Abstract
Intraneural perineuriomas are benign peripheral nerve sheath tumors that cause progressive debilitating focal extremity weakness. The etiology of perineuriomas is largely unknown. We utilized whole exome sequencing, copy number algorithm evaluation, and high-resolution whole genome microarray to investigate for a genetic causal link to intraneural perineuriomas. Ten of 16 (60%) tumor cases had mutations in the WD40 domain of TRAF7, the same location for causal mutations of meningiomas. Two additional perineurioma cases had large chromosomal abnormalities in multiple chromosomes, including chromosome 22q. This study identifies a common cause for intraneural perineuriomas and an unexpected shared pathogenesis with intracranial meningiomas.
Intraneural perineurioma is a hypertrophic peripheral nerve tumor composed of whorls of perineurial cells surrounding axons that does not metastasize outside peripheral nerves.1,2 It occurs most commonly in young patients, and presents insidiously with a motor predominant neuropathy or plexopathy affecting individual nerves and/or plexuses.3,4 The perineurial cell origin is supported by immunoreactivity to epithelial membrane antigen (EMA). The pathogenic cause is unknown, and currently no therapeutic options exist. Trauma or reactive changes have been considered as causal factors,5 but the majority of reported cases have no association with a previous trauma, suggesting a genetic tumorigenic origin.3,6 A chromosome 22q11 deletion has been reported in an intraneural perineurioma patient, suggesting a potential clonal expansion neoplasm mechanism.6 Deletions of chromosome 22 also occur in other solid tumors, including benign or malignant schwannomas, neurofibromas, and benign meningiomas.7,8 In addition, non-neural soft tissue perineuriomas have shown deletions on chromosome 13 and 22q12 identified by fluorescence in situ hybridization.9,10 To date, comprehensive, high-resolution genetic studies have not been conducted. For the majority of intraneural perineuriomas, the etiology remains unknown.
We investigated the genetic causal link of intraneural perineuriomas utilizing whole exome sequencing and copy number variation (CNV) analysis. The genetic findings of this study are integrated with our earlier published clinical and histopathological evaluations of the same group of patients.3
Patients and Methods
Patients
The study was approved by our institutional review board. We identified 16 intraneural perineurioma cases with available frozen nerve tissue sample (slurry of isopentane over liquid nitrogen stored at −80 °C) within the Mayo Peripheral Nerve Laboratory database from 1985 to December 2012. All identified cases had confirmatory pathologic review by immunostaining with epithelial membrane antibodies. The nerve tissue DNAs of 16 intraneural perineurioma cases were extracted from 25mg of frozen nerve tissue (Qiagen, Valencia, CA). We also obtained 4 blood and 3 saliva samples from 7 of these 16 patients, and extracted their genomic DNA as germline controls.
Immunohistochemistry
Immunohistochemistry on paraffin sections was performed using rabbit polyclonal TRAF7 antibody (Sigma, St Louis, MO) with the Polymer Refine Detection System (Leica Microsystems, Wetzlar, Germany).
Whole Exome Sequencing and Bioinformatics Analysis
Sixteen tumor DNA, 4 blood DNA, and 3 saliva DNA samples were prepared using an Agilent Technologies (Santa Clara, CA) SureSelect Whole Exome V5 capture kit and True-seq library kit (Illumina, San Diego, CA), then sequenced on Hi-Seq4000 (Illumina). Sequencing reads were aligned to a human reference (hg19) using in-house pipeline GenomeGPS as previously described.11 Variant-detections were conducted on exons and flanking 50 base pairs of intronic sequence. Variant data for each case was summarized in variant call format (VCF). VCF files were analyzed using Ingenuity Variant Analysis software (Qiagen). Our recently developed CNV detection algorithm (PatternCNV)11 was applied to the next generation sequencing data to assess CNV.
Whole Genome Comparative Genome Hybridization Microarray
To evaluate and validate the whole genome structural variations including deletions and duplications, we also conducted whole genome Comparative Genome Hybridization (CGH) microarray analysis (Agilent 2×400K SuperPrintG3) using DNA extracted from 16 perineurioma cases. To identify the tumor-specific changes, we used the available matching germline DNA from blood or saliva as controls for 7 cases; for the other 9 cases, we used a purchased CGH control DNA sample (Agilent). This control sample consisted of an equal molar mix of 6 normal control DNA samples, which helps reduce background noise arising from the variation of each individual.
Structural Model of TRAF7 WD40 Domain
The structure of TRAF7 WD40 domain was modeled by homology with the structure of WRD5 WD40 domain (Protein Data Bank code 2H14) using the software Modeller. PyMOL (Schrödinger, New York, NY) was used to generate the structure representations.
Results
Clinical Characteristics
Summary of clinical characteristics is in the Table. The median age at the time of our evaluation was 30 years (range = 2–57 years). The median length of presentation was 54 months (range = 6–360 months), all with slowly progressive motor weakness. Median Neuropathy Impairment Score was 10.25 points (range = 3.25–49 points).3,12 The nerves affected in order of frequency were sciatic (n = 6), tibial (n = 2), brachial plexus (n = 2), radial (n = 2), peroneal (n = 1), ulnar (n = 1), lumbosacral plexus (n = 1), and median (n = 1). Characteristic diagnostic intraneural perineurioma pathology was available in all patients (Fig 1A).
TABLE.
Perineurioma Mutation and Clinical Summary of Patients Studied
| Case | Mutation Found |
Affected Nerve(s) |
Age at Evaluation, yr |
Gender | Symptom Duration, mo |
Presenting Symptom |
Pain | Sensory Loss |
NIS |
|---|---|---|---|---|---|---|---|---|---|
| C1 | TRAF7 p.L519P | Sciatic | 14 | M | 18 | Weakness and atrophy | N | Y | 7 |
| C2 | Abnormal CGHa | Tibial | 35 | M | 180 | Weakness | N | Y | 9.5 |
| C3 | TRAF7 p.S561R | Sciatic | 12 | F | 6 | Weakness | Y | Y | 7 |
| C4 | TRAF7 p.S561R | Ulnar | 13 | F | 24 | Weakness | Y | Y | 3.25 |
| C5 | None | Sciatic | 30 | M | 78 | Weakness | Y | N | 22 |
| C6 | TRAF7 p.H521R | Brachial plexus | 35 | M | 360 | Weakness | N | Y | 49 |
| C7 | TRAF7 p.S561R | Lumbosacral plexus | 2 | F | 24 | Weakness | N | Y | 30 |
| C8 | TRAF7 p.L519P | Tibial | 37 | M | 77 | Weakness | N | Y | 9.5 |
| C9 | TRAF7 p.L519P | Radial | 30 | F | 48 | Pain | Y | Y | 6 |
| C10 | Abnormal CGHa | Radial | 57 | F | 108 | Weakness | N | Y | 11 |
| C11 | TRAF7 p.H521R | Brachial plexus | 12 | F | 48 | Weakness | N | N | 19 |
| C12 | TRAF7 p.S561R | Sciatic | 7 | M | 60 | Weakness | N | N | 14 |
| C13 | None | Sciatic | 34 | M | 48 | Weakness | Y | Y | 11.5 |
| C14 | TRAF7 p.H521R | Median | 40 | M | 108 | Weakness | Y | Y | 9 |
| C15 | None | Peroneal | 18 | M | 18 | Weakness | N | Y | 7 |
| C16 | None | Sciatic | 31 | M | 156 | Weakness and atrophy | Y | Y | 12.25 |
FIGURE 1.
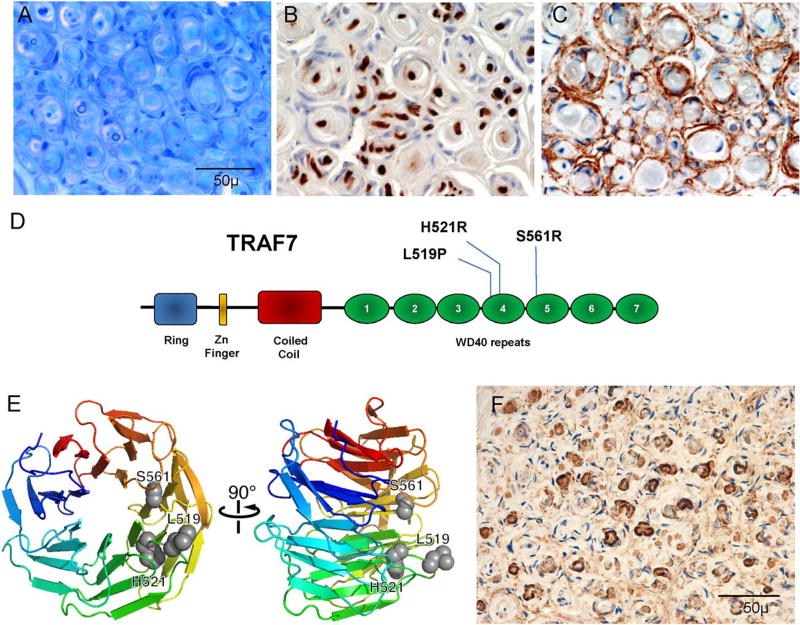
Typical histology and gene discovery in intraneural perineurioma. (A–C) Illustrative case (A) epoxy section at similar level and magnification as shown in paraffin preparations with pseudo–onion bulb formations around occasionally seen thin myelinated fibers with (B) Schwann cell preparation (S-100) demonstrating sparse reactivity of the Schwann cells at the center and (C) prominent epithelial membrane antigen (EMA)-reactive staining of the leaflets of the pseudo–onion bulbs. (D) Functional domains and location of recurrent TRAF7 mutations. The WD40 domain is the region with identified mutations in meningiomas. (E) Rainbow cartoon representation of a model of TRAF7 WD40 domain structure. The 3 residues mutated in intraneural perineuriomas are highlighted as gray spheres and cluster together on the protein predicted to interfere with TRAF7 interaction with other proteins and/or to cause instability of the WD40 domain. (F) TRAF7 immunoreactivity in perineurioma occurs in the same areas that reacted to EMA.
Exome Sequencing Identifies Somatic Novel Mutations Predicated to Alter Protein Function
Whole exome sequencing obtained an average coverage depth of ×306 for all samples. Because intraneural perineurioma is a rare disorder, we applied stringent selection criteria for mutation filtering as follows: (1) variants absent from all 4 blood and 3 saliva DNA samples, (2) absent from our in-house ~250 exome sequencing data (processed using the same analysis pipeline) from cases without perineurioma; (3) minor allele frequency < 0.0001 in ExAC databases, dbSNP147, 1000 Genomes, ESP6500, and Ingenuity-community collected exome database totaling ~100,000 samples, of which ~86,000 are exomes and 14,000 are whole genome sequencing; (4) Polyphen2 and SIFT evaluation as damaging for nonsynonymous variants; and (5) exist in at least 3 of 16 tumor cases. These criteria returned only 1 gene, tumor necrosis factor receptor-associated factor 7 (TRAF7). Specifically, 10 of 16 cases share 3 novel, heterozygous, damaging mutations in TRAF7: p.L519P (n = 3), p.H521R (n = 3), and p.S561R (n = 4), and all 3 mutations of TRAF7 are within its WD40 domain, with p.L519P and p.H521R within exon 17 and p.S561R within exon 18 (see Fig 1D). These 3 mutated sites map to a limited region of TRAF7 by our protein structure modeling (see Fig 1E).
Copy Number Variation Analysis Identified Somatic Chromosome Abnormalities
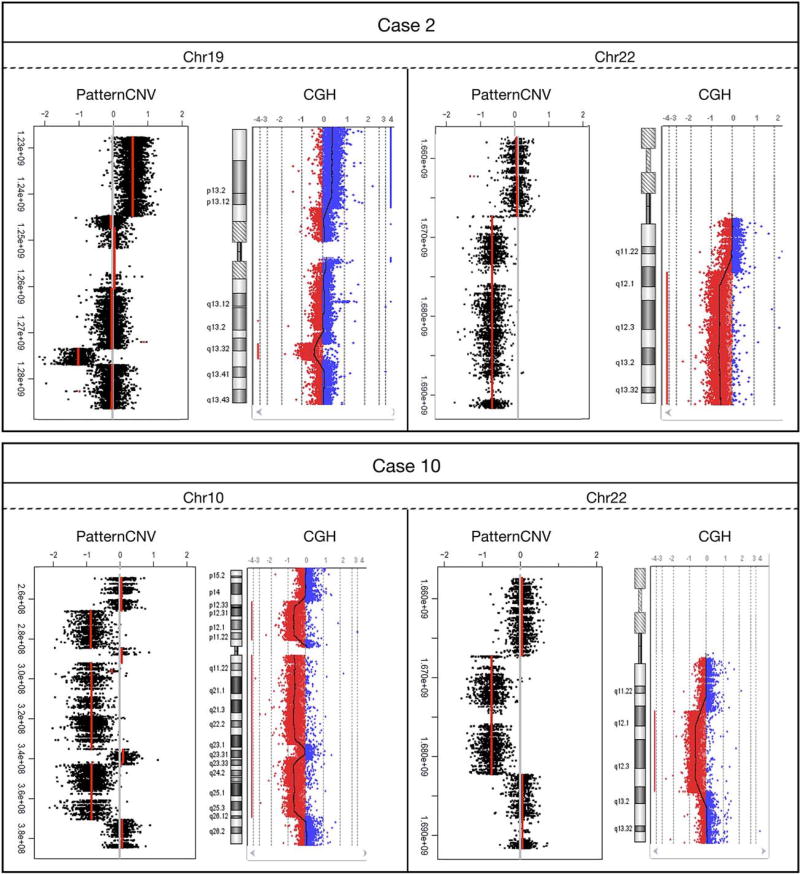
We applied CNV analysis algorithm on our exome sequencing data and found 2 cases (not overlapping with 10 cases with TRAF7 mutation) demonstrated macro duplication/deletions on multiple chromosomes (Fig 2). Specifically, Cases 2 and 10 showed large copy number abnormalities expanding sizable regions on multiple chromosomes. Case 2 demonstrated large deletions (Chr10, 11, 19, 22) and duplications (Chr16, 17, 19). Case 10 demonstrated large deletions (Chr2, 7, 10, 11, 22) and entire chromosome 13. The data from the CGH microarray analysis of 16 cases matched our CNV results obtained from exome data analysis, confirming all identified large chromosome duplication and deletions. Note both Cases 2 and 10 had chromosome 22 deletions, consistent with earlier reports.6
FIGURE 2.
Cases 2 and 10 showed large copy number abnormalities expanding sizable regions on multiple chromosomes. Shown are examples of deletions and duplications found on chromosomes 10, 19, and 22. Note both cases have large deletions on chromosome 22q. The Comparative Genome Hybridization (CGH) microarray analysis of 16 cases matched all copy number variation (CNV) results obtained from exome data analysis, providing confirmation for all deletions and duplications.
Discussion
Our comprehensive and high-resolution genomic analysis of intraneural perineuriomas identifies 3 recurrent TRAF7 somatic mutations, providing strong evidence for a specific tumor gene driver in intraneural perineurioma. The 3 novel mutations (p.L519P, p.H521R, and p.S561R) were shared among 10 cases. No clinical or histologic differences were found between patients having TRAF7 mutations (62.5%), those having large genomic deletions/duplications (12.5%), and those with no discovered mutations (25%).
The TRAF7 protein belongs to a family of signal-transducers, the tumor necrosis factor (TNF) receptor superfamily.13 In addition to having a RING finger domain and a zinc finger domain, TRAF7 is unique in this superfamily of proteins by having 7 WD40 repeats at its C-terminus (see Fig 1D).14 TRAF7 specifically interacts with MEKK3 and regulates the TNF-α/NF-κB signal transduction pathway.15 TRAF7 is a proapoptotic E3 ubiquitin-ligase, and p53 has been shown as a target of TRAF7 for ubiquitination.16 Interestingly, germline mutation in the WD40 domain of DCAF8, also an E3 ubiquitin-ligase, has been linked with giant axon neuropathy.17 Those patients have axonal swellings with dense inclusions, possibly resulting from the failure of intermediate filament breakdown. Two recent series of >300 meningioma cases showed TRAF7 mutations also frequently occur as a genetic causal factor, linked with more benign course.18,19 Thus, TRAF7 is considered as the main genetic driver for the most common primary brain tumor, and now one of the most common benign nerve tumors of young persons. All mutations identified in meningioma also occurred in the WD40 domain. Two residues, H521 and S561, mutated in both meningioma and perineurioma, but with different amino acid change.18,19 Protein modeling shows the 3 identified TRAF7 mutation sites map to a limited region of the protein (see Fig 1B). These mutations are predicted to inactivate the interaction of TRAF7 with other proteins or destabilize the WD40 domain. In grade 1 meningiomas, aside from TRAF7, 4 other genes, AKT1, SMO, NF2, and KLF4, also had mutations. We did not find any rare potential causal variants in these 4 genes.
Similar to meningiomas,18,19 chromosomal instability was found in our patients. Both cases with macro deletion/duplications had chromosome 22q deletions. We did not observe any chromosome instability in cases with TRAF7 mutation, and NF2 abnormalities were limited to the 2 patients with macro deletions of chromosome 22q. NF2 mutations tend to show greater genomic instability and malignancy potential.18,19 Of note, 1 of our cases with macro chromosomal deletion/duplication (Case 10) also has CHEK2 p.Y390C, a variant associated with lung cancer.20 That patient is without malignancy and is the oldest onset in our cohort. No patients had malignant transformation.
It will be important to investigate TRAF7 mutations in the extraneural varieties of perineuriomas, including soft tissue, sclerosing, and reticular forms. There is a greater risk of malignant transformation among them, and TRAF7 mutations may not be seen.4 Currently, local resection of intraneural perineurioma is problematic, given their endoneurial intertwined nature. Because perineurial-based tumors are now recognized to share a similar pathogenic mechanism with meningeal benign tumors, a better understanding of tumorigenic pathogenesis will be critical in discovering novel therapies.
Data Deposit
The sequencing data will be accessible through National Center for Biotechnology Information data submission SUB2189278.
Acknowledgments
Funding was provided by the Mayo Foundation and Mayo Center of Individualized Medicine.
We thank J. K. Engelstad for superior histologic preparation in review of all cases.
Footnotes
Author Contributions
Study design: C.J.K., M.L.M. Data acquisition and analysis: all authors. Drafting the manuscript and figures: C.J.K., Y.W., G.M., M.L.M.
Potential Conflicts of Interest
Nothing to report.
References
- 1.Boyanton BL, Jr, Jones JK, Shenaq SM, et al. Intraneural perineurioma: a systematic review with illustrative cases. Arch Pathol Lab Med. 2007;131:1382–1392. doi: 10.5858/2007-131-1382-IPASRW. [DOI] [PubMed] [Google Scholar]
- 2.Pina-Oviedo S, Ortiz-Hidalgo C. The normal and neoplastic perineurium: a review. Adv Anat Pathol. 2008;15:147–164. doi: 10.1097/PAP.0b013e31816f8519. [DOI] [PubMed] [Google Scholar]
- 3.Mauermann ML, Amrami KK, Kuntz NL, et al. Longitudinal study of intraneural perineurioma—a benign, focal hypertrophic neuropathy of youth. Brain. 2009;132(pt 8):2265–2276. doi: 10.1093/brain/awp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macarenco RS, Ellinger F, Oliveira AM. Perineurioma: a distinctive and underrecognized peripheral nerve sheath neoplasm. Arch Pathol Lab Med. 2007;131:625–636. doi: 10.5858/2007-131-625-PADAUP. [DOI] [PubMed] [Google Scholar]
- 5.Tsang WY, Chan JK, Chow LT, Tse CC. Perineurioma: an uncommon soft tissue neoplasm distinct from localized hypertrophic neuropathy and neurofibroma. Am J Surg Pathol. 1992;16:756–763. doi: 10.1097/00000478-199208000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Emory TS, Scheithauer BW, Hirose T, et al. Intraneural perineurioma. A clonal neoplasm associated with abnormalities of chromosome 22. Am J Clin Pathol. 1995;103:696–704. doi: 10.1093/ajcp/103.6.696. [DOI] [PubMed] [Google Scholar]
- 7.Wolff RK, Frazer KA, Jackler RK, et al. Analysis of chromosome 22 deletions in neurofibromatosis type 2-related tumors. Am J Hum Genet. 1992;51:478–485. [PMC free article] [PubMed] [Google Scholar]
- 8.Rey JA, Pestana A, Bello MJ. Cytogenetics and molecular genetics of nervous system tumors. Oncol Res. 1992;4:321–331. [PubMed] [Google Scholar]
- 9.Sato K, Ueda Y, Miwa S, et al. Low-grade malignant soft-tissue perineurioma: interphase fluorescence in situ hybridization. Pathol Int. 2008;58:718–722. doi: 10.1111/j.1440-1827.2008.02299.x. [DOI] [PubMed] [Google Scholar]
- 10.Giannini C, Scheithauer BW, Jenkins RB, et al. Soft-tissue perineurioma. Evidence for an abnormality of chromosome 22, criteria for diagnosis, and review of the literature. Am J Surg Pathol. 1997;21:164–173. doi: 10.1097/00000478-199702000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Wang C, Dawson DB, et al. Target-enrichment sequencing and copy number evaluation in inherited polyneuropathy. Neurology. 2016;86:1762–1771. doi: 10.1212/WNL.0000000000002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyck PJ, Sherman WR, Hallcher LM, et al. Human diabetic endoneurial sorbitol, fructose, and myo-inositol related to sural nerve morphometry. Ann Neurol. 1980;8:590–596. doi: 10.1002/ana.410080608. [DOI] [PubMed] [Google Scholar]
- 13.Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. 2013;8:7. doi: 10.1186/1750-2187-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Q. A network of cancer genes with co-occurring and anti-co-occurring mutations. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouwmeester T, Bauch A, Ruffner H, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Wang L, Zhang S, et al. Downregulation of ubiquitin E3 ligase TNF receptor-associated factor 7 leads to stabilization of p53 in breast cancer. Oncol Rep. 2013;29:283–287. doi: 10.3892/or.2012.2121. [DOI] [PubMed] [Google Scholar]
- 17.Klein CJ, Wu Y, Vogel P, et al. Ubiquitin ligase defect by DCAF8 mutation causes HMSN2 with giant axons. Neurology. 2014;82:873–878. doi: 10.1212/WNL.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non- NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125:351–358. doi: 10.1007/s00401-013-1093-x. [DOI] [PubMed] [Google Scholar]
- 20.Wang N, Ding H, Liu C, et al. A novel recurrent CHEK2 Y390C mutation identified in high-risk Chinese breast cancer patients impairs its activity and is associated with increased breast cancer risk. Oncogene. 2015;34:5198–5205. doi: 10.1038/onc.2014.443. [DOI] [PubMed] [Google Scholar]