Abstract
Tissue engineering aims to utilise biologic mediators to facilitate tissue regeneration. Several recombinant proteins have potential to mediate induction of bone production, however, the high production cost of mammalian cell expression impedes patient access to such treatments. The aim of this study is to produce recombinant human osteopontin (hOPN) in plants for inducing dental bone regeneration. The expression host was Nicotiana benthamiana using a geminiviral vector for transient expression. OPN expression was confirmed by Western blot and ELISA, and OPN was purified using Ni affinity chromatography. Structural analysis indicated that plant-produced hOPN had a structure similar to commercial HEK cell-produced hOPN. Biological function of the plant-produced hOPN was also examined. Human periodontal ligament stem cells were seeded on an OPN-coated surface. The results indicated that cells could grow normally on plant-produced hOPN as compared to commercial HEK cell-produced hOPN determined by MTT assay. Interestingly, increased expression of osteogenic differentiation-related genes, including OSX, DMP1, and Wnt3a, was observed by realtime PCR. These results show the potential of plant-produced OPN to induce osteogenic differentiation of stem cells from periodontal ligament in vitro, and suggest a therapeutic strategy for bone regeneration in the future.
Introduction
Tissue engineering is a technology in medical therapeutics using bioactive substitutes for functional restoration of lost tissues and impaired organs1. Various proteins are necessary for tissue or organ engineering. These proteins function in regulating cellular behavior, the structure of many signaling molecules, and the fabrication of scaffolds2. However, there are several limitations to using proteins for this objective, such as source availability and stability, batch-to-batch variability, transmission of infecting organisms and immunogenicity3,4. Recombinant DNA technology used to expression foreign genes in convenient host systems can overcome these impediments.
Many recombinant proteins were reported to function in tissue engineering, such as bone matrix protein, collagens5,6, elastin7,8, and spider silk9,10. Presently, several sources for recombinant protein production are available, including E. coli, yeast, insect cells, mammalian cells, and plants. Among these platforms, the plant system is the most recently developed. One advantage of plants over other systems is eukaryotic post-translational modification, which may be necessary for the function of proteins used in tissue engineering processes11. Other advantages include lack of human pathogens, speed, low cost, and highly scalable manufacturing11,12.
OPN has a role in bone formation. Also known as secreted phophoprotein1 (SPP1), it is a highly phosphorylated glycoprotein that is a prominent component of the mineralized extracellular matrix of bone13. OPN contains an Arg-Gly-Asp sequence that is a major integrin-binding site and functions to support adhesion of bone cells to the mineralized matrix14,15. The molecular weight of OPN varies between 45–74 kDa, depending on the level of phosphorylation and glycosylation16. OPN is a soluble protein present in most body fluids. One study suggested that OPN plays a role in adhesion and movement of osteoblasts in bone17. Besides bone cells, OPN can be secreted by mesenchymal stem cells (MSC) and can be further up-regulated during the osteogenic differentiation of these cells18.
In this study, plants are used to produce human osteopontin (hOPN) as a model for recombinant proteins for tissue engineering. The recombinant hOPN was expressed transiently in Nicotiana benthamiana using geminiviral vectors. The hOPN was purified by Ni affinity chromatography and characterized for its structural conformation compared with commercial hOPN. The effect of plant-produced hOPN on cell cytotoxicity and osteogenic differentiation were also investigated.
Results
Transient expression of hOPN in N. benthamiana leaves
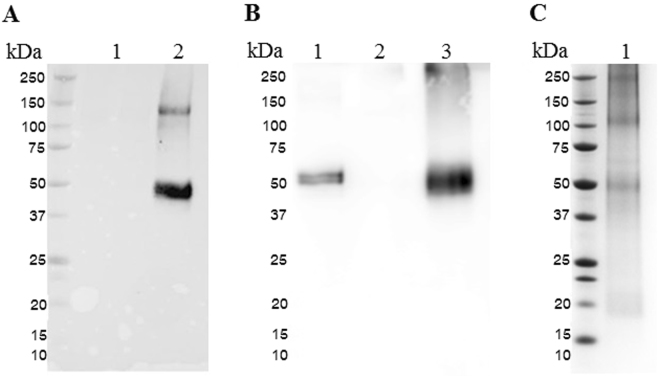
We produced hOPN by co-expression of hOPN (geminiviral vector pBY-OPN) with the gene silencing inhibitor p1919 from tomato bushy stunt virus, pPS19 (Fig. 1). The expression level of hOPN in N. benthamiana leaves was examined from 1 to 5 dpi by Western blot. The highest level of protein expression was found on day 3 (Supplementary Figure 1), up to ~100 ng hOPN per g leaf mass. Therefore, leaves were subsequently harvested on day 3 for protein extraction and purification. hOPN protein was observed at ~50 kDa by Western blot using anti-OPN antibody, while a negative control leaf extract showed no signal (Fig. 2A). The hOPN protein was purified from plant leaves using Ni affinity chromatography. The purified protein was confirmed by Western blot using an anti-OPN antibody (Fig. 2B) and SDS-PAGE (Fig. 2C).
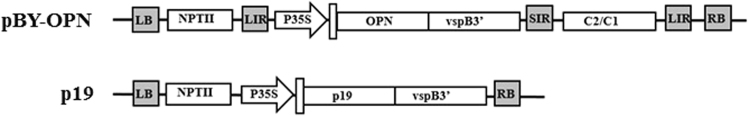
Figure 1.
Schematic representation of the T-DNA regions of the vectors used in this study P35S: Cauliflower Mosaic Virus (CaMV) 35S promoter, O. sativa leader: Oryza sativa leader sequence, OPN: human osteopontin gene with 8X histidine residues at C-terminus, vspB3’: soybean vspB gene 3′ element, C2/C1: Bean Yellow Dwaft Virus (BeYDV) ORFs C1 and C2 which encode for replication initiation protein (Rep) and RepA, LIR: long intergenic region of BeYDV genome, SIR: short intergenic region of BeYDV genome, NPTII: expression cassette encoding nptII gene for kanamycin resistance P19: P19 gene from Tomato Bushy Stunt Virus (TBSV). LB and RB: the left and right borders of the Agrobacterium T-DNA region.
Figure 2.
Western blot and SDS-PAGE of plant-produced hOPN. (A)Western blot of hOPN in crude extract of N. benthamiana leaf agroinfiltrated with pBY-OPN and pPS19 detected with mouse anti-human OPN and goat anti-mouse IgG conjugated with HRP. lane 1: wildtype N. benthamiana, lane 2: N. benthamiana agroinfiltrated with pBY-OPN and pPS19; (B) Western blot of purified hOPN from N. benthamiana detected with mouse anti-human OPN and goat anti-mouse IgG conjugated with HRP. lane 1: hOPN produced from HEK 293 cells, lane 2: wildtype N. benthamiana, lane 3: N. benthamiana agroinfiltrated with pBY-OPN and pPS19; (C) Purified plant-produced hOPN in SDS-PAGE. The numbers on the left are the size of the protein marker in kDa.
Secondary structure comparison by CD spectroscopy
To verify the secondary structure, the CD spectra of commercial hOPN expressed in HEK 293 cells and plan-produced hOPN were monitored. The secondary structure contents of both proteins are showed in Table 1. The spectrum of both proteins had peaks at wavelengths of 208 nm and 222 nm (Fig. 3). These spectra suggested that both proteins contain similar secondary structures.
Table 1.
The estimated secondary structure contents of commercial hOPN and plant hOPN.
| Secondary structure contents (%) | hOPN produced from HEK 293 cells | hOPN produced from N. benthamiana |
|---|---|---|
| α-helix | 2.5 | 8.3 |
| β-sheet | 34.3 | 28.1 |
| turn | 12.5 | 12.5 |
| random | 40.3 | 39.2 |
| sum | 89.7 | 88.1 |
Figure 3.

CD Structure analysis. Circular dichroism spectra of plant-produced hOPN and commercial hOPN (7.7 μM) in PBS (pH 7.4) were scanned by a circular dichroism spectrometer from 200–250 nm at. Both proteins exhibited maximum wavelength at 210 nm.
Tertiary structure comparison by fluorescence spectroscopy
The intrinsic tryptophan fluorescence spectra of commercial hOPN expressed in HEK 293 cells and plant-produced hOPN were characterized (Fig. 4). The maximum wavelengths of commercial hOPN and plant-produced hOPN were 340 and 320 nm, respectively. This result implied that the overall tertiary structure including the structure of glycosylation of these proteins were different.
Figure 4.
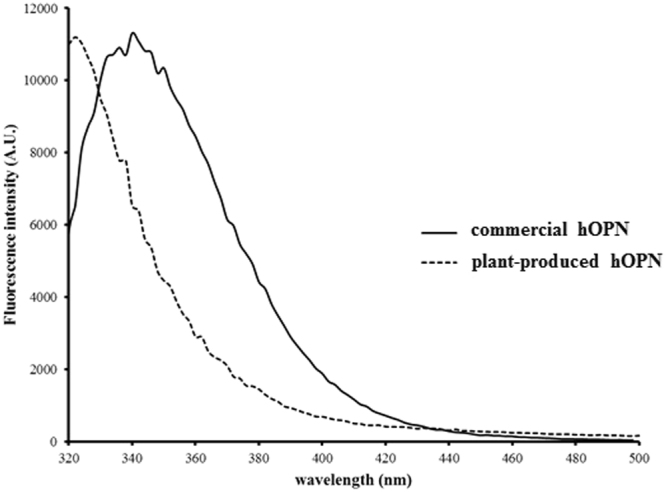
Intrinsic fluorescence spectra of commercial hOPN and plant-produced hOPN. Plant-produced hOPN showed blue shift of the maximum wavelength when compared to the commercial hOPN. The emission spectra were obtained from 320–500 nm, with the excitation wavelength at 280 nm.
Effect of plant-produced hOPN on cell proliferation
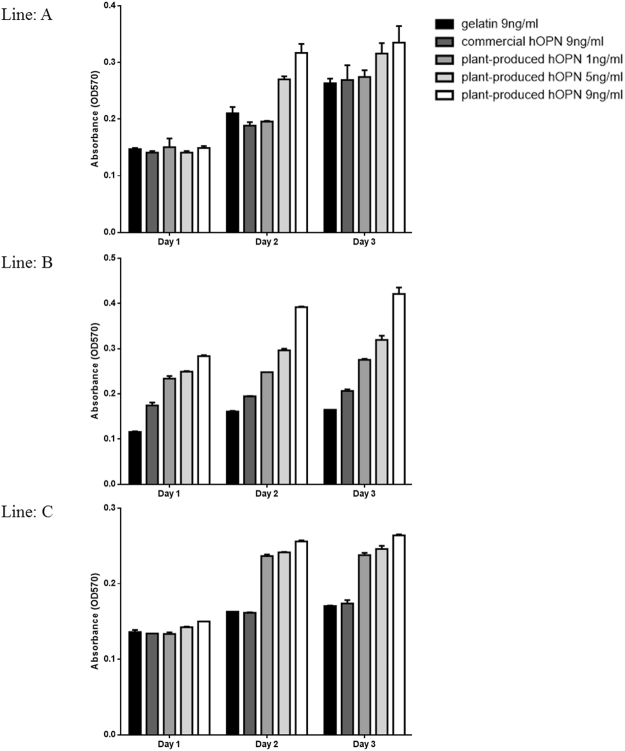
The effect of OPN on cell proliferation was examined by culturing hPDL cells established from 3 different donors on plant-produced hOPN-coated surface compared to cell culture on gelatin-coated and commercial hOPN-coated surfaces. The cell number was determined by MTT assay after cultured for 1, 2 and 3 days. The results showed that all 3 lines of hPDL cells could attach and grew normally on all surfaces tested (Fig. 5) indicating that none of the materials tested cytotoxic. However, cells cultured on 5 and 9 ng/ml of plant-produced hOPN grew significantly faster than when cultured on 9 ng/ml of gelatin on day 2 and day 3.
Figure 5.
Effect of OPN on periodontal ligament cells (PDL) proliferation. Three PDL cell lines established from three different donors were cultured on gelatin-coated (9 ng/well), commercial hOPN-coated (9 ng/well) and plant-produced hOPN-coated surface (1. 5 and 9 ng/well) for 1–3 days. The experiments were done in triplicated. Cell survival was evaluated by MTT assay at 24 hours. Data represented the absorbance at 540 nm. Data are means of 3 independent replicate samples +/−SD. Control in this experiment is the untreated cells. (*p ≤ 0.05).
Plant-produced hOPN activates osteogenesis related genes
Cells were treated with 1, 5 and 9 ng/ml of plant-produced hOPN or 9 ng/ml commercial hOPN for 72 hours. The mRNA was extracted and tested for the expression of OSX, DMP1, and Wnt3a genes by qRT-PCR. Figure 6 shows the expression level of these genes compared to cells cultured on gelatin with commercial OPN 9 ng/ml. The results from Fig. 6 indicated that commercial hOPN–coated surface could up-regulate the expression of OSX, DMP1, and Wnt3a. Interestingly, the mRNA expression of these three genes in cells seeded on plant-produced hOPN were significantly higher than those found in cells seeded on either gelatin-coated or 9 ng/ml commercial hOPN-coated surfaces.
Figure 6.
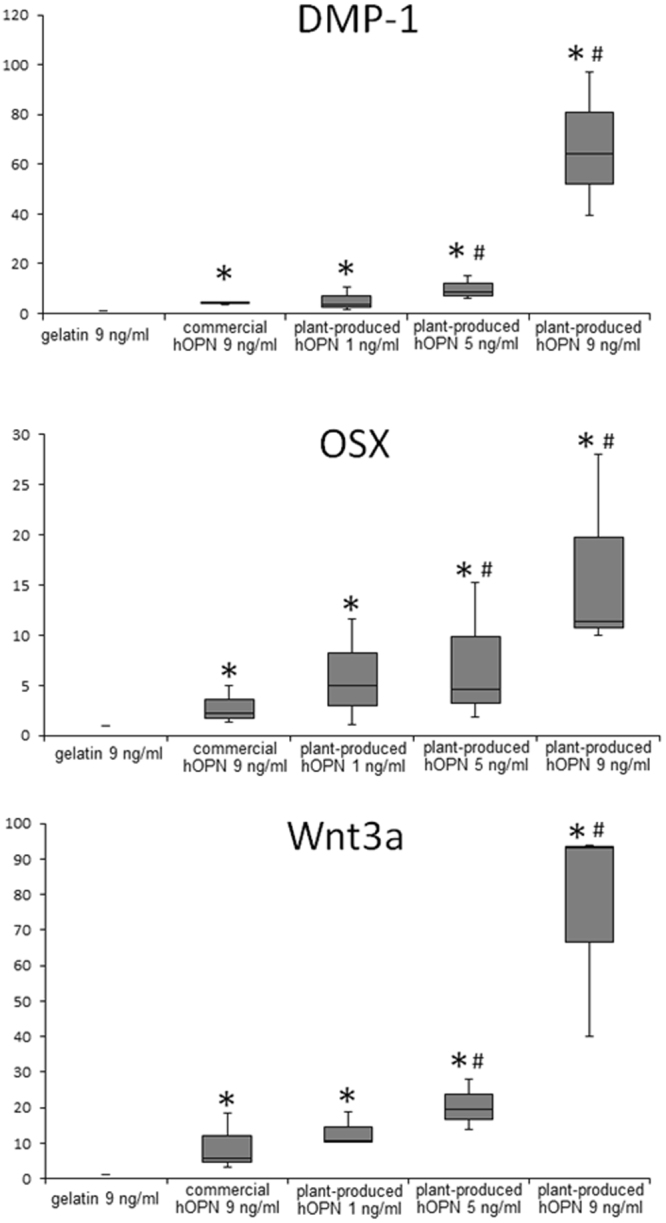
Plant-produced hOPN increased the mRNA expression of osteogenic markers. PDL cells from three different donors were treated with of 9 ng/ml gelatin, 9 ng/ml commercial hOPN, and 1, 5, 9 ng/ml plant-produced hOPN for 24 hours. Total RNA was extracted and real time PCR was performed using primer sets for human OSX, DMP1, and Wnt3a genes. Relative mRNA expression values were calculated normalized to the cells treated with gelatin. The values obtained for control gelatin were set at 1 for subsequent fold change calculation. (*p ≤ 0.05 compared to 9 ng/ml gelatin, #p ≤ 0.05 compared to 9 ng/ml commercial hOPN).
Discussion
In this study, we have demonstrated that functional matrix protein hOPN could be produced in N. benthamiana. The plant-produced hOPN was purified by Ni affinity chromatography and characterized by Western blot analysis and ELISA. The molecular weight of plant-produced hOPN was approximately 50 kDa, which is similar to that found in human tissue20,21. Our results also indicated that plant-produced hOPN contained similar epitopes as the protein synthesized by mammalian cells as judged by the ability to be recognized by a monoclonal anti-human OPN (Fig. 2B). Interestingly, plant produced hOPN showed increased biological function regarding osteogenic differentiation as compared to the same amount of recombinant hOPN from HEK cells (Fig. 6).
Recombinant hOPN was previously expressed in E. coli 22–25. Although hOPN produced from E. coli showed activity in cell adhesion, proliferation and differentiation of osteoblast cells23, it has been shown that the post-translational modifications of recombinant hOPN affects the biological activities related to the regulation of cell adhesion regulation26, which is necessary for bone regeneration. Therefore, a production platform that can provide effective post-translational modification is needed for recombinant hOPN production.
This is the first study showing that plants can be used as a platform to produce hOPN. The geminiviral replicon system has previously been used to produce different proteins such as antibody27,28, antigens28, and immune complex29. In this study, the geminiviral vector resulted in rapid transient expression - only 3 days after transfection. Although the expected mass of the unmodified hOPN protein is 33 kDa, posttranslational modifications increase the molecular weight to 45–75 kDa depending on conditions30. Our result showed that plant-produced hOPN is approximately 50 kDa (Fig. 2). OPN contains both N-linked and O-linked glycosylation sites31. Two N-glycan sites were found in OPN from bone, kidney tissues, macrophages, urinary stones and human milk32. Seven O-glycosylation regions were detected on hOPN, which are occupied by highly heterogeneous O-glycans33. A previous study demonstrated that the O-glycosylation of hOPN regulated its biological activities and affected the phosphorylation status34.
We used circular dichroism (CD) to evaluate the secondary structure of both commercial hOPN and plant-produced hOPN (Fig. 3). These data confirmed the similarity of secondary structures between hOPN produced from mammalian cells and plants. However, the intrinsic fluorescence spectra of these proteins were found to be somewhat different (Fig. 4). Since hOPN is a glycosylated protein, the different glycosylation patterns in human and plant may affect the overall protein conformation. This result is consistent with several reports that some proteins with different patterns of glycosylation showed shifted spectra35,36.
The biological activity of the purified plant-produced hOPN was further examined in human periodontal ligament stem cells. OPN has been shown to contain an RGD amino acid sequence that promotes cell attachment via integrin alphavbeta3. In this study, the ability to support cell growth and attachment was compared between gelatin, denatured type I collagen, commercial recombinant hOPN and plant-produced OPN using PDL cells. No significant differences were detected after 24 hours in culture. However, cells cultured on the plant produced hOPN-coated surface had significantly increased cell proliferation after 2 and 3 days in culture (Fig. 5). Interestingly, both types of hOPN, when immobilized on the cell surface using layer-by-layer technique, could significantly induce expression of osteogenic related genes such as OSX, DMP1, and Wnt3a (Fig. 6). Wnt3a is a the key protein to induce osteogenic differentiation of MSCs37, while osterix (OSX) is the key transcription factor that functions in osteogenic differentiation38. Dentine matrix acidic phosphoprotein-1 (DMP-1) is also one of the matrix proteins that has been recognized as a marker of mineralized tissue formation39. The ability of OPN to support osteogenic differentiation has been reported previously39. In that report, addition of a neutralizing antibody to OPN affected osteogenic differentiation of bone marrow mesenchymal stem cells. Our data showed the stimulation of these osteogenic related genes in cells isolated from three different donors. Three different donors are a minimum number for statistical analysis and were used in several reported studies40–43. Therefore, the plant-produced hOPN has clinical potential for supporting the osteogenic regeneration process.
It is also interesting to note that, at the same concentration, 9 ng/ml, the plant-produced OPN induced DMP1, OSX, and Wnt3a genes significantly better than commercial hOPN (Fig. 6) in all three established hPDL from 3 different donors, by a factor of 5–10 times. The levels of induction are different among the donor cells due to individual genetic variation. Although the exact mechanism is still unclear, it is possible that the plant-specific glycan pattern might be involved. Thus the glycan structures of plant-produced hOPN will be studied in detailed in future.
Our study firstly reported that the recombinant hOPN protein can be produced in plants by transient expression and the plant-produced hOPN can induce the genes involved in bone regeneration in cellulo. Further study of the plant-produced hOPN in an animal model is necessary to evaluate function in bone regeneration. Moreover, other proteins, which induce a different mechanism of bone regeneration, will also tested for expression in plants. The plant platform has potential to be a robust factory to produce low cost and effective recombinant proteins, which can facilitate the development of tissue engineering techniques that are affordable for patients in the future.
Methods
Expression vector construction
Human osteopontin gene44 was amplified using hOPN forward and hOPN reverse primers (Table 2). The vector containing human OPN was a gift from Professor Cecilia M. Giachelli. The PCR product was cloned into pGEM-T Easy (Promega, USA) for sequencing and cut with NcoI and SacI restriction enzymes. The geminiviral vector pBY030.2R45 was cut with NcoI and SacI and ligated with the digested hOPN gene to make pBY-OPN (Fig. 1). After verification, the plasmid was electroporated into Agrobacterium tumefaciens strain GV3101.
Table 2.
Primers list.
| Gene | Sequence (5′ to 3′) | Product size (bp) | Sequence ID |
|---|---|---|---|
| hOPN | Forward: CCATGGAACTTGGACTTTCTTGG | 950 | J04765.1 |
| Reverse: GAGCTCTTAATGATGGTGATGGTGGTGATGATG | |||
| qGAPDH | Forward: CACTGCCAACGTGTCAGTGGTG | 121 | NM_002046.5 |
| Reverse: GTAGCCCAGGATGCCCTTGAG | |||
| OSX | Forward: GCCAGAAGCTGTGAAACCTC | 160 | NM_152860.1 |
| Reverse: GCTGCAAGCTCTCCATAACC | |||
| qDMP-1 | Forward: ATGCCTATCACAACAAACC | 213 | NM_004407.3 |
| Reverse: CTCCTTTATGTGACAACTGC | |||
| Wnt3a | Forward: CTGTTGGGCCACAGTATTCC | 113 | NM_033131.3 |
| Reverse: GGGCATGATCTCCACGTAGT |
Plant inoculation and protein expression
Nicotiana benthamiana plants, 6 to 8 weeks-old, were co-infiltrated with two Agrobacterium cell lines that contained pBY-OPN or pPSp19 by vacuum infiltration27. Plants were maintained in cultured room with a 16 h light/8 h dark cycle at 28 °C after infiltration. The leaves were harvested on days 1, 2, 3, 4, and 5 post-infiltration (dpi) for expression time-course experiments. For other experiments, the leaves were harvested on day 3 dpi. Infiltrated tobacco leaves were homogenized by using a blender with extraction buffer (5 mM imidazole, 20 mM Tris-HCl pH 7.4, 50 mM NaCl). Crude extract was filtered through Miracloth and centrifuged at 26,000 g at 4 °C for 30 min. The supernatant was filtered with 0.2-micron filter and used for Ni affinity purification. Chelating SepharoseTM (GE healthcare, UK) was packed into a column and washed with 10 bed volumes of distilled water. The resin was charged with 5 column volumes (CV) of 50 mM NiSO4.6H2O. After washing with 10 CV distilled water, the resin was washed with 10 CV of binding buffer (5 mM imidazole, 20 mM Tris-HCl pH7.4, 50 mM NaCl). The plant extract solution was loaded into the column. After washing with 10 CV washing buffer (20 mM imidazole, 20 mM Tris-HCl pH7.4, 50 mM NaCl), the purified protein was eluted with eluting buffer (250 mM imidazole, 20 mM Tris-HCl pH7.4, 50 mM NaCl) and analyzed by SDS-PAGE and Western blot.
SDS-PAGE and Western blot
The proteins were denatured by boiling for 5 minutes with loading buffer (125 mM Tris-HCl, 12% (w/v) SDS, 10% (v/v) glycerol, 22% (v/v) β-mercaptoethanol, and 0.001% (w/v) bromophenol blue) and separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE). Proteins were either visualized by Coomassie blue staining or electrophoretically transferred to polyvinlidene difluoride (PVDF) membrane (Amersham Hybond-ECL; Amersham Biosciences, UK). The membrane was blocked with 5% non-fat dried milk, 0.1% Tween20 in PBS (PBST). The membrane was probed with mouse monoclonal anti-OPN antiserum (Abcam, UK) diluted 1:5,000 in 1% non-fat dried milk in PBST and goat anti-mouse IgG-HRP conjugated (Sigma, USA) diluted 1:10,000 in 1% non-fat dried milk in PBST. The membranes were developed by chemiluminescence using ECL plus detection reagent (GE Healthcare, UK).
Secondary structure characterization by circular dichroism spectroscopy
Circular dichroism (CD) spectra were recorded by Chirascan (Applied Photophysics, Ltd) to determine the secondary structure of recombinant hOPN expressed in HEK 293 cells (Sigma-Aldrich, USA) and plant-produced hOPN. The spectra were measured between 190 nm and 250 nm. The measurements were conducted using protein concentrations of 0.10 mg/mL in 10 mM potassium phosphate buffer (pH 7.4). All data presented are the means of three independent measurements. The secondary contents of both proteins were calculated using Raussens et al. method46.
Intrinsic fluorescence spectroscopy
Emission spectra of commercial hOPN and plant-produced hOPN were monitored. The spectra were scanned from 300 to 500 nm using Spark 10 M multimode microplate reader (Tecan Group Ltd., Männedorf, Switzerland), based on an excitation of intrinsic fluorescence from aromatic side chains at 280 nm. Samples containing 0.10 mg/ml of protein were analyzed. Three repetitive scans were obtained and averaged.
OPN quantification by ELISA
The purified plant-produced hOPN was quantified by ELISA. The protocol was done according to manuals of Human Osteopontin (OPN) ELISA Kit (Sigma-Aldrich, USA). The absorbance was measured using microplate reader at 450 nm.
Cells
Human periodontal ligament (hPDL) cells were isolated and maintained according to a previous report47. The protocol was approved by the Ethics Committee, Faculty of Dentistry, Chulalongkorn University and all methods were performed in accordance with the relevant guidelines and regulations. The informed consents were obtained. Briefly, periodontal tissues were scraped from the middle one-third of the root surface the extracted third molars. The explants were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 2 mM L-glutamine, 100 units/mL penicillin, 100 µg/mL streptomycin and 250 ng/mL amphotericin B at 37 °C in a humidified 5% carbon dioxide atmosphere until the cells were outgrown from the explants and routinely subcultured after reaching confluency. All cell culture reagents were purchased from Gibco BRL (Carlsbad, CA, USA). Cells were prepared from six donors, represented by line A, B, C, D, E, and F. All the experiments were done using cells from passage 3–5. To characterize the mesenchymal surface markers, flow cytometry was performed to determine the surface expression of CD45, CD73, CD90 and CD105 according to our previously published report48,49.
Cell proliferation assay
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (USB Corporation, USA) is a tetrazolium compound that will be reduced to a formazan product by mitochondrial dehydrogenase. The amount of formazan product represents the metabolic activity of viable cells at a particular time point. Thus, cell proliferation can be indirectly determined by the changes in the amount of formazan. Cells were seeded at density of 50,000 cells per wells in 24-well-plate and assay was performed at days 24, 48, or 72 hours. Cells were treated with 300 µl of 0.5 mg/ml MTT solution and the plate was incubated for 20 min at 37 °C. After that, the MTT solution was aspirated and the well washed with PBS. Then 500 µl of Glycine:DMSO (1:9) was added to each well. After the formazan crystals had dissolved, the absorbance was determined spectrophotometrically at 570 nm using a reference wavelength of 630 nm on an ELX800UV universal microplate reader (Bio-Tek Instruments Inc., Vermont, USA). The experiments were done in triplicate.
Real-time PCR analysis for osteoblast differentiation markers
Total RNA was extracted from each experiment with Isol-RNA Lysis reagent (5Prime, Gaithersburg, MD, USA) and 1 µg of RNA per sample was converted to cDNA using a reverse transcriptase kit (Promega, Madison, WI, USA). Real-time quantitative polymerase chain reaction was performed using a Lightcycler Nano realtime polymerase chain reaction machine (Roche Applied Science, Indianapolis, IN, USA) using FastStart Essential DNA Green Master (Roche Applied Science). The PCR protocol was set as; denaturation at 94 °C for 10 seconds, annealing at 60 °C for 10 seconds, and extension at 72 °C for 10 seconds for 45 cycles. The reaction product of GAPDH was used as a reference gene for the internal control. The primer sequences are shown in Table 2.
Statistical Analysis
All experiments were performed using cells isolated from three different donors. Statistical evaluation was performed using SPSS 16.0 software (SPSS, USA). The Mann Whitney U test was employed for two group comparison and the Kruskal Wallis test followed by pairwise comparison was used for comparing three or more groups. A significant difference was considered at p ≤ 0.05.
Electronic supplementary material
Acknowledgements
This study was supported by Thailand Research Fund grant No MRG5980087 and IRN59W0001. KR was supported by the Ratchadaphiseksomphot Fund, Chulalongkorn University for the Postdoctoral Fellowship. PP was supported by the 2012 Research Chair Grant, Thailand National Science and Technology Development Agency (NSTDA). DC was supported by Research grant, Faculty of Dentistry, and IRN59W0001. We would like to thank Professor Cecilia M. Giachelli, University of Washington, USA for providing the osteopontin gene.
Author Contributions
J.M., P.P., and W.P. conceived and designed the study. K.R., S.A., D.C., A.K., H.M., and W.P. performed the experiment. H.M., P.P., and W.P. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17666-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Persidis A. Tissue engineering. Nature biotechnology. 1999;17:508–510. doi: 10.1038/8700. [DOI] [PubMed] [Google Scholar]
- 2.Farajollahi MM, Hamzehlou S, Mehdipour A, Samadikuchaksaraei A. Recombinant proteins: hopes for tissue engineering. BioImpacts: BI. 2012;2:123–125. doi: 10.5681/bi.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von der Mark K, Park J, Bauer S, Schmuki P. Nanoscale engineering of biomimetic surfaces: cues from the extracellular matrix. Cell and tissue research. 2010;339:131–153. doi: 10.1007/s00441-009-0896-5. [DOI] [PubMed] [Google Scholar]
- 4.Langer R, Tirrell DA. Designing materials for biology and medicine. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Cui FZ, Hu K, Zhu XD, Fan DD. Bone regeneration by using scaffold based on mineralized recombinant collagen. Journal of biomedical materials research. Part B, Applied biomaterials. 2008;86:29–35. doi: 10.1002/jbm.b.30984. [DOI] [PubMed] [Google Scholar]
- 6.Liu W, et al. Recombinant human collagen for tissue engineered corneal substitutes. Biomaterials. 2008;29:1147–1158. doi: 10.1016/j.biomaterials.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Jordan SW, et al. The effect of a recombinant elastin-mimetic coating of an ePTFE prosthesis on acute thrombogenicity in a baboon arteriovenous shunt. Biomaterials. 2007;28:1191–1197. doi: 10.1016/j.biomaterials.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 8.Sallach RE, Conticello VP, Chaikof EL. Expression of a recombinant elastin-like protein in pichia pastoris. Biotechnology progress. 2009;25:1810–1818. doi: 10.1002/btpr.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabotyagova OS, Cebe P, Kaplan DL. Self-assembly of genetically engineered spider silk block copolymers. Biomacromolecules. 2009;10:229–236. doi: 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- 10.Agapov II, et al. Three-dimensional scaffold made from recombinant spider Silk protein for tissue engineering. Doklady. Biochemistry and biophysics. 2009;426:127–130. doi: 10.1134/S1607672909030016. [DOI] [PubMed] [Google Scholar]
- 11.Fischer R, Stoger E, Schillberg S, Christou P, Twyman RM. Plant-based production of biopharmaceuticals. Current opinion in plant biology. 2004;7:152–158. doi: 10.1016/j.pbi.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Ma JK, et al. Plant-derived pharmaceuticals–the road forward. Trends in plant science. 2005;10:580–585. doi: 10.1016/j.tplants.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Saito K, Nakatomi M, Ida-Yonemochi H, Ohshima H. Osteopontin Is Essential for Type I Collagen Secretion in Reparative Dentin. Journal of dental research. 2016;95:1034–1041. doi: 10.1177/0022034516645333. [DOI] [PubMed] [Google Scholar]
- 14.Liaw L, Lindner V, Schwartz SM, Chambers AF, Giachelli CM. Osteopontin and beta 3 integrin are coordinately expressed in regenerating endothelium in vivo and stimulate Arg-Gly-Asp-dependent endothelial migration in vitro. Circulation research. 1995;77:665–672. doi: 10.1161/01.RES.77.4.665. [DOI] [PubMed] [Google Scholar]
- 15.Denhardt DT, Guo X. Osteopontin: a protein with diverse functions. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1993;7:1475–1482. [PubMed] [Google Scholar]
- 16.Denhardt DT, Noda M. Osteopontin expression and function: role in bone remodeling. Journal of cellular biochemistry. Supplement. 1998;30–31:92–102. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<92::AID-JCB13>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 17.Ishijima M, et al. Enhancement of osteoclastic bone resorption and suppression of osteoblastic bone formation in response to reduced mechanical stress do not occur in the absence of osteopontin. The Journal of experimental medicine. 2001;193:399–404. doi: 10.1084/jem.193.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickard DJ, Sullivan TA, Shenker BJ, Leboy PS, Kazhdan I. Induction of rapid osteoblast differentiation in rat bone marrow stromal cell cultures by dexamethasone and BMP-2. Developmental biology. 1994;161:218–228. doi: 10.1006/dbio.1994.1022. [DOI] [PubMed] [Google Scholar]
- 19.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant journal: for cell and molecular biology. 2003;33:949–956. doi: 10.1046/j.1365-313X.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 20.Oldberg A, Franzen A, Heinegard D. Cloning and sequence analysis of rat bone sialoprotein (osteopontin) cDNA reveals an Arg-Gly-Asp cell-binding sequence. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:8819–8823. doi: 10.1073/pnas.83.23.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullrich O, Mann K, Haase W, Koch-Brandt C. Biosynthesis and secretion of an osteopontin-related 20-kDa polypeptide in the Madin-Darby canine kidney cell line. The Journal of biological chemistry. 1991;266:3518–3525. [PubMed] [Google Scholar]
- 22.Ashkar S, Teplow DB, Glimcher MJ, Saavedra RA. In vitro phosphorylation of mouse osteopontin expressed in E. coli. Biochemical and biophysical research communications. 1993;191:126–133. doi: 10.1006/bbrc.1993.1193. [DOI] [PubMed] [Google Scholar]
- 23.Jang JH, Kim JH. Improved cellular response of osteoblast cells using recombinant human osteopontin protein produced by Escherichia coli. Biotechnology letters. 2005;27:1767–1770. doi: 10.1007/s10529-005-3551-6. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Y, et al. Expression and purification of bioactive high-purity recombinant mouse SPP1 in Escherichia coli. Applied biochemistry and biotechnology. 2014;173:421–432. doi: 10.1007/s12010-014-0849-7. [DOI] [PubMed] [Google Scholar]
- 25.Weng S, Zhou L, Han L, Yuan Y. Expression and purification of non-tagged recombinant mouse SPP1 in E. coli and its biological significance. Bioengineered. 2014;5:405–408. doi: 10.4161/bioe.34424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangaswami H, Bulbule A, Kundu GC. Osteopontin: role in cell signaling and cancer progression. Trends in cell biology. 2006;16:79–87. doi: 10.1016/j.tcb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Huang Z, et al. High-level rapid production of full-size monoclonal antibodies in plants by a single-vector DNA replicon system. Biotechnology and bioengineering. 2010;106:9–17. doi: 10.1002/bit.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai H, He J, Engle M, Diamond MS, Chen Q. Robust production of virus-like particles and monoclonal antibodies with geminiviral replicon vectors in lettuce. Plant biotechnology journal. 2012;10:95–104. doi: 10.1111/j.1467-7652.2011.00649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phoolcharoen W, et al. Expression of an immunogenic Ebola immune complex in Nicotiana benthamiana. Plant biotechnology journal. 2011;9:807–816. doi: 10.1111/j.1467-7652.2011.00593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sodek J, Ganss B, McKee MD. Osteopontin. Critical reviews in oral biology and medicine: an official publication of the American Association of Oral Biologists. 2000;11:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 31.Miwa HE, Gerken TA, Jamison O, Tabak LA. Isoform-specific O-glycosylation of osteopontin and bone sialoprotein by polypeptide N-acetylgalactosaminyltransferase-1. The Journal of biological chemistry. 2010;285:1208–1219. doi: 10.1074/jbc.M109.035436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuda K, Takahashi N, Tsukamoto Y, Honma H, Kohri K. N-Glycan structures of an osteopontin from human bone. Biochemical and biophysical research communications. 2000;268:814–817. doi: 10.1006/bbrc.2000.2224. [DOI] [PubMed] [Google Scholar]
- 33.Li H, et al. Site-specific structural characterization of O-glycosylation and identification of phosphorylation sites of recombinant osteopontin. Biochimica et biophysica acta. 2015;1854:581–591. doi: 10.1016/j.bbapap.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 34.Kariya Y, et al. Osteopontin O-glycosylation contributes to its phosphorylation and cell-adhesion properties. The Biochemical journal. 2014;463:93–102. doi: 10.1042/BJ20140060. [DOI] [PubMed] [Google Scholar]
- 35.Arakawa T, et al. Glycosylated and unglycosylated recombinant-derived human stem cell factors are dimeric and have extensive regular secondary structure. The Journal of biological chemistry. 1991;266:18942–18948. [PubMed] [Google Scholar]
- 36.Zhu BC, Laine RA, Barkley MD. Intrinsic tryptophan fluorescence measurements suggest that polylactosaminyl glycosylation affects the protein conformation of the gelatin-binding domain from human placental fibronectin. European journal of biochemistry. 1990;189:509–516. doi: 10.1111/j.1432-1033.1990.tb15516.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, et al. Wnt3a enhances bone morphogenetic protein 9-induced osteogenic differentiation of C3H10T1/2 cells. Chinese medical journal. 2013;126:4758–4763. [PubMed] [Google Scholar]
- 38.Sinha KM, Zhou X. Genetic and molecular control of osterix in skeletal formation. Journal of cellular biochemistry. 2013;114:975–984. doi: 10.1002/jcb.24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalajzic I, et al. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. 2004;35:74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Mindaye ST, et al. Impact of Influenza A Virus Infection on the Proteomes of Human Bronchoepithelial Cells from Different Donors. Journal of proteome research. 2017;16:3287–3297. doi: 10.1021/acs.jproteome.7b00286. [DOI] [PubMed] [Google Scholar]
- 41.Dommisch H, Chung WO, Plotz S, Jepsen S. Influence of histamine on the expression of CCL20 in human gingival fibroblasts. Journal of periodontal research. 2015;50:786–792. doi: 10.1111/jre.12265. [DOI] [PubMed] [Google Scholar]
- 42.Kim DS, et al. Gene expression profiles of human adipose tissue-derived mesenchymal stem cells are modified by cell culture density. PloS one. 2014;9:e83363. doi: 10.1371/journal.pone.0083363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richert L, et al. Species differences in the response of liver drug-metabolizing enzymes to (S)-4-O-tolylsulfanyl-2-(4-trifluormethyl-phenoxy)-butyric acid (EMD 392949) in vivo and in vitro. Drug metabolism and disposition: the biological fate of chemicals. 2008;36:702–714. doi: 10.1124/dmd.107.018358. [DOI] [PubMed] [Google Scholar]
- 44.Laura L. Smith, et al. Osteopontin N-terminal Domain Contains a Cryptic Adhesive Sequence Recognized by α β Integrin . Journal of Biological Chemistry271 (45):28485-28491 (1996). [PubMed]
- 45.Huang Z, Chen Q, Hjelm B, Arntzen C, Mason H. A DNA replicon system for rapid high-level production of virus-like particles in plants. Biotechnology and bioengineering. 2009;103:706–714. doi: 10.1002/bit.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raussens, V., Ruysschaert, J. M. & Goormaghtigh, E. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: a new scaling method. Analytical biochemistry319, 114-121 (2003). [DOI] [PubMed]
- 47.Pattamapun, K., Tiranathanagul, S., Yongchaitrakul, T., Kuwatanasuchat, J. & Pavasant, P. Activation of MMP- 2 by Porphyromonas gingivalis in human periodontal ligament cells. Journal of periodontal research38, 115-121 (2003). [DOI] [PubMed]
- 48.Dae Seong Kim, et al. Gene Expression Profiles of Human Adipose Tissue-Derived Mesenchymal Stem Cells Are Modified by Cell Culture Density. PLoS ONE9 (1):e83363 (2014). [DOI] [PMC free article] [PubMed]
- 49.Lv, F., Tuan, R. S., Cheung, K. M. C. & Leung, V. Y. L. Concise review: the surface markers and identify of human mesenchymal stem cells. Stem Cells32, 1408-1419 (2014). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.