Abstract
Host genotype may be closely related to the different outcomes of Hepatitis B virus (HBV) infection. To identify the association of variants and HBV infection, we comprehensively investigated the cytokine- and immune-related gene mutations in patients with HBV associated hepatocellular carcinoma (HBV-HCC). Fifty-three HBV-HCC patients, 53 self-healing cases (SH) with HBV infection history and 53 healthy controls (HCs) were recruited, the whole exon region of 404 genes were sequenced at >900× depth. Comprehensive variants and gene levels were compared between HCC and HC, and HCC and SH. Thirty-nine variants (adjusted P<0.0001, Fisher’s exact test) and 11 genes (adjusted P<0.0001, optimal unified approach for rare variant association test (SKAT-O) gene level test) were strongly associated with HBV-HCC. Thirty-four variants were from eight human leukocyte antigen (HLA) genes that were previously reported to be associated with HBV-HCC. The novelties of our study are: five variants (rs579876, rs579877, rs368692979, NM_145007:c.*131_*130delTG, NM_139165:exon5:c.623-2->TT) from three genes (REAT1E, NOD-like receptor (NLR) protein 11 (NLRP11), hydroxy-carboxylic acid receptor 2 (HCAR2)) were found strongly associated with HBV-HCC. We found 39 different variants in 11 genes that were significantly related to HBV-HCC. Five of them were new findings. Our data implied that chronic hepatitis B patients who carry these variants are at a high risk of developing HCC.
Keywords: cytokine, gene level association, HBV-HCC, immune genes, Target Region Sequencing (TRS)
Introduction
Hepatitis B virus (HBV) affected more than 248 million individuals worldwide [1] and caused deaths of 500000–1.2 million each year [2]. The HBV infection outcomes including self-healing and persistent infection, are associated with viral factors, environmental factors, and host genetic factors [3]. Host immune responses, such as virus recognition, antigen processing and presentation, as well as immune regulation are associated with pathogen infection and clearance [4]. The innate immunity plays important roles in immunopathology and treatment of HBV infection [5]. Toll-like receptors (TLRs) and innate immune sensors of pathogen serve as immune regulators of both innate and adaptive immune responses [6]. Recent study showed that young mice with TLR4 mutation exhibit rapid HBV clearance [7]. NOD-like receptor (NLR), another immune sensor can sense and react to viral infection through the inflammasomes [8,9]. The adaptive immunity genes such as human leukocyte antigen (HLA), have been widely studied in the HBV infection by genome-wide association study (GWAS) [10–14]. Polymorphisms of some cytokine genes are also associated with the outcomes of patients with HBV infection [15–18]. With the advances in high-throughput sequencing (HTS), some rare variants have been found and associated with HBV infection. To identify the variants associated with outcomes of patients with HBV infection, we systematically sorted and sequenced 404 genes related to innate and adaptive immunity, cytokine and cytokine receptors, and immune regulator genes in Chinese population infected with/without HBV infection. The identification of variants associated with the outcome of patients with HBV infection helps physicians treat the patients more specifically.
Materials and methods
Study subjects and sample collections
Three groups were included in the present study: hepatocellular carcinoma (HCC) group individuals who had HBV infection history and self-healed (self-healing (SH)) and healthy controls (HCs). The participants were from the First, Second, Third, and Fourth Hospital of Hebei Medical University (Shijiazhuang, China) and the Fifth Hospital of Shijiazhuang (Shijiazhuang, China) between January 2015 and January 2016. HCC diagnosis was according to the guidelines of the America Association for the Study of Liver Diseases (AASLD) [19]. HCC patients were seropositive for HBsAg or had HBV infection history. Self-healing HBV patients had no previous HBV immunization and the liver enzymes were normal, seropositive for HBsAb and HBeAb, and seronegative for HBsAg, HBeAg, and HBV DNA. HCs had normal liver enzymes, seronegative for HBcAb, HBeAb, HBeAg, HBsAb, and HBsAg, no previous HBV immunization, and no endocrine, cardiovascular, and kidney diseases, and other liver diseases history. All participants had written informed consents. The protocol of the present study was in accordance with the Declaration of Helsinki, and was approved by the Human Ethic Committee of the Second Hospital of Hebei Medical University. Five milliliters of whole blood were drawn and stored at −80°C until use. DNA was extracted from whole blood samples using an Invitrogen PureLink Genomic DNA Mini Kit (Thermo Fisher, Foster City, CA, U.S.A.).
Cytokines and immune genes search and gene panel design
The cytokines and cytokine receptors, TLRs, NLRs, HLA family genes, T-cell activation, and co-stimulation genes, natural killer cell target genes, G-protein-coupled receptor genes such as free fatty acid receptor (FFAR), hydroxy-carboxylic acid receptor (HCAR) family genes were sorted according to the HUGO Gene Nomenclature Committee (HGNC) database [20]. TargetSeq™ (iGeneTech, Beijing, China), an RNA probe based liquid phase chips were used to capture the whole exome region of selected genes.
Targetted region sequencing
The concentration of DNA was determined using a Qubit dsDNA BR Assay Kit (Thermo Fisher, Foster City, CA, U.S.A.). Two microliters of sample DNA were used for sequencing. Using an automated SPRIworks System (Beckman Coulter, San Jose, CA, U.S.A.), the 200–500 bp size libraries were constructed with a TruSeq DNA Sample Preparation Kit (Illumina, San Diego, CA, U.S.A.). Then the small (200–500 bp) size libraries were further used for capturing using TargetSeq™ liquid chip capture sequencing kits (iGeneTech, Beijing, China) according to the manufacturer’s instructions. In short, before the hybrid capture, small size libraries were mixed with Hybridization Block (iGeneTech, Beijing, China) to avoid any repeating sequences to form hybrid itself. And then, melting the Hybridization Buffer at room temperature and preheating Hybridization Buffer in 65°C water bath. After the solution completely dissolved, 20 µl Hybridization Buffer was mixed with 20 μl small-size library in a PCR tube, and 5 µl RNase block (Thermo Fisher, Foster City, CA, U.S.A.) was mixed with ssRNA probe to prevent probe degradation. The purpose of liquid phase hybrid was to capture objective DNA fragments by ssRNA probe complementary pairing. The Hybridization capture was performed in ABI 2720 PCR (Thermo Fisher, Foster City, CA, U.S.A.) at 65°C overnight for incubation. After hybridization, the DNA:RNA hybrid was enriched by biotin-labeled magnetic beads (Thermo Fisher, Foster City, CA, U.S.A.). Finally, the targetted sequences were amplified in ABI 2720 PCR. PCR parameters were: 95°C for 4 min, 98°C for 20 s, 65°C for 30 s, 16 cycles, 72°C for 30 s, 72°C for 5 min, and 12°C hold. PCR reagents were KAPA Taq PCR Kits (KAPA Biosystems, Boston, Massachusetts, U.S.A.) and Nextflex primers were synthesized by Invitrogen (Thermo Fisher, Foster City, CA, U.S.A.). The quality of amplified library was determined by using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, U.S.A.), and the library DNA concentration was again determined by Qubit dsDNA BR assay kit according to the manufacturer’s instructions. Libraries with good quality and DNA concentration >3 ng/μl were sequenced using an Illumina Hiseq X-ten sequencer (Illumina, San Diego, CA, U.S.A.).
Bioinformatics analyses
Data QC
The adaptor sequences and raw reads with low quality reads were filtered using Trimmomatic software [21]. The adaptor sequences were GATCGGAAGAGCACACGTCT and AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT, the criterion of low-quality sequence was the quality value of no more than Q20 or the accuracy of no more than 99%. The bases length should be longer than 40 bp after removing the base that does not meet the criteria. Finally fastqc software (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) was used to measure data quality to make sure that 95% of the remaining reads or clean reads with quality were more than Q30.
Variants calling
The clean reads were aligned to the reference human genome (February 2009, hg19, GRCh37, downloaded from UCSC) by BWA MEM software [22] to generate BAM files. In order to improve the accuracy, the samtools [23] and picard software (http://broadinstitute.github.io/picard/) were used to remove PCR repetitive sequence. The Genome Analysis Toolkit (GATK) [24] was used to detect the variants such as SNPs and InDels in BAM files. Finally, ANNOVAR software [25] was used to annotate the variants.
Functional effect of variants
Phenolyzer [26] was used to predict the association between the genotype and phenotype of HBV infection.
Association testing
Both single-variant and gene-based tests were performed. For single-variant based test, Fisher’s exact test was used to compare the different variants between HCC/control groups or subgroup, the false discovery rate (FDR) adjusted P-value <0.001 was considered significant. For gene-based tests, optimal unified approach for rare variant association test (SKAT-O) [27], a rare-variant association test used in small-sample case–control genome study, was used to assess excess-risk mutations in HCC/controls and subgroups. Default setting was used and significance were defined at FDR adjusted P-value <0.0001.
Results
Study subjects
A total of 53 HCC patients, 53 SH cases with previous HBV infection and 53 HCs were recruited, the clinical characteristics and statistics of study subjects were listed in Table 1.
Table 1. Demographics.
| HCC | HC | SH | |
|---|---|---|---|
| Gender, male/female | 45/8 | 26/27 | 30/23 |
| Age, M (Q) | 56.0 (11.50) | 34.0 (16.0) | 55.0 (19.0) |
| Drinking, no/yes | 40/13 | 46/7 | 43/10 |
| Smoking, no/yes | 41/12 | 46/7 | 41/12 |
| HBV DNA, M (Q) | 3200 (30842) | - | - |
| HbsAg, (+/−) | 41/12 | 0/53 | 0/53 |
| HbeAg, (+/−) | 17/36 | 0/53 | 0/53 |
| Anti-Hbe, (+/−) | 7/11 | - | - |
| ALT, M (Q) | 38.0 (68.8) | - | - |
| AST, M (Q) | 58.0 (72.5) | - | - |
| AFP, M (Q) | 161.2 (1070.0) | - | - |
Abbreviations: AFP, α-fetoprotein; ALT, Alanine aminotransferase; Anti-Hbe, antibody to HBeAg; AST, aspartate aminotransferase; HbeAg, HBV E antigen; HbsAg, HBV surface antigen; M, mean; Q, quartile.
Targetted gene investigations
A total of 404 genes or coding region of ~500 kilo (K) bp, were sorted according to HGNC database, including the whole cytokine and receptor family genes, some innate immunity and adaptive immunity related genes which reported as pathogen sensors (Table 2). The cytokine and receptor family genes were: 43 interleukin (IL) family genes, 42 IL receptor genes, 21 interferon (IFN) genes, 5 IFN receptor genes, 45 chemokine ligand (CCL) genes, 24 chemokine receptor genes, 18 tumor necrosis factor genes, and 29 tumor necrosis factor receptor genes. The innate genes included 10 TLR genes, and 22 NLRs family genes. The adaptive immunity genes comprised 27 MHC-related genes, 18 T-cell inhibition and co-inhibition related genes, 28 T-cell activation and co-stimulation genes. Other immune response related genes were: 37 natural killer cell target genes, 54 G-protein-coupled receptor genes, and 16 other genes.
Table 2. Summary of genes investigated in the study.
| Group | Subgroup and gene numbers |
|---|---|
| Cytokines and receptors | ILs and receptors, n=85; IFNs and receptors, n=26; chemokines and receptors, n=69; tumor necrosis factor and receptors, n=47 |
| Innate immunity | TLRs, n=10; NLRs, n=22 |
| Adaptive immunity | MHCs, n=27; T-cell inhibitors, n=18; T-cell activators, n=28 |
| Others | GPCRs, n=54; natural killer cell targets, n=37, others, n=16; |
Abbreviation: GPCR, G-protein-coupled receptor.
Targetted region sequencing performance
TRS yielded 10.84M clean reads (averaged read length was 143.65 bp) or 1577.94M clean bp for each sample, the average base quality was 40.26 and Q30 of call clean reads was 95.64%; the coverage rate of 499.968K region was 99.48%, duplication rate was 39.84%, and align rate was 99.23%; the capture rate was 50.53%. The target average depth was 919.55X, with target 10X rate was 99.31% and target 20X rate was 99.18%.
Variants and genes related to HBV associated with HCC
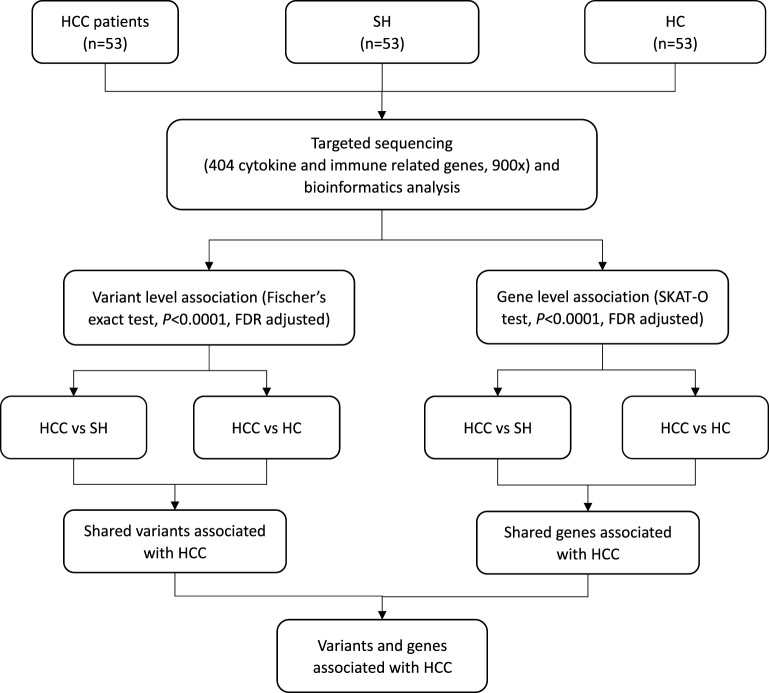
As shown in flowchart (Figure 1), both HC and self-healing groups were used as controls to identify variants and genes associated with HCC. The results of variant and gene level association test for each subgroup were listed in Table 3.
Figure 1. Flowchart of study design.
CC, hepatocellular carcinoma with hapatitis B virus infection; FDR, false discovery rate; SH, self-healing cases with previous hepatitis B virus infection; HC, healthy controls; SKAT-O, optimal unified approach for rare variant association test.
Table 3. Association of variants and genes with HBV associated with HCC.
| Gene symbol | Associated SNVs | Variant level OR | Variant level P-value (FDR adjusted) | Numbers of SNVs | SKAT-O gene level P-value (FDR adjusted) | |||
|---|---|---|---|---|---|---|---|---|
| HCC compared with HC | HCC compared with SH | HCC compared with HC | HCC compared with SH | HCC compared with HC | HCC compared with SH | |||
| HLA-B | rs2596493 | Inf | Inf | 2.25E-18 | 3.59E-18 | 106 | 1.7232E-16 | 2.67597E-10 |
| HLA-B | rs1131285 | Inf | Inf | 1.7E-13 | 1.13E-13 | 106 | 1.7232E-16 | 2.67597E-10 |
| HLA-B | rs1065386 | Inf | Inf | 6.97E-09 | 6.97E-09 | 106 | 1.7232E-16 | 2.67597E-10 |
| HLA-B | rs1050570 | Inf | Inf | 1.87E-08 | 1.83E-08 | 106 | 1.7232E-16 | 2.67597E-10 |
| HLA-B | rs1050556 | Inf | Inf | 0.0000019 | 0.0000019 | 106 | 1.7232E-16 | 2.67597E-10 |
| HLA-B | rs1131215 | Inf | Inf | 0.0000623 | 0.0000619 | 106 | 1.7232E-16 | 2.67597E-10 |
| HLA-C | rs41556617 | Inf | Inf | 7.92E-12 | 6.79E-12 | 108 | 5.33706E-07 | 3.14762E-05 |
| HLA-C | rs2308527 | Inf | Inf | 0.000000801 | 0.00000074 | 108 | 5.33706E-07 | 3.14762E-05 |
| HLA-DPA1 | rs1042174 | 16.63 | 28.69 | 0.000000899 | 3.15E-08 | 33 | 1.4521E-08 | 4.3979E-07 |
| HLA-DPA1 | rs1126543 | Inf | Inf | 2.9E-09 | 2.9E-09 | 33 | 1.4521E-08 | 4.3979E-07 |
| HLA-DPA1 | rs2308930 | Inf | Inf | 6.97E-09 | 6.97E-09 | 33 | 1.4521E-08 | 4.3979E-07 |
| HLA-DPA1 | rs2308929 | Inf | Inf | 6.97E-09 | 6.97E-09 | 33 | 1.4521E-08 | 4.3979E-07 |
| HLA-DPA1 | rs1126542 | Inf | Inf | 1.87E-08 | 1.83E-08 | 33 | 1.4521E-08 | 4.3979E-07 |
| HLA-DQA1 | rs9272896 | Inf | Inf | 0.0000116 | 0.0000113 | 81 | 3.33767E-07 | 1.30562E-05 |
| HLA-DQB1 | rs1049055 | 36.19 | 75.53 | 2.7E-11 | 6.3E-14 | 110 | 9.04603E-10 | 7.65036E-10 |
| HLA-DQB1 | rs1130430 | 19.53322682 | 11.3 | 0.00000465 | 0.0000835 | 110 | 9.04603E-10 | 7.65036E-10 |
| HLA-DQB1 | rs1049057 | Inf | Inf | 4.67E-08 | 4.18E-08 | 110 | 9.04603E-10 | 7.65036E-10 |
| HLA-DQB1 | rs1063345 | Inf | Inf | 0.00000465 | 0.0000045 | 110 | 9.04603E-10 | 7.65036E-10 |
| HLA-DQB1 | rs1140316 | Inf | Inf | 0.0000623 | 0.0000619 | 110 | 9.04603E-10 | 7.65036E-10 |
| HLA-DQB2 | rs9276572 | Inf | Inf | 2.05E-15 | 2.05E-15 | 21 | 5.04185E-06 | 0.000688766 |
| HLA-DRB1 | rs17211105 | 18.48 | 18.48 | 3.21E-08 | 3.02E-08 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs9270299 | 12.56 | 16.04 | 0.00000974 | 0.00000051 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs17883134 | 44.09 | 20.97 | 1.07E-10 | 1.83E-08 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs9270303 | 20.97 | 20.97 | 1.95E-08 | 1.83E-08 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs2308759 | 353.69 | 214.66 | 5.04E-20 | 3.59E-18 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs17211091 | 18.04 | 96.28 | 0.000000363 | 1.61E-10 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs113734598 | 48.46 | 15.62 | 0.00000159 | 0.0000619 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs9269693 | 136.13 | Inf | 9.53E-13 | 3.74E-14 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs1064697 | Inf | Inf | 0.000000124 | 0.000000107 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB1 | rs1071752 | Inf | Inf | 0.0000623 | 0.0000619 | 126 | 1.08223E-14 | 1.74002E-11 |
| HLA-DRB5 | rs17211043 | Inf | Inf | 0.000000124 | 0.000000107 | 83 | 4.77962E-10 | 2.52675E-06 |
| HLA-DRB5 | rs77853982 | Inf | Inf | 0.000000801 | 0.00000074 | 83 | 4.77962E-10 | 2.52675E-06 |
| HLA-DRB5 | rs701884 | 38.71 | Inf | 0.0000245 | 0.0000019 | 83 | 4.77962E-10 | 2.52675E-06 |
| HLA-DRB5 | rs1064587 | 35.87 | Inf | 0.000058 | 0.0000045 | 83 | 4.77962E-10 | 2.52675E-06 |
| HCAR2 | rs579877 | Inf | Inf | 0.0000277 | 0.000027 | 11 | 2.03163E-06 | 3.36441E-05 |
| HCAR2 | rs579876 | Inf | Inf | 0.0000277 | 0.000027 | 11 | 2.03163E-06 | 3.36441E-05 |
| NLRP11 | NM_145007: c.*131_*130delTG | 9.684525939 | 14.70656392 | 2.71228E-05 | 6.95927E-07 | 22 | 6.83802E-07 | 2.97726E-09 |
| NLRP11 | rs368692979 | 17.64443693 | 32.53501457 | 8.81316E-08 | 8.54585E-10 | 22 | 6.83802E-07 | 2.97726E-09 |
| RAET1E | NM_139165: exon5:c.623-2- >TT | 16.6900097 | 20.59517629 | 1.0742E-07 | 9.68061E-09 | 21 | 0.000603407 | 3.07916E-05 |
Abbreviations: Inf, infinite; HCAR2, hydroxy-carboxylic acid receptor 2; OR, odds ratio; NLRP11, NLR protein 11; RAET1E, retinoic acid early transcript 1E; SH, self-healing cases with previous HBV infection.
A total of 39 significantly different variants and 11 genes were identified. Thirty four of the variants were from eight HLA genes, including HLA-B, HLA-C, HLA-DPA1, HLA-DQA1, HLA-DQB1, HLA-DQB2, HLA-DRB1, and HLA-DRB5. We also identified five non-HLA variants in retinoic acid early transcript 1E (RAET1E), NLR protein 11 (NLRP11), and HCAR2. NM_139165:exon5:c.623-2->TT was located in RAET1E. The variants identified were significantly enriched in HCC patients, and rare or no variants were found enriched in controls. rs368692979 and NM_145007:c.*131_*130delTG were located in NLRP11, and were two of the top ten most significant variants (P-values were 6.72E-14 and 7.08E-11 in HCC compared with HC and HCC compared with self-healing, respectively). Previous studies showed that NLRP11 was related to inflammation [41]. These variants were also significantly enriched in HCC patients, and rare or no variants were found enriched in controls. rs579876 and rs579877 were located in 3′-UTR of HCAR2.
Discussion
The present study found that 39 variants in 11 genes were strongly associated with HBV associated with HCC (HBV-HCC). Amongst them, 34 variants from eight genes in HLA region were previously described and our data were consistent with previous studies [30–35]. The novelties of our study are: five variants (rs579876, rs579877, rs368692979, NM_145007:c.*131_*130delTG, NM_139165:exon5:c.623-2->TT) from three genes (REAT1E, NLRP11, HCAR2) were found to be strongly associated with HBV-HCC. Our study provided fundamental data which needs further study to confirm the roles of these new variants and genes.
HBV infection is a serious public health problem [1]. Patients with HBV infection have different outcomes, such as self-healing, HBV carrier, chronic hepatitis, or HCC [4]. It has been found that the host genotype may be closely related to the different outcomes. Some HCC-related loci have been identified based on GWAS [10–14,28]. However, these studies are mainly based on the common variants, and most of the variants are located in the non-gene region. It is difficult to verify the gene function and explore the mechanism of disease. Given that HTS methods such as whole exome sequencing (WES) or whole genome sequencing (WGS) focus on not only common but also rare variants, HTS has been widely used in complex disease genetics studies and shows promising results. WES has been successfully applied in HCC genetics research [29].
Large sample size is necessary for HCC studies. However, WES-based GWAS is very expensive. TRS, which targets the specific gene families or pathway genes, provides relatively cost-effective solution. The present study investigated the cytokines and immune genes associated with HCC. We selected 404 genes that may be associated with HBV infection. These genes are mainly involved in antigen recognition, processing and presentation, immune regulatory cytokines and receptors, as well as those included in innate immune system, such as TLRs and NLRs. We have also studied some genes that are directly related to immune checkpoint.
We included SH cases because we speculated that they may carry less risk mutations or carry more protective mutations. Previous study demonstrated that the performance of gene level associated/burden test is reliable in WES/WGS study even in a small sample size in WES/WGS study [27]. The present study performed a comprehensive variants and gene level association analysis between both HCC compared with HC and HCC compared with SH. We found that 39 variants and 11 genes were strongly associated with HBV-HCC.
Not surprisingly, 34 variants from eight genes in HLA region were found strongly associated with HBV-HCC. Our data were consistent with previous studies [30–35]. We found five new variants from three genes that are also strongly related to HBV-HCC.
RAET1E is an MHC class I related gene from the RAET1 family, which functions as a ligand for the NKG2D receptor. NKG2D receptors are expressed on the surface of several types of immune cells and are involved in both innate and adaptive immune responses [36]. Previous studies showed that reduced NKG2D ligand expression in HCC correlates with early recurrence [37], and that the immunoreceptor NKG2D promotes tumor growth in a model of HCC [38]. Consistent with these studies, we found that a splicing variant in RAET1E, NM_139165:exon5:c.623-2->TT, might lead to malfunction of RAET1E, a ligand for the NKG2D receptor. As NKG2D receptor blockade is an attractive target for HCC therapy [39], further research will be necessary to study how RAET1E may be involved in HBV-associated HCC and develop candidate drugs.
NLRP11 is a member of the NLR gene family, which is reported to be closely related to antiviral immunity [40]. Although little is known about whether NLRP11 is involved in HBV infection, another member of the NLR family, NLRP3, is involved in danger signal inflammatory responses [41]. Furthermore, the four variants identified, including rs368692979 and NM_145007:c.*131_*130delTG in NLRP11, together with rs579876 and rs579877 in HCAR2, a gene closely related to immune activation [42], were all from the 3′-UTR region, which suggests that the expression of NLRP11 and HCAR2 might be under the regulation of miRNA.
Conclusion
Our comprehensive investigation of cytokine- and immune-related gene mutations in HBV-HCC patients and controls showed that 39 different variants and 11 genes were significantly related to HBV-HCC. Our data implied that chronic hepatitis B patients who carry these variants need intensive monitoring.
Abbreviations
- BAM
Binary Alignment/Map
- FDR
false discovery rate
- GWAS
genome-wide association study
- HbsAg
Hepatitis B surface antigen
- HBeAg
Hepatitis B e antigen
- HBcAb
Hepatitis B core antibody
- HBeAb
Hepatitis B e antibody
- HBV
hepatitis B virus
- HBV-HCC
HBV associated hepatocellular carcinoma
- HC
healthy control
- HCAR2
hydroxy-carboxylic acid receptor 2
- HCC
hepatocellular carcinoma
- HGNC
HUGO Gene Nomenclature Committee
- HLA
human leukocyte antigen
- HTS
high-throughput sequencing
- IFN
interferon
- IL
interleukin
- NLR
NOD-like receptor
- NLRP11
NOD-like receptor protein 11
- NOD
nucleotide-binding oligomerization domain
- RAET
retinoic acid early transcript
- RAET1E
retinoic acid early transcript 1E
- SH
self-healing case
- SNV
Single Nucleotide Variant
- TLR
toll-like receptor
- TRS
target region sequencing
- QC
quality control
- WES
whole exome sequencing
- WGS
whole genome sequencing
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81402729]; and the Natural Science Foundation of Hebei Province [grant number H2015206152].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
Fengxue Yu, Xiaolin Zhang and Mingbang Wang conducted the research design; Suzhai Tian, Lianxia Geng, Chaojun Zhang, Lina Guo and Wenting An performed DNA extracting; Weili Xu, Yuan Jia, Xuechen Liu, Junji Ma and Yuan Quan collected the samples; Fengxue Yu, Ning Ma, Xiaolin Zhang, Mingbang Wang and Dianwu Liu analyzed the data.
References
- 1.Schweitzer A., et al. (2015) Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386, 1546–1555 [DOI] [PubMed] [Google Scholar]
- 2.Lavanchy D. (2004) Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J. Viral Hepat. 11, 97–107 [DOI] [PubMed] [Google Scholar]
- 3.Rehermann B. and Bertoletti A. (2015) Immunological aspects of antiviral therapy of chronic hepatitis B virus and hepatitis C virus infections. Hepatology 61, 712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehermann B. and Nascimbeni M. (2005) Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 5, 215–229 [DOI] [PubMed] [Google Scholar]
- 5.Maini M.K. and Gehring A.J. (2016) The role of innate immunity in the immunopathology and treatment of HBV infection. J. Hepatol. 64 (1 Suppl.), S60–S70 [DOI] [PubMed] [Google Scholar]
- 6.Akira S. and Takeda K. (2004) Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 7.Chou H.H., et al. (2015) Age-related immune clearance of hepatitis B virus infection requires the establishment of gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 112, 2175–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franchi L., Munoz-Planillo R. and Nunez G. (2012) Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 13, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanneganti T.D. (2010) Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 10, 688–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang D.K., et al. (2015) Genetic variants in five novel loci including CFB and CD40 predispose to chronic hepatitis B. Hepatology 62, 118–128 [DOI] [PubMed] [Google Scholar]
- 11.Kim Y.J., et al. (2013) A genome-wide association study identified new variants associated with the risk of chronic hepatitis B. Hum. Mol. Genet. 22, 4233–4238 [DOI] [PubMed] [Google Scholar]
- 12.Hu Z., et al. (2013) New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat. Genet. 45, 1499–1503 [DOI] [PubMed] [Google Scholar]
- 13.Mbarek H., et al. (2011) A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum. Mol. Genet. 20, 3884–3892 [DOI] [PubMed] [Google Scholar]
- 14.Kamatani Y., et al. (2009) A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat. Genet. 41, 591–595 [DOI] [PubMed] [Google Scholar]
- 15.Yucesoy B., et al. (2002) IL-1beta gene polymorphisms influence hepatitis B vaccination. Vaccine 20, 3193–3196 [DOI] [PubMed] [Google Scholar]
- 16.Hohler T., et al. (2005) A functional polymorphism in the IL-10 promoter influences the response after vaccination with HBsAg and hepatitis A. Hepatology 42, 72–76 [DOI] [PubMed] [Google Scholar]
- 17.Sodsai P., et al. (2013) Association of cytokine and cytokine receptor gene polymorphisms with the risk of chronic hepatitis B. Asian Pac. J. Allergy Immunol. 31, 277–285 [DOI] [PubMed] [Google Scholar]
- 18.Gao Q.J., et al. (2009) Polymorphisms of some cytokines and chronic hepatitis B and C virus infection. World J. Gastroenterol. 15, 5610–5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruix J., Sherman M. and (2011) Management of hepatocellular carcinoma: an update. Hepatology 53, 1020–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray K.A., et al. (2015) Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 43, D1079–D1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger A.M., Lohse M. and Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H. (2014) Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A., et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K., Li M. and Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H., Robinson P.N. and Wang K. (2015) Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat. Methods 12, 841–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S., et al. (2012) Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 91, 224–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., et al. (2016) Genome-wide association study identifies 8p21.3 associated with persistent hepatitis B virus infection among Chinese. Nat. Commun. 7, 11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Q., et al. (2012) Rare inborn errors associated with chronic hepatitis B virus infection. Hepatology 56, 1661–1670 [DOI] [PubMed] [Google Scholar]
- 30.Mathew S., et al. (2016) Host nucleotide polymorphism in hepatitis B virus-associated hepatocellular carcinoma. World J. Hepatol. 8, 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malmassari S.L., et al. (2007) Impact of hepatitis B virus basic core promoter mutations on T cell response to an immunodominant HBx-derived epitope. Hepatology 45, 1199–1209 [DOI] [PubMed] [Google Scholar]
- 32.Posuwan N., et al. (2014) Genetic association of human leukocyte antigens with chronicity or resolution of hepatitis B infection in thai population. PLoS ONE 9, e86007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., et al. (2016) Polymorphisms of HLA-DQB1 predict survival of hepatitis B virus-related hepatocellular carcinoma patients receiving hepatic resection. Clin. Res. Hepatol. Gastroenterol. 40, 739–747 [DOI] [PubMed] [Google Scholar]
- 34.Kurokohchi K., et al. (1996) Expression of HLA class I molecules and the transporter associated with antigen processing in hepatocellular carcinoma. Hepatology 23, 1181–1188 [DOI] [PubMed] [Google Scholar]
- 35.Li S., et al. (2012) GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 8, e1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muntasell A., et al. (2017) Targeting NK-cell checkpoints for cancer immunotherapy. Curr. Opin. Immunol. 45, 73–81 [DOI] [PubMed] [Google Scholar]
- 37.Kamimura H., et al. (2012) Reduced NKG2D ligand expression in hepatocellular carcinoma correlates with early recurrence. J. Hepatol. 56, 381–388 [DOI] [PubMed] [Google Scholar]
- 38.Sheppard S., et al. (2017) The immunoreceptor NKG2D promotes tumour growth in a model of hepatocellular carcinoma. Nat. Commun. 8, 13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilarinho S., et al. (2007) Blockade of NKG2D on NKT cells prevents hepatitis and the acute immune response to hepatitis B virus. Proc. Natl. Acad. Sci. U.S.A. 104, 18187–18192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupfer C. and Kanneganti T.D. (2013) The expanding role of NLRs in antiviral immunity. Immunol. Rev. 255, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baroja-Mazo A., et al. (2014) The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat. Immunol. 15, 738–748 [DOI] [PubMed] [Google Scholar]
- 42.Feingold K.R., et al. (2014) Inflammation stimulates niacin receptor (GPR109A/HCA2) expression in adipose tissue and macrophages. J. Lipid Res. 55, 2501–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]