Abstract
Cellulase production in the model cellulolytic fungus Trichoderma reesei is subject to a variety of environmental and physiological conditions involving an intricate regulatory network with multiple transcription factors. Here, we identified the mating type locus protein MAT1-2-1 as an interacting partner for the key transcriptional activator Xyr1 of T. reesei cellulase genes. Yeast two-hybrid and GST pulldown analyses revealed that MAT1-2-1 directly interacted with the putative transcription activation domain (AD, 767~940 aa) and the middle homology region (MHR2, 314~632 aa) of Xyr1. Disruption of the mat1-2-1 gene compromised the induced expression of cellulase genes with Avicel in response to light or with lactose. Chromatin immunoprecipitation (ChIP) demonstrated that MAT1-2-1 was recruited to the cbh1 (cellobiohydrolase 1-encoding) gene promoter in a Xyr1-dependent manner. These results strongly support an important role of MAT1-2-1 as a physiological cofactor of Xyr1, and suggest that MAT1-2-1 represents another regulatory node that integrates the light response with carbon source signaling to fine tune cellulase gene transcription.
Introduction
The ascomycete Trichoderma reesei (teleomorph Hypocrea jecorna), a saprophytic filamentous fungus, represents an important workhorse for industrial production of cellulases as well as other proteins. The secreted cellulase mixture synergistically decomposes the insoluble cellulosic materials to fermentable sugars1. In T. reesei, most of the cellulase genes are coordinately controlled by a suite of transcription factors2. Among others, xylanase regulator 1 (Xyr1) is absolutely necessary for activating the expression of most cellulases/hemicellulases. Lack of Xyr1 has been shown to eliminate cellulase induction by almost all inducers (such as cellulose and lactose)3. Regulation of Xyr1 expression and/or activity has thus been considered to be crucial for the production of various hydrolytic enzymes in T. reesei though the exact mechanism remains largely elusive4–6. Apart from the extensively studied carbon source-dependent expression of cellulase genes, other environmental cues, especially light, have also been found to exert a profound effect on cellulase production7–11. Our understanding on cellulase gene regulation in this filamentous fungi is thus far from complete.
Recently, T. reesei has been demonstrated to be able to carry out a heterothallic sexual cycle and successful mating has thus been achieved in this industrial fungus12,13. Similar to other filamentous ascomycete fungi, T. reesei has two mating types, MAT1-1 and MAT1-2, with MAT-idiomorphs occupying the same genomic region in the respective types. Whereas the MAT1-1 locus contains three genes, the MAT1-2 locus encodes a single protein with a high motility group (HMG) domain12. Moreover, the composition of the pheromone system in T. reesei is almost the same as other ascomycete fungi except that a novel h-type pheromone peptide (Hpp1) with characteristics of a- and α-type exists instead of the classical a-type peptide pheromone precursor14. Unlike other fungi, it has been found that neither pheromone precursor genes nor pheromone receptor genes of T. reesei are transcribed in a strictly mating type dependent manner although enhanced expression levels have been observed in the cognate mating type15. Another peculiarity in T. reesei is that sexual development is more efficient in light and upon growth on rich medium12. Potential signaling pathways transmitting these light signals in controlling development have been extensively investigated11,16,17. In particular, both the PAS domain protein ENVOY (ENV1) and the light-dependent regulator VEL1 have been found to be important for balanced regulation of pheromone precursor genes and receptor genes largely in a mating type dependent manner, and thus crucial for successful mating16,17. On the other hand, both regulators have been also found to be involved in regulation of cellulase gene expression7,11.
In this study, we screened for proteins interacting with Xyr1 using the yeast two-hybrid system and identified the mating type locus protein MAT1-2-1 as an interacting partner. We further provided insights into the role of MAT1-2-1 in regulation of cellulase gene expression in light. MAT1-2-1 may thus represent another regulatory node that integrates the light response with carbon source signaling to fine tune cellulase gene transcription.
Results
Mat1-2-1 interacts with Xyr1 in a yeast two-hybrid screen
To explore the mechanism of Xyr1 mediating the transcriptional activation of cellulase genes in T. reesei, a yeast two-hybrid (Y2H) screen was performed to isolate any protein that would interact with Xyr118. With the C-terminal putative activation domain (AD) of Xyr1 (767~940 aa) as a bait, a T. reesei cDNA expression library prepared under cellulase-inducing conditions was transformed into the reporter strain and transformants were selected on the quadruple dropout medium (QDO, SD/–Ade/–His/–Leu/–Trp) containing 100 ng/ml AbA. A positive clone with a 0.73 Kb cDNA insert was repeatedly obtained in three independent screens. Sequencing analysis of the cDNA insert revealed a full-length open reading frame of 241 amino acids corresponding to the mating type locus (MAT1-2) encoding protein, also annotated as a mating related transcription factor MAT1-2-1 (Tr_124341).
To see whether other regions of Xyr1 were also involved in the interaction with MAT1-2-1, the same Y2H assay was performed with different regions of Xyr1 as the bait. As with AD, a middle homology region of Xyr1 (MHR2, 314~632 aa) was also found to interact with MAT1-2-1 (Fig. 1A). However, an extended MHR region (MHR1, 314~772 aa) failed to support the activation of reporter genes (Fig. 1A). To further verify the interaction, we performed a pull-down assay. In accordance with the results of Y2H, His-tagged MAT1-2-1 was efficiently retained by recombinant GST-Xyr1AD and -Xyr1MHR2, whereas none of them could be detected in the GST only coupled beads (Fig. 1B). Taken together, these results indicate that MAT1-2-1 directly interacts with different regions of Xyr1 in vitro.
Figure 1.
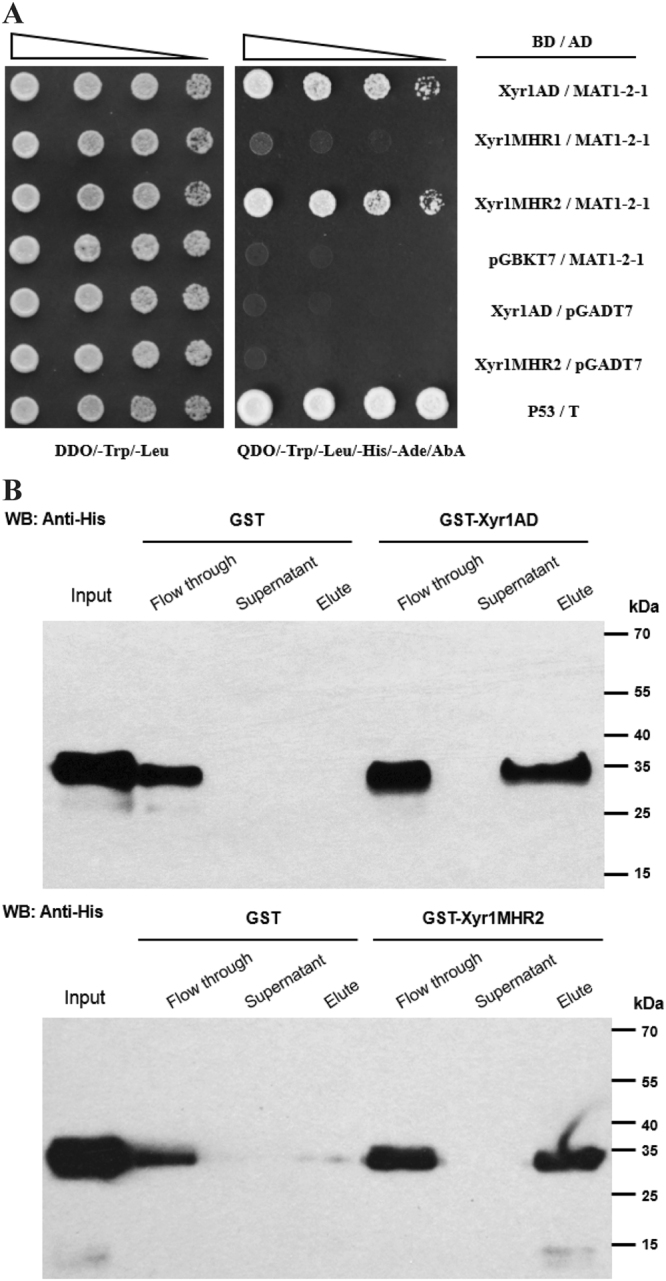
MAT1-2-1 interacts with Xyr1 in vitro. (A) Yeast two-hybrid analyses of the interaction between MAT1-2-1 and different regions of Xyr1. Serial dilutions of yeast transformant cells harboring the indicated plasmids were spotted on double dropout medium (DDO, SD/–Leu/–Trp) and quadruple dropout medium (QDO, SD/–Ade/–His/–Leu/–Trp) containing AbA plates, respectively, and were allowed grow at 30 °C for 3 days. Transformant containing p53/T pair was used as the positive control. (B) MAT1-2-1 interacts with Xyr1 AD and Xyr1 MHR2 in an in vitro GST pull-down assay. Recombinant 6 × His-MAT1-2-1 was incubated with glutathione sepharose 4B bead-coupled GST-Xyr1 AD, GST-Xyr1 MHR2 or GST as a control. MAT1-2-1 distributed in different fractions of the assay was detected by western blot with anti-His antibody.
MAT1-2-1 is a nuclear protein that co-localizes with Xyr1 and its transcription is upregulated by light or on inducing carbon sources
To investigate the subcellular localization of MAT1-2-1, a recombinant strain that simultaneously expressed EGFP-MAT1-2-1 and mCherry-Xyr1 both under the control of a copper responsive promoter Ptcu1 was constructed19. As shown in Fig. 2, EGFP-MAT1-2-1 fluorescence was readily observed to be distributed as a series of dots across the hypha and matched well with mCherry-Xyr1 fluorescence, which has been reported to be rapidly imported into the nucleus upon induced expression6. MAT1-2-1 also remained in the nucleus when transferred to medium containing inducing carbon sources including Avicel or lactose (data not shown). These results thus indicate that MAT1-2-1 is mainly a nuclear protein that co-localizes with Xyr1 regardless of the carbon source.
Figure 2.
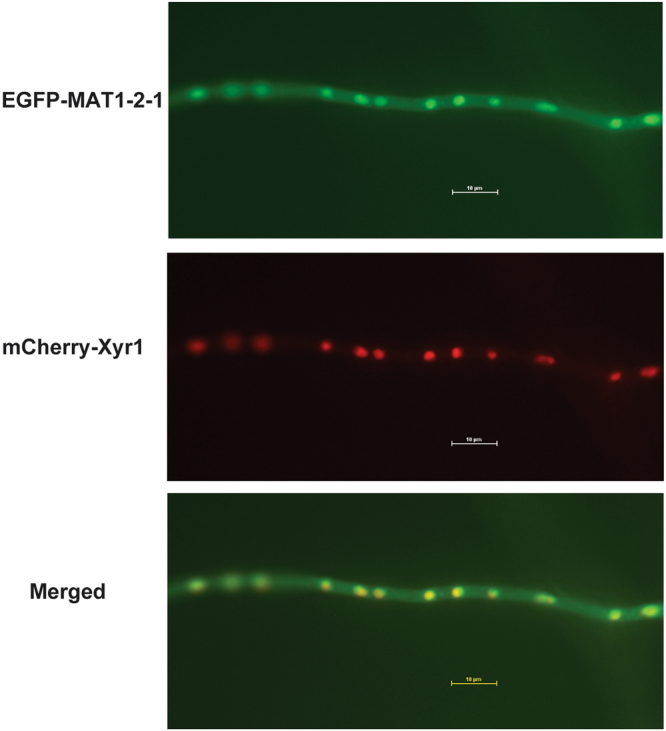
MAT1-2-1 is co-localized with Xyr1 in the nucleus. Germinated hypha of T. reesei transformant simultaneously expressing EGFP-MAT1-2-1 and mCherry-Xyr1 were analyzed for the respective fluorescence with a Nikon Eclipse 80i fluorescence microscope. The result shown represents one of at least two independent experiments.
The transcription of mat1-2-1 under different cultivation conditions was determined by RT-qPCR (Fig. 3). Whereas the transcription of mat1-2-1 was up-regulated on inducing carbon sources compared to that on repressing glucose, the upregulation was found to be more dramatic on lactose than on Avicel. In accordance with previous report20, the enhanced expression of mat1-2-1 on both carbon sources was further boosted under constant light (LL) compared to that on constant darkness (DD). These results implied that MAT1-2-1 may play a role in induced cellulase synthesis in response to nutrient and light signals.
Figure 3.
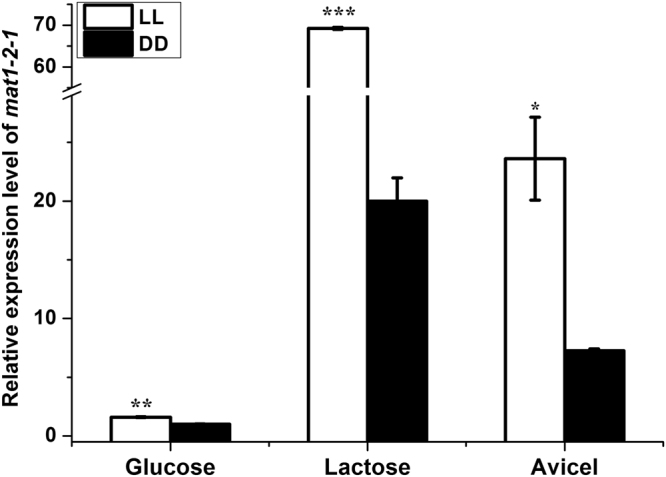
Transcription of mat1-2-1 under different cultivation conditions. qRT-PCR analysis of the relative transcription level of mat1-2-1. The mean values of each column were obtained from three independent biological replicates. Error bars indicated the standard error (SE) from three independent biological replicates. Significant differences (T-test *P < 0.05, **P < 0.01, ***P < 0.001) were detected for the transcription of mat1-2-1 when comparing light and darkness for the indicated inducing carbon sources.
MAT1-2-1 modulates the induced cellulase gene expression in response to light
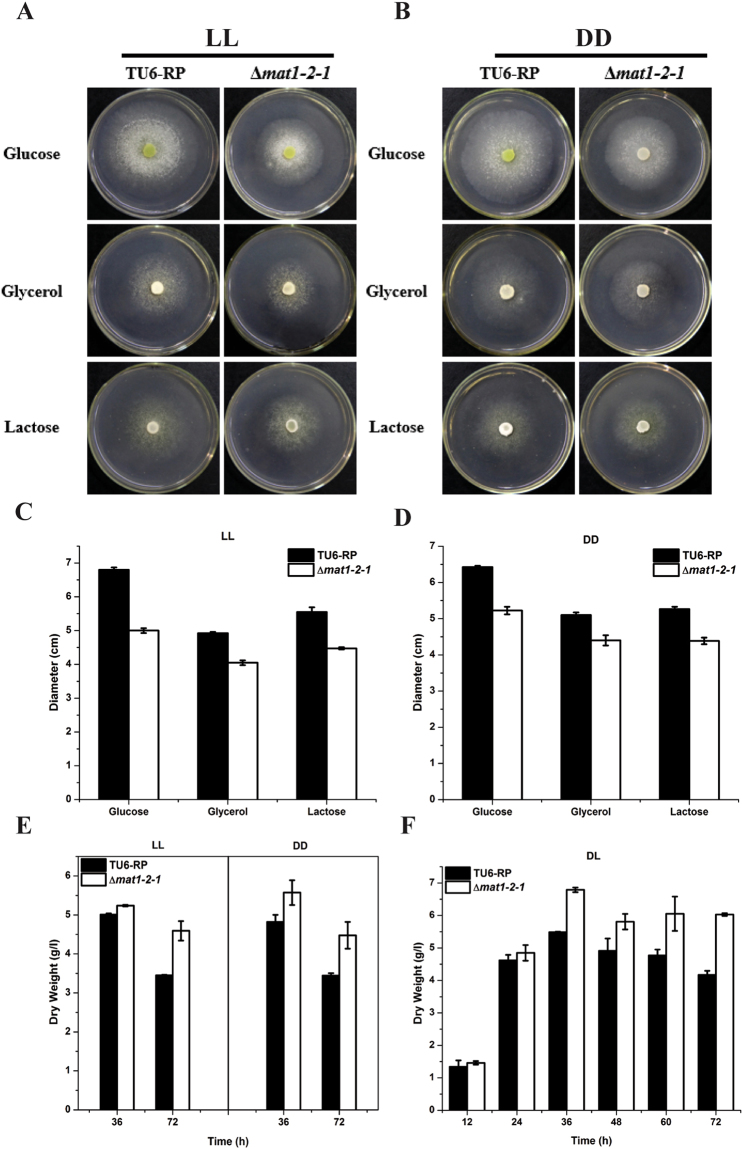
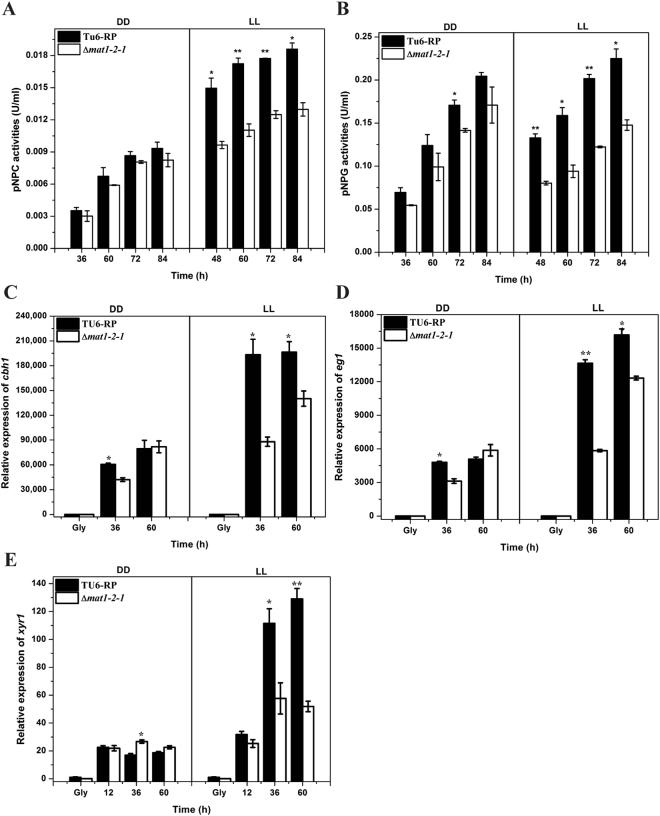
Considering the detected interaction between MAT1-2-1 and Xyr1, the role of MAT1-2-1 in cellulase gene expression was investigated. A T. reesei mutant lacking the mat1-2-1 gene was obtained by targeted gene replacement with the orotidine-5-decarboxylase gene pyr4 in TU-6 strain. PCR verification and Southern blot analysis confirmed that the deletion of mat1-2-1 occurred as predicted, integrating within the T. reesei genome resulting in the removal of the expected coding sequences (Fig. S1). Whereas deletion of mat1-2-1 resulted in a slightly slower growth rate on agar plate (Fig. 4A–D), the final biomass of the mutant strain cultured in liquid MA medium with glucose as the sole carbon source was slightly higher than that of the parental strain (Fig. 4E and F). Considering the light-responsive transcription of mat1-2-1, the induced production of cellulase activity in the mutant and the parental strains was analyzed on Avicel in either constant light or darkness by measuring the extracellular pNPC and pNPG hydrolytic activities. Consistent with previous report7, the induction of cellulases was stimulated by light compared to that in constant darkness (Fig. 5A and B). The results also revealed that the extracellular pNPC and pNPG hydrolytic activities were significantly decreased in the Δmat1-2-1 mutant compared to those in the parental strain under constant light condition, whereas hardly any effect was observed in darkness except that only a slight decrease in pNPG hydrolytic activities was observed at 72 h of induction. A similar light-dependent effect of the mat1-2-1 deletion on the induced xylanase activities was also observed in media using Avicel or xylan as the sole carbon source (Fig. S2). Further examination of the endogenous cbh1 and eg1 mRNA by RT-qPCR demonstrated that the decreased cellulase activities as observed in the mat1-2-1 deletion strain resulted from a down-regulation in the steady state transcripts of cellulase genes (Fig. 5C and D). Similarly, the transcription of the key transcriptional activator gene xyr1 was significantly decreased on Avicel in the absence of MAT1-2-1 in light but not in dark compared with that of the control strain (Fig. 5E).
Figure 4.
Growth assay of the TU6-RP and Δmat1-2-1 strains on solid agar plate or in liquid MA medium under constant light (LL) or darkness (DD) condition. (A and B) Growth of the Δmat1-2-1 and TU6-RP strains on MM agar plates with the indicated carbon sources under constant light and darkness conditions, respectively. (C and D) The diameter of the colony measured after three days of growth. (E and F) Dry weight of the Δmat1-2-1 and TU6-RP strains grown in liquid MA medium with 1% glucose (w/v) at the indicated time points in constant light/darkness or in daylight with normal light-dark cycle.
Figure 5.
Deletion of mat1-2-1 leads to down-regulation of cellulase gene expression on cellulose in light but not in darkness. (A and B) pNPC and pNPG hydrolytic activities of the extracellular supernatant of the Δmat1-2-1 and TU6-RP strains. (C to E) Relative expression levels of cbh1 (C), eg1 (D) and xyr1 (E) in these two strains cultured under the same conditions for enzymatic activity analyses were assayed by RT-qPCR. Error bars indicated the standard error (SE) from three independent biological replicates. Significant differences (T-test *P < 0.05, **P < 0.01) were detected for the extracellular activities and the transcription of the cbh1, egl1 and xyr1 genes between TU6-RP and the Δmat1-2-1 strain for the indicated time points after induction.
MAT1-2-1 plays an important role in lactose induced expression of cellulases
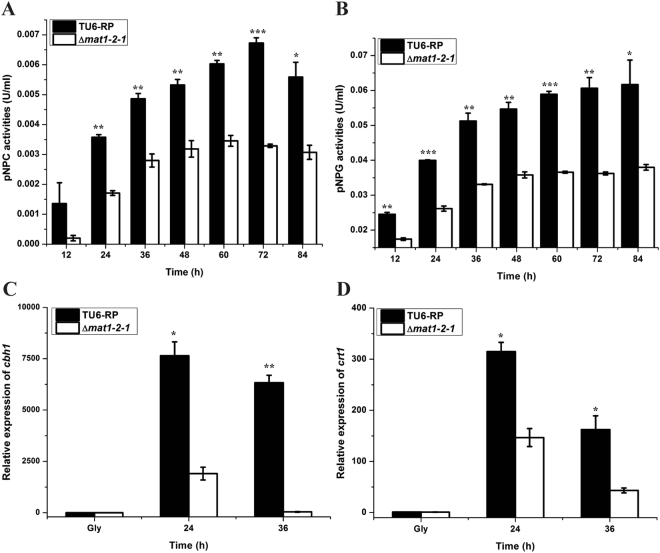
In contrast to the case on Avicel, light does not stimulate T. reesei cellulase gene expression on lactose but rather has a slightly negative effect21. Considering that transcription of mat1-2-1 also showed a significant upregulation on lactose even under constant darkness (Fig. 3), the effect of the absence of MAT1-2-1 on lactose induced cellulase expression was investigated. Cultivation of the Δmat1-2-1 and TU6-RP strains was performed under normal daylight condition (light–dark cycles) with 1% lactose (w/v) as the sole carbon source, wherein the induced cellulase synthesis in TU6-RP strain was comparable to that in darkness (data not shown). Analyses of the extracellular pNPC and pNPG hydrolytic activities revealed that deletion of mat1-2-1 resulted in a significantly decreased level of extracellular cellobiohydrolase and β-glucosidase activities (Fig. 6A and B). Transcriptional analyses by RT-qPCR further revealed that the expression level of the major cellulase gene cbh1 was dramatically decreased in the Δmat1-2-1 strain than that of the TU6-RP (Fig. 6C). Deletion of mat1-2-1 also resulted in a significant down-regulation of the MFS superfamily transporter crt1 (Fig. 6D), which has been reported to be essential for lactose uptake and cellulase induction by lactose22,23. Taken together, these results indicate that MAT1-2-1 also plays an important regulatory role in celllulase gene expression on lactose.
Figure 6.
MAT1-2-1 plays an important role in lactose induced expression of cellulases. (A and B) pNPC and pNPG hydrolytic activities of the extracellular supernatant, and (C and D) the relative transcription levels of cbh1 and crt1 of the Δmat1-2-1 and TU-6-Rp strains cultured on lactose. Error bars indicated the SE from three independent biological replicates. Significant differences (T-test, *P < 0.05, **P < 0.01, ***P < 0.001) were detected for the extracellular pNPC and pNPG activities between TU6-RP and Δmat1-2-1 for the indicated time points after induction. Significant differences also detected for the transcription of the cbh1 and crt1 genes (T-test *P < 0.05, **P < 0.01).
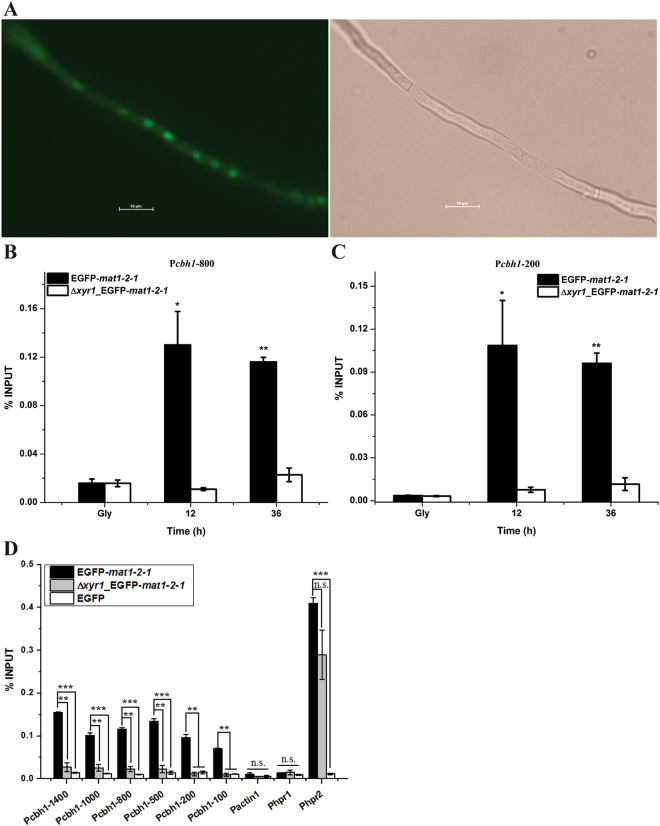
MAT1-2-1 is recruited to the cbh1 promoter in a Xyr1-dependent manner
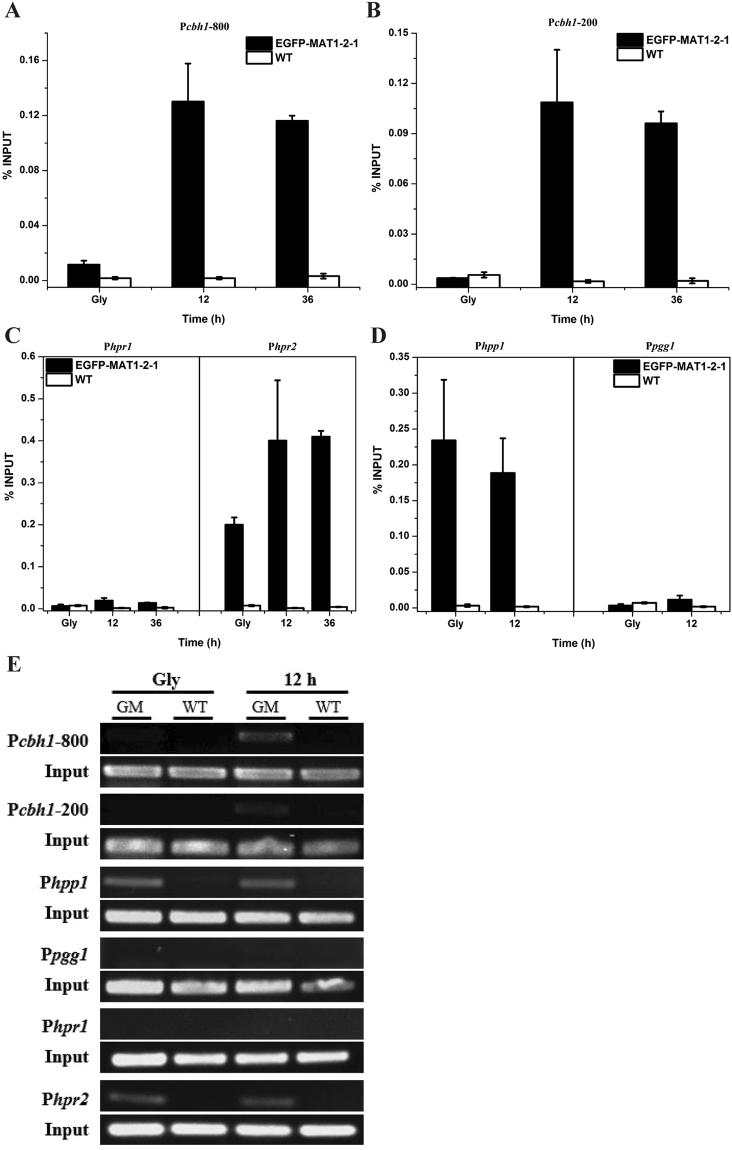
To test the possibility that MAT1-2-1 is recruited to cellulase gene promoters and thus is directly involved in regulation of cellulase gene expression upon induction, chromatin immunoprecipitation (ChIP) experiments were performed using the GFP antibody followed by quantitative real-time PCR in a T. reesei recombinant strain expressing EGFP-MAT1-2-1. The results demonstrated that, in contrast to the non-inducing conditions with glycerol as the sole carbon source, MAT1-2-1 was efficiently recruited to the −800 and −200 bp regions of the cbh1 promoter under inducing conditions (Fig. 7A,B and E). The same two regions were also found to be more efficiently occupied by Xyr1 whose expression only occurred upon induction24. As a control, relatively higher MAT1-2-1 occupancy was found on the promoters of the pheromone gene hpp1 and its cognate receptor gene hpr2 regardless of the culture conditions, whereas no such enrichment occurred on promoters of pgg1 and hpr1, thus indicating that MAT1-2-1 was also directly involved in the transcriptional regulation of hpp1 and hpr2 (Fig. 7C–E).
Figure 7.
MAT1-2-1 is recruited to the cbh1 promoter upon cellulase induction. ChIP analyses of MAT1-2-1 recruitment to the −800 and −200 regions of the cbh1 promoter (A and B), the pheromone receptor gene promoters (hpr1 and hpr2) (C), and the pheromone precursor gene promoters (hpp1 and pgg1) (D) before (Gly) and after induction with Avicel for the indicated time periods. (E) Semi-quantitative PCR products amplified from the above promoter fragments in the immunoprecipitated samples. GM and WT denoted samples from the EGFP-MAT1-2-1 and TU6-RP strains, respectively. Gly denoted pre-cultures on glycerol.
To elucidate whether the MAT12-1 recruitment to the cbh1 promoter is dependent on Xyr1, a recombinant strain expressing GFP-MAT1-2-1 but with xyr1 deleted was constructed and the same ChIP experiments were repeated. As shown in Fig. 8, whereas the appropriate expression and nuclear localization of GFP-MAT1-2-1 was hardly affected by the absence of Xyr1 (Fig. 8A), its enrichment on all tested cbh1 promoter regions was almost abolished in the absence of Xyr1 (Fig. 8B–D). On the contrary, MAT1-2-1 was still capable of being recruited to the hpr2 promoter (Fig. 8D). The same ChIP assay with a control strain expressing EGFP alone revealed that there were no recruitment signal of EGFP to all the tested promoters (Fig. 8D). Taken together, these results indicated that MAT1-2-1 may be directly involved in regulating cellulase gene expression by being specifically recruited to their promoters, and that its recruitment is most likely mediated by its interaction with Xyr1.
Figure 8.
MAT1-2-1 recruitment to the cbh1 promoter is dependent on Xyr1. (A) Expression and subcellular localization of EGFP-MAT1-2-1 in the Δxyr1 strain. (B and C) ChIP analyses of MAT1-2-1 recruitment to the −800 and −200 regions of the cbh1 promoter in the presence or absence of Xyr1 after induction with Avicel for the indicated time periods. Gly denoted pre-cultures on glycerol. (D) ChIP analyses of MAT1-2-1 recruitment to different regions of the cbh1 promoter under Avicel inducing conditions in the EGFP-MAT1-2-1, Δxyr1_EGFP-MAT1-2-1 strains and a control strain expressing EGFP. Error bars indicated the SD from two independent biological replicates. The actin, hpr1 and hpr2 promoters were used as controls. Significant differences (T-test *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant) were detected for the recruitment of EGFP-MAT1-2-1 to all the tested regions of the cbh1 promoter between the EGFP-MAT1-2-1 and the Δxyr1_EGFP-MAT1-2-1 strains. No significant recruitment was detected for MAT1-2-1 to the actin and hpr1 promoters or for GFP alone to all the tested promoters.
Discussion
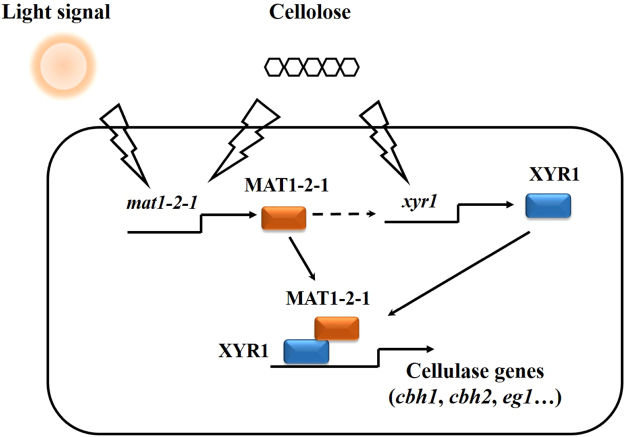
The regulation of cellulase gene expression has been extensively investigated in T. reesei, and the process has been shown to be controlled by a suit of transcription factors3,24–27. Whereas Xyr1 is the central regulator and absolutely necessary for activating the expression of most cellulases and hemicellulases3, several lines of evidence strongly suggest that fine-tuning of (hemi)-cellulase expression in T. reesei is achieved by an as-yet-unknown robust regulatory network involving the combinatorial and synergistic actions of different repressors and activators bound at multiple sites of the promoter27,28. In this study, we identified the mating type locus protein MAT1-2-1 of T. reesei that interacts with Xyr1, binds to the cbh1 promoter and modulates cellulase gene expression. As a conserved transcription factor (TF), MAT1-2-1 contains a high mobility group (HMG) domain that has no similarity to S. cerevisiae mating-type TFs, but is found in TFs of diverse lower or higher eukaryotes. Functional analyses in Penicillium chrysogenum provided evidence that MAT1-2-1 has functions beyond sexual development though the precise underlying mechanism is unclear29. In fact, HMG protein has been implicated in the regulation of transcription through interacting both with the basal transcription machinery and with individual TFs30,31. Once localized for binding, HMG proteins have been also reported to play an important role in maintaining chromatin structure and as a co-regulator of transcription through its strong DNA bending activity32. In our in vitro studies, we found that MAT1-2-1 directly interacted with the putative AD and MHR2 regions of Xyr1. Further ChIP assay revealed that MAT1-2-1 was recruited to the endogenous cbh1 promoter in an induction and Xyr1-dependent manner. The exact reason for the almost evenly recruitment of MAT1-2-1 to all the tested cbh1 promoter regions was not clear at present. One possible explanation is that, once MAT1-2-1 was recruited to some specific regions bound preferentially by Xyr1, it could somehow spread to the neighboring regions of the promoter. Nevertheless, the significant down-regulation of the induced cellulase gene expression in the absence of MAT1-2-1 under conditions wherein the endogenous mat1-2-1 was otherwise up-regulated, strongly supports an important role of MAT1-2-1 as a physiological cofactor of Xyr1. The negligible effect of mat1-2-1 deletion on the induced expression of cellulase genes in darkness is largely due to the fact that mat1-2-1 was expressed at a much lower level compared to that in light.
In addition to the type of carbon source, additional environmental conditions are known to affect T. reesei cellulase production. Among others, light is a crucial environmental factor for fungi wherein the light-induced changes in gene expression impact various physiological processes including growth, asexual and sexual reproduction, pigment formation, carbon metabolism, and circadian rhythms33–35. Interestingly, it has been found that the induction of cellulase gene transcription in T. reesei by cellulose is enhanced by light7. Although the effect of light on cellulase expression is likely not a direct effect, possibly an indirect one through general effects on metabolism, evidence exists that this light-dependent regulation is impacted by the PAS domain light-regulatory protein ENVOY (ENV1)7,10. Besides representing an important node in integrating signals from light and nutrients via heterotrimeric G-protein and cyclic AMP (cAMP) pathways, ENV1 has been also shown to be important for successful mating by balancing the levels of genes in light at least in part by exerting a negative effect on MAT1-2-1 16. In the absence of env1, a much dramatic up-regulation of MAT1-2-1 was observed in daylight16. Although possibility that ENV1 participates in regulating cellulase gene expression in response to light via alternative signal effectors could not be excluded, the observation that MAT1-2-1 acts as a cofactor of Xyr1 promoted us to speculate that the abnormally higher up-regulation of MAT1-2-1 than that observed in WT could otherwise in part account for the retarded cellulase induction as displayed in the Δenv1 mutant7 by otherwise interfering with Xyr1 function. Our data thus supports a regulatory model where MAT1-2-1 functions during the cellulolytic response in T. reesei (Fig. 9). Upon cellulose induction in light, both Xyr1 and an appropriate level of MAT1-2-1 are induced and then synergize in binding to cellulase gene promoters, thus leading to the efficient induction of cellulolytic genes. The mating-type-dependent effect on cellulase gene expression thus suggests that MAT1-2-1 may represent another regulatory node that integrates the light response with carbon source signaling to fine tune cellulase gene transcription.
Figure 9.
The putative regulatory effect of MAT1-2-1 on the cellulolytic response in T. reesei.
Materials and Methods
Strains, medium, and cultivations
The uridine-auxotrophic strain T. reesei TU-6 (ATCC MYA-256) was used as a parental strain for targeted gene knock-out. A recombinant TU-6 strain with a retransformed pyr4 gene termed TU6-RP, was used as control strain for all the phenotypic analysis experiments24. T. reesei strains were maintained on malt extract agar (Sigma-Aldrich, Madison, USA) supplemented with 10 mM uridine when necessary. For the transcription and cellulase production analyses, T. reesei strains were pre-grown in 1-liter Erlenmeyer flasks on a rotary shaker (200 rpm) at 30 °C in 250 ml Mandels-Andreotti medium with 1% glycerol (v/v) as the carbon source for 48 h. Mycelia were harvested by filtration and washed twice with Mandels-Andreotti medium with no carbon source. Equal amounts of mycelia were then transferred to a fresh medium containing 1% Avicel (w/v) (Sigma-Aldrich) or other carbon sources as indicated, and the incubation was continued for the indicated time periods.
The Saccharomyces cerevisiae strain Y2H Gold (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS–Gal1TATA–His3, GAL2UAS–Gal2TATA–Ade2 URA3::MEL1UAS–Mel1TATA AUR1-C MEL1) (Clontech) was used as the host for the two-hybrid screen. Yeast cells were routinely cultivated at 30 °C in YPD medium (1% yeast extract, 2% peptone and 2% glucose). Synthetic complete (SC) medium lacking tryptophan and leucine with 100 ng/ml of Aureobasidin A (AbA, Clontech) was used for transformant selection. For plate growth assays, yeast cells with serial dilutions were spotted onto a selective quadruple dropout medium (QDO, SD/–Ade/–His/–Leu/–Trp) containing 100 ng/ml of AbA.
Escherichia coli DH5α cells were used for plasmid construction and amplification. Escherichia coli BL21 (DE3) cells were used as a host for expression of the recombinant proteins.
cDNA library construction and yeast-based two-hybrid screening
Preparation of the cDNA library from mycelia of the strain QM9414 induced on Avicel was carried out as described previously by Cao Y et al.24.
For constructing the bait vector for yeast two-hybrid screening, the Xyr1 coding sequences either for its putative activation domain (AD, aa 767~940) or the middle homology domains (MHR1, aa 314~772 and MHR2, aa 314~632) were amplified from T. reesei cDNA with primers containing NdeI and BamHI sites. The digested DNA fragments were ligated into pGBKT7 (Clontech) to obtain pGBKT7-Xyr1AD, pGBKT7-MHR1 and pGBKT7-MHR2, respectively.
The bait plasmid pGBKT7-Xyr1AD was transformed into the yeast Y2H Gold strain by a PEG-LiAC method to obtain the bait strain36. Ten micrograms of T. reesei cDNA library plasmid was then transformed into the bait strain and transformants were selected on SC plates lacking tryptophan and leucine but containing 100 ng ml-1 AbA. Growing colonies were picked and the harbored plasmids with cDNA insert were verified by DNA sequencing after retransformation.
Plasmids and recombinant T. reesei strain construction
To delete mat1-2-1 in TU-6, two DNA fragments corresponding to approximately 2-kb of mat1-2-1 up- and downstream of the open reading frame (ORF) regions were amplified from T. reesei TU-6 genomic DNA and ligated into the pUCpyr4 plasmid37 to yield the knock-out vector pUCpyr4-delMAT1-2-1, which was used to transform T. reesei TU-6 after linearization with EcoRI.
Transformation of T. reesei protoplast was carried out essentially as previously described37. Transformants were selected on minimal medium either for uridine prototroph or for resistance to hygromycin B (120 µg ml-1) after at least three rounds of single conidium purification.
To determine the subcellular localization of MAT1-2-1 and Xyr1, the tcu1 gene (Tr_52315) promoter and the cbh2 terminator were first amplified from QM9414 genomic DNA and inserted into the HindIII/SalI and SpeI/SalI sites in the pUC19-hph and pUC19-pyr4 plasmids37 to obtain Ptcu1-hph and Ptcu1-pyr4, respectively. The EGFP and mCherry coding genes were then ligated into Ptcu1-hph and Ptcu1-pyr4 after digestion with EcoRV and NotI to generate Ptcu1-EGFP-hph and Ptcu1-mCherry-pyr4, respectively. The MAT1-2-1 and Xyr1 coding sequences were finally amplified from T. reesei cDNA and inserted into the NotI and SpeI sites in the above two vectors to obtain Ptcu1-EGFP-MAT1-2-1 and Ptcu1-mCherry-Xyr1, respectively. These two intact vectors were co-transformed into TU-6 and Ptcu1-EGFP-MAT1-2-1 was also transformed into the Δmat1-2-1 and Δxyr1 strains3.
Recombinant protein production in E. coli and GST pull down
For the expression of Xyr1 MHR2, Xyr1 AD and MAT1-2-1 in E. coli, the DNA fragments coding for Xyr1 MHR2 and Xyr1 AD were amplified from TU-6 cDNA and ligated into the pGEX 4T-1 vector after digestion with EcoRI and NotI to obtain plasmids pGEX 4T-1-MHR2 and pGEX 4T-1-AD, respectively. The MAT1-2-1 coding sequence was amplified from TU-6 cDNA with primers harboring NdeI and EcoRI sites and ligated into pET22b (+) to obtain pET22b-MAT1-2-1.
The indicated expression constructs were transformed into CaCl2-treated competent E. coli BL21 (DE3) cells38. Protein purification was carried out essentially as described by Cao Y et al.24. All of the protein preparations were stored at −80 °C in the presence of 20% (v/v) glycerol.
For GST pull-down assay, the purified 6 × His MAT1-2-1 was incubated with glutathione sepharose bead-coupled GST, GST-Xyr1 AD, and GST-Xyr1 MHR2, respectively, with rotation at room temperature for half an hour before being washed three times with PBST buffer (137 mmol/L NaCl, 2.7 mmol/L KCl,10 mmol/L Na2HPO4,2 mmol/L KH2PO4, 0.5% Triton X-100, pH 7.4). The proteins in the flow through as well as those retained on the beads were resolved by SDS-PAGE and detected by Western blot with anti-His antibody (Santa Cruz, sc-8036).
Fluorescence microscopy
The subcellular localization of EGFP-MAT1-2-1 and mCherry-Xyr1 was analyzed using fluorescence microscopy essentially as described previously37.
Enzyme activity analysis
Extracellular cellobiohydrolase and β-glucosidase activities were determined by measuring the amount of released p-nitrophenol using p-nitrophenyl-D-cellobioside (pNPC; Sigma) and p-nitrophenyl-β-D- glucopyranoside (pNPG; Sigma) as the substrates, respectively. The enzymatic reactions were performed essentially as described previously39. One unit (U) of pNPCase or pNPGase activity corresponds to the conversion of 1 μmol substrate per minute under the test conditions.
Quantitative RT-PCR (qRT-PCR)
To determine mat1-2-1 transcription under different cultivation conditions, the parental strain TU-6 was pre-grown in Mandels-Andreotti (MA) medium with 1% glycerol (v/v) for 36 h and then transferred into a fresh batch of MA medium supplemented with 1% glucose (w/v), 1% lactose (w/v) or 1% Avicel (w/v) as the sole carbon source. The culture was continued under constant light (LL) or darkness (DD) condition for another 24 h. As for the transcription of cellulase genes, the Δmat1-2-1 and TU6-RP strains were pre-grown in MA medium with 1% glycerol (v/v) for 48 h and then transferred into MA medium supplemented with 1% Avicel (w/v) or 1% lactose (w/v) as the sole carbon source under constant light (LL) or darkness (DD).
RNA extraction and qRT-PCR analyses were performed as described previously37. Data analysis was performed using the relative quantitation/comparative CT (ΔΔCT) method and was normalized to an endogenous control (rpl6e 40). Transcript level from cultures on glycerol was used as the reference and set to 1. Three biological replicates were performed for each analysis. The results and the error bars are the mean of and SE (standard error) from these replicates, respectively. Statistical analysis was performed using the student’s t-test when determining whether significant differences exist between TU6-RP and the Δmat1-2-1 strain for the extracellular activities and the transcription of the cbh1 and egl1 genes. Similar statistical analysis was also performed for the recruitment signal of EGFP-MAT1-2-1 to each regions of the cbh1 promoter and the control promoter between recombinant strains with or without Xyr1.
Chromatin immunoprecipitation (ChIP)
To analyze the binding of indicated proteins to specific promoter regions, the recombinant strain expressing EGFP-MAT1-2-1 was cultivated in liquid MA medium containing 1% Avicel for 12 h and 36 h, respectively. ChIP assays were performed according to a previously described protocol41,42. Briefly, the mycelia were fixed in minimal medium containing 1% formaldehyde at 30 °C for 10 min with shaking and followed with 125 mM glycine shaking for an additional 5 min, which were then collected, ground in liquid N2 and broken in lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF (phenylmethanesulfonyl fluoride), 1 μg ml-1 leupeptin, and 1 μg ml−1 pepstatin A) with ceramic beads (0.5 mm). Chromatin DNA was further sonicated to obtain an average DNA fragment size of approximately 500 bp. Immunoprecipitation was then performed with anti-GFP (#ab290, Abcam) or anti-Xyr1 antibody24 with equal amounts of extract proteins (2 mg) at 4 °C for 5 h. Quantitative PCR was finally performed with the precipitated chromatin DNAs using a Bio-Rad IQ2 thermocycler (Bio-Rad) (Takara). Relative enrichment of the DNAs was calculated as a percentage of the input DNA. Primers used in quantitative RT-PCR and ChIP were listed in Supplementary Table S1.
Electronic supplementary material
Acknowledgements
We thank Dr. Robert L. Mach for kindly providing the Δxyr1 strain. We also thank Prof. Qun He for technical help of the ChIP experiments. This work is supported by grants from the National Natural Science Foundation of China (31270116, 31470162 and 31670040) and National Key Technology Support Program (2015BAD15B05). Zhang WX would also like to acknowledge the support from the Scientific Research Foundation for Excellent Young and Middle-Aged Scientists of Shandong Province of China (BS2013NJ021), and China Postdoctoral Science Foundation (2012M521325). The authors declare no conflict of interest with the content of this article.
Author Contributions
F.L.Z., Y.L.C., L.W. and X.X.L. performed the experiments. W.F.L., W.X.Z., X.F.M. and G.J.C. performed data analysis. F.L.Z. and W.F.L. designed the project. F.L.Z., and W.F.L. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17439-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiology and Molecular Biology Reviews. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels. 2009;2:19. doi: 10.1186/1754-6834-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stricker AR, Grosstessner-Hain K, Wurleitner E, Mach RL. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryotic cell. 2006;5:2128–2137. doi: 10.1128/EC.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mach-Aigner AR, et al. Transcriptional regulation ofxyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Applied and environmental microbiology. 2008;74:6554–6562. doi: 10.1128/AEM.01143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derntl C, et al. Mutation of the Xylanase regulator 1 causes a glucose blind hydrolase expressing phenotype in industrially used Trichoderma strains. Biotechnol Biofuels. 2013;6:62. doi: 10.1186/1754-6834-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichius, A., Seidl-Seiboth, V., Seiboth, B. & Kubicek, C. P. Nucleo-cytoplasmic shuttling dynamics of the transcriptional regulators XYR1 and CRE1 under conditions of cellulase and xylanase gene expression in Trichoderma reesei. Mol Microbiol, 10.1111/mmi.12824 (2014). [DOI] [PMC free article] [PubMed]
- 7.Schmoll M, Franchi L, Kubicek CP. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot Cell. 2005;4:1998–2007. doi: 10.1128/EC.4.12.1998-2007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seibel C, et al. Light-dependent roles of the G-protein alpha subunit GNA1 of Hypocrea jecorina (anamorph Trichoderma reesei) BMC Biol. 2009;7:58. doi: 10.1186/1741-7007-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmoll M, Schuster A, Silva Rdo N, Kubicek CP. The G-alpha protein GNA3 of Hypocrea jecorina (Anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light. Eukaryot Cell. 2009;8:410–420. doi: 10.1128/EC.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellanos F, et al. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet Biol. 2010;47:468–476. doi: 10.1016/j.fgb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Karimi Aghcheh R, et al. The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PloS one. 2014;9:e112799. doi: 10.1371/journal.pone.0112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidl V, Seibel C, Kubicek CP, Schmoll M. Sexual development in the industrial workhorse Trichoderma reesei. Proc Natl Acad Sci USA. 2009;106:13909–13914. doi: 10.1073/pnas.0904936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linke R, et al. Restoration of female fertility in Trichoderma reesei QM6a provides the basis for inbreeding in this industrial cellulase producing fungus. Biotechnol Biofuels. 2015;8:155. doi: 10.1186/s13068-015-0311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmoll M, Seibel C, Tisch D, Dorrer M, Kubicek CP. A novel class of peptide pheromone precursors in ascomycetous fungi. Mol Microbiol. 2010;77:1483–1501. doi: 10.1111/j.1365-2958.2010.07295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seibel C, Tisch D, Kubicek CP, Schmoll M. The role of pheromone receptors for communication and mating in Hypocrea jecorina (Trichoderma reesei) Fungal Genet Biol. 2012;49:814–824. doi: 10.1016/j.fgb.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seibel C, Tisch D, Kubicek CP, Schmoll M. ENVOY is a major determinant in regulation of sexual development in Hypocrea jecorina (Trichoderma reesei) Eukaryot Cell. 2012;11:885–895. doi: 10.1128/EC.05321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bazafkan, H. et al. Mating type dependent partner sensing as mediated by VEL1 in Trichoderma reesei. Mol Microbiol, 10.1111/mmi.12993 (2015). [DOI] [PMC free article] [PubMed]
- 18.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 19.Lv X, et al. Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction-independent production of cellulases in Trichoderma reesei. Biotechnol Biofuels. 2015;8:67. doi: 10.1186/s13068-015-0249-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tisch D, Kubicek CP, Schmoll M. The phosducin-like protein PhLP1 impacts regulation of glycoside hydrolases and light response in Trichoderma reesei. BMC Genomics. 2011;12:613. doi: 10.1186/1471-2164-12-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuster A, Tisch D, Seidl-Seiboth V, Kubicek CP, Schmoll M. Roles of protein kinase A and adenylate cyclase in light-modulated cellulase regulation in Trichoderma reesei. Appl Environ Microbiol. 2012;78:2168–2178. doi: 10.1128/AEM.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanova C, Baath JA, Seiboth B, Kubicek CP. Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PloS one. 2013;8:e62631. doi: 10.1371/journal.pone.0062631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, et al. Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. The Journal of biological chemistry. 2013;288:32861–32872. doi: 10.1074/jbc.M113.505826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao, Y. et al. Rce1, a novel transcriptional repressor, regulates cellulase gene expression by antagonizing the transactivator Xyr1 in Trichoderma reesei. Mol Microbiol, 10.1111/mmi.13685 (2017). [DOI] [PubMed]
- 25.Saloheimo A, Aro N, Ilmen M, Penttila M. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. Journal of Biological Chemistry. 2000;275:5817–5825. doi: 10.1074/jbc.275.8.5817. [DOI] [PubMed] [Google Scholar]
- 26.Aro N, Saloheimo A, Ilmen M, Penttila M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. Journal of Biological Chemistry. 2001;276:24309–24314. doi: 10.1074/jbc.M003624200. [DOI] [PubMed] [Google Scholar]
- 27.Hakkinen M, et al. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels. 2014;7:14. doi: 10.1186/1754-6834-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dos Santos CL, et al. Understanding the Role of the Master Regulator XYR1 in Trichoderma reesei by Global TranscriptionalAnalysis. Frontiers in microbiology. 2016;7:175. doi: 10.3389/fmicb.2016.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boehm J, Dahlmann TA, Guemueser H, Kueck U. A MAT1-2 wild-type strain from Penicillium chrysogenum: functional mating-type locus characterization, genome sequencing and mating with an industrial penicillin-producing strain. Molecular Microbiology. 2015;95:859–874. doi: 10.1111/mmi.12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Critical reviews in biochemistry and molecular biology. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 31.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci. 2001;26:167–174. doi: 10.1016/S0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 32.Dowell NL, Sperling AS, Mason MJ, Johnson RC. Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription. Genes Dev. 2010;24:2031–2042. doi: 10.1101/gad.1948910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrera-Estrella A, Horwitz BA. Looking through the eyes of fungi: molecular genetics of photoreception. Mol Microbiol. 2007;64:5–15. doi: 10.1111/j.1365-2958.2007.05632.x. [DOI] [PubMed] [Google Scholar]
- 34.Tisch D, Schmoll M. Light regulation of metabolic pathways in fungi. Appl Microbiol Biotechnol. 2010;85:1259–1277. doi: 10.1007/s00253-009-2320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS biology. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q, et al. Differential involvement of beta-glucosidases from Hypocrea jecorina in rapid induction of cellulase genes by cellulose and cellobiose. Eukaryotic cell. 2012;11:1371–1381. doi: 10.1128/EC.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, et al. Intracellular beta-glucosidases CEL1a and CEL1b are essential for cellulase induction on lactose in Trichoderma reesei. Eukaryotic cell. 2014;13:1001–1013. doi: 10.1128/EC.00100-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tisch D, Kubicek CP, Schmoll M. New insights into the mechanism of light modulated signaling by heterotrimeric G-proteins: ENVOY acts on gna1 and gna3 and adjusts cAMP levels in Trichoderma reesei (Hypocrea jecorina) Fungal Genet Biol. 2011;48:631–640. doi: 10.1016/j.fgb.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Chen G, Liu W. Multiple metabolic signals influence GAL gene activation by modulating the interaction of Gal80p with the transcriptional activator Gal4p. Molecular microbiology. 2010;78:414–428. doi: 10.1111/j.1365-2958.2010.07343.x. [DOI] [PubMed] [Google Scholar]
- 42.Xin Q, Gong Y, Lv X, Chen G, Liu W. Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression. Curr Microbiol. 2013;67:580–589. doi: 10.1007/s00284-013-0396-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.