Abstract
Background:
Alterations in GABAnergic system are implicated in the pathophysiology of schizophrenia. Available antipsychotics that target GABA receptor form a desirable therapeutic strategy in the treatment regimen of schizophrenia, unfortunately, suffer serious setback due to their prolonged side effects. The present investigation focuses on developing QSAR models from the biological activity of herbal compounds and their derivatives that promise to be alternative candidates to GABA uptake inhibitors.
Methods:
Three sets of compounds were undertaken in the study to develop QSAR models. The first set consisted of nine compounds which included Magnolol, Honokiol and other GABA acting established compounds. The second set consisted of 16 derivatives of N-diarylalkenyl-piperidinecarboxylic acid. The third QSAR dataset was made up of thirty two compounds which were Magnolol and Honokiol derivatives. Multiple linear regressions (MLR) and support vector machine (SVM) supervised quantitative structure-activity relationship (QSAR) models were developed to predict the biological activity of these three sets. The purpose of taking three QSAR sets of diverse chemical structures but identical in their GABA targeting and pharmacological action was to identify common chemical structure features responsible for structure-activity relationship (SAR).
Results:
Linear and non-linear QSAR models confirmed that the three sets shared common structural descriptors derived from WHIM (Weighted Holistic Invariant Molecular descriptors), 3D-MoRSE and Eigenvalue classes.
Conclusion:
It was concluded that properties like electro negativity and polarizability play a crucial role in controlling the activity of herbal compounds against GABA receptor.
Keywords: Schizophrenia, Linear and non-linear QSAR models, MLR and SVM
1. Introduction
Over the past decade, much of the attention regarding the treatment for schizophrenia and related psychotic disorders has focused on a new class of antipsychotic medications. The therapeutic strategy for the treatment of schizophrenia has seen considerable growth in the past half century [1-4] by the advent of drugs targeting GABAnergic system which has marked the beginning of the pharmacologic era in psychiatry [5-7]. In spite of the tremendous progress that has been made in confronting the disease, the pharmacological properties that confer the therapeutic effects on GABAnergic system have remained elusive, and certain side effects can still impact patient health and quality of life [8]. In addition, the efficacy of antipsychotic drugs is limited prompting the clinical use of adjunctive pharmacy to augment the effects of treatment [9, 10]. Moreover, the search for novel GABAnergic antipsychotic drugs has not been successful to date, though numerous development strategies continue to be pursued [11].
Quantitative structure activity relationship (QSAR) has proved its usefulness in predicting the biological response of compounds in class as a function of their structure by adopting mathematical and statistical tools. Generally, structural properties are expressed in numerical magnitudes as molecular descriptors derived from chemical structures. QSAR studies facilitate to relate structural features in terms of molecular descriptors with biological activity, which further assist drug design community to synthesize new molecules with optimized structures of desired biological activity [12-14]. These studies have its remarkable application in medicinal chemistry to investigate new drugs or optimizing the existing ones [15, 16]. QSAR employs regression statistics using algorithms like support vector machine (SVM), artificial neural network (ANN), partial least square (PLS), regression trees and ensembles, etc. [17, 18]. Multiple linear regressions are the most simple and significant approach used to identify linear relationship among molecular structures and their biological responses. Structure activity relationship often being non-linear which cannot be identified using MLR (Multiple Linear Regression) analysis to overcome this hurdle, SVM was introduced which is an accurate, robust and fast statistical tool [19] and efficient in identifying non-linear Structure activity relationships. Furthermore, development of kernel functions like Gaussian and polynomial made SVM even more an applicable and alternative tool in QSAR studies. SVM developed for classification was further optimized and applied to achieve regression in exploring non-linear QSAR models [20].
Present studies aim to identify common chemical structural feature insights which describe SAR of GABA acting compounds which are derivatives of compounds originally derived from natural sources. There are three sets of compounds which are treated as three QSAR datasets along with their experimental biological activities targeting GABA with special reference to schizophrenia treatment as pharmacological action.
2. Methodology
2.1. Dataset Selection
Established potent GABAA and GABAB inhibitors like Acacetin, Saikosaponin A, Saikogenin G, Cimicidanol, Rutaecarpine, flunitrazepam, honokiol, magnolol, 6-methylflavone along with sixteen (16) compounds belonging to N-diarylalkenyl-piperidinecarboxylic acid derivatives designed by Zheng et al., 2006 [21], and thirty two (32) plant compound derivatives of magnolol and honokioldesigned by Fuchs et al., 2014 [22] were considered for the study. Three sets of compounds were subjected to MLR (Linear) and SVM (Non-linear) QSAR studies, so as to derive an individual QSAR model for each set and finally, to extract common chemical structure features responsible for SAR with reference to their action on GABA receptor.
2.2. Descriptor Calculation
Molecular descriptors are numerical representations to evaluate and establish the structural activity relationship. All the structures belonging to each series were generated and optimized in Marvin Sketch version 5.6.0.2 [23] which was then converted into their SMILES (Simplified Molecular Line Entry Specification). SMILES were used to calculate descriptors using E-Dragon (version 5.4) [24-26], an online server. In total, 2074 descriptors belonging to various classes were imported to data analysis package of Microsoft Excel for MLR analysis and GIST server was employed for Support Vector Machine aided non-linear analysis [27].
2.3. Model Preparation (MLR Aided Linear and SVM Aided Non-linear Models)
Descriptor-screening methods were employed to select the most significant descriptors to establish the models. Pruning of descriptors was performed by considering the parameters (standard deviation ≤0, and missing values greater than equal to 1) which drops aside constant and missing set of descriptors that are considered insignificant in statistical analysis [28]. Correlation coefficient of molecular descriptors with biological responses (endpoint) was calculated using Pearson’s correlation coefficient and ranked in the descending order. Chances of redundancy in regression models are thoroughly inspected and removed using correlation matrix [29]. A method of variable selection is required in order to find the optimal subset of the descriptors which may play a determining role in quantitative relationship of structures and their biological responses. Forward selection wrapper was introduced to select molecular descriptor subsets. Multiple linear regression (MLR), being the most popular and conventional statistical tool, was used to develop linear QSAR models [30]. SVM is the system based on structural risk minimization (SRM) principle, which provides a separating hyperplane with minimum expected generalization error. It was used in forward selection algorithm to generate non-linear QSAR models [28]. QSAR models were generated from one-variable to four-variable descriptor models for Linear (MLR) and non-linear (Gaussian kernel function aided SVM) [31]. Models were validated using internal validation tools like cross validated R2CV).
3. Results and Discussion
After pruning and dropping highly correlated descriptors, forward selection for feature selection was used to pick significant descriptors and their sets ranging from uni-variable to tetra variable models. Present QSAR studies are an attempt to obtain QSAR models for established GABA ligands (Magnolol, Honokiol and other candidates). Linear (MLR) and non-linear (Gaussian kernel function aided SVM) QSAR models obtained on a QSAR dataset of 9 molecules suggest new insights into structure-activity relationship for these structurally different, naturally derived and GABA acting compounds. Multiple linear regression (MLR) used in forward selection ended with various sets of molecular descriptors from one-variable to tetra variable variable QSAR models whereas similar but non-linear models with different molecular descriptor were produced by Gaussian kernel function aided Support Vector Machine (SVM).
A good rule of thumb allows us stretching variable selection from uni-variable to bi-variable with nine (9) compounds in QSAR dataset though it was extended to tetra variable in order to compare the obtained linear and non-linear QSAR models with other datasets. Nevertheless, QSAR models were found statistically fit and predictive even with bi-variable model in case of QSAR dataset of main compounds consisting of nine (9) compounds.
with corresponding values in linear (R2CV=0.7684) and non-linear (R2CV=0.8455) bi-variable QSAR models.
A similar forward selection method was applied to QSAR dataset 2 (16 compounds) to retrieve the structure information in terms of molecular descriptor which could further be subjected to analyze structure-activity relationship. Table 3 shows selected descriptors and corresponding statistical fitness parameters of QSAR models staring from uni variable to tetra variable. In the case of QSAR dataset 2, linear models appeared to be more fit than non-linear models with the same number of descriptors.
Table 3.
Molecular descriptors and forward selection statistics for linear (MLR) and non-linear (SVM) QSAR dataset 2 (16 Compounds).
| Model | Descriptors | Variables | R2 | Max. Abs. Error | Mean Abs. Error | R2CV (N-Fold) |
|---|---|---|---|---|---|---|
|
Linear (MLR) |
H0m | 1 | 0.4827 | 1.1753 | 0.4315 | 0.2908 |
| H0m, C-025 | 2 | 0.7114 | 1.0899 | 0.3089 | 0.5508 | |
| H0m, C-025, nBnz | 3 | 0.8670 | 0.5661 | 0.2180 | 0.7736 | |
| H0m, C-025, nBnz, Mor17m | 4 | 0.9274 | 0.4868 | 0.1542 | 0.8547 | |
|
Non-linear (SVM) |
GGI9 | 1 | 0.6459 | 1.1049 | 0.3207 | 0.4738 |
| GGI9, R7v+ | 2 | 0.7902 | 0.7553 | 0.2370 | 0.6831 | |
| GGI9, R7v+, G(O..S) | 3 | 0.8970 | 0.7558 | 0.1135 | 0.7834 | |
| GGI9, R7v+, G(O..S), HATSe | 4 | 0.8803 | 0.7725 | 0.1255 | 0.8155 |
Statistical fitness derived from various statistical parameters of linear and non-linear QSAR models show that models were acceptable in the current form. R2 values indicate a strong confidence level even in bi-variable linear (R2=0.8634) and non-linear (R2=0.9747) QSAR models. R2CV values further confirm the stability of QSAR models
Further in Dataset-3 (32 Compounds) and their QSAR models derived after forwards selection, Table 3 illustrates acceptable tetra variable models in both linear and non-linear relationships. Since the activity has been expressed in terms of percentage (%) and discrete values, the models have suffered a rough training and therefore have got reported in comparatively low statistical profile.
For QSAR dataset-1, Multi linear regression (MLR) aided Equation for tetra variable model is presented below as equation 1. In addition to R2 and R2CV, Adjusted regression coefficient R2A (0.959) values, Standard error estimate (S.E.) 0.144 and F-stat values (47.967) approve and allow the use of tetra variable models even with limited compounds (9).
QSAR Dataset-1: Linear QSAR Model Equation (Tetra variable model)
pIC50 = 35.277 + 1.610[nR09] + 6.892[E1u] - 18.772[G2e] -16.721[BELp2] [Eq. 1]
N=9 R2 = 0.979 R2A= 0.959 S.E. = 0.144 F-statistics=47.967
Moving to dataset-2 with 16 compounds the tetra variable model came out to be competitive with that obtained for dataset-1. Standard Error and adjusted R2were also found comparative to equation 1. Thought significance test from F-stat showed that instead of identical statistical fitness tetra variable model of dataset-1 (F stat = 47.967) was more significant than that obtained for dataset-2 (F-stat = 35.144).
QSAR Dataset-2: Linear QSAR Model Equation (Tetra variable model)
pIC50 = 0.198 + 3.281[H0m] + 0.608[nBnz] - 0.868 [Mor17m]-0.778 [C-025] [Eq. 2]
N=16 R2 = 0.927 R2A= 0.901 S.E. = 0.244 F-statistics=35.144
Dataset-3 was found to be statistically less confident in regression with R2 (0.833) when compared to confidence received in dataset-1 and dataset-3, respectively. Equation 3 presents linear model based on multiple linear regression analysis for a set of 32 compounds,although the significance (F-stat = 39.733) is found to be equivalent to dataset-1 and dataset-2.
QSAR Dataset-3: Linear QSAR Model Equation (Tetra variable model)
Potential % = 0.996 - 0.014[W] + 805.159[R8v+] - 9.627 [EEig05x] + 12.939 [EEig07r] [Eq. 3]
N=32 R2 = 0.854 R2A= 0.833 S.E. = 1.553 F-statistics=39.733
Tetra variable models using the above equations from linear (MLR) QSAR models and Gaussian kernel function aided SVM models were, thereafter, used to check the predictive powers of QSAR models. The endpoint values (pIC50) of compounds were predicted using molecular descriptor values and corresponding coefficients. A graphical correlation of experimental (actual) and predicted (estimated) end point values (biological activities) is presented below in Figs. 1-3.
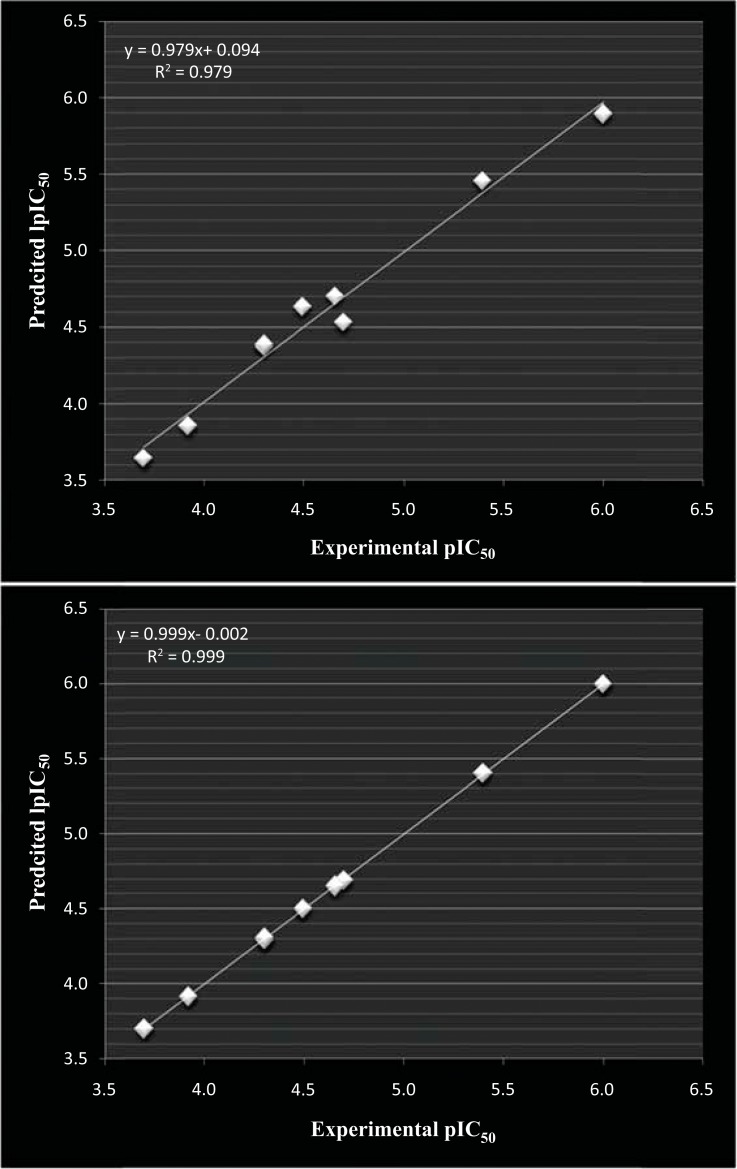
Fig. (1).
(A): correlation of experimental and predicted pIC50 calculated from linear (MLR) aided tetra variable model for dataset -1 and (B) correlation of experimental and predicted pIC50 calculated from non-linear (SVM) aided tetra variable model for dataset-1.
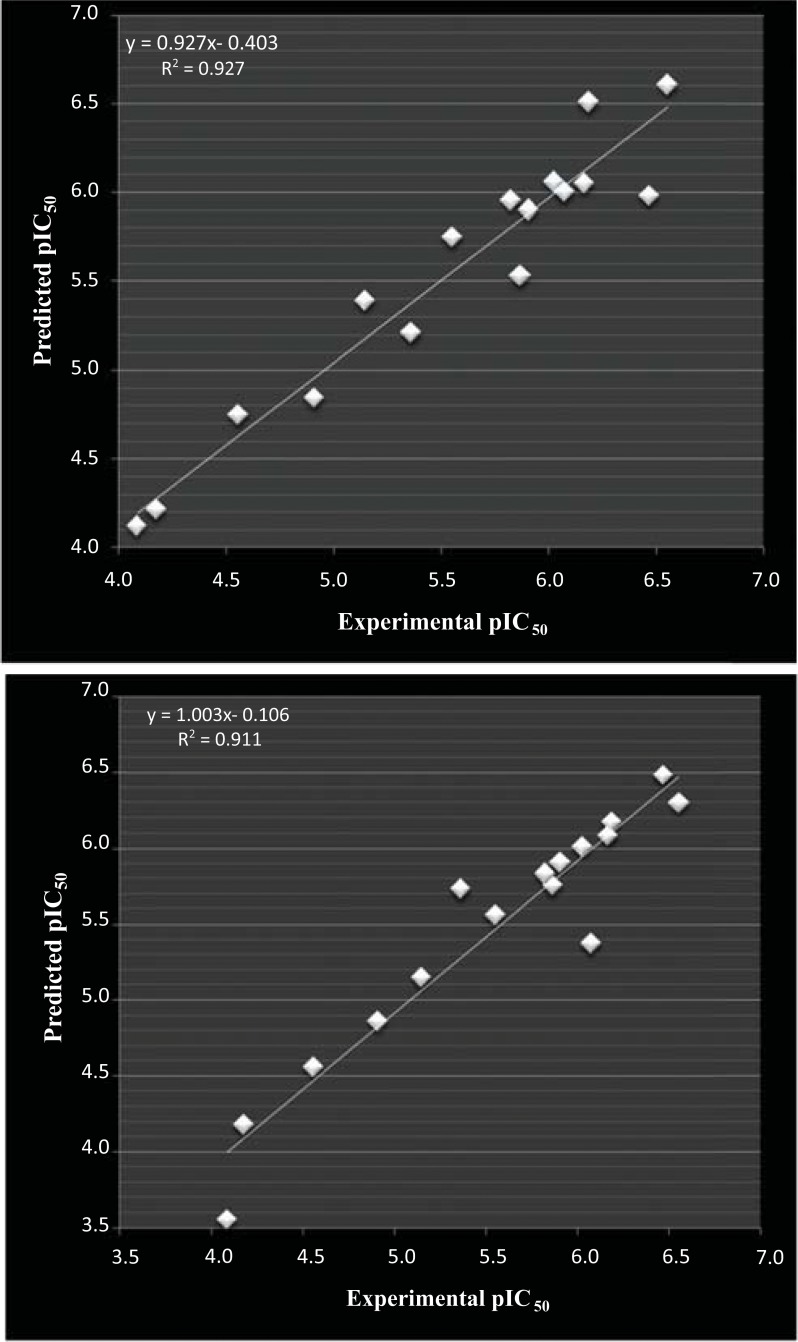
Fig. (3).
(A): correlation of experimental and predicted pIC50 calculated from linear (MLR) aided tetra variable model for dataset -3 and (B) correlation of experimental and predicted pIC50 calculated from non-linear (SVM) aided tetra variable model for dataset-3.
In Fig. (1A and 1B) graphical correlation of experimental pIC50 and predicted pIC50 is compared for 9 compounds. Correlation declares high degree of predictive powers of QSAR models obtained hereby. The R2 metric values reached near to 1 in tetra variable model based on non-linear a (SVM) model which is pretty clear in graphical correlation of experimental and predicted values of pIC50.
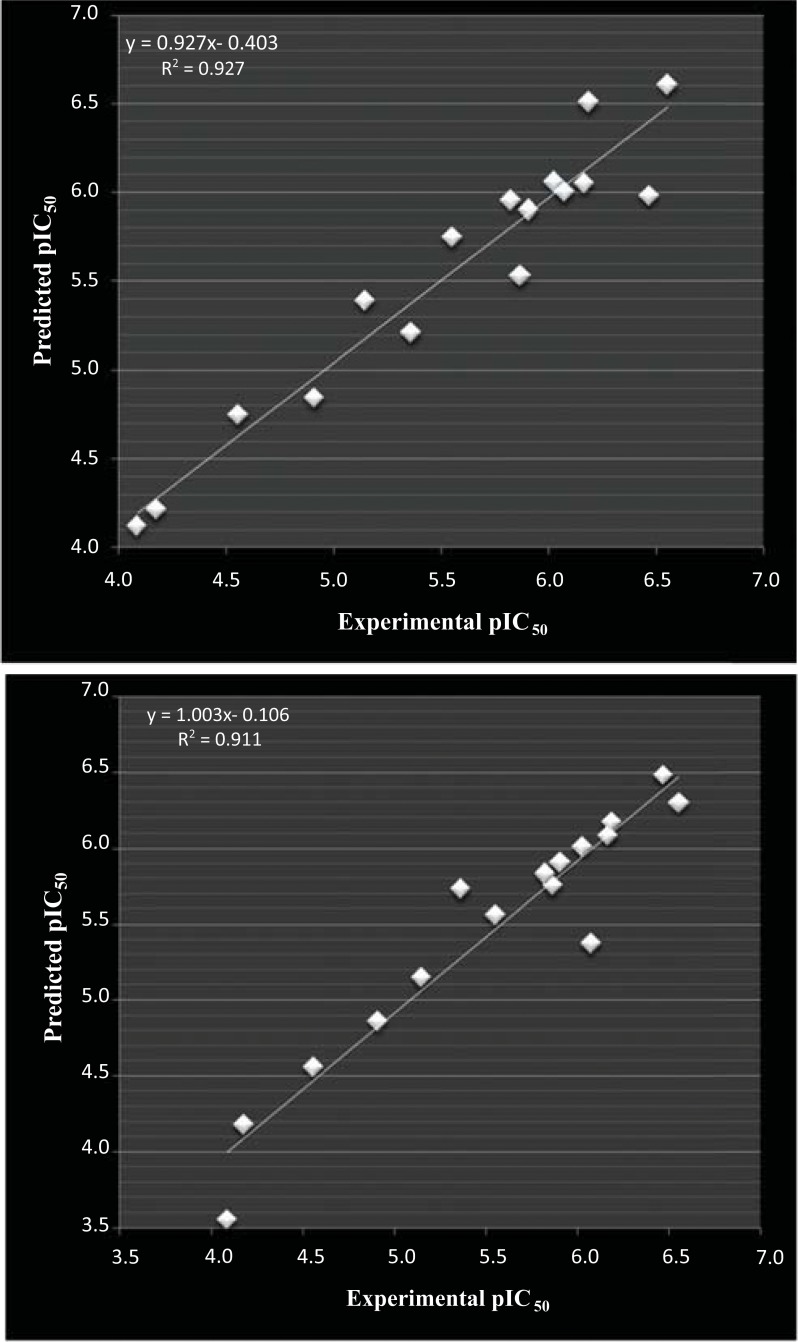
For dataset-2, graphical correlation of experimental with predicted pIC50 using tetra variable linear (MLR) and non-linear (SVM) QSAR models is given below in Fig. (2A and B), respectively. First graphical look confirms the predictive powers of QSAR models wherein values are found in close vicinity to regression line.
Fig. (2).
(A): correlation of experimental and predicted pIC50 calculated from linear (MLR) aided tetra variable model for dataset -2 and (B) correlation of experimental and predicted pIC50 calculated from non-linear (SVM) aided tetra variable model for dataset-2.
Graphical correlation of experimental and predicted binding potential percentage (%) for dataset-3 with 32 compounds using tetra variable linear (MLR) and non-linear (SVM) models appeared non-smooth around regression line. The most probable reason could be due to non-smooth nature of end point values (biological activity) in terms of percentage (%) binding of compounds to GABA receptor. SVM aided non-linear models appeared superior than the respective MLR aided linear QSAR models in the case of dataset-3. The graphical correlation is presented in Fig. (3A and B).
Descriptors selected in the above model can thereby be used to understand and illustrate the underlying SAR of compounds towards GABA receptor. There are various classes of descriptors selected in forward selection of linear and non-linear QSAR models in dataset-1, dataset-2 and dataset-3. Interestingly, there are four classes of descriptors (Topological charge indices, WHIM descriptors, 3D-MoRSE and Eigenvalue based descriptors) which frequently repeated in linear and non-linear QSAR models described above. When compared with respect to chemical structure features derived from these identified various classes of descriptors, mapped on frequency of occurrence, it can be concluded that electro-negativities, polarizabilities, van der Waals volume, resonance integrals and number of rings are found to be decisive in structure-activity relationship of compounds targeting GABA receptor.
Conclusion
The present QSAR studies successfully obtained QSAR models on three different QSAR datasets which consist of chemically dispersed molecules but acting on GABA receptor evaluated for their pharmacological action against schizophrenia. Attempts to identify underlying common chemical structure features which are responsible for their SAR towards GABA receptor included MLR aided linear QSAR models and Gaussian kernel function aided non-linear QSAR models. Descriptors identified in linear and non-linear QSAR models could assist medicinal chemists to synthesize analogues of magnolol and hankiol based compounds. Statistical fitness and predictive powers for all QSAR models received are acceptable. Mechanistic analysis on QSAR models identified chemical structural features based on van der Waals volumes, electronegativities, polarizability and number of rings available in compounds included in QSAR datasets. These structural properties are derived as the most repetitive properties in WHIM, 3D-MoRSE and Eigenvalue sets of descriptors. Linear and non-linear QSAR models also confirm this observation by selecting various descriptors in forwards selection but they belong to the same class and, more relatively, the same structure-activity relationship.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
Table 1.
Molecular descriptors and forward selection statistics for linear (MLR) and non-linear (SVM) for QSAR dataset 1 (9 Compounds).
| Model | Descriptors | Variables | R2 | Max. Abs. Error | Mean Abs. Error | R2CV (N-Fold) |
|---|---|---|---|---|---|---|
|
Linear (MLR) |
nR09 | 1 | 0.5097 | 0.6185 | 0.4564 | -0.0114 |
| nR09, BELp2 | 2 | 0.8634 | 0.6036 | 0.1688 | 0.7684 | |
| nR09, G2e, BELp2 | 3 | 0.9012 | 0.5066 | 0.1338 | 0.7860 | |
| nR09, E1u, G2e, BELp2 | 4 | 0.9796 | 0.1682 | 0.0876 | 0.8607 | |
|
Non-linear (SVM) |
Mor24m | 1 | 0.8686 | 0.5820 | 0.1453 | 0.5183 |
| Mor24m, Se1C3C3ad | 2 | 0.9747 | 0.2414 | 0.0576 | 0.8455 | |
| Mor24m, Se1C3C3ad, Mp | 3 | 0.9984 | 0.0611 | 0.0140 | 0.9441 | |
| Mor24m, Se1C3C3ad, Mp, Hnar | 4 | 1.0000 | 0.0094 | 0.0029 | 0.9250 |
Table 2.
Observed and predicted pIC50 values for tetra- variable model using SVM and MLR dataset 1 (9 Compounds).
| Molecule | Experimental pIC50 | Predicted(pIC50) | Predicted(pIC50) |
|---|---|---|---|
| Linear (MLR) | Non-Linear (SVM) | ||
| Acacetin | 4.699 | 4.531 | 4.687 |
| Saikosaponin A | 4.301 | 4.368 | 4.289 |
| Saikogenin G | 4.301 | 4.388 | 4.307 |
| Cimicidanol | 5.398 | 5.455 | 5.402 |
| Rutaecarpine | 6.000 | 5.895 | 5.994 |
| flunitrazepam | 3.699 | 3.645 | 3.699 |
| honokiol | 4.658 | 4.703 | 4.652 |
| magnolol | 4.495 | 4.633 | 4.498 |
| 6-methylflavone | 3.921 | 3.854 | 3.915 |
Table 4.
Observed and predicted pIC50 values for tetra- variable model using SVM and MLR dataset 2 (16 Compounds).Nipecotic Acid Tiagabine
| Molecule | pIC50 | Predicted(pIC50) | Predicted(pIC50) |
|---|---|---|---|
| Linear (MLR) | Non-linear (SVM) | ||
| 1_NIPECOTIC ACID | 4.089 | 4.121 | 3.555 |
| 2_TIAGABINE | 6.553 | 6.606 | 6.299 |
| 1a | 5.866 | 5.529 | 5.756 |
| 1b | 4.910 | 4.843 | 4.862 |
| 1d | 6.027 | 6.062 | 6.010 |
| 1e | 6.187 | 6.510 | 6.173 |
| 1f | 4.559 | 4.745 | 4.559 |
| 2b | 5.553 | 5.749 | 5.560 |
| 2c | 5.149 | 5.390 | 5.149 |
| 2d | 6.167 | 6.051 | 6.082 |
| 2e | 6.469 | 5.982 | 6.482 |
| 2f | 6.076 | 6.004 | 5.372 |
| 2g | 4.178 | 4.213 | 4.179 |
| 3d | 5.907 | 5.903 | 5.907 |
| 3e | 5.824 | 5.955 | 5.835 |
| 3f | 5.363 | 5.211 | 5.732 |
Table 5.
Molecular descriptors and forward selection statistics for linear (MLR) and non-linear (SVM) QSAR dataset 3 (32 Compounds).
| Model | Descriptors | Variables | R2 | Max. Abs. Error | Mean Abs. Error | R2CV (N-Fold) |
|---|---|---|---|---|---|---|
|
Linear (MLR) |
W | 1 | 0.3422 | 4.5701 | 1.8949 | 0.2516 |
| W, EEig07r | 2 | 0.5222 | 4.1811 | 1.5139 | 0.4614 | |
| W, EEig07r, EEig05x | 3 | 0.7109 | 2.9664 | 1.2478 | 0.6503 | |
| W, EEig07r, EEig05x, R8v+ | 4 | 0.8548 | 2.3028 | 0.8479 | 0.8054 | |
|
Non-linear (SVM) |
EEig09r | 1 | 0.3134 | 5.8419 | 1.6281 | 0.3524 |
| EEig09r, Mor08u | 2 | 0.7250 | 3.8783 | 1.0052 | 0.6296 | |
| EEig09r, Mor08u, HATS5e | 3 | 0.8169 | 3.0712 | 0.7513 | 0.7578 | |
| EEig09r, Mor08u, HATS5e, JGI9 | 4 | 0.8973 | 2.3043 | 0.5929 | 0.7947 |
Series 1.Magnolol analogues (1-21) Series 2.Honokiol analogues (1-11).
Table 6.
Observed and predicted Experimental Potential %values for tetra- variable model using SVM and MLR dataset 3 (32 Compounds).
|
Experimental
Potential % |
Predicted
Potential % |
Predicted
Potential % |
||||
|---|---|---|---|---|---|---|
| Linear (MLR) | Non-linear (SVM) | |||||
| Series I. Magnolol Analogues | ||||||
| R1 | R2 | R3 | ||||
| 1 | H | pentyl | H | 5 | 4.4 | 0.9 |
| 2 | H | hexyl | H | 7 | 6.1 | 5.1 |
| 3 | methyl | butyl | H | 5 | 4.8 | 3.5 |
| 4 | methyl | pentyl | H | 3 | 3.5 | 2.6 |
| 5 | methyl | hexyl | H | 7 | 6.4 | 7.0 |
| 6 | ethyl | propyl | H | 5 | 4.4 | 5.0 |
| 7 | ethyl | butyl | H | 3 | 4.6 | 3.0 |
| 8 | ethyl | pentyl | H | 3 | 3.1 | 3.0 |
| 9 | propyl | pentyl | H | 5 | 5.3 | 3.9 |
| 10 | propyl | hexyl | H | 5 | 2.7 | 4.8 |
| 11 | propyl | heptyl | H | 1 | 1.1 | 3.2 |
| 12 | propyl | octyl | H | 1 | -0.3 | 1.0 |
| 13 | butyl | pentyl | H | 5 | 6.2 | 3.9 |
| 14 | butyl | hexyl | H | 3 | 2.5 | 3.0 |
| 15 | ethyl | pentyl | CH3 | 7 | 7.5 | 6.6 |
| 16 | ethyl | hexyl | CH3 | 5 | 5.0 | 5.0 |
| 17 | propyl | pentyl | CH3 | 1 | 3.3 | 3.0 |
| 18 | propyl | hexyl | CH3 | 1 | 0.5 | 1.0 |
| 19 | pentyl | ethyl | CH3 | 3 | 2.6 | 3.6 |
| 20 | pentyl | propyl | CH3 | 3 | 3.5 | 3.0 |
| 21 | hexyl | propyl | CH3 | 1 | -0.1 | 0.8 |
| Series II. 4’-O-methyl Honokiol Analogues | ||||||
| 1 | methyl | methyl | - | 1 | 1.1 | 1.0 |
| 2 | ethyl | methyl | - | 3 | 2.8 | 3.0 |
| 3 | propyl | methyl | - | 10 | 9.2 | 10.0 |
| 4 | butyl | methyl | - | 10 | 8.9 | 10.0 |
| 5 | pentyl | methyl | - | 7 | 8.2 | 7.0 |
| 6 | hexyl | methyl | - | 10 | 8.5 | 9.6 |
| 7 | heptyl | methyl | - | 7 | 6.4 | 7.1 |
| 8 | octyl | methyl | - | 1 | 3.1 | 3.2 |
| 9 | hexyl | ethyl | - | 3 | 4.5 | 3.3 |
| 10 | hexyl | propyl | - | 1 | 2.6 | 1.0 |
| 11 | hexyl | isopropyl | - | 1 | 0.5 | 1.0 |
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Gottesman I.I. Schizophrenia genesis: The origins of madness.WH Freeman/Times Books/Henry Holt. 1991. 0026 Co. [Google Scholar]
- 2.Endicott J., Spitzer R.L. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch. Gen. Psychiatry. 1978;35(7):837–844. doi: 10.1001/archpsyc.1978.01770310043002. [http://dx.doi.org/10.1001/archpsyc.1978. 01770310043002]. [PMID: 678037]. [DOI] [PubMed] [Google Scholar]
- 3.Kane J.M., McGlashan T.H. Treatment of schizophrenia. Lancet. 1995;346(8978):820–825. doi: 10.1016/s0140-6736(95)91630-x. [http://dx.doi.org/10.1016/S0140-6736(95)91630-X]. [PMID: 7545770]. [DOI] [PubMed] [Google Scholar]
- 4.Green A.I. Treatment of schizophrenia and comorbid substance abuse: pharmacologic approaches. J. Clin. Psychiatry. 2006;67(Suppl. 7):31–35. [http://dx.doi.org/10.4088/JCP.0906e08]. [PMID: 16961422]. [PubMed] [Google Scholar]
- 5.Simpson M.D., Slater P., Deakin J.F., Royston M.C., Skan W.J. Reduced GABA uptake sites in the temporal lobe in schizophrenia. Neurosci. Lett. 1989;107(1-3):211–215. doi: 10.1016/0304-3940(89)90819-7. [http://dx.doi.org/10. 1016/0304-3940(89)90819-7]. [PMID: 2616032]. [DOI] [PubMed] [Google Scholar]
- 6.Wassef A.A., Dott S.G., Harris A., Brown A., O’Boyle M., Meyer W.J., III, Rose R.M. Critical review of GABA-ergic drugs in the treatment of schizophrenia. J. Clin. Psychopharmacol. 1999;19(3):222–232. doi: 10.1097/00004714-199906000-00004. [http://dx.doi.org/10.1097/00004714-199906000-00004]. [PMID: 10350028]. [DOI] [PubMed] [Google Scholar]
- 7.Javitt D.C., Spencer K.M., Thaker G.K., Winterer G., Hajós M. Neurophysiological biomarkers for drug development in schizophrenia. Nat. Rev. Drug Discov. 2008;7(1):68–83. doi: 10.1038/nrd2463. [http:// dx.doi.org/10.1038/nrd2463]. [PMID: 18064038]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore T.R. A primer of drug action: A concise nontechnical guide to the actions, uses, and side effects of psychoactive drugs. Am. J. Health Syst. Pharm. 2002;59(11):1130–1131. [Google Scholar]
- 9.Cousins M.S., Roberts D.C., de Wit H. GABA(B) receptor agonists for the treatment of drug addiction: a review of recent findings. Drug Alcohol Depend. 2002;65(3):209–220. doi: 10.1016/s0376-8716(01)00163-6. [http://dx. doi.org/10.1016/S0376-8716(01)00163-6]. [PMID: 11841892]. [DOI] [PubMed] [Google Scholar]
- 10.Johnson B.A., Swift R.M., Addolorato G., Ciraulo D.A., Myrick H. Safety and efficacy of GABAergic medications for treating alcoholism. Alcohol. Clin. Exp. Res. 2005;29(2):248–254. doi: 10.1097/01.alc.0000153542.10188.b0. [http://dx. doi.org/10.1097/01.ALC.0000153542.10188.B0]. [PMID: 15714047]. [DOI] [PubMed] [Google Scholar]
- 11.Macdonald R.L., McLean M.J. Anticonvulsant drugs: mechanisms of action. Adv. Neurol. 1986;44:713–736. [PMID: 2871724]. [PubMed] [Google Scholar]
- 12.Yadav M., Joshi S., Nayarisseri A., Jain A., Hussain A., Dubey T. Global QSAR modeling of logP values of phenethylamines acting as adrenergic alpha-1 receptor agonists. Interdiscip. Sci. 2013;5(2):150–154. doi: 10.1007/s12539-013-0162-0. [http://dx.doi.org/10.1007/s12539-013-0162-0]. [PMID: 23740397]. [DOI] [PubMed] [Google Scholar]
- 13.Ojha M., Yadav M., Nayarisseri A., Jyoti P., Neetesh P., Payal C. Overlapping structure features selection in linear and non-linear QSAR. J. Pharm. Res. 2013;6(1):183–187. [http://dx.doi.org/10. 1016/j.jopr.2012.11.038]. [Google Scholar]
- 14.Sharma N., Ethiraj K.R., Yadav M., Nayarisseri S.A., Chaurasiya M., Vankudavath R.N., Rao K.R. Identification of LOGP values and Electronegativities as structural insights to model inhibitory activity of HIV-1 capsid inhibitors - a SVM and MLR aided QSAR studies. Curr. Top. Med. Chem. 2012;12(16):1763–1774. [http://dx.doi.org/10.2174/1568026611209061763]. [PMID: 23030611]. [PubMed] [Google Scholar]
- 15.Mbarki S., Dguigui K., Hallaoui M.E. Construction of 3D-QSAR models to predict antiamoebic activities of pyrazoline and dioxazoles derivatives. J. Mater. Environ. Sci. 2011;2(1):61–70. [Google Scholar]
- 16.Jhanwar B., Sharma V., Singla R.K., Shrivastava B. QSAR - Hansch analysis and related approaches in drug design. Pharmacologyonline. 2011;1:306–344. [Google Scholar]
- 17.Deeb O., Shaik B., Agrawal V.K. Exploring QSARs of the interaction of flavonoids with GABA (A) receptor using MLR, ANN and SVM techniques. J. Enzyme Inhib. Med. Chem. 2014;29(5):670–676. doi: 10.3109/14756366.2013.839557. Epub ahead of print [PMID: 24102524]. [DOI] [PubMed] [Google Scholar]
- 18.Suykens J.A., Vandewalle J.P. Least squares support vector machine classifiers. Neural Process. Lett. 1999;(9):293–300. [http://dx.doi.org/10.1023/A:1018628609742]. [Google Scholar]
- 19.Pavlidis P., Wapinski I., Noble W.S. Support vector machine classification on the web. Bioinformatics. 2004;20(4):586–587. doi: 10.1093/bioinformatics/btg461. [http://dx.doi.org/10.1093/bioinformatics/btg461]. [PMID: 14990457]. [DOI] [PubMed] [Google Scholar]
- 20.Corinna C., Vladimir V. Support-vector networks. Mach. Learn. 1995;20(3):273–297. [http://dx.doi.org/10.1007/BF00994018]. [Google Scholar]
- 21.Zheng J., Wen R., Luo X., Lin G., Zhang J., Xu L., Guo L., Jiang H. Design, synthesis, and biological evaluation of the N-diarylalkenyl-piperidinecarboxylic acid derivatives as GABA uptake inhibitors (I). Bioorg. Med. Chem. Lett. 2006;16(1):225–227. doi: 10.1016/j.bmcl.2005.09.004. [http://dx.doi.org/10.1016/j.bmcl.2005.09.004]. [PMID: 16246548]. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs A., Baur R., Schoeder C., Sigel E., Müller C.E. Structural analogues of the natural products magnolol and honokiol as potent allosteric potentiators of GABA(A) receptors. Bioorg. Med. Chem. 2014;22(24):6908–6917. doi: 10.1016/j.bmc.2014.10.027. [http://dx.doi.org/10.1016/j.bmc.2014.10. 027]. [PMID: 25456080]. [DOI] [PubMed] [Google Scholar]
- 23.Csizmadia F. John Willy & Sons. Java applets and modules supporting chemical database handling from web browsers. J. Chem. Inf. Comput. Sci. 2000;40(2):323–324. doi: 10.1021/ci9902696. [http://dx.doi.org/ 10.1021/ci9902696]. [PMID: 10761134]. [DOI] [PubMed] [Google Scholar]
- 24.Xue L., Bajorath J. Molecular descriptors in chemoinformatics, computational combinatorial chemistry, and virtual screening. Comb. Chem. High Throughput Screen. 2000;3(5):363–372. doi: 10.2174/1386207003331454. [http://dx.doi.org/10.1002/9783527628766]. [DOI] [PubMed] [Google Scholar]
- 25.Tetko I.V., Gasteiger J., Todeschini R., Mauri A., Livingstone D., Ertl P., Palyulin V.A., Radchenko E.V., Zefirov N.S., Makarenko A.S., Tanchuk V.Y., Prokopenko V.V. Virtual computational chemistry laboratory--design and description. J. Comput. Aided Mol. Des. 2005;19(6):453–463. doi: 10.1007/s10822-005-8694-y. [http://dx.doi.org/ 10.1007/s10822-005-8694-y]. [PMID: 16231203]. [DOI] [PubMed] [Google Scholar]
- 26.Tetko I.V. Computing chemistry on the web. Drug Discov. Today. 2005;10(22):1497–1500. doi: 10.1016/S1359-6446(05)03584-1. [http://dx.doi.org/10.1016/S1359-6446(05)03584-1]. [PMID: 16257371]. [DOI] [PubMed] [Google Scholar]
- 27.Pavlidis P., Wapinski I., Noble W.S. Support vector machine classification on the web. Bioinformatics. 2004;1(20(4)):586–587. doi: 10.1093/bioinformatics/btg461. [DOI] [PubMed] [Google Scholar]
- 28.Smola A.J., Schölkopf B.A. Tutorial on Support Vector Regression. Statistics and Computing. Stat. Comput. 2004;14:199–222. [http://dx.doi.org/10.1023/B:STCO.0000035301.49549.88]. [Google Scholar]
- 29.Sharma B.K., Shekhawat M., Singh P.A. QSAR study on Pyrazole and Triazole Derivatives as Selective Canine COX-2 Inhibitors. IJRAP. 2011;2(1):186–197. [Google Scholar]
- 30.Liu P., Long W. Current mathematical methods used in QSAR/ QSPR studies. Int. J. Mol. Sci. 2009;10(5):1978–1998. doi: 10.3390/ijms10051978. [http://dx. doi.org/10.3390/ijms10051978]. [PMID: 19564933]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schölkopf B., Smola A.J. Learning with kernels: Support vector machines, regularization, optimization and beyond. 2002 [Google Scholar]