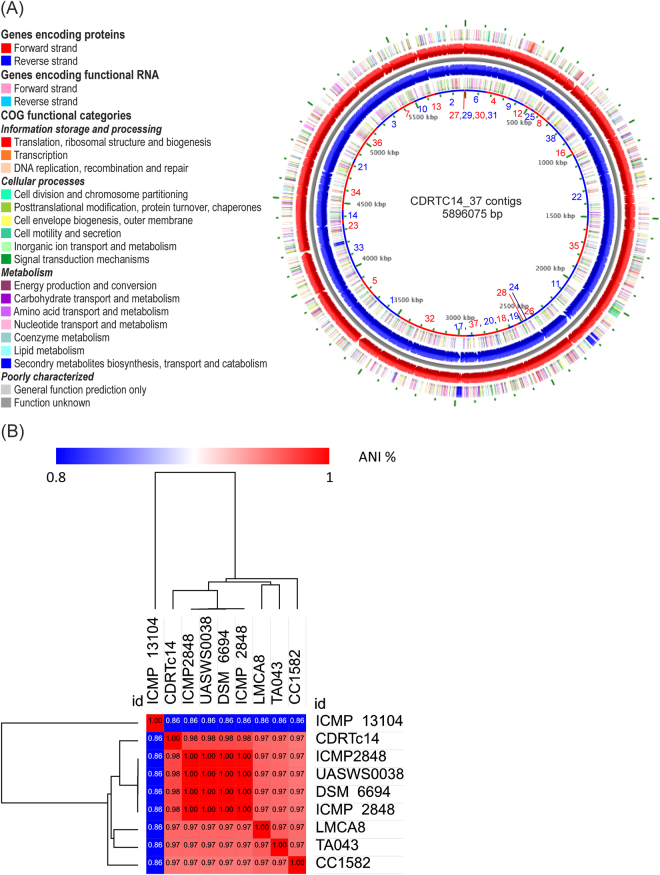
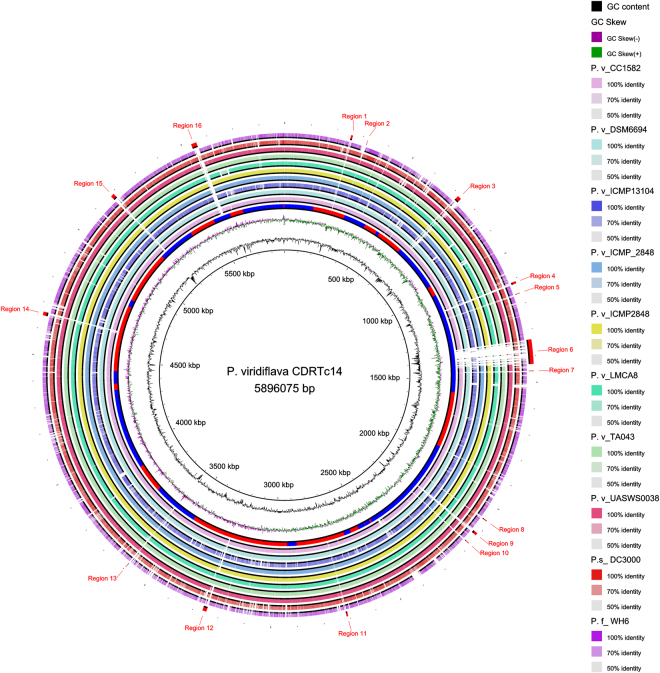
The CDRTc14 genome was sequenced using Illumina HiSeq and then assembled with SPAdes. The details about genome sequencing were reported earlier19 and genomic features are summarized in Tables S1 and S2. The whole genome of CDRTc14 has a total length of 5.96 Mb, a GC content of 59.3%, contains one chromosome and one plasmid (67,392 bp). The map of CDRTc14 genome with its annotation is represented in Fig. 2A. Genome relatedness of the strain CDRTc14 was analyzed on the basis of average nucleotide identity (ANI). Strains with ANI values >96% are considered to be the same species24. As shown in Fig. 2B, strain CDRTc14 has the greatest nucleotide identity (98.06%) with strain UASWS0038, which has been reported as a biocontrol agent against postharvest disease of pip fruits25. Other P. viridiflava genomes showed ANI values between 96.72% and 98.04% with CDRTc14 except ICMP_13104, which has only 85.90% ANI with CDRTc14. Low ANI similarity (less than 86%) of ICMP_13104 to all whole genome sequenced strains question its postulated taxonomic position as P. viridiflava 26.
Figure 2.
(A) Graphical circular map of P. viridiflava CDRTc14 with annotation. The innermost ring shows contig boundaries as alternating red and blue color where contig accession numbers MBPF01000001.1 to MBPF010000037.1 show the each contig position. (B) Heat-map of average nucleotide identity (ANI) values amongst different strains of Pseudomonas viridiflava revealing two major groups.
Identification of genes potentially involved in herbicide production
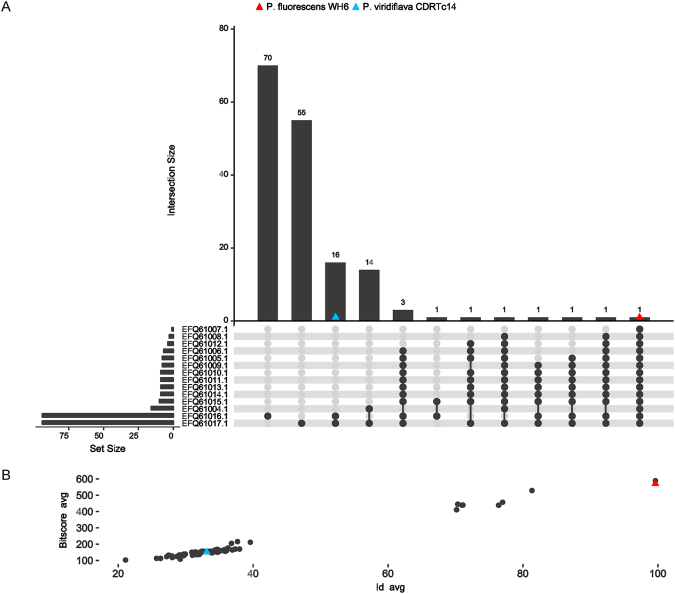
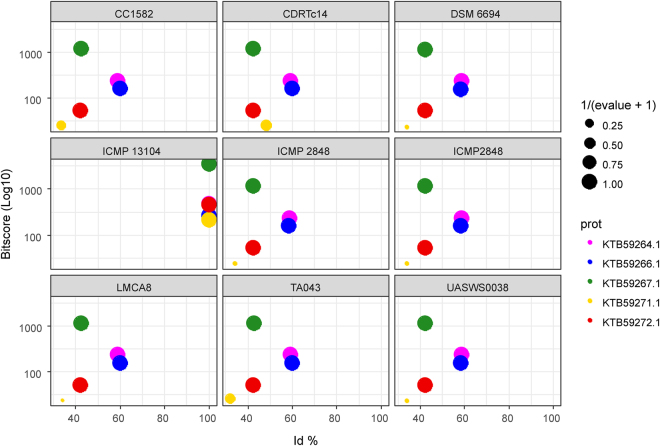
Strain CDRTc14 showed similar phytotoxic effects on L. draba and lettuce seedlings in vitro as reported earlier for P. fluorescens strain WH6, which was correlated with the biosynthesis of 4-formylaminooxyvinylglycine (FVG) responsible for germination arrest of grasses10. The whole genomes of these two herbicidal strains (P. viridiflava CDRTc14 and P. fluorescens WH6) shared 183 genes (which correspond to 3.5% and 3.1% of all genes in CDRTc14 and WH6, respectively), while 5,043 and 5,644 genes were found specifically in CDRTc14 and WH6, respectively (Fig. 3B, Table S9). At the molecular level, the germination arrest factor of WH6 was reported to be encoded by the gvg cluster containing fourteen genes9,11. Eight genes (gvgR, gvgA-C, gvgF-I) of this cluster are involved in the FVG production and two genes (gvgJ and gvgK) are involved in the export of FVG while other genes (gvgJ, gvgK, tam, ssb) were shown not to be important for FVG production11. We investigated how commonly the gvg cluster (13–15 kbp) occurs in 185 sequenced genomes of the genus Pseudomonas and also compared the protein encoded by the cluster using BLAST analysis against the whole NCBI non-redundant protein database (see Methods for details). This bioinformatics analysis revealed that the complete gvg cluster (14 genes) is absent in any of the Pseudomonas genomes, apart from the P. fluorescens WH6 genome (in which it was originally found) (Fig. 4, Table S8). Two strains, P. syringae pv. maculicola strain ES4326 and P. antarctica strain PAMC 27494, contained genes which matched most of the cluster genes (12 out of 14 genes displayed in 8th and 11th intersection bar from left in Fig. 3, respectively). Both strains (ES4326 and PAMC 27494) harbor all genes involved in the production of the FVG except gvgB (EFQ61007.1) gene. The gvgB gene codes for a small hypothetical protein and it was exclusively found in WH6. In CDRTc14, only two genes of this cluster were detected, more precisely gvgK (EFQ61016.1), a LysE family transporter, which is involved in the transport of the FVG out of the cell11, and ssb (EFQ61017.1) encoding a ssDNA binding protein. However, the genes involved in FVG production were not found in CDRTc14 (Fig. 4, Table S5). Overall, the genes responsible for FVG production were found very rarely in the Pseudomonas genomes investigated, for example only 4.8% of the investigated genomes harboured any gene involved in the production of FVG. Also at the protein level, the products encoded by the gvg genes appeared to be very rare (Fig. S8, Table S7). For example, at 60% protein identity threshold, hits (Nhits) for eight FVG producer genes varied between 260 for the gvgF gene (EFQ61011.1) to three for gvgB (EFQ61007.1). The gvgB gene that was not detected in the genus Pseudomonas (except WH6) showed similarity with genes found in other genera including Streptomyces albulus (62% identity) and Burkholderia cenocepacia (89% identity). However, the two FVG transporter genes (gvgJ and gvgK) were found more commonly showing 449 and 1033 hits at the 60% protein identity threshold, respectively. Overall, it seems that multiple genes of the gvg cluster have been horizontally transferred, however, our analysis does not indicate transfer of the complete cluster among the strains/genomes analysed. In particular, the eight genes within the gvg cluster supposed to be involved in FVG production occur to be very rare and among them, the small CDS (gvgB) consisting of only 28 amino acids seems to be very specific for WH6. FVG belongs to a class of non-proteinogenic amino acids oxyvinylglycine. Some other oxyvinylglycines such as L-2-amino-4-methoxy-trans-3-butenoic acid (AMB), aminoethoxyvinylglycine and rhizobitoxin are also known for their role in plant-microbe interactions but the prediction of their biosynthetic pathways is difficult using bioinformatic tools because their biosynthetic routes might vary considerably between different species27. More detailed genetic and metabolomic analysis is needed to obtain further information on the genes involved in the production of oxyvinylglycine or other unknown herbicidal compounds.
Figure 3.
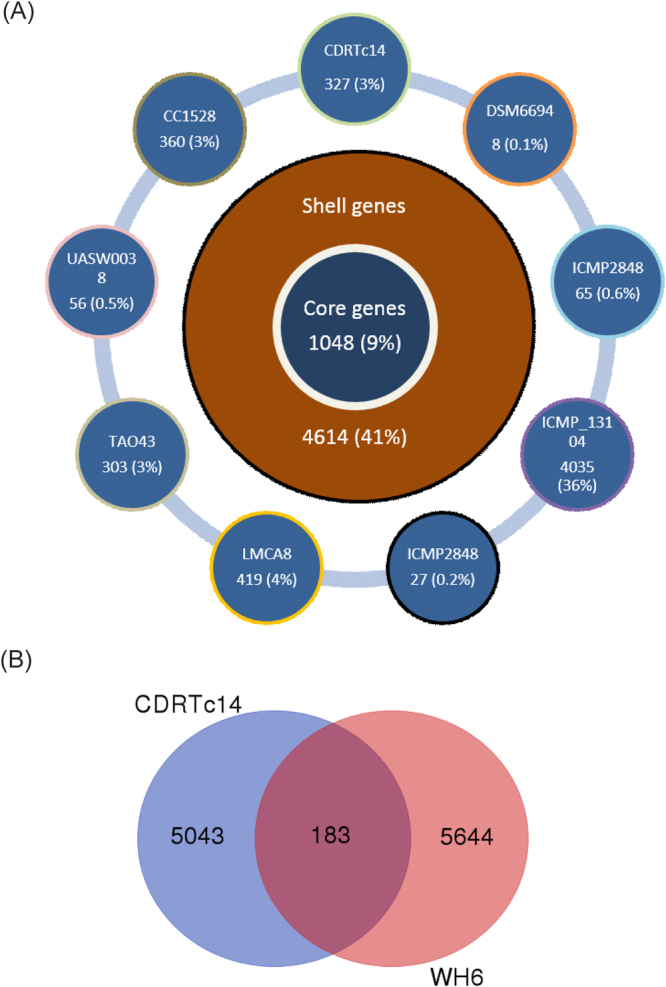
(A) Pan-genome of P. viridiflava strains calculated in Roary (with blastp at 95%). The inner ring shows the total number of the core genes (present in >99% of the genomes); the middle ring shows the number of shell genes (present in 15–95% of the genomes); the outer rings show the number of genes in the cloud of the pan-genome (present in 1–15% of the genomes), gene annotation of all genes is provided in Table S10. (B) Representing core and unique genes between P. viridiflava CDRTc14 and P. fluorescens WH6, annotation of core genes is provided in Table S9.
Figure 4.
Occurrence of formylaminooxyvinylglycine (FVG) biosynthetic genes (gvg gene cluster of WH6) across tested genomes are shown by UpSet composite plot: (A) Intersection size vs set size: this plot shows the number of genomes (vertical barplots) sharing the same combination of gvg cluster elements (black dots read vertically) whereas the horizontal barplots report how many genomes have one specific element of the gvg cluster, gene by gene (black dots read horizontally). As expected, all the genes of the gvg cluster are present in P. fluorescens WH6 genome (red triangle). None of the other Pseudomonas genomes has the complete set of elements of the cluster (at least with an identity percentage ≥ 75%). P. viridiflava CDRTc14 shows only two genes and these are also the most abundant among Pseudomonas genomes. (B) Scatterplot showing the identity % average (x-axis) and the bitscore average (y-axis) for all the genes in the gvg cluster in each genome. P. fluorescens WH6 has an id avg of 100% and bitscore avg of 590, P. viridiflava CDRTc14 has an id avg of 32% and bitscore avg of 136.
Bacteria with herbicidal activity (except WH6) are hardly explored for their herbicidal mechanism at the molecular and genetic level and also bacterial herbicides against broadleaf plants are less often studied than those against grasses8. However, auxin-producing bacteria have often been shown effective in inhibiting the growth of broadleaf plants28,29. It has been also shown that broadleaf weeds are more prone to certain auxinic herbicides than grassy weeds30. In our experiment, we observed that all strains showing herbicidal activity (on broadleaf plants) produced IAA in vitro 31 (Table S6). Strain CDRTc14 produces indole acetic acid (IAA) up to 42 μg ml−1 and harbors genes for IAA production (Table 2). However, different plants generally show different responses to exogenous IAA depending on the concentration perceived by tissues. Rather low levels of IAA stimulate plant growth whereas high concentration of IAA can cause a negative impact on plants32. For example, it has been shown that low quantities of IAA produced by P. putida GR12–2 increased plant growth, while an elevated level of IAA inhibited root growth33. Moreover, IAA increases the level of ethylene by stimulating the production of aminocyclopropane-1-carboxylate (ACC) synthase34.
Table 2.
Functional characters tested in vitro and their related genes found in CDRTc14.
| Functional characters | In vitro test | Putative Genes/pathway |
|---|---|---|
| HCN | − | hcnB_1, hcnB_2, hcnC |
| Siderophores | + | entS |
| P solubilization | + | PqqB, PqqC, PqqD, PqqE, PqqF |
| IAA | + | indole−3−pyruvate (IPyA) (iaaM, iaaH gene) |
| ACC deaminase | − | acdS |
| Antifungal | − | ubiC, gabP, gapT, gapD, PhzF1–2 |
Cyanogenic bacteria have been also reported for their herbicidal activity since hydrogen cyanide (HCN) is known to inhibit plant growth by taking part in the metabolism (inhibiting respiratory path, CO2 and blocking photosynthetic electron transport) and by finally causing plant death due to cellular hypoxia (cells suffering from lack of oxygen)35. CDRTc14 is equipped with genes related to HCN production but did not show HCN activity in vitro (Table 2). Bacterial cyanogenesis can be influenced by many factors under laboratory conditions, such as the growth media, cell density, aerobic growth conditions and quorum sensing36–38.
Identification of genes involved in herbicide metabolism and metal resistance
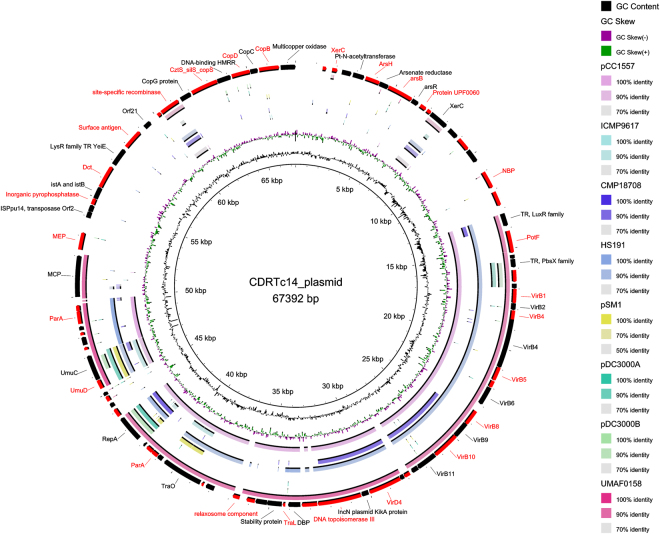
Plant‐associated bacteria are often described for their potential to degrade commonly used herbicides39. In the CDRTc14 genome we found several genes and/or molecular pathways known for the metabolism of commonly used herbicides such as N-(phosphonomethyl) glycine (glyphosate), atrazine, 2,4-dichlorophenoxyacetic acid (2,4 D), alachlor and the complete KEGG pathway of atrazine degradation (predicted by KEGG Automatic Annotation Server) (Table 3). Interestingly, we also found a glufosinate resistance gene (phosphinothricin N-acetyltransferase/RePAT), both on the chromosome and the plasmid of CDRTc14 (Fig. 5 and Table 3). The gene was used to develop glufosinate resistant transgenic rice40. In vitro testing confirmed (Fig. S9) that CDRTc14 can grow in the presence of glyphosate (20 mM), glufosinate (8 mM), and 2,4 D (1200 mg/L). Strain CDRTc14 could have adapted to herbicides commonly used in vineyards, possibly by acquiring resistance genes through horizontal gene transfer. A wide range of mechanisms for microbial degradation of herbicides has been reported, especially through hydrolytic, bond cleavage, oxidation and reduction reactions39. Rhizobacteria and endophytes particularly belonging to the Proteobacteria (α, β and γ classes) have been often described to degrade herbicides and to protect plants from herbicides41–43. P. viridiflava isolates from agricultural fields have also been reported to degrade pesticides16,17. Recently, it has been suggested that plant-associated bacteria contribute to herbicide resistance in plants39, for example pea (Pisum sativum L.) inoculated with an endophytic strain Pseudomonas putida POPHV6 showed a higher removal of 2,4 D from soil in the presence of endophytic strain without the translocation of the herbicide into aerial plant parts44. Another study in Australia has shown that herbicide resistance of the annual ryegrass (Lolium rigidum) is correlated with the colonization of the endophytic fungus Neotyphodium spp.45. However, the introduction of herbicide resistance traits into plants via microbial engineering has so far been hardly explored. BLAST analysis of the CDRTc14 plasmid and comparison with eight closely related plasmids revealed even more unique features in CDRTc14 (Fig. 6). Many genes found on the CDRTc14 plasmid code for features such as motility, chemotaxis, heavy metal tolerance, herbicidal metabolism (phosphinothricin N-acetyltransferase), DNA repair system, type IV secretion system, and tyrosine recombinase XerC (Fig. 5, Table S12). We also identified putative tyrosine recombinases at the left end of the RePAT (phosphinothricin N-acetyltransferase), a well-known herbicide resistance gene. Tyrosine recombinases have been reported to mediate the DNA integration/excision that might be involved in horizontal gene transfer46.
Table 3.
Molecular pathways or genes found in CDRTc14 related to herbicide metabolism.
| Herbicide | In vitro test | Putative genes/pathway/gene products | Reference |
|---|---|---|---|
| 2,4-dichlorophenoxyacetate (2,4-D) and | + | tfdA (Alpha-ketoglutarate-dependent 2,4 dichlorophenoxyacetate dioxygenase) | 80,81 |
| 2-methyl-4-chlorophenoxyacetic (MCPA) | NA | cadA | 77 |
| Atrazine | NA | atzE (biuret hydrolase), atzF1–2 (allophanate hydrolase) | 82 |
| Glyphosate | + | Enzyme EPSPS (5-enol-pyruvylshikimate-3-phosphate synthase), aroA1–2 aroA_2 (3-phosphoshikimate 1-carboxyvinyltransferase)argF, sdhA, speA, ahpC (shikimate pathway) | 83–85 |
| Glufosinate | + | bar_1, bar_2 (phosphinothricin N-acetyltransferase) | 40 |
| Alachlor | NA | gstB_1–5 (glutathione S-transferase)) | 86 |
Figure 5.
Circular visualization of the comparison of CDRTc14 plasmid with other related (Top 8 blast hits was taken from NCBI using BLASTn, plasmid accession numbers: CP007015.1, NZ_CM002754.1, NZ_CP012180.1, NZ_CP006257.1, CM001987.1, NC_004633.1, NC_004632.1, NZ_CP005971.1). The figure was designed using BRIG. The interesting features are labelled in the outer most circle and annotation is provided in Table S12.
Figure 6.
Circular visualization of the whole genome comparison of CDRTc14 chromosome with other related genomes. The figure was designed using BRIG. The gaps in the circles represent regions of low or no similarity provided in Table S11 with annotation.
Further, we calculated a pan-genome of all available P. viridiflava genomes. The P. viridiflava pan-genome has a total of 11,262 genes: 1,048 represent the core genes (genes present in >99% of isolates within this study), whereas 4,614 and 5,600 form the shell (genes present in 15–95% of isolates analyzed) and cloud genes (genes present in 0–15% isolates analyzed), respectively (Fig. 3A). The genome of DSM_6694 shared the highest number of genes (4,827) while ICMP_13104 shared the least number of genes (1,084) with CDRTc14. Several unique genes (327) were found in the CDRTc14 genome, particularly several genes related to heavy metal resistance including copper (copB, copC, copD) and arsenic (arsB, arsC, arsR, arsH) homeostasis, herbicidal resistance (phosphinothricin N-acetyltransferase), type IV secretion system, such as virB1, virB4, virB6, virB8, virB10, virB11, chemotaxis and motility (Table S10). Altogether, comparative analysis between genomes using both BRIG and Roary showed that the CDRTc14 genome is equipped with several unique features located on the chromosome as well on the plasmid. Many of these features correlate with its host L. draba and the vineyard soil environment. For instance, L. draba is a noxious weed species, known for the hyperaccumulation of heavy metals47,48. It has been shown that metal hyperaccumulating plants host metal-resistant endophytes due to local adaptation to the metal-rich environment49. A recent study revealed that long-term association with metal-hyperaccumulator plant leads to local adaptation by Pseudomonas endophytes50. Copper and arsenic resistant Pseudomonas strains have been frequently isolated from the vineyard environment51,52. Indeed, vineyards have been treated with copper-containing pesticides to control plant pathogens53 as well as with sodium arsenate till the end of the last century, which may still be present in vineyard soils54. The fact that CDRTc14 carries heavy metal resistance genes on its plasmid suggests that these genes have been obtained by horizontal gene transfer53.
Identification of genes involved in plant interaction
Endophytes use multiple strategies to interact with their host and their various types of genes have been described for endophytic colonization55. A set of known mechanisms, which endophytes often use to interact with plants, are found in the genome of CDRTc14 such as genes coding for type IV pili, motility and chemotaxis (90 genes for flagellar motility, 59 for chemotaxis), transporters (i.e. 80 genes for ABC transporters, 11 for antiporter membrane transporters, six for TRAP transporters, 23 for cation transporters) and protein secretion systems (type I-II & type IV-VII), plant adaptation and protection (i.e. four genes for plant hormones, 15 for siderophores and 93 for stress response), quorum sensing (synthase gene: acuI, recipient genes: 14 for autoinducer receptors of LuxR family, lasR, rhtB, acuR) and quorum quenching (pvdQ, quiP, aiiA) (Table S13). To gain in-depth knowledge of the CDRTc14 genome, we performed a comparative genome analysis using all available P. viridiflava whole genomes (November 11th, 2016), P. syringae DC3000 and P. fluorescens WH6 (listed in Table 4). We included P. syringae DC3000, because it is the most closely related complete genome to CDRTc14 and is a well-studied plant pathogen. Comparison of ten genomes revealed many regions in the CDRTc14 genome showing less (<70%) or no similarity to the other analyzed genomes. These differences are shown in the circular map designed in BRIG (Fig. 6). Interestingly, 16 unique regions were found in CDRTc14 and their annotation revealed 235 coding sequences (CDS) and three coding for RNA (Table S11). Most of these annotated unique regions appear to be similar to the ones predicted by the Roary pipeline during the pan genome analysis (described above in detail).
Table 4.
General features of the P. viridiflava CDRTc14 and related organisms used for comparative studies.
| Species/strain | Size | % G+C | Scaffolds | Plasmid | Genes | % coding | tRNA | rRNA | GenBank | Isolation source | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P. viridiflava CDRTc14 | 5.96 | 59.3 | 38 | 1 | 5,330 | 97.9 | 58 | 9 | MBPF00000000 | L. draba | Austria |
| P. viridiflava TA043 | 5.98 | 59.2 | 218 | — | 5,282 | 98.1 | 29 | 2 | AVDV00000000 | P. officinalis | France |
| P. viridiflava CC1582 | 6.02 | 59.2 | 211 | — | 5,350 | 98.0 | 28 | 2 | AVDW00000000 | epilithon | France |
| P. viridiflava ICMP2848 | 5.90 | 59.4 | 254 | — | 5,278 | 97.0 | 53 | 4 | LJRS00000000 | P. vulgaris | Switzerland |
| P. viridiflava UASWS0038 | 5.91 | 59.4 | 201 | — | 5,278 | 97.1 | 56 | 10 | AMQP00000000 | Rhododendron sp. | Switzerland |
| P. viridiflava LMCA8 | 5.99 | 59.3 | 73 | — | 5,358 | 96.9 | 69 | 19 | JXQO00000000 | A. thaliana | USA |
| P. viridiflava DSM 6694 | 5.91 | 59.4 | 188 | — | 5,256 | 97.8 | 54 | 3 | JRXH00000000 | P. coccineus | Switzerland |
| P. viridiflava ICMP 13104 | 5.55 | 58.9 | 204 | — | 4,916 | 89.4 | 58 | 4 | LKEJ00000000 | A. deliciosa | France |
| P. viridiflava ICMP 2848 | 5.92 | 59.4 | 48 | — | 5,261 | 97.8 | 52 | 6 | LKEH00000000 | P. vulgaris | Switzerland |
| P. syringae DC3000 | 6.4 | 58.4 | 1 | 2 | 5,721 | 85.6 | 15 | 63 | AE016853.1 | S. lycopersicum | UK |
| P. fluorescens WH6 | 6.3 | 60.6 | 1 | — | 5,724 | 93.8 | 4 | 53 | CM001025.1 | T. aestivum | USA |
Strain CDRTc14 harbors several genes (such as siderophore production, ACC deaminase, and IAA and phosphate solubilization, Table 2) potentially involved in plant growth promotion and conferring enhanced stress tolerance in plants, although under controlled conditions the strain negatively affects its host plant L. draba. It might be that the herbicidal activity of this strain is bacterial cell-dependent as phytotoxicity can be regulated by quorum sensing56. Through quorum sensing bacteria regulate the expression of several phenotypic characteristics such as motility, antibiotics production, phytohormones production in a cell-density dependent manner56. Also bacterial autoinducers regulate the expression of virulence genes57, a phenomenon which is not restricted to pathogenic interactions. For example, quorum sensing plays a role in the symbiotic interaction between rhizobia and legumes58. Furthermore, the expression of virulence genes in a group of (otherwise non-pathogenic) fluorescent pseudomonads has been shown to correlate with their population level inside leatherleaf ferns59. Additionally, the amount of IAA produced may reach toxic levels when the population size is high60. This correlates with our results obtained in testing dose dependency where we did not observe any negative effect on lettuce growth upon CDRTc14 inoculation with a low inoculum dose (<105 CFU). However, CDRTc14 significantly reduced germination and root length when applied at a higher concentration (CFU = 107) (Figs 1F, S7). Altogether, endophytes may establish very complex and multilayered interactions with their hosts, which range from mutualistic to pathogenic under specific conditions61.
Identification of genes for pathogenicity
The pathogenicity of the known plant pathogen P. viridiflava has been correlated with the presence of complete pathogenic islands of type III secretion system62,63. We analysed the genomes of all available P. viridiflava strains using RAST and found altogether seven potential protein secretion systems under membrane transport subsystem category (Table 5). However, the type III secretion system containing thirty genes was only found in strain ICMP 13104, a strain found in kiwi fruits infected with stem cankers (Actinidia spp.), while this strain lacks the type I secretion system. In the case of other membrane transport systems, no substantial differences regarding the number of genes were observed among all the strains except for strain ICMP 13104, which was found to be quite different in almost all secretions systems compared to other strains. Furthermore, we screened (using TBLASTN) all P. viridiflava strains for the presence of the pathogenicity island of type III secretion, using the five pathogenicity related gene products of strain ICMP 13104 as sequence queries. All tested genomes, except ICMP 13104, showed a similarity between 32% and 60% to the five pathogenicity related genes (Figs 7, S7). Moreover, we could not detect the presence of the complete pathogenicity island in CDRTc14 using three different annotation pipelines (PGAAP, Prokka, and RAST). In agreement with this, strain CDRTc14 did not show any pathogenicity symptoms on tomato, A. thaliana, common bean and N. benthamiana while other P. syringae pathovars (DC3000, 1448 A, ATCC11528) showed clearly visible symptoms on all tested hosts species in the greenhouse experiment (Table S3, Fig. S4). Colonization of each inoculated strain was confirmed by 16 S rRNA gene sequencing followed by high resolution melting curve analysis with qPCR (Figs S5 and S6). Moreover, the phylogenetically closest strain (UASWS38) to CDRTc14 also lacks the complete pathogenicity island25, whereas strain ICMP 13104 carries the complete pathogenicity island but is phylogenetically only distantly related to CDRTc14 (Fig. 2B). Further research is needed to explore in depth the molecular mechanism underlying virulence-related plant-microbe interactions of CDRTc14.
Table 5.
P. viridiflava membrane transport subsystem prediction as estimated by RAST.
| Protein family/ Strain (No of total proteins) | CDRTc14 | TA043 | CC1582 | ICMP2848 | UASWS0038 | LMCA8 | DSM 6694 | ICMP 13104 | ICMP 2848 |
|---|---|---|---|---|---|---|---|---|---|
| Protein secretion system, Type I | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 0 | 3 |
| Protein secretion system, Type II | 28 | 30 | 30 | 30 | 30 | 30 | 30 | 17 | 30 |
| Protein secretion system, Type III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 30 | 0 |
| Protein and nucleoprotein secretion system, Type IV | 23 | 23 | 23 | 23 | 23 | 23 | 23 | 23 | 23 |
| Protein secretion system, Type V | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 12 | 2 |
| Protein secretion system, Type VI | 40 | 46 | 52 | 41 | 41 | 45 | 40 | 26 | 40 |
| Protein secretion system, Type VII (Chaperone/Usher pathway, CU) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABC transporters | 80 | 84 | 83 | 80 | 79 | 76 | 79 | 73 | 79 |
| Uni- Sym- and Antiporters | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 10 | 11 |
| Membrane Transport - no subcategory | 54 | 59 | 59 | 59 | 58 | 54 | 59 | 41 | 59 |
| TRAP transporters | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 3 | 6 |
| Protein translocation across cytoplasmic membrane | 7 | 7 | 6 | 7 | 7 | 8 | 7 | 7 | 7 |
| Cation transporters | 23 | 18 | 18 | 19 | 18 | 19 | 19 | 15 | 19 |
Figure 7.
BLAST plot of P. viridiflava strains tested for the presence of the pathogenicity island of type III secretion system, where strain ICMP 13104 was used as a reference. For this purpose, a tBLASTn search was performed using the gene products of the pathogenicity cluster against the P. viridiflava genome nucleotide sequences. The scatter plot shows the similarity percentages on the x-axis and the bitscores on the y-axis for each genome, whereas the e-values were transformed as 1/(evalue + 1) so that the size of each point representing a gene product could correlate positively with the e-value significance.
Conclusions
Functional assays in combination with an in-depth comparative genome analysis revealed that strain CDRTc14, which was isolated as a root endophyte from a perennial weed in a vineyard, can be assigned to P. viridiflava. Although this species comprises many plant pathogens strain CDRTc14 did not induce disease symptoms on the plant species tested in this study and lacks known virulence genes. Moreover, strain CDRTc14 shows a number of features, which are well known from beneficial plant-associated bacteria. However, this strain showed herbicidal activity against its host and to other related plant species, while it showed neutral or beneficial effects on the growth of others. These findings refute the centrifugal phylogenetic method (CPM), which is used as a standard in host-specificity testing of biological control agents. Strain CDRTc14 shows similar herbicidal activities as strain WH6, but the gene cluster responsible for herbicidal activity in this strain was absent in CDRTc14, suggesting that both strains employ different mechanisms. Although genetic and metabolomics analyses are needed to further elucidate the mechanisms responsible for and metabolites involved in herbicidal activity, we found activities and responsible genes indicating the adaptation of this strain to conditions typically encountered in vineyards such as heavy metal resistance and herbicide/pesticide degradation. The amount of pesticide application in vineyards is in comparison to other crops extremely high. Furthermore, pesticides based on copper are still widely used and products with arsenic were used in the past. Features like herbicide degradation and heavy metal resistance reflect the adaptation of strain to prevailing soil environmental stressors.
Methods
Germination assay in growth chamber
Germination tests were carried out to evaluate the herbicidal effect of bacterial inoculation on different plant species. All isolates used in this assay were previously isolated from a vineyard field (L. draba or grapevine)31 and CDRTc14 was isolated from surface sterilized roots of the vineyard weed L. draba L.31 (Table S6). The seeds of all tested plants (L. draba lettuce and A. thaliana Col-1) were surface sterilized by immersion in 3.25% (v/v) sodium hypochlorite (NaOCl) for 1 min, followed by 70% (v/v) ethanol for 1 min, rinsed five times in sterile distilled water and blotted on sterilized filter paper. Bacterial cultures, grown for overnight at 28 °C in R2A broth (Lab M limited, Lancashire, United Kingdom), were adjusted to an OD600 of 0.2 (∼107 CFU ml−1). Then 10 ml of each bacterial culture were centrifuged at 4.000 x g for 20 min at 10 °C and after discarding the supernatants bacterial pellets were dissolved into 10 ml PBS (phosphate-buffered saline). Five ml of the bacterial culture was used for seed imbibement for 30 min. Each treatment contained three replicates, for a mock treatment PBS alone was used. Fifteen to twenty surface-sterilized seeds were plated on a 145 mm Petri dish containing approximately 60 mL of 1% water agar medium and placed in a plant culturing room under conditions of 16/8 hours of day/night (20 °C, 50% air humidity) for 12–18 days depending on the growth of the various plant species.
Greenhouse experiment with Lepidium draba
Six isolates (Table S6) were further tested on L. draba under greenhouse conditions (an average temperature of 29 °C, a light regime of 12:12 h L: D (light: dark), and 68–80% relative humidity). In the greenhouse high pressure sodium lamp with clear outer bulb (MASTER Agro 400 W E40 1SL/12, Philips) was used as a source of light energy. Ten seeds of L. draba were sown in small plastic pots (500 g substrate containing 3 parts Einheitserde classic and 1 part premium perlite of Gramoflor, Germany) in triplicates for each treatment. Bacterial pellets from five ml of each bacterial culture (OD600 = 0.2) were resuspended in PBS and dispensed onto seeds in the pots just after sowing, and control was treated with 5 ml PBS (pH = 7.4). Plants were harvested after 9 weeks. Germination percentage, root length, shoots length and fresh biomass of seedlings were recorded.
Plant host range test of CDRTc14
To test the host range of strain CDRTc14, seven different plant species, closest relative of L. draba (Lepidium meyenii, Hesperis matronalis, Isatis tinctoria, Erysimum odoratum, Sisymbrium officinale, Lepidium sativum, Brassica n. var. Cleopatra, B. napus var. Pharao) (Fig. S3) were tested under greenhouse conditions using the same protocol as mentioned above, but instead of pots 145 mm petri dishes were used (with 10 seeds per petri dish). Plants were harvested after 3–4 weeks depending on growth of different plant species. The number of seeds germinated was counted daily, root and shoot length of all the germinated seedlings were recorded in cm and fresh biomass of all seedlings were recorded in g.
Dose dependent test of CDRTc14 on lettuce
To evaluate if the phytotoxicity of CDRTc14 is concentration dependent we tested 101 to 107 CFU ml−1 of CDRTc14 on lettuce seeds. Lettuce was chosen due to its faster and higher germination rate than L. draba. All the experimental conditions were the same as described above for growth chamber experiment with the exception of plate type and number of replicates, 12 wells plates were used instead of 145 mm Petri dish and each treatment contained five replicates.
Genome sequencing, annotation and comparative analysis
The information about the CDRTc14 whole genome sequencing, genome assembly and quality check was reported earlier16. The CDRTc14 genome was annotated using the NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) as well as Prokka64,65. RAST (Rapid Annotation using Subsystem Technology) was used to compare different P. viridiflava strains at the genome level66.
All genomes used in this analysis were downloaded in FASTA format using the NCBI-genome-download python script (available at https://github.com/kblin/ncbi-genome-download). The gvg biosynthetic gene cluster products were downloaded in FASTA format using the NCBI E-utils (available at ftp://ftP.ncbi.nlm.nih.gov/entrez/entrezdirect/). Then an Average Nucleotide Analysis (ANI) was calculated using the pyani Python3 module (available at https://github.com/widdowquinn/pyani) with both MUMmer (NUCmer) alignment and BLAST search methods used as confirmation.
Pangenome analysis of all strains was performed with Roary67 after genome annotation in Prokka. Functional annotation of core genes of CDRTc14 and WH6 was performed using the EggNOG 4.5 resource68. Genome wide search for the gvg genes cluster was done by adopting two different approaches: (1) a tBLASTn search of each gene of the gvg cluster against the whole set of Pseudomonas complete genomes available in NCBI (November 11th, 2016) including also four draft genomes of P. viridiflava CDRTc14, P. viridiflava UASWS0038, P. syringae ES4326 and P. fluorescens WH6. The best hit of each gene against each nucleotide genome sequence was used to generate a table reporting bitscores, similarity percentages and e-values. The table was processed further and binary values generated assuming 75% of sequence similarity as a threshold for putative homology. The table was imported in R and an UpSet technique69 applied using the UpSetR package. (2) A BLASTp search was conducted for each gvg cluster gene product against the entire NCBI nr database (November 25th, 2016) in R, using the Bio3D package70. Normalized scores were clustered to partition hits in groups by similarity to query and e-value, bitscore, identity and length values of the hits showed in a plot for each accession. BLAST Ring Image Generator (BRIG) was used for visualisation of genome comparisons71 and bacterial annotation system (BASys) was used to prepare CDRTc14 circular image with annotation72.
Pathogenicity test
Plant pathogenicity tests were performed in the greenhouse to test compatible (disease) and incompatible (resistance) responses due to strain CDRTc14 on tomato (var. Cobra), A. thaliana (Col-0), common bean (var. Roma II) and Nicotiana benthamiana which are known susceptible host of P. syringae 73,74 and P. viridiflava 13,75. Bacterial cultures (OD600 = 0.2) in PBS buffer with 0.01% Silwet L-77 (OSI Specialties Inc., Danbury, CT, U.S.A.) were sprayed to runoff on the abaxial and adaxial leaf surfaces of about 3-week-old plants. Control plants were treated with PBS buffer containing 0.01% Silwet L-77. N. benthamiana plants were also infiltrated with a needleless syringe to find out if cells produce a hypersensitive response. The pathogenicity test was based on a completely randomized design with three replications in the greenhouse conditions as mentioned above. The plants were scored for bacterial speck severity one week after inoculation using the scale of 0–3 (0 = free of symptoms). Pathogenic strains of each plant species were used as reference and are listed in Table S3.
Re-isolation of bacteria from plant tissues and high resolution melting PCR (HRMA)
Leaves were detached 3 weeks after the inoculation and surface sterilized (70% ethanol for 30 s, followed by rinsing with sterile distilled water 3 times). The surface sterilization was checked by plating the last washing on R2A. Leaf discs from three different leaves were ground in 10 mM MgCl2 with a glass rod into test tubes. Then the samples were thoroughly vortex-mixed and 1:10 serially diluted. Samples were plated on King’s B medium and incubated at 28 °C for 2 days. Resulting colonies were picked and subjected to high resolution melting analysis (HRMA)76. Pure cultures of each reference strain (DC3000, 1448 A, ATCC11528, listed in Table S3) was also run in parallel for melting curve comparison. The PCR was carried out on a CFX96 cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA), with 10 µl reaction mixture containing 0.5 µl (10 µM) of each primer (Gamma395F 5′-CMATGCCGCGTGTGTGAA-3′ and Gamma871r 5′-ACTCCCCAGGCGGTCDACTTA-3′) targeting Gammaproteobacteria77, 5 µl of 2 × SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Inc.), 3 µl water and 1 µl (10 ng) genomic DNA. Cycling conditions were one cycle at 95 °C for 5 min, followed by 40 cycles at 95 °C for 5 s and 60° for 5 s. Melting curves of PCR amplicons were obtained with temperatures increasing from 65 °C to 95 °C. Each single DNA batch was done in triplicates, and analyzed by high-resolution melting analysis software (Bio-Rad Laboratories, Inc.), which automatically clusters the samples per their melting profiles and assigns a confidence score to each sample (Fig. S5). Pure genomic DNA of each inoculated strain was used as reference to confirm the identity of each sample tested by comparing its melting profile to the reference. The confidence level threshold for a sample to be included in a cluster was 99.5%. The HRM data were confirmed by sequencing the amplicons of each cluster using the Gamma395F primer and phylogenetic analysis was performed using MEGA678.
Herbicide resistance assay
CDRTc14 was grown in R2A broth (Lab M, UK) at 28 °C for overnight. The culture was collected by centrifugation and washed twice using liquid M9 minimal medium79. Bacteria in the culture (OD600 = 0.2) were then grown with shaking at 28 °C in liquid M9 minimal medium supplemented with either glyphosate (at the concentrations of 0, 5, 10, 15, 20 mM), glufosinate (2, 4, 6, 8 mM) or 2, 4 D (at the concentrations of 200, 400, 800 and 1200 mg/l). The OD600 values of the cultures were determined at 1-h intervals to record the growth rates of the strains till 48 h. Alternatively, bacteria were plated on M9 agar79 containing herbicide as mentioned above. After 5 days of incubation at 28 °C, the colonies growth was observed on the plates.
Availability of data
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Supplementary Material and Methods and Figures
Acknowledgements
The authors are extremely grateful to Helmut Gangl (Gangl wines) for the permission and help with plant sampling. We gratefully acknowledge the Higher Education Commission (HEC) of Pakistan for financial support for A. Samad.
Author Contributions
A. Samad performed the experiments, wrote the whole manuscript and prepared figures and tables, L. Antonielli performed the bioinformatics analysis, S. Compant A. Sessitsch and F. Trognitz designed and supervised the study, give advices for the analysis and made correction in the manuscript
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16495-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oerke E-C. Crop losses to pests. J. Agric. Sci. 2006;144:31–43. doi: 10.1017/S0021859605005708. [DOI] [Google Scholar]
- 2.Delye C, Jasieniuk M, Le Corre V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013;29:649–658. doi: 10.1016/j.tig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Chauvel, B., Guillemin, J.-P., Gasquez, J. & Gauvrit, C. History of chemical weeding from 1944 to 2011 in France: Changes and evolution of herbicide molecules. Crop Prot. 42, 320–326 (2012).
- 4.Cordeau S, Triolet M, Wayman S, Steinberg C, Guillemin J-P. Bioherbicides: Dead in the water? A review of the existing products for integrated weed management. Crop Prot. 2016;87:44–49. doi: 10.1016/j.cropro.2016.04.016. [DOI] [Google Scholar]
- 5.Charudattan, R. P. Use of tobacco mild green mosaic virus (TMGMV) mediated lethal hypersensitive response (HR) as a novel method of weed control. US Patent 7494955 B2 filed 8 Jan. 2004, and issued 24 Feb. 2009.
- 6.Johnson, D. R., Wyse, D. L. & Jones, K. J. Controlling weeds with phytopathogenic bacteria. Weed Technol. 621–624 (1996).
- 7.Li Y, et al. Research progress on microbial herbicides. Crop Prot. 2003;22:247–252. doi: 10.1016/S0261-2194(02)00189-8. [DOI] [Google Scholar]
- 8.Harding DP, Raizada MN. Controlling weeds with fungi, bacteria and viruses: a review. Front. Plant Sci. 2015;6:659. doi: 10.3389/fpls.2015.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halgren A, et al. Genetics of germination-arrest factor (GAF) production by Pseudomonas fluorescens WH6: identification of a gene cluster essential for GAF biosynthesis. Microbiology. 2013;159:36–45. doi: 10.1099/mic.0.062166-0. [DOI] [PubMed] [Google Scholar]
- 10.McPhail KL, Armstrong DJ, Azevedo MD, Banowetz GM, Mills DI. 4-Formylaminooxyvinylglycine, an herbicidal germination-arrest factor from Pseudomonas rhizosphere bacteria. J. Nat. Prod. 2010;73:1853–1857. doi: 10.1021/np1004856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okrent RA, Trippe K, Maselko M, Manning VA. Functional analysis of a biosynthetic cluster essential for production of 4-formylaminooxyvinylglycine, a germination-arrest factor from Pseudomonas fluorescens WH6. Microbiology. 2017;163:207–217. doi: 10.1099/mic.0.000418. [DOI] [PubMed] [Google Scholar]
- 12.Bartoli C, et al. The Pseudomonas viridiflava phylogroups in the P. syringae species complex are characterized by genetic variability and phenotypic plasticity of pathogenicity‐related traits. Environ. Microbiol. 2014;16:2301–2315. doi: 10.1111/1462-2920.12433. [DOI] [PubMed] [Google Scholar]
- 13.Sarris PF, Trantas EA, Mpalantinaki E, Ververidis F, Goumas DE. Pseudomonas viridiflava, a multi host plant pathogen with significant genetic variation at the molecular level. PloS ONE. 2012;7:e36090. doi: 10.1371/journal.pone.0036090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller C, et al. Ecomycins, unique antimycotics from Pseudomonas viridiflava. J. Appl. Microbiol. 1998;84:937–944. doi: 10.1046/j.1365-2672.1998.00415.x. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Miller, C., Miller, R. V. & Strobel, G. A. Antifungal compounds from Pseudomonas viridiflava. US Patent 6,103,875 filed 26 Nov. 1997, and issued 15 Aug. 2000.
- 16.Selvam ADG, Thatheyus A, Vidhya R. Biodegradation of the synthetic pyrethroid, fenvalerate by Pseudomonas viridiflava. Am. J. Microbiol. Res. 2013;1:32–38. doi: 10.12691/ajmr-1-2-4. [DOI] [Google Scholar]
- 17.Ning J, et al. Functional assembly of bacterial communities with activity for the biodegradation of an organophosphorus pesticide in the rape phyllosphere. FEMS Microbiol. Lett. 2010;306:135–143. doi: 10.1111/j.1574-6968.2010.01946.x. [DOI] [PubMed] [Google Scholar]
- 18.Bezza FA, Chirwa EMN. Pyrene biodegradation enhancement potential of lipopeptide biosurfactant produced by Paenibacillus dendritiformis CN5 strain. J. Hazard. Mater. 2017;321:218–227. doi: 10.1016/j.jhazmat.2016.08.035. [DOI] [PubMed] [Google Scholar]
- 19.Samad, A., Trognitz, F., Antonielli, L., Compant, S. & Sessitsch, A. High-quality draft genome sequence of an endophytic Pseudomonas viridiflava strain with herbicidal properties against its host, the weed Lepidium draba L. Genome Announc. 4 (2016). [DOI] [PMC free article] [PubMed]
- 20.Wapshere AJ. A strategy for evaluating the safety of organisms for biological weed control. Ann. Appl. Biol. 1974;77:201–211. doi: 10.1111/j.1744-7348.1974.tb06886.x. [DOI] [Google Scholar]
- 21.Zhang, W. & Sulz, M. Control of chickweed using Burkholderia andropogonis as a bioherbicide. US Patent 7141407 B2 filed 5 Mar. 2004, and issued 28 Nov. 2006.
- 22.Kennedy AC, Johnson BN, Stubbs TL. Host range of a deleterious rhizobacterium for biological control of downy brome. Weed Sci. 2001;49:792–797. doi: 10.1614/0043-1745(2001)049[0792:HROADR]2.0.CO;2. [DOI] [Google Scholar]
- 23.Briese D. Translating host-specificity test results into the real world: the need to harmonize the yin and yang of current testing procedures. Biol. Control. 2005;35:208–214. doi: 10.1016/j.biocontrol.2005.02.001. [DOI] [Google Scholar]
- 24.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefort, F., Calmin, G., Crovadore, J., Osteras, M. & Farinelli, L. Whole-genome shotgun sequence of Pseudomonas viridiflava, a bacterium species pathogenic to Ararabidopsis thaliana. Genome Announc. 1 (2013). [DOI] [PMC free article] [PubMed]
- 26.Visnovsky, S. B. et al. Draft genome sequences of 18 strains of Pseudomonas Isolated from kiwifruit plants in New Zealand and overseas. Genome Announc. 4 (2016). [DOI] [PMC free article] [PubMed]
- 27.Lee X, et al. Identification of the biosynthetic gene cluster for the Pseudomonas aeruginosa antimetabolite L-2-amino-4-methoxy-trans-3-butenoic acid. J. Bacteriol. 2010;192:4251–4255. doi: 10.1128/JB.00492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarwar M, Kremer RJ. Enhanced suppression of plant growth through production of L-tryptophan-derived compounds by deleterious rhizobacteria. Plant Soil. 1995;172:261–269. doi: 10.1007/BF00011328. [DOI] [Google Scholar]
- 29.Park J-M, Radhakrishnan R, Kang S-M, Lee I-J. IAA producing Enterobacter sp. I-3 as a potent bio-herbicide candidate for weed control: a special reference with lettuce growth inhibition. Indian J. Microbiol. 2015;55:207–212. doi: 10.1007/s12088-015-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McSteen P. Auxin and monocot development. Cold Spring Harb. Perspect. Biol. 2010;2:a001479. doi: 10.1101/cshperspect.a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samad, A., Trognitz, F., Compant, S., Antonielli, L. & Sessitsch, A. Shared and host-specific microbiome diversity and functioning of grapevine and accompanying weed plants. Environ. Microbiol. (2016). [DOI] [PubMed]
- 32.Sarwar M, Frankenberger WT. Influence of L-tryptophan and auxins applied to the rhizosphere on the vegetative growth of Zea mays L. Plant Soil. 1994;160:97–104. doi: 10.1007/BF00150350. [DOI] [Google Scholar]
- 33.Xie H, Pasternak JJ, Glick BR. Isolation and characterization of mutants of the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2 that overproduce indoleacetic Acid. Curr. Microbiol. 1996;32:67–71. doi: 10.1007/s002849900012. [DOI] [Google Scholar]
- 34.Kende H. Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993;44:283–307. doi: 10.1146/annurev.pp.44.060193.001435. [DOI] [Google Scholar]
- 35.Kremer RJ, Souissi T. Cyanide production by rhizobacteria and potential for suppression of weed seedling growth. Curr. Microbiol. 2001;43:182–186. doi: 10.1007/s002840010284. [DOI] [PubMed] [Google Scholar]
- 36.Blumer C, Haas D. Iron regulation of the hcnABC genes encoding hydrogen cyanide synthase depends on the anaerobic regulator ANR rather than on the global activator GacA in Pseudomonas fluorescens CHA0. Microbiology. 2000;146:2417–2424. doi: 10.1099/00221287-146-10-2417. [DOI] [PubMed] [Google Scholar]
- 37.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michelsen CF, Stougaard P. Hydrogen cyanide synthesis and antifungal activity of the biocontrol strain Pseudomonas fluorescens In5 from Greenland is highly dependent on growth medium. Can. J. Microbiol. 2012;58:381–390. doi: 10.1139/w2012-004. [DOI] [PubMed] [Google Scholar]
- 39.Tétard-Jones C, Edwards R. Potential roles for microbial endophytes in herbicide tolerance in plants. Pest Manag. Sci. 2016;72:203–209. doi: 10.1002/ps.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui Y, et al. Application of a novel phosphinothricin N-acetyltransferase (RePAT) gene in developing glufosinate-resistant rice. Sci. Rep. 2016;6:21259. doi: 10.1038/srep21259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobsen CS. Plant protection and rhizosphere colonization of barley by seed inoculated herbicide degrading Burkholderia (Pseudomonas) cepacia DBO1 (pRO101) in 2, 4-D contaminated soil. Plant Soil. 1997;189:139–144. doi: 10.1023/A:1004296615446. [DOI] [Google Scholar]
- 42.Ngigi AN, Getenga ZM, Boga HI, Ndalut PK. Biodegradation of s-triazine herbicide atrazine by Enterobacter cloacae and Burkholderia cepacia sp. from long-term treated sugarcane-cultivated soils in Kenya. J. Environ. Sci. Health Part B. 2012;47:769–778. doi: 10.1080/03601234.2012.676364. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y-J, Liu S-J, Drake HL, Horn MA. Alphaproteobacteria dominate active 2-methyl-4-chlorophenoxyacetic acid herbicide degraders in agricultural soil and drilosphere. Environ. Microbiol. 2011;13:991–1009. doi: 10.1111/j.1462-2920.2010.02405.x. [DOI] [PubMed] [Google Scholar]
- 44.Germaine KJ, et al. Bacterial endophyte-enhanced phytoremediation of the organochlorine herbicide 2, 4-dichlorophenoxyacetic acid. FEMS Microbiol. Ecol. 2006;57:302–310. doi: 10.1111/j.1574-6941.2006.00121.x. [DOI] [PubMed] [Google Scholar]
- 45.Vila-Aiub MM, Martinez-Ghersa MA, Ghersa CM. Evolution of herbicide resistance in weeds: vertically transmitted fungal endophytes as genetic entities. Evol. Ecol. 2003;17:441–456. doi: 10.1023/B:EVEC.0000005580.19018.fb. [DOI] [Google Scholar]
- 46.Hirano N, Muroi T, Takahashi H, Haruki M. Site-specific recombinases as tools for heterologous gene integration. Appl. Microbiol. Biotechnol. 2011;92:227. doi: 10.1007/s00253-011-3519-5. [DOI] [PubMed] [Google Scholar]
- 47.DiTomaso, J. M. et al. Weed control in natural areas in the Western United States. (University of California Weed Research and Information Center, 2013).
- 48.Georgieva S, Atanassova J, Dinev N. Metal hyperaccumulation in Cardaria draba (L.) Desv. (Brassicaceae) and heavy metal effects on the nematodes and a weevil associated with the plant roots in sites near a non-ferrous metal smelter in Bulgaria. Soil Sci. Agrochem. Ecol. 2015;XLIX:55–64. [Google Scholar]
- 49.Sessitsch A, et al. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013;60:182–194. doi: 10.1016/j.soilbio.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fones, H. N., McCurrach, H., Mithani, A., Smith, J. A. C. & Preston, G. M. Local adaptation is associated with zinc tolerance in Pseudomonas endophytes of the metal-hyperaccumulator plant Noccaea caerulescens. Proc. Biol. Sci. 283 (2016). [DOI] [PMC free article] [PubMed]
- 51.Chong TM, et al. Heavy-metal resistance of a France vineyard soil bacterium, Pseudomonas mendocina strain S5.2, revealed by whole-genome sequencing. J. Bacteriol. 2012;194:6366. doi: 10.1128/JB.01702-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chong TM, et al. Comprehensive genomic and phenotypic metal resistance profile of Pseudomonas putida strain S13.1.2 isolated from a vineyard soil. AMB Express. 2016;6:95. doi: 10.1186/s13568-016-0269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andreazza R, et al. Characterization of copper bioreduction and biosorption by a highly copper resistant bacterium isolated from copper-contaminated vineyard soil. Sci. Total Environ. 2010;408:1501–1507. doi: 10.1016/j.scitotenv.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 54.Frank, A. C. The genomes of endophytic bacteria. In Endophytes of Forest Trees 107–136 (Springer, 2011).
- 55.Robinson B, et al. Leaching of copper, chromium and arsenic from treated vineyard posts in Marlborough, New Zealand. Sci. Total Environ. 2006;364:113–123. doi: 10.1016/j.scitotenv.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 56.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 57.Antunes LCM, Ferreira RB, Buckner MM, Finlay BB. Quorum sensing in bacterial virulence. Microbiology. 2010;156:2271–2282. doi: 10.1099/mic.0.038794-0. [DOI] [PubMed] [Google Scholar]
- 58.Sanchez-Contreras M, Bauer WD, Gao M, Robinson JB, Downie JA. Quorum-sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007;362:1149–1163. doi: 10.1098/rstb.2007.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kloepper JW, McInroy JA, Liu K, Hu C-H. Symptoms of fern distortion syndrome resulting from inoculation with opportunistic endophytic fluorescent Pseudomonas spp. PloS ONE. 2013;8:e58531. doi: 10.1371/journal.pone.0058531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Preston GM. Plant perceptions of plant growth-promoting. Pseudomonas. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2004;359:907–918. doi: 10.1098/rstb.2003.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brader, G. et al. Ecology and genomic insights on plant-pathogenic and-nonpathogenic endophytes. Annu. Rev. Phytopathol (2017). [DOI] [PubMed]
- 62.Jakob K, et al. Pseudomonas viridiflava and P. syringae-natural pathogens of Arabidopsis thaliana. Mol. Plant Microbe Interact. 2002;15:1195–1203. doi: 10.1094/MPMI.2002.15.12.1195. [DOI] [PubMed] [Google Scholar]
- 63.Araki, H. et al. Presence/absence polymorphism for alternative pathogenicity islands in Pseudomonas viridiflava, a pathogen of Arabidopsis. Proc. Natl. Acad. Sci. USA. 103 5887–5892 (2006). [DOI] [PMC free article] [PubMed]
- 64.Angiuoli SV, et al. Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. Omics. 2008;12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinforma. Oxf. Engl. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 66.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huerta-Cepas J, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–93. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: Visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 2014;20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 71.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Domselaar GH, et al. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33:W455–9. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 2000;64:624–653. doi: 10.1128/MMBR.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buell CR, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilkie JP, Dye D, Watson D. Further hosts of Pseudomonas viridiflava. N. Z. J. Agric. Res. 1973;16:315–323. doi: 10.1080/00288233.1973.10421110. [DOI] [Google Scholar]
- 76.Gori A, Cerboneschi M, Tegli S. High-resolution melting analysis as a powerful tool to discriminate and genotype Pseudomonas savastanoi pathovars and strains. PloS ONE. 2012;7:e30199. doi: 10.1371/journal.pone.0030199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mühling M, Woolven-Allen J, Murrell JC. & Joint, I. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2008;2:379–392. doi: 10.1038/ismej.2007.97. [DOI] [PubMed] [Google Scholar]
- 78.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yi, S. et al. A novel naturally occurring class i 5-enolpyruvylshikimate-3-phosphate synthase from Janibacter sp. Confers high glyphosate tolerance to rice. Sci. Rep. 6 (2016). [DOI] [PMC free article] [PubMed]
- 80.Itoh K, et al. tfda-like genes in 2,4-dichlorophenoxyacetic acid-degrading bacteria belonging to the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster in α-Proteobacteria. Appl. Environ. Microbiol. 2002;68:3449–3454. doi: 10.1128/AEM.68.7.3449-3454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nielsen TK, et al. Novel insight into the genetic context of the cadAB genes from a 4-chloro-2-methylphenoxyacetic acid-degrading Sphingomonas. PloS ONE. 2013;8:e83346. doi: 10.1371/journal.pone.0083346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Govantes F, Porrua O, Garcia-Gonzalez V, Santero E. Atrazine biodegradation in the lab and in the field: enzymatic activities and gene regulation. Microb. Biotechnol. 2009;2:178–185. doi: 10.1111/j.1751-7915.2008.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Green JM. Review of glyphosate and ALS-inhibiting herbicide crop resistance and resistant weed management. Weed Technol. 2007;21:547–558. doi: 10.1614/WT-06-004.1. [DOI] [Google Scholar]
- 84.Rogers SG, Brand LA, Holder SB, Sharps ES, Brackin MJ. Amplification of the aroA gene from Escherichia coli results in tolerance to the herbicide glyphosate. Appl. Environ. Microbiol. 1983;46:37–43. doi: 10.1128/aem.46.1.37-43.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fei Y-Y, Gai J-Y, Zhao T-J. Identification of regulated genes conferring resistance to high concentrations of glyphosate in a new strain of Enterobacter. FEMS Microbiol. Lett. 2013;349:135–143. doi: 10.1111/1574-6968.12306. [DOI] [PubMed] [Google Scholar]
- 86.Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. FEBS J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material and Methods and Figures