Abstract
Understanding the biological factors that are characteristic of metastasis in melanoma remains a key approach to improving treatment. In this study, we seek to identify a gene signature of metastatic melanoma. We configured a new network-based computational pipeline, combined with a machine learning method, to mine publicly available transcriptomic data from melanoma patient samples. Our method is unbiased and scans a genome-wide protein-protein interaction network using a novel formulation for network scoring. Using this, we identify the most influential, differentially expressed nodes in metastatic as compared to primary melanoma. We evaluated the shortlisted genes by a machine learning method to rank them by their discriminatory capacities. From this, we identified a panel of 6 genes, ALDH1A1, HSP90AB1, KIT, KRT16, SPRR3 and TMEM45B whose expression values discriminated metastatic from primary melanoma (87% classification accuracy). In an independent transcriptomic data set derived from 703 primary melanomas, we showed that all six genes were significant in predicting melanoma specific survival (MSS) in a univariate analysis, which was also consistent with AJCC staging. Further, 3 of these genes, HSP90AB1, SPRR3 and KRT16 remained significant predictors of MSS in a joint analysis (HR = 2.3, P = 0.03) although, HSP90AB1 (HR = 1.9, P = 2 × 10−4) alone remained predictive after adjusting for clinical predictors.
Introduction
Malignant melanoma, a cancer arising from the melanocytes is reported to have one of the largest rates of increase in incidence worldwide1,2. According to the World Health Organization, current statistics indicate that 132,000 cases occur globally each year3. The majority of primary tumours are cured by local excision4 but the trend towards increased numbers of tumours in older males (age and male sex5 being risk factors for melanoma death) suggests that metastatic AJCC stage IV melanoma will continue to increase in incidence. Although the advent of targeted therapies, such as BRAF inhibitors and checkpoint therapies have for the first time produced a survival advantage, long term survival is still only seen in around 20% of patients6. Improvement is therefore necessary both in its detection and in its treatment. Computational methods are necessary for an unbiased comprehensive analysis so as to identify the characterizing genes of the metastatic phenotype.
Genome sequencing and analysis by The Cancer Genome Atlas (TCGA) has led to the identification of driver mutations in around 70% of tumours and a classification of patients into BRAF, NRAS, NF1 and tripleWT subtypes7. In addition, other studies have identified a series of mutations or copy number changes8. These have provided insights into the underlying molecular mechanisms but have little prognostic or diagnostic significance. Many of the currently available markers (TYR, HMGA2, TRIB2, MITF and PMEL) depend on the differential expression of these markers in the diseased state9–13. Although there was previously little agreement between transcriptomic studies in melanoma, we recently replicated signatures described by Jonsson’s group14,15 giving rise to the view that increasing study size, better platforms and bioinformatics may now lead to the identification of better biomarkers.
The most common analytical tools used for biomarker identification are clustering methods16, classification using a support vector machine17, decision trees and random forest classifiers18,19, artificial neural networks20 and simple differential expression based analysis13,21. These methods are typically data-driven and do not consider any biological information of the component genes as an input, but have an advantage of identifying distinguishing features even when no information is available about that feature. On the other hand, biological networks, constructed on the basis of the known functions and interactions of individual molecules, offer alternate approaches that are superior to blind learning approaches. Networks have the added advantage of combining condition specific transcriptome data and allow understanding of the functional role of the individual genes capable of discriminating disease from healthy or between different disease stages22–24. Machine learning methods on the other hand are capable of providing a quantitative picture of the classification efficiencies of the individual genes25,26. They fail when the number of features is higher than the number of samples. To get the best of both approaches, we have combined the two and used network analysis which facilitates the usage of machine learning methods by reducing the number of features to be tested for classification efficiency and derive the final signature. This type of a combination approach has been suggested earlier to yield the best classification as compared to individual methods alone27. Initially, a genome-scale molecular interaction network was rendered condition-specific by integrating transcriptome data. Next, we mined the networks to identify a shortlist of key components that would define the state of tumour, progression stages and key points of perturbation. We then used a machine learning method to derive different signatures with an optimal length to discriminate primary and metastatic melanoma, respectively. We then went on to validate the signature genes based on Melanoma specific survival (MSS) analysis from an independent cohort.
Results and Discussion
Biomarker identification strategy
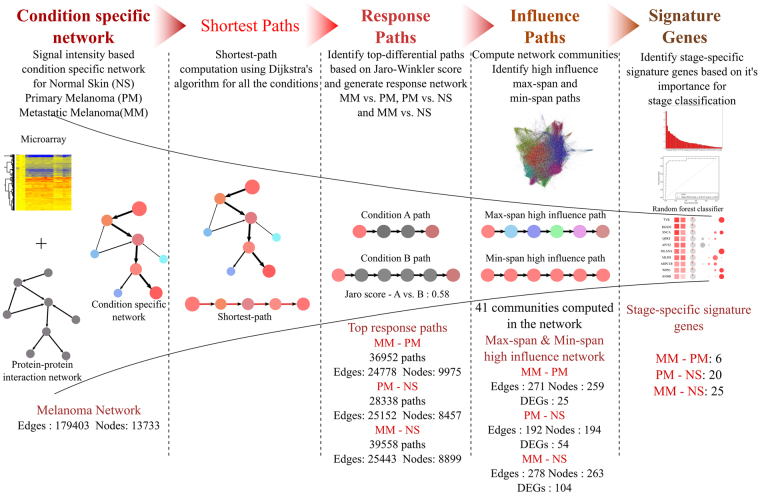
We configured a pipeline to identify RNA based biomarker candidates distinguishing metastatic melanoma from primary melanoma in an unbiased fashion using well established methods at each step. As illustrated in Fig. 1, the pipeline (a) begins with the reconstruction of knowledge-based protein-protein interaction networks. (b) These are then rendered condition-specific by weighing the network based on fitted signal intensity values resulting in three different networks for NS, PM and MM respectively, using methods previously established by us28,29. (c) The PM and MM networks were either compared with each other or compared with NS to generate disease response networks. For this, the highest activity paths were compared for a given pair of conditions and the top-ranked perturbed paths shortlisted for inclusion in further steps. The selected set of such paths in each comparison were found to be well connected with each other and hence form the corresponding response networks. (d) This is followed by identification of high-influence paths based on the paths impact on the network by constructing network communities using standard graph theory approaches. (e) Highest ranked influential paths and genes in them were then used as an input into a machine learning classifier that yields a final signature that discriminated metastatic from primary melanoma and predicts the risk of disease progression in primary melanoma.
Figure 1.
A schematic representation of the biomarker identification pipeline. The pipeline involves 5 major steps. Condition specific network: Construction of weighted network using protein-protein interaction network and gene expression data. Shortest Path analysis: Identification of all-vs.-all nodes shortest paths using Dijkstra’s algorithm. Response Paths: Paths with highest differential activity in diseased condition identified using string matching metric. Influence paths: Prioritizing paths based on influence on the network. Signature genes: Feature ranking of genes to obtain minimal set classifying conditions. The conditions considered for study: Normal Skin (NS), Primary Melanoma (PM) and Metastasis Melanoma (MM). Analysis results in numbers is shown in bottom section of the image
Among the publicly available transcriptome repositories for cutaneous melanoma, we initially selected a transcriptional profiling dataset that contained data for tissue samples of primary melanoma, metastatic melanoma along with adjacent normal skin10. We used this to identify biomarker signatures to distinguish between (a) metastatic and primary melanoma (b) metastatic melanoma and normal skin (c) primary melanoma and normal skin. The shortlist of possible biomarker candidates was obtained and evaluated for the performance of the signatures on an independent dataset for the first phase of validation and used it for pruning the candidate set, thereby deriving an optimal signature. For the next phase of fully independent validation, we evaluated the performance of the optimized biomarker panel using a large dataset from the Leeds Melanoma30 cohort for which survival information was available.
Response networks capture disease stage-specific variations in an unbiased fashion
We utilized a comprehensive master network of interactions between human proteins previously constructed in the laboratory28 (Methods). The master network comprises 13733 proteins (nodes) connected by 179403 interactions (edges) and includes both structural as well as functional interactions, belonging to several signaling, metabolic and regulatory processes, thus providing a global coverage of the human protein interactome. We rendered the master network condition-specific by weighting the individual nodes proportional to their respective fitted gene expression intensities from the transcriptomes of 46 and 12 samples of primary melanoma (PM) and metastatic melanoma (MM), respectively10. The dataset also contained 16 normal skin (NS) samples. A transcriptome comparison of MM vs. PM indicated 925 differentially expressed genes (DEGs, adjusted p-value ≤ 0.05, fold change ≥ 2). The same comparison for PM vs. NS is in the order of 2739 DEGs while that in MM vs. NS are 4262. A majority of DEGs (72% of MM vs. PM) were present in the initial network, indicating that the network has high coverage of the variations in melanoma and a similar trend was observed in other comparisons as well. From the three condition-specific networks reflecting conditions of NS, PM and MM, we obtained shortest paths by computing paths for all-vs.-all node pairs in each weighted network.
The paths abstracted as strings were compared (MM vs. PM, PM vs. NS, MM vs. NS), using a string similarity metric, that provided a measure of dissimilarity among the three conditions (see Methods). Highest scoring paths in each comparison reflect the set of highest perturbations in the network. We use the term ‘highest perturbations’ to describe the top ranked difference paths in the given pair of conditions (Supplementary Figure S1). A total of about 188 million paths were computed for each condition, of which about 19.5% paths of MM were unique to paths of PM. Similarly, 15% paths of PM and 21% paths of MM were unique when compared to NS. Higher the dissimilarity score, higher are the differential activities and hence the paths were sorted on this basis. We selected only the top ranked 0.001% of paths consisting of ~50% DEGs for further analysis amounting to 36952, 28338 and 39558 paths in the three comparisons MM vs. PM, PM vs. NS and MM vs. NS, respectively. A stringent threshold of 0.001% was used to obtain a shortlist containing sufficient number of promising candidates for taking them further in the pipeline, while minimising chances of false-positives. In each case, although only a small fraction of paths was selected, we observed that these paths form a well-connected subnet. The fact that they are connected subnets strongly suggests that the perturbations are not random in nature and appear to be orchestrated as a system’s response to melanoma. Thus these paths of highest perturbations in a MM vs PM comparison defines the systems’ response to progression of disease from a primary melanoma to a metastatic form and referred to as response paths. Likewise, the paths for PM vs NS and MM vs NS represent the systems’ response for primary melanoma with respect to normal skin or metastatic melanoma versus normal skin respectively. The response paths can also be viewed as highest differences in ‘flows’ in the network, where a ‘flow’ implies a transfer of effect through the path containing differentially regulated genes. The paths, in addition to differentially expressed genes, contain bridging genes that may be constitutively expressed at high levels, and also hub nodes that serve as the main link to multiple flows. The response networks were constructed using the response paths and consisted of 9975 (MM vs. PM), 8457 (PM vs. NS) and 8899 (MM vs. NS) nodes. The potential of such response networks to identify top perturbations and a common core in disease-specific networks has been explored previously28,29. A response network of PM vs. NS is shown in Fig. 2A. This exercise resulted in elimination of 49% of DEGs, resulting in a list of 472 DEGs between MM and PM for further processing. Similarly, in the other two comparisons 56% (PM vs. NS) and 53% (MM vs. NS) of DEGs were eliminated. A point to note is that the protein-protein interactome is likely to be incomplete, since many interactions may not even be characterized in any system and it is therefore possible to miss some promising DEGs at this step. This however is not a major limitation in our study, as our goal is to identify biomarkers with high discriminative power rather than evaluate all possible markers.
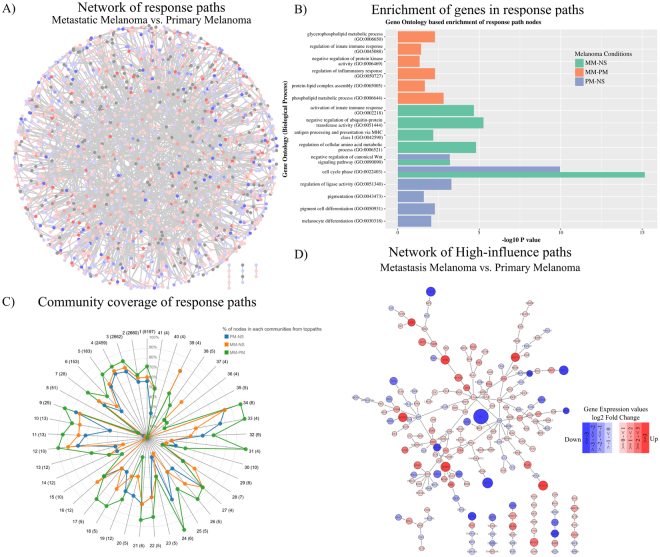
Figure 2.
(A) A network view of response paths identified using the Jaro-Winkler metric for the MM vs. PM comparison. (B) Functional enrichment of differentially regulated genes in top-response paths of MM vs. PM, MM vs. NS and PM vs. NS. (C) Percentage coverage of genes in 41 communities by genes of top-response paths from 3 comparisons. (D) A subnetwork of response paths of MM vs. PM prioritized based on influence score.
Functional enrichment analysis of the response networks
To gain insights about the functional categories of the genes in the response networks (foreground set), a gene enrichment analysis was carried out against all human genes (background set). The predominant biological processes of each of the response networks are illustrated in Fig. 2B (Supplementary Table S2). For the MM vs. PM set, several processes linked to metastasis such as the phospholipid metabolic process (P = 1.5 × 10−3), protein-lipid complex assembly (P = 2.1 × 10−2), regulation of inflammatory response (P = 5.1 × 10−3), negative regulation of protein kinase activity (P = 4.5 × 10−2) and regulation of innate immune response (P = 3.7 × 10−2) were enriched. On the other hand, the processes of melanocyte differentiation (P = 8.8 × 10−3), pigment cell differentiation (P = 5.2 × 10−3), pigmentation (P = 2.5 × 10−2), regulation of ligase activity (P = 5.08 × 10−4), and cell cycle phase (P = 1.9 × 10−10) are the most enriched processes in the PM vs. NS set. Overall, the enrichment analysis indicated that the cell cycle, immune process and processes related to metastasis are prominent in metastatic melanoma, whereas the pigmentation processes was predominantly only in the case of PM but not in MM, as reported earlier by Raskin et al.10 Another GO process, the negative regulation of the canonical Wnt signaling pathway (P = 6.4 × 10−4) was also present in both PM vs. NS and MM vs. NS enrichment. In addition, the metastatic condition has a high enrichment of genes related to lipid synthesis (P = 2.1 × 10−2), consistent with the report of Baenke et al. for various cancers31. SPP1 (osteopontin), a gene involved in melanoma invasion and tumour progression32 is increased by 8-fold in MM compared to PM and is present in MM vs. PM response paths. In an earlier work, we have reported that SPP1 differential expression increased hazard of death33. MITF, RAC1, PTEN and Jak-Stat pathway proteins (STAT1 and STAT3) are some of other proteins involved in invasive and metastatic behaviour of malignant melanoma that are part of top-response networks34.
Screening for High influence genes in the response networks
The genes in the response network were further prioritized based on the extent of influence they wielded in the whole network. The network communities are densely connected subnets of the whole network and are involved in performing similar or interrelated biological functions35,36. Functional perturbations to the nodes percolate effectively due to high connectedness within a community. Based on the network topology, we identified 41 communities in the master network. We tested if the nodes in each response network showed good coverage of the communities and observed that most communities (87%) were indeed well covered (Fig. 2C), and hence it was meaningful to use community-spanning to identify the most-influential nodes in the response networks. We then score the paths in each top-response paths based on the number of communities they span using two scoring schemes: (a) the paths consisting of nodes that belong to maximum communities - max-span paths (the top 1% of paths spanning the largest number of communities) and (b) the paths with nodes belonging to a single community - min-span paths (the top 1% of paths within a community). Max-span paths are enriched with differential genes across multi-function communities while min-span paths are enriched with genes that are most important within a community.
The next filter in the biomarker identification pipeline retains only the highly influential paths and eliminates the rest. For this, we computed an influence score (equation 1) for each node to capture the extent of its topological importance in the network and the gene expression variation in the given condition, and thus obtained a measure of a consolidated influence score (equation 2) for each path. Top 1% of max-span and min-span paths ranked based on consolidated influence score in each condition were selected and high-influence networks were built. 72 paths (271 edges, 259 nodes) make the high-influence network of MM vs. PM (Fig. 2D). Similarly, 56 paths (192 edges, 194 nodes) for PM vs. NS and 78 paths (278 edges, 263 nodes) for MM vs. NS form the high-influence networks (Supplementary Figure S3 and Table S3). 25 DEGs from the high-influence network were identified as possible candidates to discriminate MM from PM. Similarly, 54 and 104 DEGs were identified for PM vs. NS and MM vs. NS, respectively. These form the first version of the signature panels in each case (Supplementary Table S4).
Optimization of the panel length and performance evaluation
The signature genes were derived based on median expression values which are oblivious to the heterogeneity of the disease. Given that high extents of heterogeneity are typically observed among patients with the same clinical presentations, it becomes necessary to use a panel of genes. The next question therefore is to identify how many and which genes should constitute the panel to achieve high discrimination in multiple datasets. Towards this, the relative importance of each gene, when treated as a feature was computed in the present dataset (GSE15605). The feature ranking and the receiver operating curves (ROC) from a random forest classifier are shown in Supplementary Figure S4. KRT16, a regulator of innate immunity in the skin, significantly downregulated in MM was found to be the highest discriminator between MM and PM. ALDH1A1, IRX4, REST, WNT3A and SPRR3 were the other top ranked genes (full list of all condition comparisons in Supplementary Table S5). Further, we retained only those genes that showed consistent differential expression in another independent transcriptome dataset of 14 PM, 40 MM and 4 NS samples (GSE7553).
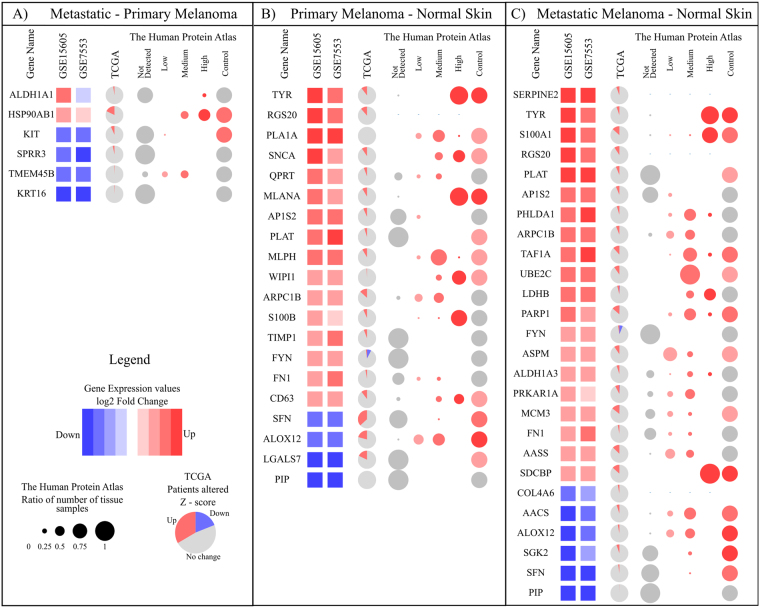
We thus identified a final panel of 6 genes (ALDH1A1, HSP90AB1, KIT, SPRR3, TMEM45B and KRT16) which achieved a classification of 87% for MM vs. PM, a panel of 20 genes achieved a classification of 95% for PM vs. NS and 96% by a panel of 25 genes for MM vs. NS. Figure 3 provides a comprehensive illustration of how each gene fared in the two datasets based on gene expression values. In addition, we compared the gene-expression fold change patterns for each gene with the available protein expression levels in melanoma tissue (no stage-specific data was available) and normal skin (Fig. 3), which showed reasonable agreement for many genes. The protein abundances were obtained from the human protein atlas, which were based on antibody staining of the melanoma tissue. The panel is intended as a RNA-signature and hence differential proteomic data is not directly relevant. However, understanding the trend in protein abundances can provide insights towards a mechanistic understanding of the role of the individual gene products, in disease progression and lend support for the selection of biomarkers. Of the 6 markers, ALDH1A1 (P = 4.3 × 10−7) and HSP90AB1 (P = 4 × 10−3) are upregulated in 75% and 83% of patients respectively, while the other 4 genes KIT (P = 2 × 10−3), SPRR3 (P = 3.2 × 10−6), TMEM45B (P = 4 × 10−3) and KRT16 (P = 3.2 × 10−6) are downregulated in around 80% patients. We compared the discriminatory power of our panel with that of a similar-sized panel identified without the use of networks, based on only machine learning approach (Supplementary information, Table S1 and Figure S2), which showed that the network based methods have a distinct advantage in identifying the best panel and also contains biologically meaningful genes.
Figure 3.
Final signature genes for 3 condition comparisons. (A) 6 genes of MM vs. PM (B) 20 genes of PM vs. NS (C) 25 of MM vs. NS. The first two columns after gene name show the differential expression level in cohort GSE15605 and GSE7553, respectively. Third column show Venn diagrams indicating percentage of patients, the gene is differentially regulated in TCGA based on z-score. In the human protein atlas section, first four columns show the antibody stain levels observed in melanoma tissue. The size of each circle is based on a ratio of the number of patients showing particular expression to the total patients and the colouring is based on the intensity of expression. The last column is stain intensity in control tissue.
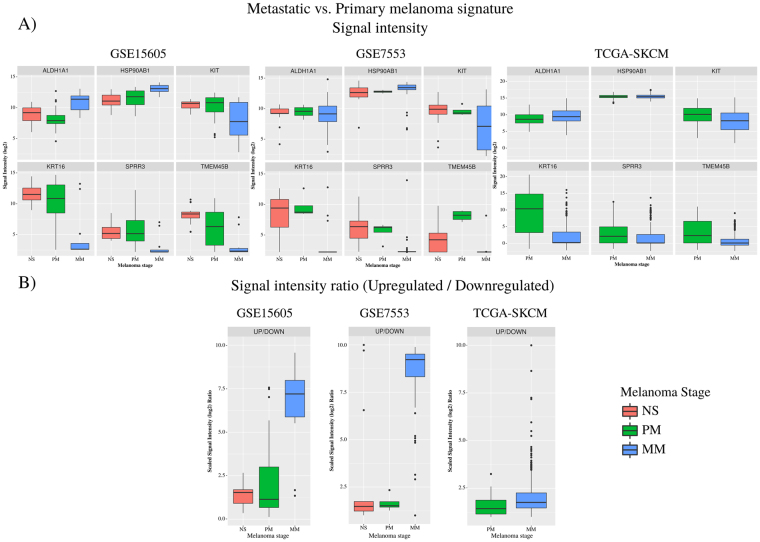
Figure 4A shows the log2 intensity values of these 6 genes for each condition in GSE15605, GSE7553 and TCGA. Figure 4B is ratio of upregulated genes expression product (HSP90AB1 and ALDH1A1) to the downregulated genes expression product (KIT, KRT16, SPRR3 and TMEM45B) among the 6 gene signature for the 3 cohorts and shows a good separation between the MM and PM. The combined effect size of the panel is seen to be very high in the first two datasets. A clear interpretation is difficult from the TCGA dataset, although the combined score is still higher in MM as compared to PM, because the dataset that is publicly accessible is a pool of samples of known primaries and metastatic samples of unknown primaries, and those collected from different tissues including from lymph nodes, but not individually annotated beyond the broad classification of ‘primary’ and ‘metastatic’ conditions’. The first two datasets on the other hand are more clearly annotated and the samples are all from the skin samples with known primaries.
Figure 4.
(A) log2 intensity values of the 6 genes for each condition in GSE15605, GSE7553 and TCGA. (B) The combined score which is computed as a ratio of product of the signal intensity (log2) of upregulated genes (HSP90AB1 and ALDH1A1) to the product of the signal intensity (log2) downregulated genes (KIT, KRT16, SPRR3 and TMEM45B).
Biological significance of the identified panel
To understand the significance of the identified genes, we first analysed how our signature fares with respect to expression of genes known to be differentially expressed in melanoma: tyrosinase (TYR), S100 family proteins, PMEL, MLANA, MITF, FN1, LDH, S100B, MIA and CSG4 37,38. From our analysis, we identified that 10 such genes are found either in our PM vs. NS or MM vs. NS signatures. We provide a full list of genes in Table 1, along with their functional categories and their role characterised in melanoma or other cancers. 16 genes in our signatures such as QPRT, ALOX12 and PIP are seen to be either known or potential markers of other cancers, but not previously identified in melanoma.
Table 1.
Biological significance of PM vs. NS and MM vs. NS signature.
| Gene symbol | Description | Association with cancer | GO biological process | Remarks | |||
|---|---|---|---|---|---|---|---|
| A | B | C | |||||
| Genes in PM vs. NS | |||||||
| TYR* | ↑ | Tyrosinase | ✓ | pigmentation | Well established biomarker of melanoma9 | ||
| RGS20* | ↑ | Regulator Of G-Protein Signaling 20 | ✓ | cell differentiation | Involved in cancer cell aggregation, migration, invasion and adhesion in other cancers51 | ||
| PLA1A | ↑ | Phospholipase A1 Member A | ✓ | metabolic process | Identified to be related to short survival of melanoma patients52. | ||
| SNCA | ↑ | Synuclein Alpha | ✓ | metabolic process | Protein of many diseases and also reported as biomarker of malignant melanoma53. | ||
| QPRT | ↑ | Quinolinate Phosphoribosyltransferase | ✓ | metabolic process | A potential marker for follicular thyroid carcinoma54. | ||
| MLANA | ↑ | Melan-A | ✓ | An established melanoma biomarker37 | |||
| AP1S2* | ↑ | Adaptor Related Protein Complex 1 Sigma 2 Subunit | ✓ | ✓ | intracellular protein transport | Upregulated in expression profile of 20 cancer types55 | |
| PLAT* | ↑ | Plasminogen Activator, Tissue Type | ✓ | cell mobility | Plasminogen activation system studied in uveal melanoma56 | ||
| MLPH | ↑ | Melanophilin | ✓ | intracellular protein transport | Differentially expressed in melanoma57 | ||
| WIPI1 | ↑ | WD Repeat Domain, Phosphoinositide Interacting 1 | ✓ | metabolic process | Coordinates Melanosome Formation and Melanogenic Gene Transcription58 | ||
| ARPC1B* | ↑ | Actin Related Protein 2/3 Complex Subunit 1B | ✓ | ✓ | cell mobility | Prediction marker for choroidal malignant melanoma and lung cancer59 | |
| S100B | ↑ | S100 Calcium Binding Protein B | ✓ | cell proliferation | An established melanoma biomarker37,60 | ||
| TIMP1 | ↑ | TIMP Metallopeptidase Inhibitor 1 | ✓ | cell proliferation | Timp1 interacts with CD63 to activate PI3-K signaling pathway in melanoma61,62 | ||
| CD63 | ↑ | CD63 Molecule | ✓ | cell mobility | |||
| FYN* | ↑ | FYN Proto-Oncogene, Src Family Tyrosine Kinase | ✓ | ✓ | cell mobility | Potential biomarker for melanoma and other cancers63,64. | |
| FN1* | ↑ | Fibronectin 1 | ✓ | cell mobility | Used in a diagnostic assay of metastatic melanoma65 | ||
| SFN* | ↓ | Stratifin | ✓ | ✓ | cell death | Downregulated in melanoma and other cancers12,66 | |
| ALOX12* | ↓ | Arachidonate 12-Lipoxygenase, 12S Type | ✓ | ✓ | skin development | Biomarker for prostate cancer and also downregulated in melanoma11,67 | |
| LGALS7 | ↓ | Galectin 7 | ✓ | ✓ | apoptotic process | Dual role observed in melanoma. Downregulation studied in cervical cancer and gastric cancer68–70 | |
| PIP* | ↓ | Prolactin Induced Protein | ✓ | regulation of immune system process | Biomarker for Breast Cancer71 | ||
| Genes in MM vs. NS | |||||||
| SERPINE2 | ↑ | Serpin Family E Member 2 | ✓ | cell differentiation | Therapeutic target for colorectal cancer52,72 | ||
| S100A1 | ↑ | S100 Calcium Binding Protein A1 | ✓ | cell proliferation | Established melanoma marker37 | ||
| PHLDA1 | ↑ | Pleckstrin Homology Like Domain Family A Member 1 | ✓ | cell differentiation | Expression involved in intestinal tumorigenesis73 | ||
| TAF1A | ↑ | TATA Box-Binding Protein-Associated Factor 1A | Regulation of transcription | ||||
| UBE2C | ↑ | Ubiquitin Conjugating Enzyme E2 C | ✓ | cell proliferation | Therapeutic target for melanoma74 | ||
| LDHB | ↑ | Lactate Dehydrogenase B | ✓ | metabolic process | Established biomarker of melanoma75 | ||
| PARP1 | ↑ | Poly(ADP-Ribose) Polymerase 1 | ✓ | cell differentiation | Associated with poor survival of melanoma patients76 | ||
| ASPM | ↑ | Abnormal Spindle Microtubule Assembly | ✓ | cell differentiation | Has a pro-invasion role in metastasis77 | ||
| ALDH1A3 | ↑ | Aldehyde Dehydrogenase 1 Family Member A3 | ✓ | metabolic process | Identified as marker and target of melanoma therapeutics78 | ||
| PRKAR1A | ↑ | Protein Kinase A Type 1a Regulatory Subunit | ✓ | cell differentiation | Overexpression studied in cholangiocarcinoma79 | ||
| MCM3 | ↑ | Minichromosome Maintenance Complex Component 3 | ✓ | metabolic process | Is a possible independent prognostic marker for melanoma80 | ||
| AASS | ↑ | Aminoadipate-Semialdehyde Synthase | ✓ | metabolic process | Is an oncogene81 | ||
| SDCBP | ↑ | Syndecan Binding Protein | ✓ | cell mobility | Involved in cancer development and progression82 | ||
| COL4A6 | ↓ | Collagen Type IV Alpha6 Chain | ✓ | ✓ | cell adhesion | Involved in aggressiveness and metastasis of melanoma and other cancers83 | |
| AACS | ↓ | Acetoacetyl-CoA Synthetase | ✓ | cell differentiation | Low expression studied in tumor tissues84 | ||
| SGK2 | ↓ | SGK2,Serine/ThreonineKinase 2 | ✓ | regulation of cell growth | Dowregulated in melanoma85 | ||
*Genes also present in MM vs. NS signature. A: Melanoma biomarker B: Studies related to melanoma C: Studies related to other cancers.
Of the 6 gene MM vs. PM panel, HSP90 (heat shock protein 90) is a well-known marker for melanoma and its expression increases with disease progression39, ALDH1A1 is also a previously suggested marker and also potential target to decrease growth, tumorigenicity and metastasis of melanoma40. SPRR family and Keratin family genes were described to be downregulated in metastatic melanoma as compared to primary melanoma41, consistent with the trend that we observe, for two members of the family, KRT16 and SPRR3. KIT, a downregulated gene in this set has been linked to disease progression and is also being explored as a therapeutic target42. Overall, as listed in Table 1, we observe that genes belonging to the following gene ontology categories are upregulated in the PM vs. NS and MM vs. NS panels, (a) pigmentation, (b) cell differentiation, (c) cell proliferation and cell mobility (d) metabolic processes, while some genes related to (e) cell death and (f) skin development are downregulated.
Assessing disease severity and prognosis in a retrospective study of 703 primary melanoma samples from the Leeds melanoma Cohort
To validate the significance of the MM vs. PM signature, expressions of key genes from the identified network were analysed in the Leeds Melanoma cohort (703 primary tumours) to assess their individual and joint effect on melanoma-specific survival (MSS) as well as their association with melanoma histological characteristics: AJCC stage, Breslow thickness, ulceration and mitotic rate (See Methods).
Melanoma specific survival analysis (MSS)
In a univariable Cox model, elevated HSP90AB1 expression significantly predicted increased hazard of dying from melanoma (HR = 1.9, P = 0.0002) while higher expression of KRT16, KIT and TMEM45B reduced the death hazard (HR = 0.9 and P ≤ 0.05 for all three) (see Table 2). In unadjusted multivariable analysis three genes showed independent effects: HSP90AB1, KRT16 and SPRR3 (Table 2). In multivariable analysis adjusted for sex, tumour site, age at diagnosis and AJCC stage, only HSP90AB1 remained significant with unchanged death hazard ratio estimate (HR = 2.0, P = 10−4, see Table 2).
Table 2.
Hazard ratios for MSS for individual genes in the whole dataset&.
| Gene | Univariable MSS | Multivariable unadjusted MSS | Multivariable adjusted MSS | |||
|---|---|---|---|---|---|---|
| HR | P value | HR | P value | HR | P value | |
| ALDH1A1 | 0.9 | 0.1 | 0.9 | 0.2 | 0.9 | 0.3 |
| HSP90AB1 | 1.9 | 2 × 10−4 | 1.7 | 0.002 | 2.0 | 10−4 |
| KIT | 0.9 | 0.05 | 0.9 | 0.3 | 1.0 | 0.2 |
| SPRR3 | 1.03 | 0.5 | 1.1 | 0.03 | 1.0 | 0.5 |
| TMEM45B | 0.9 | 0.005 | 0.96 | 0.4 | 1.0 | 0.5 |
| KRT16 | 0.9 | 0.001 | 0.93 | 0.04 | 0.9 | 0.07 |
&Death hazard ratio (HR) reflects the change from the baseline of 1.0 each time the gene expression is doubled.
Association with melanoma histology
The 4 genes that were associated with MSS in univariable analysis (Table 2) were also significantly correlated with ulceration, mitotic rate and Breslow thickness, and concordantly, AJCC stage (see Table 3). KIT, KRT16 and THEN45B are known to be expressed by normal skin appendages or stromal tissue, and hence it is possible that the differential expression of these genes in primary compared with metastatic tissue may represent sampling of those normal tissues in primary disease. Among these 4 genes, as expected, expression of HSP90AB1 increased with tumour thickness and higher mitotic rate, while the other 3 were negatively correlated.
Table 3.
Association between each gene and histological features of melanoma.
| Gene | AJCC (Pvalue) | Ulceration (Pvalue) | Mitotic rate correlation (P-value) | Breslow thickness correlation(P-value) |
|---|---|---|---|---|
| ALDH1A1 | 0.4 | 0.4 | −0.04 (0.4) | −0.01(0.7) |
| HSP90AB1 | 0.03 | 9 × 10−4 | 0.13 (0.001) | 0.2 (1.6 × 10−5) |
| KIT | 5 × 10−5 | 10−5 | −0.11 (0.005) | −0.2 (2.3 × 10−10) |
| SPRR3 | 0.7 | 0.6 | −0.08 (0.06) | −0.06 (0.1) |
| TMEM45B | 5.5 × 10−13 | 2.2 × 10−11 | −0.2 (7.9 × 10−8) | −0.3 (3.1 × 10−16) |
| KRT16 | 4.8 × 10−12 | 2 × 10−5 | −0.2 (3.1 × 10−7) | −0.3, (1.3 × 10−20) |
Composite gene expression score and MSS
A composite score was created using expressions of ALDH1A1, HSP90AB1, KIT, SPRR3, TMEM45B, KRT16 in the training dataset (random 2/3 of the total dataset, see Methods). When dichotomized on median and applied to the test data (remaining 1/3 of the data), higher values of score predicted worse prognosis with HR = 2.3, P = 0.003 (Fig. 5), remaining prognostic upon adjustment of sex, tumour site, age at diagnosis and AJCC stage with HR = 2.0, P = 0.01.
Figure 5.
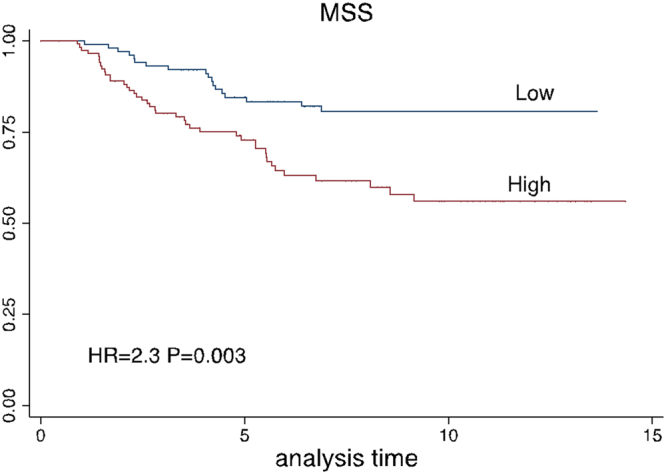
Survival curves according to the combined 6-gene score (unadjusted) in test data (1/3 of total sample). The score was dichotomised by the median.
Composite gene expression score and tumour histology
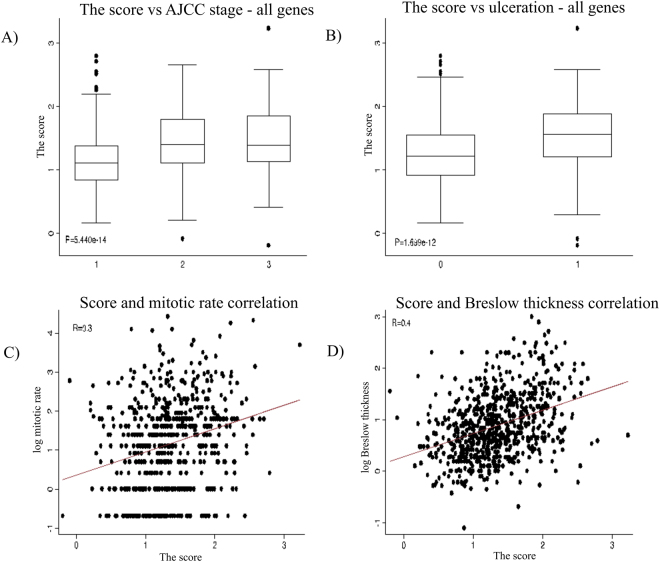
The score was significantly lower in samples derived from patients with stage 1 (P = 5.4 × 10−14) but there was no difference between stages 2 and 3 (Fig. 6A). Tumours with the higher scores were more likely to be ulcerated (P = 1.7 × 10−12, Fig. 6B). Mitotic rate and Breslow thickness positively correlated with the score (see Fig. 6C and D).
Figure 6.
The 6-gene score distribution by AJCC stage (A), ulceration status (B), mitotic rate (C) and Breslow thickness (D). Note the log scale for mitotic rate and Breslow thickness.
The scores obtained by removing up to 3 genes (firstly ALDH1A1, then ALDH1A1 and KIT and lastly these two and TMEM45B) did not significantly change the MSS results (Supplementary Figure S5). All the scores were comparable to the initial score and remained significant after adjustment. The score utilizing the three genes (HSP90AB1, SPRR3 and KRT16) was shown to be as strong as the initial 6-gene score in terms of predicting MSS.
Because KRT16 is highly expressed in the epidermis and its expression may not be entirely from tumours in our data, we further eliminated it and recalculated a score combining only the 2 remaining genes (HSP90AB1, SPRR3). This new score remained associated with MSS in the test data (Supplementary Figure S5D).
Thus, from the survival analysis, HSP90AB1 expression significantly predicted reduced survival (HR = 1.9, P = 2 × 10−4 in multivariable analysis and the result remained significant after the adjustment of confounders (HR = 2, P = 10−4). Expression of this gene was associated with higher likelihood of ulceration (P = 9 × 10−4), higher AJCC stage (P = 0.03) and it was positively correlated with mitotic rate (R = 0.13) and Breslow thickness (R = 0.2), which is concordant with MSS results.
Higher fold change (downregulated) of KIT, KRT16 and TMEM45B predicted better prognosis in univariable analysis, however the result did not remain significant in multivariable analysis adjusting confounders (Table 2). The expression of those genes negatively correlated with ulceration, thickness, mitotic rate and ultimately AJCC stage, which is consistent with positive prognostic value. The lack of an independent effect of these genes on MSS is explained by this correlation with these other prognostic tumour characteristics. Although HSP90AB1 correlated with these tumour characteristics as well (see Table 3), its residual effect on MSS remained significant, suggesting a putatively more potent role.
From the second approach, we see that the 6-gene score trained in 2/3 of the data was a strong predictor of MSS in the remaining 1/3, independent of AJCC stage; hence it might be explored as a prognostic biomarker. This independent prognostic effect was observed in spite of the 6-gene score being also associated with classical melanoma prognostic factors (AJCC stage, ulceration, mitotic rate and Breslow thickness). Interestingly, even after eliminating 3 of the 6 genes, the new 3-gene score (HSP90AB1, SPRR3, KRT16) remained strongly predictive of MSS. This suggests that 3 of the 6 genes identified in the protein network analysis may play a key role in melanoma progression. We note that the combined effect of the 3 genes is roughly similar to that of HSP90AB1 alone, which is consistent with the results from multivariable analysis which singled out this gene as the only significant when melanoma characteristics are adjusted (Table 2). Therefore, the results from our two analysis approaches highlight the importance of HSP90AB1 in progression of melanoma.
Conclusions
We developed a pipeline that combines a network approach with machine learning, through which we identified a biomarker signature capable of discriminating metastatic from primary melanoma tumours. The approach is based on constructing condition-specific genome-wide molecular interaction networks that are specific to each condition and subsequently mining the networks to identify nodes most influential in differentiating between disease stages. The signature genes identified by this network approach have been previously suggested as melanoma markers. In addition, our approach also identifies new potential markers. For many of these, there are studies reported in literature, supporting their roles in the pathophysiology. The discriminatory signature between MM and PM comprises a panel of 6 genes, which exhibit a 6 to 7 fold difference in their combined score between MM and PM. Melanoma specific survival (MSS) analysis for these 6 genes showed 3 genes HSP90AB1, SPRR3 and KRT16 to be strongly predictive of survival, of which HSP90AB1 by itself remained significant for predicting risk of disease progression, even after adjusting for confounding variables and hence has an added prognostic value. In addition to the 6-gene panel, our approach also identified two panels of 20 and 25 genes that can discriminate PM from NS and MM from NS, respectively.
Materials and Methods
Datasets
Microarray datasets (i) GSE15605, that contain expression profiles of 16 normal skin, 46 primary melanoma and 12 metastatic melanoma samples10, and (ii) GSE7553, that contains expression profiles of 14 primary melanomas, 40 metastases, taken from tumor samples from patients and 4 normal skin samples as controls12, were obtained from the NCBI Gene Expression Omnibus (GEO) and used for the discovery phase. Additional datasets used for validation are: (iii) Transcriptomic data of 703 primary melanoma patients from the Leeds Melanoma Cohort generated from formalin fixed primaries using the Illumina DASL array (iv) TCGA dataset – 104 primary melanoma and 367 metastatic melanoma, as available through the cBio Cancer Genomics Portal43, and (v) The Human Protein Atlas44 containing measurements of proteins based on antibody staining from a few melanoma patients.
Leeds-cohort: 2184 participants with primary melanoma were recruited to the Leeds Melanoma Cohort. As previously described, whole genome transcriptomes were generated from formalin fixed samples taken using a tissue microarray needle14. Normalization and analysis was carried out in Leeds as described previously.
Transcriptome analysis
The microarray analysis was carried out using Bioconductor-R (http://www.bioconductor.org/). The raw intensity values for each tissue sample were normalized using the method GCRMA in Bioconductor package affy. eBayes function was used to identify differential gene expression on linear fitted model generated by lmFit from the package LIMMA. The P-values obtained were adjusted for multiple tests using Benjamini-Hochberg false discovery rate method. Genes with adjusted P-value ≤ 0.05 and a fold change of ±2 were considered as differentially expressed genes (DEGs).
Protein-protein interaction network
A master human protein-protein interaction network curated earlier in the laboratory28 was used. The master network, where proteins were considered as nodes and interactions as edges, consists of 17015 nodes and 200361 edges, of which nearly 80% were directed edges and the rest taken as bi-directional edges (Supplementary information). The available transcriptome data for melanoma GSE15605, mapped onto 21209 genes and finally the corresponding protein-protein interaction network of melanoma genes consisted of 13733 proteins and 179403 interactions.
Construction of condition-specific interaction networks
The network was rendered condition-specific by integrating it with the gene-expression profile of that condition. The nodes in the network were assigned weights based on the fitted normalized signal intensity values of all genes of NS, PM and MM condition, thus obtaining three networks. The edge between two nodes was weighted as the inverse of the product of the node weights making it compliant with Dijkstra’s algorithm.
Identification of response paths for each comparison
Response paths are the paths that are highly perturbed between two conditions. Between all-vs-all nodes, high-activity paths were computed using Dijkstra’s algorithm implemented in python-igraph45, on condition-specific interaction network of both conditions. The high-activity paths between any two nodes (source and target) in a network were modelled as the linear combination of genes through which information flows with minimal resistance. A path between two nodes is termed ‘perturbed’ if the nodes used to transmit information between the source and target was altered between the two conditions being compared. The paths were considered as strings and the path deregulation was captured using the Jaro-Winkler (JW) distance, a string matching metric46 (Supplementary File). The Jaro-Winkler score was normalized between 0 and 1 with 0 indicating an exact match. The top 0.001% perturbed (dissimilar) paths between two conditions were selected using this metric and considered further as response paths.
Functional enrichment analysis was carried out using web-based tool PANTHER – Protein Analysis Through Evolutionary Relationships. The P-values are FDR corrected using Bonferroni correction and considered significant if adjusted P-value < 0.0547.
Identification of high-influence paths
Identification of paths that have the highest influence in the network involved two steps, the first to detect communities or clusters in the network and the second to compute the influence of paths based on the span of these paths across communities and influence wielded by each node in these paths on the entire network.
Community detection for the network
Communities were computed using an unweighted, master PPI network to identify the span of paths and reduce the total number of paths based on their efficiency to percolate effect of differential expression. For detecting communities, the Fast-greedy algorithm48 implemented in igraph45 was used as it is proven to reflect biological network properties efficiently over other community detection methods49. A minimal node size ≥4 was imposed to consider a community, which yielded 41 communities, which were taken through further steps in the pipeline.
Max-span and Min-span high influence paths
The response paths overlapping on maximum communities were classified as max-span paths, which reflect high inter-community influence and the response paths assigned to a single community were classified as min-span paths and reflect high intra-community influence. Further, an influence score was computed for each node in the max-span and min-span paths. The score is a combination of the differential expression of the node and its topological position in the network. The influence score of node v is given as:
| 1 |
Where,
DC (Degree conserved), E (eccentricity) and BC (betweeness centrality) values were computed using functions in python-igraph (See supplementary file).
Consolidated Influence score
After obtaining an influence score of each node in the max-span and min-span paths, a consolidated influence score was computed for each path.
| 2 |
Where, n = number of nodes in path P.
It is the sum of influence scores of all nodes in the path, normalized by the path length. The max-span paths and min-span paths with high score were finally shortlisted to generate a high-influence network for each condition. The DEGs from these high influence networks were further validated for their ability to classify the conditions in a larger dataset.
Identifying discriminatory genes
From the set of high influence nodes, those that are the most discriminatory between the two conditions in a comparison are then identified. This module involves two steps, the first determines the feature importance and the second prunes the list to finally identify a signature with the best classifying power and the least length.
Feature Importance
The ‘extra trees’ classifier was employed to rank important features. The number of estimates was set to 500 while the class weights were automatically assigned so that proportional weights were given to undersampled/oversampled class labels. The criteria for selection was based on entropy (i.e. information gain) and the maximum number of features selected for finding the best split was taken as the sqrt of n_features. The maximum depth was disabled and the nodes were expanded until all leaves are pure.
Classification accuracy of the final signature
An AdaBoost classifier wrapper was used around the random forest algorithm to compute the classification accuracy of the signature gene set. The number of estimators was set to 500 with a learning rate of 0.08 and the criterion for selection was based on entropy. The boosting function is implemented using the Stagewise Additive Modeling with a Multiclass Exponential loss function (SAMME)50. A stratified K-fold was used for cross validation (see supplementary file).
Validation in the Leeds Melanoma Cohort
Three types of analysis were performed in transcriptomic data in the Leeds Melanoma Cohort using the top ranked network genes:
Univariable and multivariable association with melanoma-specific survival (MSS) in a Cox proportional hazards regression, adjusted and unadjusted for patient age at diagnosis, gender, AJCC stage and tumour site.
Univariable association of each gene with melanoma histological features using the Kruskall-Wallis test for the categorical variables (AJCC stage, ulceration) and the Spearman correlation coefficient for continuous variables (Breslow thickness and mitotic rate).
A composite gene expression score was created by calculating a weighted sum of the expression of each gene. The weight was the univariable log hazard-ratio from MSS analysis in a random selection of 2/3 of the dataset (training set). This score was then applied to the remaining 1/3 (test set) to assess its prediction of MSS and association with tumour histology (AJCC, ulceration, Breslow thickness and mitotic rate). The score’s stability was assessed by removing one by one the genes that had earlier shown the smallest independent effect on MSS in multivariable analyses.
Electronic supplementary material
Acknowledgements
We acknowledge financial support from the Department of Biotechnology, Govt. of India and partial support from Department of Science & Technology, Government of India. The University of Leeds work was supported by Cancer Research UK Programme grant C588/A19167 and Project Grant C8216/A6129 and NIH Award CA83115. JP has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 641458.
Author Contributions
R.M. analysed the gene expression data, formulated and carried out network analysis. A.M. developed method for identification of response network and carried out machine learning analysis. N.C. conceptualized the pipeline and supervised all the analysis. J.P. and J.N. conducted analyses of the Leeds Melanoma Cohort transcriptomic data under supervision of J.N.-B. and T.B. C.M.P. provided valuable inputs for the project. All authors read and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17330-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Forschner A, et al. Melanoma staging: facts and controversies. Clin Dermatol. 2010;28:275–80. doi: 10.1016/j.clindermatol.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Macdonald JB, et al. Malignant melanoma in the elderly: different regional disease and poorer prognosis. J Cancer. 2011;2:538–43. doi: 10.7150/jca.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Skin Cancers. Available at: http://www.who.int/uv/faq/skincancer/en/index1.html (2016).
- 4.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 5.Joosse A, et al. Gender differences in melanoma survival: female patients have a decreased risk of metastasis. J Invest Dermatol. 2011;131:719–26. doi: 10.1038/jid.2010.354. [DOI] [PubMed] [Google Scholar]
- 6.Larkin J, Hodi FS, Wolchok JD. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373:1270–1. doi: 10.1056/NEJMoa1504030. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas, N. Genomic classification of cutaneous melanoma. Cell161, 1681–1696 %@ 0092–8674 (2015). [DOI] [PMC free article] [PubMed]
- 8.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quaglino P, et al. Prognostic relevance of baseline and sequential peripheral blood tyrosinase expression in 200 consecutive advanced metastatic melanoma patients. Melanoma Res. 2007;17:75–82. doi: 10.1097/CMR.0b013e328054c667. [DOI] [PubMed] [Google Scholar]
- 10.Raskin L, et al. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. J Invest Dermatol. 2013;133:2585–92. doi: 10.1038/jid.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill R, et al. TRIB2 as a biomarker for diagnosis and progression of melanoma. Carcinogenesis. 2015;36:469–77. doi: 10.1093/carcin/bgv002. [DOI] [PubMed] [Google Scholar]
- 12.Riker AI, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haqq C, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci UA. 2005;102:6092–7. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nsengimana J, et al. Independent replication of a melanoma subtype gene signature and evaluation of its prognostic value and biological correlates in a population cohort. Oncotarget. 2015;6:11683–93. doi: 10.18632/oncotarget.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbst K, et al. Molecular profiling reveals low- and high-grade forms of primary melanoma. Clin Cancer Res. 2012;18:4026–36. doi: 10.1158/1078-0432.CCR-12-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emmert-Streib F, et al. Collectives of diagnostic biomarkers identify high-risk subpopulations of hematuria patients: exploiting heterogeneity in large-scale biomarker data. BMC Med. 2013;11:12. doi: 10.1186/1741-7015-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrauder MG, et al. Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS One. 2012;7:e29770. doi: 10.1371/journal.pone.0029770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Z, Li J, Xiong Y, Xu W, Zheng G. Identification of candidate colon cancer biomarkers by applying a random forest approach on microarray data. Oncol Rep. 2012;28:1036–42. doi: 10.3892/or.2012.2055. [DOI] [PubMed] [Google Scholar]
- 19.Tung CW, et al. Identification of biomarkers for esophageal squamous cell carcinoma using feature selection and decision tree methods. ScientificWorldJournal. 2013;2013:782031. doi: 10.1155/2013/782031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, et al. Combining multiple serum tumor markers improves detection of stage I epithelial ovarian cancer. Gynecol Oncol. 2007;107:526–31. doi: 10.1016/j.ygyno.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Peng Y, Tobin DJ. A new 12-gene diagnostic biomarker signature of melanoma revealed by integrated microarray analysis. PeerJ. 2013;1:e49. doi: 10.7717/peerj.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YC, Chen BS. A network-based biomarker approach for molecular investigation and diagnosis of lung cancer. BMC Med Genomics. 2011;4:2. doi: 10.1186/1755-8794-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang L, et al. A network biology approach to discover the molecular biomarker associated with hepatocellular carcinoma. Biomed Res Int. 2014;2014:278956. doi: 10.1155/2014/278956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Gene selection from microarray data for cancer classification–a machine learning approach. Comput Biol Chem. 2005;29:37–46. doi: 10.1016/j.compbiolchem.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Glaab E, Bacardit J, Garibaldi JM, Krasnogor N. Using rule-based machine learning for candidate disease gene prioritization and sample classification of cancer gene expression data. PLoS One. 2012;7:e39932. doi: 10.1371/journal.pone.0039932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azencott, C.-A. In Machine Learning for Health Informatics (ed. Holzinger, A.) 9605, 319–336 (Springer International Publishing, 2016).
- 28.Sambarey A, et al. Meta-analysis of host response networks identifies a common core in tuberculosis. NPJ Syst. Biol. Appl. 2017;3:4. doi: 10.1038/s41540-017-0005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambarey, A., Prashanthi, K. & Chandra, N. Mining large-scale response networks reveals ‘topmost activities’ in Mycobacterium tuberculosis infection. Sci. Rep. 3 (2013). [DOI] [PMC free article] [PubMed]
- 30.Newton-Bishop JA, et al. Serum 25-hydroxyvitamin D3 levels are associated with breslow thickness at presentation and survival from melanoma. J Clin Oncol. 2009;27:5439–44. doi: 10.1200/JCO.2009.22.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Model Mech. 2013;6:1353–63. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, et al. Osteopontin expression correlates with melanoma invasion. J Invest Dermatol. 2005;124:1044–52. doi: 10.1111/j.0022-202X.2005.23680.x. [DOI] [PubMed] [Google Scholar]
- 33.Conway C, et al. Gene expression profiling of paraffin-embedded primary melanoma using the DASL assay identifies increased osteopontin expression as predictive of reduced relapse-free survival. Clin Cancer Res. 2009;15:6939–46. doi: 10.1158/1078-0432.CCR-09-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orgaz JL, Sanz-Moreno V. Emerging molecular targets in melanoma invasion and metastasis. Pigment Cell Melanoma Res. 2012;26:39–57. doi: 10.1111/pcmr.12041. [DOI] [PubMed] [Google Scholar]
- 35.Lewis AC, Jones NS, Porter MA, Deane CM. The function of communities in protein interaction networks at multiple scales. BMC Syst Biol. 2010;4:100. doi: 10.1186/1752-0509-4-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rives AW, Galitski T. Modular organization of cellular networks. Proc Natl Acad Sci UA. 2003;100:1128–33. doi: 10.1073/pnas.0237338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinstein D, Leininger J, Hamby C, Safai B. Diagnostic and prognostic biomarkers in melanoma. J Clin Aesthet Dermatol. 2014;7:13–24. [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer SR, Erickson LA, Ichetovkin I, Knauer DJ, Markovic SN. Circulating serologic and molecular biomarkers in malignant melanoma. Mayo Clin Proc. 2011;86:981–90. doi: 10.4065/mcp.2011.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy MM, et al. HSP90 as a marker of progression in melanoma. Ann Oncol. 2008;19:590–4. doi: 10.1093/annonc/mdm545. [DOI] [PubMed] [Google Scholar]
- 40.Yue L, et al. Targeting ALDH1 to decrease tumorigenicity, growth and metastasis of human melanoma. Melanoma Res. 2015;25:138–48. doi: 10.1097/CMR.0000000000000144. [DOI] [PubMed] [Google Scholar]
- 41.Kavak E, Unlu M, Nister M, Koman A. Meta-analysis of cancer gene expression signatures reveals new cancer genes, SAGE tags and tumor associated regions of co-regulation. Nucleic Acids Res. 2010;38:7008–21. doi: 10.1093/nar/gkq574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slipicevic A, Herlyn M. KIT in melanoma: many shades of gray. J Invest Dermatol. 2015;135:337–8. doi: 10.1038/jid.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 45.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695:1–9. [Google Scholar]
- 46.Winkler W. E. String Comparator Metrics and Enhanced Decision Rules in the Fellegi-Sunter Model of Record Linkage. (1990).
- 47.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clauset A, Newman MEJ, Moore C. Finding community structure in very large networks. Phys. Rev. E. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 49.Tripathi, S., Moutari, S., Dehmer, M. & Emmert-Streib, F. Comparison of module detection algorithms in protein networks and investigation of the biological meaning of predicted modules. BMC Bioinformatics17, (2016). [DOI] [PMC free article] [PubMed]
- 50.Zhu J, Zou H, Rosset S, Hastie T. Multi-class adaboost. Stat. Interface. 2009;2:349–360. doi: 10.4310/SII.2009.v2.n3.a8. [DOI] [Google Scholar]
- 51.Yang L, Lee MM, Leung MM, Wong YH. Regulator of G protein signaling 20 enhances cancer cell aggregation, migration, invasion and adhesion. Cell Signal. 2016;28:1663–72. doi: 10.1016/j.cellsig.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 52.Eriksson J, et al. Gene expression analyses of primary melanomas reveal CTHRC1 as an important player in melanoma progression. Oncotarget. 2016;7:15065–92. doi: 10.18632/oncotarget.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welinder C, et al. Analysis of alpha-synuclein in malignant melanoma - development of a SRM quantification assay. PLoS One. 2014;9:e110804. doi: 10.1371/journal.pone.0110804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinsch N, Frank M, Doring C, Vorlander C, Hansmann ML. QPRT: a potential marker for follicular thyroid carcinoma including minimal invasive variant; a gene expression, RNA and immunohistochemical study. BMC Cancer. 2009;9:93. doi: 10.1186/1471-2407-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Y, et al. Common human cancer genes discovered by integrated gene-expression analysis. PLoS One. 2007;2:e1149. doi: 10.1371/journal.pone.0001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Vries TJ, et al. Components of the plasminogen activation system in uveal melanoma–a clinico-pathological study. J Pathol. 1995;175:59–67. doi: 10.1002/path.1711750110. [DOI] [PubMed] [Google Scholar]
- 57.Soikkeli J, et al. Systematic search for the best gene expression markers for melanoma micrometastasis detection. J Pathol. 2007;213:180–9. doi: 10.1002/path.2229. [DOI] [PubMed] [Google Scholar]
- 58.Ho H, Kapadia R, Al-Tahan S, Ahmad S, Ganesan AK. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J Biol Chem. 2011;286:12509–23. doi: 10.1074/jbc.M110.200543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumagai K, et al. Arpc1b gene is a candidate prediction marker for choroidal malignant melanomas sensitive to radiotherapy. Invest Ophthalmol Vis Sci. 2006;47:2300–4. doi: 10.1167/iovs.05-0810. [DOI] [PubMed] [Google Scholar]
- 60.Harpio R, Einarsson R. S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma. Clin Biochem. 2004;37:512–8. doi: 10.1016/j.clinbiochem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 61.Lugowska I, et al. Serum markers in early-stage and locally advanced melanoma. Tumour Biol. 2015;36:8277–85. doi: 10.1007/s13277-015-3564-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toricelli M, Melo FH, Peres GB, Silva DC, Jasiulionis MG. Timp1 interacts with beta-1 integrin and CD63 along melanoma genesis and confers anoikis resistance by activating PI3-K signaling pathway independently of Akt phosphorylation. Mol Cancer. 2013;12:22. doi: 10.1186/1476-4598-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang C, Sheng Y, Jia J, Chen L. Identification of melanoma biomarkers based on network modules by integrating the human signaling network with microarrays. J Cancer Res Ther. 2014;10(Suppl):C114–24. doi: 10.4103/0973-1482.145816. [DOI] [PubMed] [Google Scholar]
- 64.Fleuren ED, Zhang L, Wu J, Daly RJ. The kinome ‘at large’ in cancer. Nat Rev Cancer. 2016;16:83–98. doi: 10.1038/nrc.2015.18. [DOI] [PubMed] [Google Scholar]
- 65.Kashani-Sabet M, et al. A multi-marker assay to distinguish malignant melanomas from benign nevi. Proc Natl Acad Sci U A. 2009;106:6268–72. doi: 10.1073/pnas.0901185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jaeger J, et al. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;13:806–15. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 67.Filella X, Foj L. Emerging biomarkers in the detection and prognosis of prostate cancer. Clin Chem Lab Med. 2015;53:963–73. doi: 10.1515/cclm-2014-0988. [DOI] [PubMed] [Google Scholar]
- 68.Biron-Pain K, Grosset AA, Poirier F, Gaboury L, St-Pierre Y. Expression and functions of galectin-7 in human and murine melanomas. PLoS One. 2013;8:e63307. doi: 10.1371/journal.pone.0063307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Higareda-Almaraz JC, et al. Systems-level effects of ectopic galectin-7 reconstitution in cervical cancer and its microenvironment. BMC Cancer. 2016;16:680. doi: 10.1186/s12885-016-2700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim SJ, Hwang JA, Ro JY, Lee YS, Chun KH. Galectin-7 is epigenetically-regulated tumor suppressor in gastric cancer. Oncotarget. 2013;4:1461–71. doi: 10.18632/oncotarget.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ihedioha OC, Shiu RP, Uzonna JE, Myal Y. Prolactin-Inducible Protein: From Breast Cancer Biomarker to Immune Modulator-Novel Insights from Knockout Mice. DNA Cell Biol. 2016;35:537–541. doi: 10.1089/dna.2016.3472. [DOI] [PubMed] [Google Scholar]
- 72.Bergeron S, et al. The serine protease inhibitor serpinE2 is a novel target of ERK signaling involved in human colorectal tumorigenesis. Mol Cancer. 2010;9:271. doi: 10.1186/1476-4598-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakthianandeswaren A, et al. PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res. 2011;71:3709–19. doi: 10.1158/0008-5472.CAN-10-2342. [DOI] [PubMed] [Google Scholar]
- 74.Hong JJ, Gong K, Kaufman D, Chen H, Essner R. Abstract B026: Ubiquitin-conjugating enzyme E2C: a potential therapeutic target for primary and metastatic melanoma by microarray gene expression. Cancer Immunol. Res. 2016;4:B026–B026. doi: 10.1158/2326-6074.CRICIMTEATIAACR15-B026. [DOI] [Google Scholar]
- 75.Ho J, et al. Importance of glycolysis and oxidative phosphorylation in advanced melanoma. Mol Cancer. 2012;11:76. doi: 10.1186/1476-4598-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies JR, et al. Inherited variation in the PARP1 gene and survival from melanoma. Int J Cancer. 2014;135:1625–33. doi: 10.1002/ijc.28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kabbarah O, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5:e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luo Y, et al. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30:2100–13. doi: 10.1002/stem.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Loilome W, et al. PRKAR1A is overexpressed and represents a possible therapeutic target in human cholangiocarcinoma. Int J Cancer. 2011;129:34–44. doi: 10.1002/ijc.25646. [DOI] [PubMed] [Google Scholar]
- 80.Nodin B, et al. High MCM3 expression is an independent biomarker of poor prognosis and correlates with reduced RBM3 expression in a prospective cohort of malignant melanoma. Diagn Pathol. 2012;7:82. doi: 10.1186/1746-1596-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:D721–6. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Philley JV, Kannan A, Dasgupta S. MDA-9/Syntenin Control. J Cell Physiol. 2016;231:545–50. doi: 10.1002/jcp.25136. [DOI] [PubMed] [Google Scholar]
- 83.Pasco S, Brassart B, Ramont L, Maquart FX, Monboisse JC. Control of melanoma cell invasion by type IV collagen. Cancer Detect Prev. 2005;29:260–6. doi: 10.1016/j.cdp.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 84.Tisdale MJ. Role of acetoacetyl-CoA synthetase in acetoacetate utilization by tumor cells. Cancer Biochem Biophys. 1984;7:101–7. [PubMed] [Google Scholar]
- 85.Capra M, et al. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res. 2006;66:8147–54. doi: 10.1158/0008-5472.CAN-05-3489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.