Abstract
Background
Blastocystis sp. is a unicellular eukaryote that is commonly found in the human intestine. Its ability to cause disease is debated and a subject for ongoing research. In this study, faecal samples from 35 Swedish university students were examined through shotgun metagenomics before and after travel to the Indian peninsula or Central Africa. We aimed at assessing the impact of travel on Blastocystis carriage and seek associations between Blastocystis and the bacterial microbiota.
Results
We found a prevalence of Blastocystis of 16/35 (46%) before travel and 15/35 (43%) after travel. The two most commonly Blastocystis subtypes (STs) found were ST3 and ST4, accounting for 20 of the 31 samples positive for Blastocystis. No mixed subtype carriage was detected. All ten individuals with a typable ST before and after travel maintained their initial ST. The composition of the gut bacterial community was not significantly different between Blastocystis-carriers and non-carriers. Interestingly, the presence of Blastocystis was accompanied with higher abundances of the bacterial genera Sporolactobacillus and Candidatus Carsonella. Blastocystis carriage was positively associated with high bacterial genus richness, and negatively correlated to the Bacteroides-driven enterotype. These associations were both largely dependent on ST4 – a subtype commonly described from Europe – while the globally prevalent ST3 did not show such significant relationships.
Conclusions
The high rate of Blastocystis subtype persistence found during travel indicates that long-term carriage of Blastocystis is common. The associations between Blastocystis and the bacterial microbiota found in this study could imply a link between Blastocystis and a healthy microbiota as well as with diets high in vegetables. Whether the associations between Blastocystis and the microbiota are resulting from the presence of Blastocystis, or are a prerequisite for colonization with Blastocystis, are interesting questions for further studies.
Electronic supplementary material
The online version of this article (10.1186/s12866-017-1139-7) contains supplementary material, which is available to authorized users.
Keywords: Blastocystis, Subtype, Persistence, Travel, Microbiota, Sporolactobacillus, Candidatus Carsonella, Transmission
Background
Blastocystis sp. is an anaerobic unicellular eukaryote that can reside in the gastrointestinal tract of humans and a wide variety of animals [1]. Taxonomically it belongs to the phylum Stramenopiles [2] and it is the only member of this phylum found in the human intestine. Human carriage of Blastocystis is common worldwide, and recent studies using molecular tools report detection rates of 22–56% in European countries [3–5] and 37–100% in Asian and African countries [6–8]. There is a high degree of genetic diversity within Blastocystis sp., and 17 genetic subtypes, named ST1–17, are recognized. ST1–9 and ST12 have so far been found in humans of which ST3 is the most commonly detected subtype [9, 10]. The most notable geographic difference in subtype distribution is that while ST4 is the second most common subtype in Europe it is rarely found in South America, Africa and Asia [7, 9, 10]. The effect of international travel on subtype carriage is not known. Blastocystis has been associated with diarrhoea, abdominal pain, and vomiting, but its role as a causative agent in human diseases is unclear and debated [11]. Research on subtype-dependent pathogenicity is ongoing but there is to date no widely accepted distinction between pathogenic and non-pathogenic subtypes [11, 12]. When Blastocystis is detected by microscopy or PCR in diagnostic examinations of faecal samples from humans with suspected disease, it is often not possible to determine if the finding represents acute infection or intestinal colonization. This prompts further research to better understand and characterize the nature of human Blastocystis carriership. In prolonged cases of diarrhoea or other gastrointestinal complaints after travel, findings of Blastocystis as the sole possible pathogen may be interpreted as infection and treated with metronidazole, trimethroprim- sulfamethoxazole and/or other antimicrobial drugs [11]. Treatment failure is common and eradication of Blastocystis can be difficult to achieve [13]. It is unknown whether this is due to poor drug efficacy, a failure of the host defence in eliminating a pathogen despite an appropriate drug effect, or if persistent Blastocystis findings in symptomatic disease signify that this organism constitutes a normal and stable part of the gut microbiota. To be able to determine the factors that govern the presence of Blastocystis in the human intestine, an essential step is to examine the relationships between Blastocystis and the bacterial gut microbiota. The aim of this study was to use metagenomics to investigate the prevalence and temporal stability of Blastocystis subtypes and its relation to the bacterial intestinal microbiota of Swedish university students travelling to the Indian peninsula or Central Africa.
Methods
Data collection
Participants in this study were part of a larger study in which Swedish students from the Universities in Umeå, Stockholm and Gothenburg travelling for international exchange studies submitted faecal samples for laboratory analyses before and after travel. The material has previously been used in a study on the acquisition of extended spectrum beta-lactamase (ESBL) producing bacteria through travel [14]. The present metagenomic study was planned and executed together with authors of a study in which faecal samples were examined for antibiotic resistance genes [15]. Included in these metagenomic studies were 35 health care students, 26/35 female, age 23–34 years, who travelled to Central Africa (17/35) or the Indian peninsula (18/35) with a median travel duration of 34 days (range 14 to 150). The students included reported no antibiotic treatment during travel or within six months prior to sampling. Faecal samples were frozen on arrival to the laboratory and DNA was extracted from frozen faeces using the QIAamp DNA stool mini kit (Qiagen). Metagenomic sequencing was performed on a HiSeq 2000 (Illumina). The sequence data, consisting of 40 to 178 million filtered read pairs per sample, was deposited in the European Nucleotide Archive as project PRJEB7369 [15].
Bioinformatic analysis
Quality filtering of the reads and removal of host sequences was performed as previously described [15]. The read pairs of the sequence data were searched for small subunit ribosomal DNA (SSU-rDNA) sequences using Metaxa2 version 2.0 [16] (additional option “--align none”). The genus rDNA counts for each library were normalized to the total number of rDNA counts in that library, yielding relative abundances for each genus. Partial Blastocystis SSU-rDNA sequences and their associated subtype classifications were downloaded from the Blastocystis Sequence Typing website (http://pubmlst.org/blastocystis/) on 2014–10-15. SSU-rDNA sequences in the gut metagenomes classified as deriving from the Blastocystis genus by Metaxa2 were extracted and used as queries for BLAST searches [17] against the Blastocystis Sequence Typing database (options “-p blastn -e 1e-5 -m 8 -F F”), and thereby classified to subtypes. Sequence reads that could have originated from more than one subtype were noted as non-discriminatory matches and were not used in the majority of subsequent analysis. To avoid noise, subtypes with less than three reads assigned to them were removed from each library. The detection of the Blastocystis SSU-rDNA by the Metaxa2 tool was confirmed by testing with real-time PCR (qPCR) for Blastocystis [7] in which 30 of 31 samples positive according to the metagenomic analysis were positive also by qPCR.
Detection of intestinal parasites
All samples were tested for the presence of intestinal parasites by light microscopy after formol-ether-concentration and by qPCR for the detection of Giardia intestinalis, Cryptosporidium spp., Entamoeba histolytica and Entamoeba dispar [7]. Detection of bacterial and viral enteropathogens was not performed.
Statistical analysis
Alpha diversity (richness, Shannon and Simpson indices) was assessed using the R package Vegan (http://cran.r-project.org/web/packages/vegan/). Vegan was also used for rarefaction analysis. To determine if there was a connection between human gut bacterial composition and the Blastocystis subtypes present in the gut, we performed a principal component analysis (PCA) of the relative abundances of bacterial genera, as well as a principal coordinates analysis (PCoA) of the Bray-Curtis dissimilarity matrix, in the software R [18]. Prior to PCA a variance stabilizing transform (square-root transformation) of the relative abundances of bacterial genera was applied, and then the R command prcomp was used for the PCA. Spearman correlation was used to identify relationships between the relative abundances of individual bacterial genera and the total Blastocystis abundance or the abundance of the respective subtypes. P-values for correlations were adjusted for multiple testing using the Benjamini-Krieger-Yekuteli modification of the false discovery rate (FDR) [19] and tests with an FDR < 0.05 were considered statistically significant. These correlations were calculated both before and after travel, as well as on all samples together, and only correlations that were significant in all three cases have been reported, as those seem to be stable both between individuals and over time. The association between Blastocystis presence and bacterial diversity was assessed by comparing the mean bacterial genus richness of Blastocystis positive and negative samples using a two-tailed Student’s t-test assuming unequal variance. Relationships between Blastocystis positivity/negativity and the relative abundance of the Bacteroides, Sporolactobacillus and Candidatus Carsonella genera were tested in the same manner. Those tests were also carried out separately for individuals positive or negative for subtypes ST3 and ST4. Furthermore, the effect of Blastocystis and/or specific subtype presence or absence on total community composition was assessed on the family and phylum levels using the metaxa2_uc tool, part of the Metaxa2 Diversity Tools [20] (options “-g auto --table T --matrix T” and “-g auto --table T --matrix T -c groups”). This tool iteratively calculates Bray-Curtis dissimilarity between subsamples of communities to assess whether they are significantly dissimilar (i.e. if the distribution of taxa in a sample could have been drawn from the collective pool of taxa of a group of samples).
Results
Sixteen out of the 35 subjects (46%) were positive for Blastocystis before travel and 15 (43%) were positive after travel based on the detection of Blastocystis SSU-rDNA in the metagenomic data. Eighteen (51%) were positive for Blastocystis either before or after travel and 13 (37%) were positive both before and after travel (Fig. 1). The relative abundance of Blastocystis SSU-rDNA reads varied substantially between individuals, and also between time points, ranging from 0.08 to 18.8 per million reads. Between 25% and 71% of the Blastocystis SSU-rDNA reads in each sample matched a specific subtype. Other Blastocystis SSU-rDNA reads were not specific to any single subtype because of inter-subtype sequence identity in some parts of the SSU-rDNA, and were considered as non-discriminatory with regard to subtyping. After filtering out non-discriminatory reads we did not find any evidence of co-occurrence of several different subtypes within the same individual. Instead, all SSU-rDNA reads that could be assigned to a subtype corresponded to a single variant in each individual. Subtyping was not successful in three samples in which only a few reads of non-discriminatory sequences were detected. In total, we found ST1 in 3 samples from 2 individuals, ST2 in 4 samples from 2 individuals, ST3 in 10 samples from 7 individuals, ST4 in 10 samples from 6 individuals, and ST8 in a single sample (Fig. 1; Additional file 1: Figure S1). For the ten participants that had their Blastocystis subtyped both before and after travel, the same subtype was detected in both samples. The median travel duration for these individuals was 41 days (range 22 to 150). Among all participants, travellers’ diarrhoea was reported by 69% (24/35), for a median of four days [15], and among those positive for Blastocystis both before and after travel the corresponding figure was 77% (10/13). Parasitological analysis by light microscopy revealed no pathogenic intestinal parasites, and all samples were negative in the qPCR for Giardia intestinalis (using a cycle treshold value cut-off of 36), Cryptosporidium spp., Entamoeba histolytica and Entamoeba dispar.
Fig. 1.
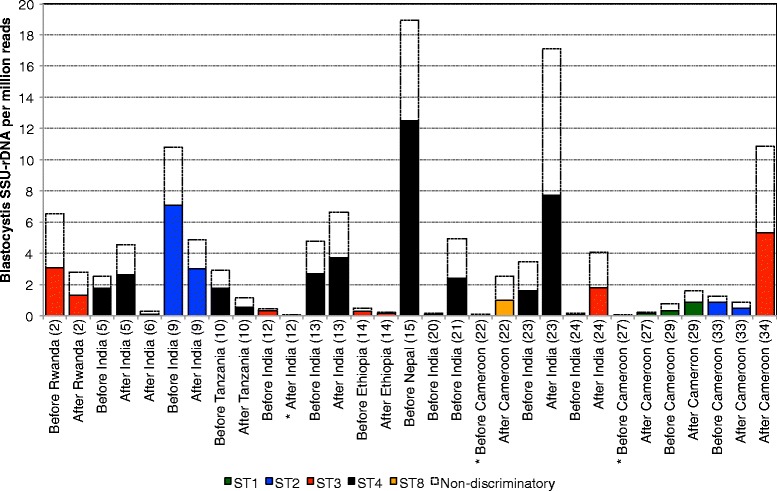
Abundance of Blastocystis SSU rDNA in positive individuals, before and after travel. Labels show the time point of sampling, destination, and participants’ number of inclusion. Asterisks indicate samples in which no subtype specific reads were detected
The metagenomic analysis revealed high interindividual variability of the bacterial community, but the microbiota was only moderately affected by travel (Additional file 2: Figure S2). This was previously presented by Bengtsson-Palme et al. [15] (see the supplemental material), which shows the taxonomic composition at the phylum level before and after travel to Central Africa and the Indian peninsula, respectively. In the present study, the relative abundance of Blastocystis was significantly positively correlated to the relative abundance of bacteria in the genera Sporolactobacillus and Candidatus Carsonella (Table 1). In addition, we found significant positive correlations between certain bacterial genera and the relative abundance of certain Blastocystis subtypes both before and after travel. Considering the relatively few individuals carrying each subtype we have chosen to present these findings in Additional file 3: Table S1, but have refrained from further interpretation of their importance due to small sample sizes at the subtype level. When comparing Blastocystis positive samples with negative samples we found positive samples to be associated with an increased relative abundance of Sporolactobacillus (p = 0.00021; Fig. 2a). An increased relative abundance of Candidatus Carsonella in Blastocystis-carriers was also noted in the total material (p = 0.03795; Fig. 2b), however, when samples taken before and after travel were analysed separately, this relationship was not significant (p = 0.1855, and p = 0.097, respectively). The relative abundance of Bacteroides was significantly lower in faecal samples containing Blastocystis (p = 0.00027; Fig. 2c). For the two most commonly found subtypes, ST3 and ST4, the negative association with Bacteroides was significant for ST4 (p = 0.005655) but not for ST3 (p = 0.7293). Presence of Blastocystis was also associated with significantly higher richness of bacterial genera (p = 0.0022; Fig. 3; Additional file 4: Figure S3), with this effect also being largely driven by ST4, which was significantly associated with high richness of bacterial genera (p = 3.65E-05), in contrast to what was found for ST3 (p = 0.7093).
Table 1.
Bacterial genera with significant correlations to the abundance of overall Blastocystis SSU-rDNA
| Before travel | After travel | In total | ||||
|---|---|---|---|---|---|---|
| Bacteria correlated to Blastocystis sp. (n = 31) | Correlation coefficient | Adjusted p-value | Correlation coefficient | Adjusted p-value | Correlation coefficient | Adjusted p-value |
| Firmicutes; Bacilli; Bacillales; Sporolactobacillaceae; Sporolactobacillus | 0.9062 | < 0.0001 | 0.8448 | < 0.0001 | 0.8731 | < 0.0001 |
| Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Candidatus Carsonella | 0.6148 | 0.0811 | 0.6617 | 0.0172 | 0.6430 | < 0.0001 |
Fig. 2.

Box-plot for the relative abundance of the bacterial genera Sporolactobacillus (a), Candidatus Carsonella (b), and Bacteroides (c) in Blastocystis positive and negative individuals. The differences were assessed statistically through Students t-test
Fig. 3.
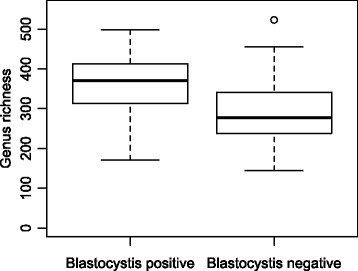
Box-plot for the bacterial genus richness in Blastocystis positive and negative individuals. The differences were assessed statistically through Students t-test
The total bacterial community compositions in the stool samples showed no systematic significant differences, neither between Blastocystis positive and negative individuals, nor between individuals with different subtypes (all p-values >0.17 and >0.89, respectively). However, our data for ST3 and ST4 suggest that a relationship to different bacteria still may exist (Fig. 4; Additional file 5: Figure S4). According to the stratification of the human intestinal bacterial flora by enterotypes suggested by Arumugam et al. [21], the PCA in Fig. 4 is shown in relation to Bacteroides and Prevotella, the genera suggested to drive the human gut enterotypes. Interestingly, the microbiota composition of individuals with different Blastocystis subtypes seems not to be equally distributed with regard to the enterotypes. Most of the faecal samples containing Blastocystis ST3 clustered more closely to the Bacteroides genus in the PCA than did the samples with ST4. Additionally, the bacterial communities of individuals carrying Blastocystis ST2 and ST8 differed the most from the Blastocystis negative individuals, and ST2 communities stood out as being highly dissimilar to those associated with ST1, ST3 and ST4 (Fig. 5; Additional file 6: Figure S5 and Additional file 7: Figure S6). For most subtypes, however, the variability in bacterial community composition was high also within groups, with none of the differences indicated above being statistically significant at the overall community level (p > 0.17).
Fig. 4.

Principal component analysis of bacterial genus composition (square-root transformed abundances). Samples are colour coded by the subtype present (ST1: green; ST2: blue; ST3: red; ST4: black; ST8: orange; Ambiguous: yellow; No Blastocystis detected: grey). The genera Bacteroides and Prevotella, indicative of the human enterotypes, are shown as drivers of the PCA separation. In addition, the two genera significantly correlated to Blastocystis abundance (Candidatus Carsonella and Sporolactobacillus) are indicated in light blue
Fig. 5.
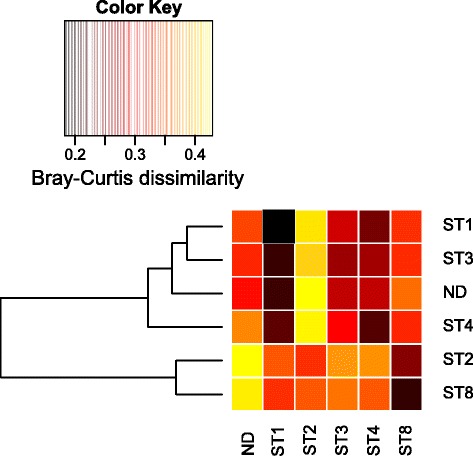
Similarity between the bacterial community compositions of individuals carrying different Blastocystis subtypes, or no Blastocystis at all (ND). Rows indicate the dissimilarity of the average community of each sample group to the average community of the sample groups of the columns, as assessed by repeated random subsampling of the communities at the family level. The dendrogram represents the overall similarity of the sample groups. Yellow colour corresponds to high average Bray-Curtis dissimilarity and black corresponds to the average communities of the sample groups being very similar sample groups. Note that the similarity within subsamples drawn from each group is also indicated in the figure
Discussion
This metagenomic study of travellers offers new insights with regard to the stability of Blastocystis carriership in the intestine, and the relationship between Blastocystis and the bacterial microbiota. In a population of 35 university students, the prevalence of Blastocystis was high both before and after intercontinental travel. The distribution of Blastocystis subtypes, with representation of ST1–4 and a dominance of ST3 and ST4, was unremarkable for a European setting [9, 22]. Apparently, there was little impact of intercontinental travel on the subtype distribution. Among the ten individuals where a reliable subtype designation could be determined from the metagenomic data before and after travel, each carried the same Blastocystis subtype on both occasions, implying a persistence of a specific subtype for three weeks to five months. This persistence was seen despite the common occurrence of travellers’ diarrhoea. No evidence of mixed subtype carriage was found, which is in contrast to a recent study from Ireland where mixed STs were found in 22% of Blastocystis-carriers [5]. However, most previous studies have found a rather low frequency of mixed STs [9]. Although the occurrence of mixed STs might be underreported in some studies because of inherent methodological limitations [5], we are confident that the method we have used is a sensitive tool to detect the occurrence of more than one ST. Our findings, with a high prevalence of Blastocystis and an absence of mixed subtype carriage raise the question if competition between STs may occur, preventing colonization of a second subtype when one is already present.
The presence of travellers’ diarrhoea and possible, non-examined, co-existing bacterial or viral enteropathogens could act as confounding factors in our metagenomic study of the intestinal microflora. However, the participants of the study reported no antibiotic use and we found that the overall composition of the bacterial gut microbiota of the study participants was only moderately affected by travel. The most notable change was an increase in abundance of families within the phylum Proteobacteria in participants returning from Africa [15]. Comparisons of the bacterial composition at the family level showed no significant systematic differences between Blastocystis-carriers and non-carriers or between carriers of different Blastocystis subtypes. We did, however, find a positive association between Blastocystis-carriage and high bacterial genus richness in the gut (Fig. 3). This strengthens the association between Blastocystis and high bacterial diversity that has also been reported in a retrospective metagenomic study by Andersen et al. [23], which included both healthy controls and patients with inflammatory bowel disease, and in a metagenomic study by Audebert et al. [24] that investigated the intestinal microbiota in individuals whose faecal samples had previously been sent for diagnostic parasitological exams. Since a gut microbiota with high diversity is generally considered healthy, Blastocystis might be seen as an indicator of gastrointestinal health, an opinion also voiced by other researchers [23–25]. Because we lack dietary data and detailed medical history of the participants in our study we cannot substantiate this assumption any further. It is interesting that the association between Blastocystis and genus richness was largely driven by ST4 in our study, and we suggest that subtype analysis is performed in further metagenomic studies of Blastocystis.
We also found interesting correlations between Blastocystis and certain bacterial genera. Firstly, the relative abundance of bacterial members of the genus Bacteroides was lower in Blastocystis-carriers compared to non-carriers (Fig. 2c). This was especially prominent for those individuals harbouring Blastocystis ST4. A negative correlation between Blastocystis and Bacteroides was also described in the study by Andersen et al. [23] and in another study using qPCR to quantify major bacterial taxa in the intestine of Blastocystis carriers and non-carriers [26]. Such a negative correlation was however not reported in the study by Audebert et al. [24], nor in a previous qPCR-based study [27]. There are several possible explanations to the inverse relationship between Blastocystis and Bacteroides, including competition for nutrients, competition for a specific ecological niche in the gut, discrepancies in the ability to utilize nutrients in the diet and/or a direct negative effect on Bacteroides by Blastocystis predation of these bacteria. The Bacteroides-driven enterotype has previously been linked to diets high in protein and animal fat, typical of a Western diet [28, 29]. However, our study specifically pinpoints Blastocystis ST4 to be associated with low Bacteroides abundance, and the high prevalence of ST4 found in Europe and Australia [9] seems contradictory to the theory of the Bacteroides-enterotype driven by a typical Western diet. Additional explanatory factors have to be considered to better understand the regional patterns of Blastocystis subtype distribution and microbiota composition.
We also observed a strong correlation between Blastocystis and the genus Sporolactobacillus. These anaerobic bacteria are gram positive, spore-forming, and lactic acid-producing rods [30, 31]. The type species, S. inulinus, was first isolated from chicken feed in 1963 [30]. Other species have since been isolated from soil or the rhizosphere of plants [32–35]. In recent years, novel species of Sporolactobacillus have been cultured from decaying foodstuffs such as spoiled orange juice (S. putidus) [36] and spoiled jelly (S. pectinovorans) [37], but to our knowledge there are no publications on the role of Sporolactobacillus in the gut microbiota. Because of its lactic acid properties S. inulinus has been suggested as a possible probiotic [38]. In general, intestinal lactic acid bacteria are favoured by diets high in non-metabolizable sugars which are typically found in vegetables [39].
Another correlation was found between Blastocystis and Candidatus Carsonella of the Gammaproteobacteria. Candidatus Carsonella ruddii, the only described species of this genus, has a tiny genome and seems to lack genes essential for bacterial life on its own [40]. It is a strict endosymbiont of sap-feeding insects of the family Psyllidae, jumping plant-lice [41], which in turn are monophagous, i.e. have a strict host plant specificity [42]. The correlation to Blastocystis is enigmatic, and could have several explanations. First of all, Blastocystis may be associated with some other, yet undescribed, species of the Candidatus Carsonella genus. Indeed, endosymbionts of Blastocystis have been described [43], and since the rDNA sequence similarity of species within the Candidatus Carsonella is, naturally, not known, we cannot exclude that these matches represent other endosymbiotic species. Another possibility is that the bacteria have been ingested through food contaminated with insect cells.
Several findings in this study raise intriguing questions about the role of diet in acquiring and sustaining Blastocystis colonization. Both the positive correlation to Sporolactobacillus and the negative correlation to Bacteroides could indirectly imply a link between Blastocystis-carriership and intake of vegetables. The differences in relative abundance of Bacteroides and bacterial diversity in carriers of ST3 and ST4 may indicate that these subtypes are favoured by different diets, or different ecological conditions in the gut, which in turn might explain the rarity of mixed subtype carriage. Because the life cycle of Blastocystis is still largely unknown [44, 45] and the transmission to humans in high-income countries with high sanitation standards is enigmatic, an interesting topic for future research is the detection of Blastocystis subtypes in different foodstuffs.
Conclusions
In this metagenomic study of Blastocystis and the intestinal microbiota in Swedish travellers, we found that persistent carriage of a specific Blastocystis subtype was common during intercontinental travel to low resource regions. Blastocystis was positively associated with high intestinal bacterial genus richness and the bacterial genus Sporolactobacillus while negatively associated with the bacterial genus Bacteroides, possibly linking Blastocystis to a healthy microbiota and diets high in vegetables.
Additional files
Distribution of Blastocystis subtypes overall (A), before travel to Africa (B), before travel to the Indian peninsula (C), after travel to Africa (D) and after travel to the Indian peninsula (E). Note that individuals are remarkably stable before and after travel. ND = not detected, ST = Blastocystis subtype. (PDF 52 kb)
Diversity of the gut communities across individual samples. (A) Genus richness, (B) Shannon diversity, (C) Simpson diversity. Samples are colour coded by the subtype present (ST1: green; ST2: blue; ST3: red; ST4: black; ST8: orange; Not detected: white). (PDF 58 kb)
Bacterial genera with significant correlations (overall adjusted p-value <0.001) to the abundance of specific Blastocystis subtype (ST) SSU-rDNA (PDF 45 kb)
Rarefaction curves of 50,000 reads corresponding to the bacterial 16S SSU rRNA gene (as detected by Metaxa2) in each sample. Samples are colour coded by the subtype present (ST1: green; ST2: blue; ST3: red; ST4: black; ST8: orange). (PDF 111 kb)
Principal Coordinates Analysis (PCoA) of the Bray-Curtis dissimilarities between the bacterial genus composition of the samples. Samples are colour coded by the subtype present (ST1: green; ST2: blue; ST3: red; ST4: black; ST8: orange; Not detected: white). Note the similarity of this pattern to the pattern shown in the PCA of Fig. 4. (PDF 40 kb)
Similarity between the bacterial community compositions of individuals carrying different Blastocystis subtypes, or no Blastocystis at all (ND). As in Fig. 5, rows indicate the dissimilarity of the average community of each sample group to the average community of the sample groups of the columns, as assessed by repeated random subsampling of the communities at the phylum level. The dendrogram represents the overall similarity of the sample groups. Yellow colour corresponds to high average Bray-Curtis dissimilarity and black corresponds to the average communities of the sample groups being very similar sample groups. Note that the similarity within subsamples drawn from each group is also indicated in the figure. (PDF 66 kb)
Pairwise Bray-Curtis dissimilarity of the communities at the phylum (A), family (B) and genus (C) levels. Yellow/white corresponds to more dissimilar communities, while red corresponds to near-identical communities. (PDF 798 kb)
Acknowledgements
We would like to thank Maria Casserdahl for her contribution in sample collection and Helén Edebro for her work with DNA-extraction. We acknowledge support from the Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure.
Funding
This work was financially supported by grants from Umeå University, Västerbotten County Council, the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), the Adlerbertska Research Foundation, and the scholarship fund Stiftelsen J C Kempes Minnes Stipendiefond. The funding bodies had no role in the design of the study, in data collection, analysis or interpretation, or in writing the manuscript.
Availability of data and materials
The sequence data used in this study has been deposited in the European Nucleotide Archive, project number PRJEB7369.
Abbreviations
- BLAST
Basic local alignment search tool
- ESBL
Extended spectrum beta-lactamase
- FDR
False discovery rate
- PCA
Principal component analysis
- PCoA
Principal coordinates analysis
- qPCR
Real-time polymerase chain reaction
- SSU-rDNA
Small subunit ribosomal DNA
- ST
Subtype
Authors’ contributions
All authors contributed to the study design. JF coordinated the study, collected data, interpreted data, drafted parts of the manuscript and co-wrote the manuscript. JBP allocated funds, curated the metagenomic data, performed data analysis, interpreted the data, drafted parts of the manuscript and co-wrote the manuscript. MA collected data and critically analysed the manuscript. AJ allocated funds and co-wrote the manuscript. BE allocated funds and critically analysed the manuscript. MG allocated funds and co-wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the regional ethical board in Umeå, Sweden (Dnr 2011–357-32 M). Informed written consent was obtained from all participants prior to inclusion in the study.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12866-017-1139-7) contains supplementary material, which is available to authorized users.
Contributor Information
Joakim Forsell, Phone: +46-0-90-785-11-93, Email: joakim.forsell@umu.se.
Johan Bengtsson-Palme, Email: johan.bengtsson-palme@gu.se.
Martin Angelin, Email: martin.angelin@umu.se.
Anders Johansson, Email: anders.f.johansson@umu.se.
Birgitta Evengård, Email: birgitta.evengard@umu.se.
Margareta Granlund, Email: margareta.granlund@umu.se.
References
- 1.Alfellani MA, Taner-Mulla D, Jacob AS, Imeede CA, Yoshikawa H, Stensvold CR, Clark CG. Genetic diversity of Blastocystis in livestock and zoo animals. Protist. 2013;164:497–509. doi: 10.1016/j.protis.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Silberman JD, Sogin ML, Leipe DD, Clark CG. Human parasite finds taxonomic home. Nature. 1996;380:398. doi: 10.1038/380398a0. [DOI] [PubMed] [Google Scholar]
- 3.Krogsgaard LR, Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: a population-based case-control study. Clin Gastroenterol Hepatol. 2015;13:507–513. doi: 10.1016/j.cgh.2014.07.065. [DOI] [PubMed] [Google Scholar]
- 4.Bart A, Wentink-Bonnema EM, Gilis H, Verhaar N, Wassenaar CJ, van Vugt M, Goorhuis A, van Gool T. Diagnosis and subtype analysis of Blastocystis sp. in 442 patients in a hospital setting in the Netherlands. BMC Infect Dis. 2013;23:389. doi: 10.1186/1471-2334-13-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scanlan PD, Stensvold CR, Cotter PD. Development and application of a Blastocystis subtype-specific PCR assay reveals that mixed-subtype infections are common in a healthy human population. Appl Environ Microbiol. 2015;15:4071–4076. doi: 10.1128/AEM.00520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popruk S, Udonsom R, Koompapong K, Mahittikorn A, Kusolsuk T, Ruangsittichai J, Palasuwan A. Subtype distribution of Blastocystis in Thai-Myanmar border, Thailand. Korean J Parasitol. 2015;53:13–19. doi: 10.3347/kjp.2015.53.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsell J, Granlund M, Samuelsson L, Koskiniemi S, Edebro H, Evengård B. High occurrence of Blastocystis sp. subtypes 1-3 and Giardia intestinalis assemblage B among patients in Zanzibar, Tanzania. Parasit Vectors. 2016;9:370. doi: 10.1186/s13071-016-1637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Safadi D, Gaayeb L, Meloni D, Cian A, Poirier P, Wawrzyniak I, Delbac F, Dabboussi F, Delhaes L, Seck M, et al. Children of Senegal River basin show the highest prevalence of Blastocystis sp. ever observed worldwide. BMC Infect Dis. 2014;14:164. doi: 10.1186/1471-2334-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ES, Fagbenro-Beyioku AF, Clark CG. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126:11–18. doi: 10.1016/j.actatropica.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Ramírez JD, Sánchez A, Hernández C, Flórez C, Bernal MC, Giraldo JC, Reyes P, López MC, García L, Cooper PJ, et al. Geographic distribution of human Blastocystis subtypes in South America. Infect Genet Evol. 2016;41:32–35. doi: 10.1016/j.meegid.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Roberts T, Stark D, Harkness J, Ellis J. Update on the pathogenic potential and treatment options for Blastocystis sp. Gut Pathog. 2014;6:17. doi: 10.1186/1757-4749-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stensvold CR, Clark CG. Current status of Blastocystis: a personal view. Parasitol Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Roberts T, Ellis J, Harkness J, Marriott D, Stark D. Treatment failure in patients with chronic Blastocystis infection. J Med Microbiol. 2014;63:252–7. [DOI] [PubMed]
- 14.Angelin M, Forsell J, Granlund M, Evengård B, Palmgren H, Johansson A. Risk factors for colonization with extended-spectrum beta-lactamase producing Enterobacteriaceae in healthcare students on clinical assignment abroad: a prospective study. Travel Med Infect Dis. 2015;13:223–229. doi: 10.1016/j.tmaid.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A. The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrob Agents Chemother. 2015;59:6551–6560. doi: 10.1128/AAC.00933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH. METAXA2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour. 2015;15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ, Gapped BLAST. PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011.
- 19.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507. doi: 10.1093/biomet/93.3.491. [DOI] [Google Scholar]
- 20.Bengtsson-Palme J, Thorell K, Wurzbacher C, Sjöling Å, Nilsson RH. Metaxa2 diversity tools: easing microbial community analysis with Metaxa2. Ecological Informatics. 2016;33:45–50. doi: 10.1016/j.ecoinf.2016.04.004. [DOI] [Google Scholar]
- 21.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forsell J, Granlund M, Stensvold CR, Clark CG, Evengård B. Subtype analysis of Blastocystis isolates in Swedish patients. Eur J Clin Microbiol Infect Dis. 2012;31:1689–1696. doi: 10.1007/s10096-011-1416-6. [DOI] [PubMed] [Google Scholar]
- 23.Andersen LO, Bonde I, Nielsen HB, Stensvold CR. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol Ecol. 2015;91:fiv072. doi: 10.1093/femsec/fiv072. [DOI] [PubMed] [Google Scholar]
- 24.Audebert C, Even G, Cian A, the Blastocystis investigation group. Loywick A, Merlin S, Viscogliosi E, Chabé M. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep. 2016;5:25255. doi: 10.1038/srep25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanlan PD, Stensvold CR, Rajilić-Stojanović M, Heilig HG, de Vos WM, O'Toole PW, Cotter PD. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol Ecol. 2014;90:326–330. doi: 10.1111/1574-6941.12396. [DOI] [PubMed] [Google Scholar]
- 26.O'Brien Andersen L, Karim AB, Roager HM, Vigsnæs LK, Krogfelt KA, Licht TR, Stensvold CR. Associations between common intestinal parasites and bacteria in humans as revealed by qPCR. Eur J Clin Microbiol Infect Dis. 2016;35:1427–1431. doi: 10.1007/s10096-016-2680-2. [DOI] [PubMed] [Google Scholar]
- 27.Nourrisson C, Scanzi J, Pereira B, NkoudMongo C, Wawrzyniak I, Cian A, Viscogliosi E, Livrelli V, Delbac F, Dapoigny M, et al. Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS One. 2014;9:e111868. doi: 10.1371/journal.pone.0111868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GD W, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitahara K, Suzuki J. Sporolactobacillus nov. subgen. J Gen Appl Microbiol. 1963;9:59–71. doi: 10.2323/jgam.9.59. [DOI] [Google Scholar]
- 31.Yanagida F, Suzuki K, Kaneko T, Kozaki M, Komagata K. Morphological, biochemical, and physiological characteristics of spore-forming lactic acid bacteria. J Gen Appl Microbiol. 1987;33:33–45. doi: 10.2323/jgam.33.33. [DOI] [Google Scholar]
- 32.Yanagida F, Suzuki K, Kozaki M, Komagata K. Proposal of Sporolactobacillus nakayamae subsp. nakayamae sp. nov., subsp. nov., Sporolactobacillus nakayamae subsp. racemicus subsp. nov., Sporolactobacillus terrae sp. nov., Sporolactobacillus kofuensis sp. nov., and Sporolactobacillus lactosus sp. nov. Int J Syst Bacteriol. 1997;47:499–504. doi: 10.1099/00207713-47-2-499. [DOI] [Google Scholar]
- 33.Nakayama O, Yanoshi M. Spore-bearing lactic acid bacteria isolated from rhizosphere. I. Taxonomic studies on Bacillus laevolacticus nov. sp. and Bacillus racemilacticus nov. sp. J Gen Appl Microbiol. 1967;13:139–153. doi: 10.2323/jgam.13.139. [DOI] [Google Scholar]
- 34.Hatayama K, Shoun H, Ueda Y, Nakamura A. Tuberibacillus calidus gen. Nov., sp. nov., isolated from a compost pile and reclassification of Bacillus naganoensis Tomimura et al. 1990 as Pullulanibacillus naganoensis gen. Nov., comb. nov. and Bacillus laevolacticus Andersch et al. 1994 as Sporolactobacillus laevolacticus comb. nov. Int J Syst Evol Microbiol. 2006;56:2545–2551. doi: 10.1099/ijs.0.64303-0. [DOI] [PubMed] [Google Scholar]
- 35.Chang YH, Jung MY, Park IS, Oh HM. Sporolactobacillus vineae sp. nov., a spore-forming lactic acid bacterium isolated from vineyard soil. Int J Syst Evol Microbiol. 2008;58:2316–2320. doi: 10.1099/ijs.0.65608-0. [DOI] [PubMed] [Google Scholar]
- 36.Fujita R, Mochida K, Kato Y, Goto K. Sporolactobacillus putidus sp. nov., an endospore-forming lactic acid bacterium isolated from spoiled orange juice. Int J Syst Evol Microbiol. 2014;60:1499–1503. doi: 10.1099/ijs.0.002048-0. [DOI] [PubMed] [Google Scholar]
- 37.Lan QX, Chen J, Lin L, Ye XL, Yan QY, Huang JF, Liu CC, Yang GW. Sporolactobacillus pectinivorans sp. nov., an anaerobic bacterium isolated from spoiled jelly. Int J Syst Evol Microbiol. 2016;66:4323–4328. doi: 10.1099/ijsem.0.001351. [DOI] [PubMed] [Google Scholar]
- 38.Huang HY, Huang SY, Chen PY, King VA, Lin YP, Tsen JH. Basic characteristics of Sporolactobacillus inulinus BCRC 14647 for potential probiotic properties. Curr Microbiol. 2007;54:396–404. doi: 10.1007/s00284-006-0496-5. [DOI] [PubMed] [Google Scholar]
- 39.Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol. 2012;2:86. doi: 10.3389/fcimb.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 41.Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol. 2000;66:2898–2905. doi: 10.1128/AEM.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodkinson ID. The biology of the Psylloidea (Homoptera): a review. Bull Entomol Res. 1974;64:325–339. doi: 10.1017/S0007485300031217. [DOI] [Google Scholar]
- 43.Stenzel DJ, Boreham PF. Bacteria-like endosymbionts in Blastocystis sp. Int J Parasitol. 1994;24:147–149. doi: 10.1016/0020-7519(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 44.Tan KS. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin Microbiol Rev. 2008;21:639–665. doi: 10.1128/CMR.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh M, Suresh K, Ho LC, Ng GC, Yap EH. Elucidation of the life cycle of the intestinal protozoan Blastocystis hominis. Parasitol Res. 1995;81:446–450. doi: 10.1007/BF00931510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of Blastocystis subtypes overall (A), before travel to Africa (B), before travel to the Indian peninsula (C), after travel to Africa (D) and after travel to the Indian peninsula (E). Note that individuals are remarkably stable before and after travel. ND = not detected, ST = Blastocystis subtype. (PDF 52 kb)
Diversity of the gut communities across individual samples. (A) Genus richness, (B) Shannon diversity, (C) Simpson diversity. Samples are colour coded by the subtype present (ST1: green; ST2: blue; ST3: red; ST4: black; ST8: orange; Not detected: white). (PDF 58 kb)
Bacterial genera with significant correlations (overall adjusted p-value <0.001) to the abundance of specific Blastocystis subtype (ST) SSU-rDNA (PDF 45 kb)
Rarefaction curves of 50,000 reads corresponding to the bacterial 16S SSU rRNA gene (as detected by Metaxa2) in each sample. Samples are colour coded by the subtype present (ST1: green; ST2: blue; ST3: red; ST4: black; ST8: orange). (PDF 111 kb)
Principal Coordinates Analysis (PCoA) of the Bray-Curtis dissimilarities between the bacterial genus composition of the samples. Samples are colour coded by the subtype present (ST1: green; ST2: blue; ST3: red; ST4: black; ST8: orange; Not detected: white). Note the similarity of this pattern to the pattern shown in the PCA of Fig. 4. (PDF 40 kb)
Similarity between the bacterial community compositions of individuals carrying different Blastocystis subtypes, or no Blastocystis at all (ND). As in Fig. 5, rows indicate the dissimilarity of the average community of each sample group to the average community of the sample groups of the columns, as assessed by repeated random subsampling of the communities at the phylum level. The dendrogram represents the overall similarity of the sample groups. Yellow colour corresponds to high average Bray-Curtis dissimilarity and black corresponds to the average communities of the sample groups being very similar sample groups. Note that the similarity within subsamples drawn from each group is also indicated in the figure. (PDF 66 kb)
Pairwise Bray-Curtis dissimilarity of the communities at the phylum (A), family (B) and genus (C) levels. Yellow/white corresponds to more dissimilar communities, while red corresponds to near-identical communities. (PDF 798 kb)
Data Availability Statement
The sequence data used in this study has been deposited in the European Nucleotide Archive, project number PRJEB7369.


