Abstract
Introduction
There is still a clear need for a widely available, inexpensive and reliable method to diagnose Alzheimer’s disease (AD) and monitor disease progression. Liquid chromatography–mass spectrometry (LC-MS) is a powerful analytic technique with a very high sensitivity and specificity.
Objectives
The aim of the present study is to measure concentrations of 20 bile acids using the novel Kit from Biocrates Life Sciences based on LC-MS technique.
Methods
Twenty bile acid metabolites were quantitatively measured in plasma of 30 cognitively healthy subjects, 20 patients with mild cognitive impairment (MCI) and 30 patients suffering from AD.
Results
Levels of lithocholic acid were significantly enhanced in plasma of AD patients (50 ± 6 nM, p = 0.004) compared to healthy controls (32 ± 3 nM). Lithocholic acid plasma levels of MCI patients (41 ± 4 nM) were not significantly different from healthy subjects or AD patients. Levels of glycochenodeoxycholic acid, glycodeoxycholic acid and glycolithocholic acid were significantly higher in AD patients compared to MCI patients (p < 0.05). All other cholic acid metabolites were not significantly different between healthy subjects, MCI patients and AD patients. ROC analysis shows an overall accuracy of about 66%. Discriminant analysis was used to classify patients and we found that 15/23 were correctly diagnosed. We further showed that LCA levels increased by about 3.2 fold when healthy subjects converted to AD patients within a 8–9 year follow up period. Pathway analysis linked these changes to a putative toxic cholesterol pathway.
Conclusion
In conclusion, 4 bile acids may be useful to diagnose AD in plasma samples despite limitations in diagnostic accuracy.
Electronic supplementary material
The online version of this article (10.1007/s11306-017-1297-5) contains supplementary material, which is available to authorized users.
Keywords: Alzheimer, Diagnosis, Plasma, Bile acid, Lithocholic acid, Biomarkers
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease that gradually leads to severe cognitive deterioration and premature death. The main neuropathological hallmarks of AD concurrently include accumulation of amyloid-β (Aβ) plaques, neurofibrillary tangles (NFT), inflammation and glial responses, synaptic and neuronal loss and vascular alterations. To date, the diagnosis of AD is based on a time-consuming combination of psychological testing, imaging and the analysis of three well-established biomarkers [amyloid-β42 (Aβ42), total tau and phospho-tau-181] in cerebrospinal fluid (Blennow 2005; Humpel 2011a). The increasing number of early dementia patients forces the identification of economic and reliable biomarkers in blood, urine, or saliva since cerebrospinal fluid collection is an invasive procedure and an expensive analysis (Humpel 2011a; Blennow et al. 2010; Song et al. 2009; Shad et al. 2013; Hu et al. 2012).
Targeted metabolomic analysis in different laboratories including ours allowed to identify several lipid metabolites in plasma being suitable to differentiate AD patients from healthy controls (Mapstone et al. 2014; Olazaran et al. 2015; Klavins et al. 2015). These preliminary data suggested that also bile acids may be altered in AD (Olazaran et al. 2015). Bile acids are major components of bile formed from cholesterol through various enzymatic reactions in hepatocytes. Primary bile acids synthesized in the liver (i.e., cholic acid and chenodeoxycholic acid) in humans are mainly conjugated with taurine or glycine amino acids through the terminal side-chain carboxylic group present in the bile acid structure (Hofmann and Hagey 2008). Bile acids are transported back to the liver via portal blood for re-secretion, a process called enterohepatic bile acid circulation (Li and Chang 2014). Bile acids have multiple roles in humans. Bile acids have hormone-like functions and interact with membrane bound G-proteins coupled receptors of various tissues (Chiang 2002; Lefebvre et al. 2009; Thomas et al. 2008). Bile acids are involved in regulating drug efficiency and toxicity via cytochrome P450 metabolism, and are increased after liver injury (Neale et al. 1971). Bile acids show effects on the gastrointestinal system, and gut microbiota are involved in the transformation of bile acids through deconjugation, dehydroxylation and reconjugation. In fact bile acids can also induce reactive oxygen species and DNA damage (Bernstein et al. 2005). And finally, bile acids are of particular interest because they are the end product of the cholesterol metabolism, which directly link bile acids to AD as cholesterol has been implicated to play a role in progression of AD (Puglielli et al. 2003). In addition to the role in cholesterol elimination, bile acids exert effects on glucose and lipid metabolism via activation of bile acid receptors nuclear-farnesoid-X receptor (FXR) and G-protein-coupled plasma-membrane bound receptors (TGR5), directly linking bile acids with progression of AD with diabetes type II (Schilling 2016) or with cerebrovascular dysfunction (Humpel 2011b).
The aim of the present study is to apply the novel Bile Acid kit from Biocrates to analyze 20 bile acid metabolites. Our data will reveal that plasma levels of the bile acid lithocholic acid (LCA) are significantly different between healthy controls, MCI patients and patients suffering from AD. In a follow up experiment, we demonstrated that LCA levels significantly increased when healthy subjects converted to AD.
Materials and methods
Patients
Cognitively healthy subjects and patients suffering from MCI and AD were recruited at Hall/Tirol State Hospital, Austria. For the test set, 80 persons were included in this study, all of whom were older than 70 years (Table 1). For the classification blinded study 23 persons were included. For a conversion study blood was taken from the same of 80 initial patients (n = 8) after 8–9 years and verified that they converted from controls to AD. The procedure for diagnosis has been described by us in detail elsewhere (Hochstrasser et al. 2011, 2012). A panel including a neurologist, psychiatrist and neuropsychologist examined all clinical and diagnostic features. Following a discussion, participants were assigned a diagnosis of MCI or AD according to published criteria (Petersen et al. 2001; McKhann et al. 1984). None of the MCI patients had a score higher than 0.5 in the global score of the Clinical Dementia Rating (Morris 1993). Cognitive functions were assessed with the German version of the CERAD plus neuropsychological battery (Welsh et al. 1994). This test battery provides a reliable profile of cognitive impairments based on demographically adjusted z scores. In addition to eight cognitive domains of CERAD battery, depressive symptoms were assessed using the 30-items version of Geriatric Depression Scale (GDS). All study participants were also assessed by magnetic resonance imaging using a 1.5 T Siemens Symphony MRI scanner with a T1-weighted FLASH 3D sequence and a repetition time (TR) of 9.7 ms, an echotime (TE) of 4 ms, a matrix size of 256 × 256 and a field of view of 230 mm, yielding sagittal slices with a thickness of 1.5 mm and an in-plane resolution of 0.98 × 0.98 mm. Imaging findings were evaluated by a trained radiologist who was blinded to diagnosis and cognitive performance of the patients. Medial temporal atrophy (MTA) was assessed using a standardized scale (Scheltens et al. 1992). Subjects were excluded from the study when suffering from acute viral hepatitis or chronic hepatitis, cirrhosis of the liver, liver damage from alcohol abuse or alcoholic fatty liver; elevations of liver enzymes alanine transaminase (ALT) and aspartate transaminase (AST), other decompensated metabolic diseases and history of drug or alcohol addiction. Participants underwent continuous statin or ezetimibe treatment for at least 3 months before study entry. No patient had a cholesterol level > 240 mg/dL that was not treated with a statin or ezetimibe.The study was approved by the Ethics Committee of the Medical University of Innsbruck and was performed in accordance with the Helsinki Declaration. All subjects gave written informed consent.
Table 1.
Characteristics of healthy subjects, MCI patients and patients suffering from AD
| Diagnostic group | CO | MCI | AD |
|---|---|---|---|
| Sample size | 30 | 20 | 30 |
| Female/male | 22 /8 | 21/9 | 11/9 |
| Age (years) | 77 ± 1.2 | 78 ± 1.2 | 79 ± 2.0 |
| MMSE | 29 ± 0.2 | 28 ± 0.3 | 20 ± 1.0 *** |
| GDS | 3.3 + 0.5 | 2.6 ± 0.4 | 3.1 ± 0.7 |
Values are given as mean ± SEM
MMSE mini-mental state examination (scale 0–30), GDS geriatric depression scale (30-items version), CO healthy controls, MCI mild cognitive impairment, AD Alzheimer’s disease
Statistical analysis was performed by One Way ANOVA with a subsequent Fisher LSD posthoc test (***p < 0.001)
Collection of plasma
Ten mL of EDTA blood was collected, centrifuged (400×g, 10 min), and the upper plasma phase was immediately frozen at − 80 °C. Blood processing time was 4.2 ± 0.2 h and did not differ between groups.
Bile acid analysis and quantification
Analysis was performed using the novel Bile Acids Kit (Biocrates Life Sciences) as described (Pham et al. 2017). The basic parameters are described as follows. The Kit is specified as to measure 16 specific human bile acids and 19 specific rodent bile acids. The total number of bile acids in its panel is 20 as many human and rodent bile acids overlap. In this study the entire panel of the Bile acid Kit is reported. The measurements were carried out on a Shimadzu Nexera X2 UHPLC system coupled with the SCIEX QTRAP 5500 mass spectrometer equipped with an ESI ion source. To ensure accuracy and precision, the Kit provides seven calibration standards, a mixture of 10 isotope-labeled internal standards, and three levels of quality control samples. The calibration range of individual compounds and their assignment of internal standards are given in Supplementary Table 1.
The sample preparation process is as follows. 10 μL of internal standards mixture was pipetted onto the filter spots suspended in the wells of the 96-well filter plate. This filter plate was fixed on top of a deep-well plate serving as a receiving plate for the extract (a combi-plate structure). Subsequently, 10-μL plasma samples were pipetted on the spots, followed by nitrogen drying. Then 100 μL methanol was added to the wells, and the combi-plate was shaken for 20 min. The combiplate was centrifuged to elute the methanol extract into the lower receiving deep-well plate, which was then detached from the upper filter plate. After adding 60 μL Milli-Q® water to the extracts and shaking briefly, the plate was ready for LC-MS/MS analysis. All target isobaric bile acids were baseline separated under ultrahigh pressure liquid chromatography (UHPLC) conditions. UHPLC systems were used at a flow rate of 0.5 mL/min, enabling a short runtime of 5 min. A proprietary reversed- phased UHPLC column (Biocrates Life Sciences) was used. Chromatographic conditions (e.g., mobile phase compositions, gradients, column temperature) were described in details in the provided user manual. Mass spectrometric detection was accomplished with electrospray ionization in negative ion mode. Two MRM transitions are used for each target compound, whereas the more intensive signals are used for quantitation. The weaker MRM transitions are used to confirm the identity of target bile acids, which, in combination with the chromatographic retention times offer a near absolute certainty for target compound identification. Supplementary Fig. 1 shows typical extracted ion chromatograms of a Bile Acids Kit calibrator (upper panel A) and a representative pooled human plasma sample (lower panel B). For the quantification, a calibration set with 7 concentration levels and a mixture of 10 internal standards were used. The calibration regression was set to quadratic with 1/x^2 weighting. An example of a calibration curve for CDCA on for different days (interday experiment), demonstrating the robustness of the assay and stability of (instrument) calibration, is shown in Supplementary Fig. 1C.
The assay has been rigorously validated in accordance with European Medicines Agency (EMA) guidelines. All performance parameters such as selectivity, dilution and spiking integrity through the intra- and interday (n = 6 and n = 4, respectively) precision and accuracy of calibrators and diluted, unspiked as well as spiked human plasma samples, matrix effect, carry-over and short-term stability have been investigated on multiple (U)HPLC-MS platforms. All acceptance criteria laid out in the EMA guideline have been fulfilled after this extensive validation process. The EMA guideline does not require explicitly the determination of the recovery rate of extraction. The recovery rate does not have to be 100% but needs to be constant to guarantee the reproducibility of measurements. This requirement has been indirectly fulfilled through the determination of the accuracy of the spiked human plasma samples. Matrix effects typically occur in MS-based analyses and generally lead to ion suppression of the analyte signals. The matrix effect was investigated in “post-extraction spiking” experiement. Six individual plasma samples have been used, including one lipaemic sample (triglyceride content > 200 mg/dL and cholesterol > 200 mg/dL) and one hemolytic sample (hemoglobin content > 20 g/dL). The samples were spiked after the extraction with bile acid standard solution at two concentration levels. Internal standards were also spiked into the extracts to compensate the matrix effects. Ideally, the suppression of analyte signals is the same as for internal standard signals, so that the ratio of their peak areas remains unchanged. By using 10 different stable isotope-labeled internal standards, it has been proven that matrix effects for the target bile acids in human samples can be effectively compensated. More importantly a proficiency test carried out in 12 different laboratories has shown excellent accuracy (within 85–115%) and precision (CV < 20% for all target bile acids) of the Kit (Pham et al. 2017).
Statistical Analysis
Bile acid concentrations were compared between groups (control, MCI, AD) by One Way ANOVA and subsequent Fisher LSD post-hoc tests. Bile acids whose concentration differed significantly between groups were resulted into a receiver operating characteristic (ROC) analysis to derive optimal cut-off levels as well as estimates of sensitivity, specificity and the area under the ROC curve. Moreover, linear discriminant analysis with forward stepwise variable selection and equal a priori probabilities for the three groups was used to obtain a classification rule based on bile acid concentrations. Selection of predictor variables was based on Wilk’s lambda and the corresponding F-statistic, entering all variables with a p value < 0.05 into the model. Predicted group membership was determined using Fisher’s linear discriminant functions. Bias-corrected classification rates were obtained by means of cross validation.
Results
Patients
The demographic and clinical characteristics for the test dataset of the healthy subjects, MCI and AD patients are given in Table 1. All individuals in the study population are in the same age group between 77 and 79 years and do not differ between groups (Table 1). All groups included more females (Table 1). The MMSE score was 29.0 for controls, not different in MCI but significantly lower in AD patients (Table 1). The GDS values were around 3 and did not differ between groups (Table 1).
Plasma bile acids analysis
Levels of lithocholic acid were significantly enhanced in plasma of AD patients (50 ± 6 nM, p = 0.004) compared to healthy controls (32 ± 3 nM; Table 2). Lithocholic acid plasma levels of MCI patients (41 ± 4 nM) were not significantly different from healthy subjects or AD patients. Levels of glycochenodeoxycholic acid, glycodeoxycholic acid and glycolithocholic acid were significantly higher in AD patients compared to MCI patients (p < 0.05; Table 2). All other cholic acid metabolites were not significantly different between healthy subjects, MCI patients and AD patients (Table 2). Muricholic acid was below detection limit in all samples (data not shown). The sum of all bile acids was significantly increased in AD patients (Table 2).
Table 2.
Levels of different bile acids in plasma of cognitively healthy subjects, MCI patients and patients suffering from Alzheimer´s disease
| Analyte | DL | Control (n = 30) | MCI (n = 20) | AD (n = 30) | CO versus MCI | CO versus AD | MCI versus AD |
|---|---|---|---|---|---|---|---|
| CA, cholic acid | 10 | 215 ± 51 | 155 ± 43 | 224 ± 161 | ns | ns | ns |
| CDCA, chenodeoxycholic acid | 6 | 361 ± 84 | 284 ± 61 | 1245 ± 1009 | ns | ns | ns |
| DCA, deoxycholic acid | 6 | 455 ± 68 | 291 ± 50 | 723 ± 236 | ns | ns | ns |
| GCA, glycocholic acid | 10 | 423 ± 145 | 207 ± 24 | 332 ± 51 | ns | ns | ns |
| GCDCA, glycochenodeoxycholic acid | 6 | 1326 ± 223 | 1156 ± 105 | 1951 ± 301 | ns | p = 0.06 | p = 0.035 |
| GDCA, glycodeoxycholic acid | 3 | 640 ± 90 | 504 ± 105 | 956 ± 180 | ns | p = 0.09 | p = 0.034 |
| GLCA, glycolithocholic acid | 3 | 41 ± 8 | 27 ± 4 | 66 ± 15 | ns | ns | p = 0.026 |
| GUDCA, glycoursodeoxycholic acid | 3 | 160 ± 29 | 137 ± 17 | 137 ± 24 | ns | ns | ns |
| LCA, lithocholic acid | 3 | 32 ± 3 | 41 ± 4 | 50 ± 6 | ns | p = 0.004 | ns |
| TCA, taurocholic acid | 6 | 134 ± 64 | 40 ± 9 | 99 ± 25 | ns | ns | ns |
| TCDCA, taurochenodeoxycholic acid | 3 | 211 ± 58 | 170 ± 37 | 347 ± 71 | ns | ns | ns |
| TDCA, taurodeoxycholic acid | 3 | 97 ± 23 | 72 ± 20 | 152 ± 36 | ns | ns | ns |
| TLCA, taurolithocholic acid | 3 | 8 ± 2 | 5 ± 1 | 11 ± 2 | ns | ns | ns |
| TMCA(a + b), tauromuricholic acid (sum) | 3 | 15 ± 3 | 17 ± 5 | 24 ± 5 | ns | ns | ns |
| TUDCA, tauroursodeoxycholic acid | 3 | 8 ± 1 | 8 ± 1 | 11 ± 2 | ns | ns | ns |
| UDCA, ursodeoxycholic acid | 6 | 88 ± 23 | 62 ± 17 | 209 ± 149 | ns | ns | ns |
| Sum of all bile acids | 4214 ± 419 | 3176 ± 324 | 6537 ± 1163 | ns | p = 0.031 | ns |
Values represent nM in mean ± SEM. Statistical analysis was performed by One Way ANOVA with a subsequent Fisher LSD posthoc test where p < 0.05 was considered as significant
DL detection limit, ns not significant
Findings of ROC analyses and discriminant analysis
Findings of the ROC analyses are summarized in Table 3. LCA levels differentiated only moderately between AD patients and controls, with an AUC of 0.689 under the ROC curve. For the cut-off level of 42.5 (the statistically best value) a fairly high specificity (0.833) was achieved in connection with rather modest sensitivity (0.5). GDCA and GLCA concentrations discriminated only moderately well between MCI and AD patients with AUC values of 0.688 and 0.654, respectively, and sensitivity and specificity values between 0.6 and 0.7. With an AUC of 0.639, GLDCA showed an even poorer discrimination between MCI and AD patients.
Table 3.
ROC analysis: discrimination between healthy controls (CO) and patients with Alzheimer’s disease (AD) or mild cognitive impairment (MCI)
| Discrimination between CO and AD | ||||||
|---|---|---|---|---|---|---|
| Parameter | Area under ROC curve | Optimal cutoff | Sensitivity | Specificity | Overall accuracya | |
| Value | 95% CI | |||||
| LCA | 0.689 | 0.556–0.822 | 42.5b | 0.500 | 0.833 | 0.667 |
| Discrimination between MCI and AD | ||||||
|---|---|---|---|---|---|---|
| Parameter | Area under ROC curve | Optimal cutoff | Sensitivity | Specificity | Overall accuracya | |
| Value | 95% CI | |||||
| GDCA | 0.688 | 0.536–0.839 | 555b | 0.667 | 0.700 | 0.687 |
| GLCDA | 0.639c | 0.487–0.791 | 1300d | 0.600 | 0.650 | 0.630 |
| GLCA | 0.654 | 0.503–0.805 | 31b | 0.600 | 0.700 | 0.660 |
aProportion of correctly classified cases
bMaximizes the sum of sensitivity and specificity
cNot significantly different from 0.5 (i.e., not significantly larger than values that may arise from guessing)
dMaximizes the sum of sensitivity and specificity under the constraint that both sensitivity and specificity are ≥ 0.6
Results of the discriminant analysis are displayed in Table 4. Of the four bile acids identified as potential predictors only two were entered into the model, LCA (Wilk’s λ = 0.876; d.f. = 1, 2, 77; F 5.45; p = 0.006) and GDCA (Wilk’s λ = 0.797 d.f. = 2, 2, 77, F = 4.56, p = 0.002); GLCA and GLDCA were not entered as they did not significantly improve prediction of group membership. With a total of 47.5% correctly classified cases (45% after bias-correction) overall prediction was only modest. The proportion of slightly misclassified cases (e.g., MCI classified as control or as AD) reached rather high levels (31.3% after bias-correction) while the percentage of completely misclassified cases (AD classified as control or vice versa) was somewhat lower (23.7%). Using discriminant analysis we aimed to classify blinded samples and correctly classified 15 out of 23 (Table 5).
Table 4.
Discriminant analysis using log-transformed LCA and GDCA as a model (GCDCA and GLCA were not significant and hence not entered into the model)
| Classification results | |||
|---|---|---|---|
| Group | Predicted group membership | ||
| Control | MCI | AD | |
| Control (N = 30) | 12 (40.0%) | 9 (30.0%) | 9 (30.0%) |
| MCI (N = 20) | 3 (15.0%) | 11 (55.0%) | 6 (30.0%) |
| AD (N = 30) | 10 (33.3%) | 5 (16.7%) | 15 (50.0%) |
| Goodness of prediction (in brackets: bias-correction by means of cross-classification) | |||
|---|---|---|---|
| Correct | One category offa | Completely misclassifiedb | |
| Control (N = 30) | 40.0% (40.0%) | 30.0% (30.0%) | 30.0% (30.0%) |
| MCI (N = 20) | 55.0% (50.0%) | 45.0% (50.0%) | – (–) |
| AD (N = 30) | 50.0% (46.7%) | 16.7% (20.0%) | 33.3% (33.3%) |
| Total (N = 80) | 47.5% (45.0%) | 28.8% (31.3%) | 23.7% (23.7%) |
Classification formula
Control f0 = 6.576* ln(GDCA) + 8.673* ln(LCA) – 35.996
MCI f1 = 5.636* ln(GDCA) + 10.451* ln(LCA) – 36.547
AD f2 = 6.528* ln(GDCA) + 10.356* ln(LCA) – 41.679
Rule Classify as control if f0 > f1 and f0 > f2, classify as MCI if f1 > f0 and f1 > f2, classify as AD if
f2 > f0 and f2 > f1
aActual group and predicted group in adjacent categories, e.g., actual group = healthy, predicted group = MCI
bActual group and predicted group are completely different, e.g., actual group = healthy, predicted group = AD
Table 5.
Classification of blinded samples using discriminant analysis
| Nr | Age | Gender | MMSE | GDS | Clinical diagnosis | LCA | GDCA | f0 | f1 | f2 | Classification | Correct |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 84 | M | 29 | 4 | Healthy | 32 | 206 | 29.10 | 29.70 | 28.99 | f1 | No |
| 2 | 76 | F | 28 | 16 | MCI | 43 | 430 | 36.50 | 36.94 | 36.86 | f1 | Yes |
| 3 | 85 | M | 21 | 2 | Alzheimer | 110 | 5211 | 61.05 | 60.81 | 62.87 | f2 | Yes |
| 4 | 76 | M | 30 | 6 | Healthy | 32 | 482 | 34.69 | 34.49 | 34.54 | f0 | Yes |
| 5 | 77 | M | 29 | 1 | MCI | 30 | 0 | 15.87 | 18.17 | 15.75 | f1 | Yes |
| 6 | 87 | M | 17 | 2 | Alzheimer | 170 | 3220 | 61.66 | 62.65 | 64.23 | f2 | Yes |
| 7 | 81 | M | 30 | 6 | Healthy | 44 | 380 | 35.89 | 36.48 | 36.29 | f1 | No |
| 8 | 71 | F | 27 | 2 | MCI | 33 | 460 | 34.65 | 34.55 | 34.56 | f0 | No |
| 9 | 89 | F | 19 | 14 | Alzheimer | 120 | 3200 | 58.60 | 58.97 | 60.59 | f2 | Yes |
| 10 | 79 | F | 29 | 3 | Healthy | 33 | 271 | 31.17 | 31.57 | 31.10 | f1 | No |
| 11 | 71 | M | 27 | 2 | MCI | 32 | 70 | 22.00 | 23.62 | 21.95 | f1 | Yes |
| 12 | 91 | F | 11 | 2 | Alzheimer | 71 | 929 | 45.92 | 46.52 | 47.08 | f2 | Yes |
| 13 | 83 | M | 29 | 3 | MCI | 32 | 515 | 35.12 | 34.87 | 34.97 | f0 | No |
| 14 | 88 | F | 23 | 4 | Alzheimer | 109 | 624 | 47.02 | 48,76 | 48.92 | f2 | Yes |
| 15 | 80 | F | 16 | 3 | Alzheimer | 83 | 595 | 44.34 | 45.64 | 45.79 | f2 | Yes |
| 16 | 82 | F | 29 | 1 | Healthy | 30 | 259 | 30.04 | 30.32 | 29.82 | f1 | No |
| 17 | 79 | F | 28 | 7 | Healthy | 32 | 682 | 36.97 | 36.45 | 36.81 | f0 | Yes |
| 18 | 82 | F | 29 | 18 | Healthy | 31 | 653 | 36.41 | 35.87 | 36.20 | f0 | Yes |
| 19 | 75 | F | 25 | 4 | Alzheimer | 92 | 557 | 44.80 | 46.34 | 46.42 | f2 | Yes |
| 20 | 90 | F | 16 | 3 | Alzheimer | 75 | 161 | 34.86 | 37.21 | 36.20 | f1 | No |
| 21 | 85 | M | 21 | 3 | Alzheimer | 56 | 551 | 40.42 | 41.09 | 41.21 | f2 | Yes |
| 22 | 80 | F | 23 | 20 | Alzheimer | 42 | 729 | 39.77 | 39.67 | 40.06 | f2 | Yes |
| 23 | 77 | M | 13 | 0 | Alzheimer | 33 | 312 | 32.10 | 32.36 | 32.02 | f1 | No |
GCDCA glycochenodeoxycholic acid, GDCA glycodeoxycholic acid, GLCA glycolithocholic acid, LCA lithocholic acid, M male, F female, age in years, MMSE minimental state examination, GDS geriatric depression scale
Verfication is based on discriminant analysis as given in Table 4 (f0 = control, f1 = MCI, f2 = AD) using LCA and GDCA. Note that 15/23 samples were correctly diagnosed
Conversion study
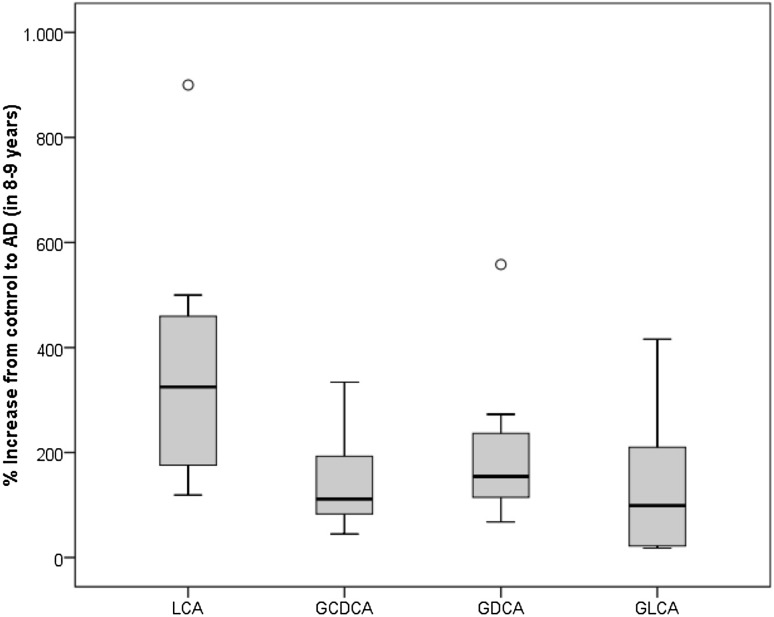
In a follow up experiment, we demonstrated changes of LCA levels in subjects converting from a cognitively healthy status to AD. We included 8 healthy controls. Blood had been collected and had been stored at − 80 °C. After 8–9 years blood was recollected from the same patients who converted to AD. Our data showed that LCA levels increased to 327 ± 100% of the baseline value (t = 0), while all 3 bile acid metabolites (GCDCA, GDCA, GLCA) increased 1.3–2.0-fold in average (Fig. 1).
Fig. 1.
Box-and Whisker Plot showing percent change of LCA, GCCA, GDCA and GLCA plasma levels in 8 converters to AD. Eight healthy control patients were included. Blood samples were recollected after 8–9 years (age 77 ± 3 years, 3 male)
Discussion
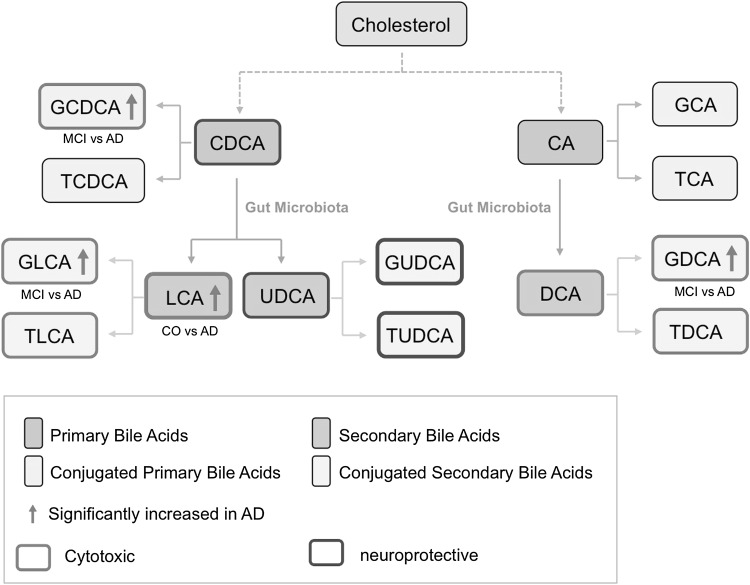
In the present study we show that the plasma bile acid lithocholic acid (LCA), as secondary bile acid, differentiates healthy controls from AD patients with a p-value of 0.004. In addition three glycine-conjugated bile acids i.e. GCDCA (conjugated primary bile acid) and GLCA and GDCA (two conjugated primary bile acids) are significantly differentiated AD patients with p values < 0.05. Figure 2 illustrates the appropriate bile acid pathway marked with these up-regulated bile acids.
Fig. 2.
Pathway analysis showing the putative role of the bile acid metabolites in the cholesterol-pathway. Abbreviations see Table 2
Bile acids and AD
Due to their broad physiological function of bile acids in the human body, and their implication in cholesterol, glucose and lipid metabolism there is a clear need to investigate bile acid metabolites in AD. The major bile acid is cholic acid, which can be conjugated or unconjugated forming different isoforms: deoxy-glyco-cheno-lith-hyo-tauro or urso. The nomeclature has been proposed by Hofmann et al. (1992). The novel Bile Acid kit developed by Biocrates allows to measure 20 bile acid metabolites with very high sensitivity (up to 3 nM). Our data not only verify previous preliminary reports on a few bile acid metabolites (Mapstone et al. 2014; Olazaran et al. 2015) but shows for the first time a detailed quantification of up to 20 bile acid metablites in human plasma. Out of these 20 analytes, four metabolites were not detectable, but our data clearly show that bile acids, and especially LCA, are altered in plasma of AD patients. The 3 metabolites GCDCA, GDCA and GLCA were slightly altered, demonstrating the effects on glycine-conjugated bile acids in AD.
Diagnosis of AD and MCI from plasma by measuring bile acids
Our data show that LCA is highly useful to separate AD patients from healthy controls. Lithocholic acid (3α-Hydroxy-5β-cholan-24-acid) is synthesized in the colon from chenodeoxycholic acid and is only reabsorbed in little amounts. LCA has been implicated in carcinogenesis and preliminary data suggests that LCA kills neuroblastoma cells and other malignant cells possibly protecting against colon cancer. LCA (and LCA acetate and LCA propionate) can also activate the vitamin D receptor. In fact, LCA is a very hydrophobic bile acid and is mostly bound to albumin and lipoproteins and may not easily interact with host cells. In addition, the 3 glycine-conjugated bile acids metabolites GCDCA, GDCA and GLCA differentiated AD from MCI patients. Although we cannot give a detailed mechanistic insight into the pathological processes of bile acid metabolism in AD patients, alterations in bile acids may be correlated to the diet of AD patients, reduced physical activity and/or gut microbial composition. Indeed such a gut microbiome may correlate with dehydroxylating gut bacteria, colonic transit time and possibly pH of the colon. A detailed characterization of the gut microbiome of these patients may yield some useful insights, but was out of focus in this study.
We used discriminant analysis for classification of patients. In our verification blinded study with 23 patients of unknown diagnosis we indeed correctly diagnosed 15 subjects (65%). This goes is line with our ROC analysis. Looking at Table 5 it becomes evident that 4 healthy controls, 2 MCI and 2 AD were incorrectly classified. In fact, all healthy controls were classified as MCI cases. This indeed may also point to the problem of incorrect clinical diagnosis. We also are aware that it is not always possible to clearly find good age-matched (70 year old) controls, as we were not able to follow up any comborbidity and additional lifestyle factors, which could influence the clinical diagnosis.
Our conversion study clearly verifies the increase in LCA when controls converted to AD. The plasma LCA increased 3.2-fold within 8–9 years during conversion to AD. There was only 1 patient where we could not measure any LCA in the plasma. The other 3 bile acid metabolites (GCDCA, GDCA and GLCA) only moderately increased during the conversion to AD within the 8–9 years. This again verifies our findings that bile acids are increased in AD. However, when examining bile acid levels within 8–9 years in a longitudinal study it will be of importance to consider what factors changed in their lifestyle. So far, we can exclude that those patients developed another disease, however, we did not evaluate detailed lifestyle parameters. This should be done in a follow-up study.
Might there be a role of bile acids in AD?
Bile acids are found endogenously in the CSF and brain (Mano et al. 2004; Ogundare et al. 2010). Bile acids are pleiotropic signaling molecules in the body, and the presence of receptors and synthesis enzymes in the CNS indicates that they could be an endogenous signaling system present in the brain. However, clinical studies investigating the dysregulation of the bile acid signaling system in the CNS during stages of Alzheimer´s disease are lacking. There is clear evidence that bile acids play an important role in cardiovascular function: they can modify the vascular tone and can interact with different receptors and transcription factors. It is well known that some cholic acids modulate endothelial cells, cerebral arteries, or aortic smooth muscle cells (Khurana et al. 2011). There is clear evidence that vessels are damaged in AD and that beta-amyloid is deposited in brain vessels (known as cerebral amyloid angiopathy). Although the reasons for development of AD are not know, there is strong evidence that a long lasting chronic vascular impairment (starting decades before onset of AD) plays a role in development of AD (Humpel 2011b). Thus, it is also very likely that such vascular alterations also affect bile acids. A damaged blood–brain barrier causes influx of toxic blood compounds and also entry of blood cells into the brain, but possibly may also result in influx or efflux of bile acids into or from the brain, respectively. Recent evidence also links bile acids with the tumor suppressor gene p53 indicating a role in anti-cancer and apoptosis (Vogel et al. 2012). Indeed, we have shown in our lab, that the tumor suppressor p21 is markedly reduced in monocytes of AD patients (Hochstrasser et al. 2011). Further studies to link bile acids with a damaged blood–brain barrier in AD are necessary.
Bile acids and the cholesterol-connection
There are several papers published suggesting a link between cholesterol and Alzheimer’s disease (see reviews Anstey et al. 2008; Ribeiro et al. 2015; Reiss and Voloshyna 2012). Also many experiments in rodents have been performed investigating if cholesterol may indeed play a role in development of this disease, including our own research group (Ehrlich and Humpel 2012). This piece of data again supports ideas that indeed the cholesterol pathway may be affected or may play a role in AD. We performed a pathway analysis and our data clearly point to a link between bile acids and cholesterol processing (see Fig. 2). Cholesterol is metabolized via different processes into 2 major primary bile acids (CDCA and CA). Both can be conjugated, leading to a putative cytotoxic GCDCA (which is increased in AD in our present study). Modulation of both primary bile acids (CDCA and CA) by the gut microbiota leads to secondary bile acids (LCA, UDCA and DCA), where again the putative cytotoxic LCA is increased in AD in our present study. Conjugation of those secondary bile acids can generate several conjugated forms, including the putative cytotoxic GLCA and GDCA forms, which were both increased in AD in our study. Thus, our data clearly shows that putative cytotoxic bile acids were altered but not putative neuroprotective bile acids (see Fig. 2), pointing to a cholesterol-mediated toxicity.
Limitation of the study
This study can be considered as a pilot study and has definitely clear limitations: A main limitation of the present study is the small size of the samples. This study should therefore be followed up by large-scale multicenter studies that should also include other types of dementia, especially vascular dementia or frontotemporal lobe dementia. In addition, a detailed correlation of ApoE genotype, plasma levels of glucose and insulin, medication, liver enzymes and/or post-prandial intervals may help to follow up the changes at a more mechanistic level, which was, however, not in focus in this study. Another limitation is that we have not yet proven the stability of the bile acids in blood. As we have processed most samples within 4 h, we probably can exclude degradation, because we recently showed that lipid metabolites are at least stable for 24 h (Klavin et al. 2015). This is also true for samples which have been stored for 8 years at − 80 °C and where we cannot exclude linear time-dependent degradation. It has been recognized that nutritional status can influence blood levels of several metabolites, and should be considered when evaluating metabolomic data. Therefore, some authors recommend fasting before blood collection. We have not yet tested the effects of nutrition in this study. More detailed investigation will be necessary in further studies. The ethnicity of patients may have an effect on metabolism; the present study used samples from a group of persons of the same ethnicity. And finally, we observed during the analysis, that the analytical limit of detection of LCA may vary between different sample batches at different days on mass spectrometry analysis due to instrumental variation in performance over the time. Thus, much care must be taken to ensure that the assays are very sensitive and robust.
Conclusion
In conclusion our data show that the secondary plasma bile acid lithocholic acid differentiates healthy controls from AD patients with a p value of 0.004. Three glycine-conjugated bile acids significantly differentiated AD with p values < 0.05. Thus, these 4 bile acids may be useful to routinely diagnose AD in plasma samples. Our discrimination and ROC analysis revealed, however, a limited specificity, sensitivity and accuracy compared to other published plasma-markers. Thus, these 4 bile acid markers (and more specifically only LCA and GDCA) may be helpful to further diagnose AD in plasma along with a broader pattern of biomarkers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Open access funding provided by University of Innsbruck and Medical University of Innsbruck. This study was supported by Österreichische Nationalbank Jubiläumsfonds (Nr. 15887). We thank Karin Albrecht and Monika Greil for assistance with blood processing. We thank Dr. Hai Pham Tuan (Biocrates) for text corrections regarding the analytical part and supplementory Fig. 1 and Table 1 and Dr. Udo Müller (Biocrates) for Fig. 2.
Abbreviations
- AD
Alzheimers disease
- MCI
Mild cognitive impairment
- GCDCA
Glycochenodeoxycholic acid
- GDCA
Glycodeoxycholic acid
- GLCA
Glycolithocholic acid
- LCA
Lithocholic acid
- MMSE
Minimental state examination
- GDS
Geriatric depression scale
Author Contributions
JM is a clincian, responsible for patient blood collection and clinical diagnosis. IB is a clinician and responsible for the conversions study. GK has done the complete statistical analysis. TK contributed to analytical part and pathway analysis. CH designed the study, analyzed the data and wrote the MS.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest. TK is employed by Biocrates.
Ethical approval
The study was approved by the Ethics Committee of the Medical University of Innsbruck and was performed in accordance with the Helsinki Declaration. All subjects gave written informed consent. Data and material are available on request.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s11306-017-1297-5) contains supplementary material, which is available to authorized users.
References
- Anstey KJ, Lipnicki DM, Low LF. Cholesterol as a risk factor for dementia and cognitive decline: A systematic review of prospective studies with meta-analysis. The American Journal of Geriatric Psychiatry. 2008;16(5):343–354. doi: 10.1097/01.JGP.0000310778.20870.ae. [DOI] [PubMed] [Google Scholar]
- Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastroinestinal cancers. Mutation Research. 2005;89(1):47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Blennow K. CSF biomarkers for Alzheimer’s disease: Use in early diagnosis and evaluation of drug treatment. Expert Review of Molecular Diagnostics. 2005;5:661–672. doi: 10.1586/14737159.5.5.661. [DOI] [PubMed] [Google Scholar]
- Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nature Reviews Neurology. 2010;6:131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- Chiang JY. Bile acid regulation of gene expression: Roles of nuclear hromone receptors. Endocrine Reviews. 2002;23(4):443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- Ehrlich D, Humpel C. Chronic vascular risk factors (cholesterol, homocysteine, ethanol) impair spatial memory, decline cholinergic neurons and induce blood-brain barrier leakage in rats in vivo. Journal of the Neurological Sciences. 2012;322(1–2):92–95. doi: 10.1016/j.jns.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser T, Ehrlich D, Marksteiner J, Sperner-Unterweger B, Humpel C. Matrix metalloproteinase-2 and epidermal growth factor are decreased in platelets of Alzheimer patients. Current Alzheimer Research. 2012;9:982–989. doi: 10.2174/156720512803251156. [DOI] [PubMed] [Google Scholar]
- Hochstrasser T, Marksteiner J, Defrancesco M, Deisenhammer EA, Kemmler G, Humpel C. Two blood monocytic biomarkers (CCL15 and p21) combined with Mini-Mental State Examination discriminates Alzheimer’s disease patients from healthy controls. Dementia Geriatric Cognitive Disorders. 2011;1(1):297–309. doi: 10.1159/000330468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Hagey LR. Bile acids: Chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cellular and Molecular Life Sciences. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Sjövall J, Kurz G, Radominska A, Schteingart CD, Tint GS, et al. A proposed nomenclature for bile acids. The Journal of Lipid Research. 1992;33:599–604. [PubMed] [Google Scholar]
- Hu WT, Holtzman DM, Fagan AM, Shaw LM, Perrin R, Arnold SE, et al. Alzheimer’s disease neuroimaging initiative. Plasma multianalyte profiling in mild cognitive impairment and Alzheimer disease. Neurology. 2012;79:897–905. doi: 10.1212/WNL.0b013e318266fa70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpel C. Identifying and validating biomarkers for Alzheimer’s disease. Trends in Biotechnology. 2011;29:26–32. doi: 10.1016/j.tibtech.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpel C. Chronic mild cerebrovascular dysfunction as a cause for Alzheimer’s disease? Experimental Gerontology. 2011;46:225–232. doi: 10.1016/j.exger.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Raufman JP, Pallone TL. Bile Acid regulate cardiovascular function. Clinical and Translational Science. 2011;4(3):210–218. doi: 10.1111/j.1752-8062.2011.00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klavin K, Koal T, Dallmann G, Marksteiner J, Kemmler G, Humpel ,C. The ratio of phosphatidylcholines to lysophosphatidylcholines in plasma differentiates healthy controls from patients with Alzheimer’s disease and mild cognitive impairment. Alzheimer’s and Dementia: Diagnosis, Assessment and Disease Monitoring. 2015;1:295–302. doi: 10.1016/j.dadm.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre P, Cariou B, Lien F. Role of bile acis and bile acid receptors in metabolic regulation. Physiological Reviews. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- Li T, Chang JY. Bile Acid signaling in metabolic disease and drug therapy. Pharmacological Reviews. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano N, Goto T, Uchida M, Nishimura K, Ando M, Kobayashi N, Goto J. Presence of protein-bound unconjugated bile acids in the cytoplasmic fraction of rat brain. Journal of Lipid Research. 2004;45:295–300. doi: 10.1194/jlr.M300369-JLR200. [DOI] [PubMed] [Google Scholar]
- Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Natural Medicines. 2014;20(4):415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of alzheimer’s disease: Report of the NINCDS-ADRDA work group under the auspices of department of health and human services task force on alzheimer’s disease. Neurology. 1984;34:939–944. doi: 10.1212/WNL.34.7.939. [DOI] [PubMed] [Google Scholar]
- Morris JC. The clinical dementia rating (CDR): Current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/WNL.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Neale G, Lewis B, Weaver V, Panveliwalla D. Serum bile acids in liver disease. Gut. 1971;12(2):145–152. doi: 10.1136/gut.12.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogundare M, Theofilopoulos S, Lockhart A, Hall LJ, Arenas E, Sjövall J, et al. Cerebrospinal fluid steroidomics: Are bioactive bile acids present in brain? The Journal of Biological Chemistry. 2010;285:4666–4679. doi: 10.1074/jbc.M109.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olazarán J, Gil-de-Gómez L, Rodríguez-Martín A, Valentí-Soler M, Frades-Payo B, Marín-Muñoz J, et al. A blood-based, 7-metabolite signature for the early diagnosis of Alzheimer’s disease. Journal of Alzheimer’s Disease: JAD. 2015;45(4):1157–1173. doi: 10.3233/JAD-142925. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, et al. Current concepts in mild cognitive impairment. Archives of Neurology. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Pham HT, Arnhard K, Asad YJ, Deng L, Felder TK, Williams LS, et al. Inter-laboratory robustness of next-generation bile acid study in mice and humans: international ring trial involving 12 laboratories. JALM. 2017 doi: 10.1373/jalm.2016.020537. [DOI] [PubMed] [Google Scholar]
- Puglielli L, Tanzi RE, Kovacs DM. Alzheimer’s disease: The cholesterol connection. Nature Neuroscience. 2003;6:345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- Reiss AB, Voloshyna I. Regulation of cerebral cholesterol metabolism in Alzheimer disease. Journal of Investigative Medicine: The Official Publication of the American Federation for Clinical Research. 2012;60(3):576–582. doi: 10.2310/JIM.0b013e318246d973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SM, Luz Sdos S, Aquino Rde C. The role of nutrition and physical activity in cholesterol and aging. Clinics in Geriatric Medicine. 2015;31(3):401–416. doi: 10.1016/j.cger.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Scheltens P, Leys D, Barkhof F, Huglo D, Weinstein HC, Vermersch P, et al. Atrophy of medial temporal lobes on MRI in probable Alzheimers disease and normal aging—diagnostic-value and neuropsychological correlates. Journal of Neurology, Neurosurgery and Psychiatry. 1992;55:967–972. doi: 10.1136/jnnp.55.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling MA. Unraveling Alzheimer’s: Making sense of the relationship between diabetes and Alzheimer´s disease. Journal of Alzheimer’s Disease: JAD. 2016;51(4):961–977. doi: 10.3233/JAD-150980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shad KF, Aghazadeh Y, Ahmad S, Kress B. Peripheral markers of Alzheimer’s disease: Surveillance of white blood cells. Synapse (New York, N. Y.) 2013;67:541–543. doi: 10.1002/syn.21651. [DOI] [PubMed] [Google Scholar]
- Song F, Poljak A, Smythe GA, Sachdev P. Plasma biomarkers for mild cognitive impairment and Alzheimer’s disease. Brain Research Reviews. 2009;61:69–80. doi: 10.1016/j.brainresrev.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile acid signaliing for metabolic diseases. Nature Reviews, Drug Discovery. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- Vogel SM, Bauer MR, Joerger AC, Wilcken R, Brandt T, Veprintsev DB, et al. Lithocholic acid is an endogenous inhibitor of MDM4 and MDM2. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(42):16906–16910. doi: 10.1073/pnas.1215060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh KA, Butters N, Mohs RC, Beekly D, Edland S, Fillenbaum G, Heyman A. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part V. A normative study of the neuropsychological battery. Neurology. 1994;44:609–614. doi: 10.1212/WNL.44.4.609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.