Abstract
It is faster and therefore cheaper to acquire magnetic resonance images (MRI) with higher in-plane resolution than through-plane resolution. The low resolution of such acquisitions can be increased using post-processing techniques referred to as super-resolution (SR) algorithms. SR is known to be an ill-posed problem. Most state-of-the-art SR algorithms rely on the presence of external/training data to learn a transform that converts low resolution input to a higher resolution output. In this paper an SR approach is presented that is not dependent on any external training data and is only reliant on the acquired image. Patches extracted from the acquired image are used to estimate a set of new images, where each image has increased resolution along a particular direction. The final SR image is estimated by combining images in this set via the technique of Fourier Burst Accumulation. Our approach was validated on simulated low resolution MRI images, and showed significant improvement in image quality and segmentation accuracy when compared to competing SR methods. SR of FLuid Attenuated Inversion Recovery (FLAIR) images with lesions is also demonstrated.
Keywords: super-resolution, MRI, self-generated training data
1 Introduction
Spatial resolution is one of the most important imaging parameters for magnetic resonance imaging (MRI). The choice of spatial resolution in MRI is dictated by many factors such as imaging time, desired signal to noise ratio, and dimensions of the structures to be imaged. Spatial resolution—or simply, the resolution—of MRI is decided by the extent of the Fourier space acquired. Digital resolution is decided by the number of voxels that are used to reconstruct the image; it can be changed by upsampling via interpolation. However, interpolation by itself does not add to the frequency content of the image. Super-resolution (SR) is the process by which we can estimate high frequency information that is lost when a low resolution image is acquired.
MRI images are typically acquired with an anisotropic voxel size; usually with a high in-plane resolution (small voxel lengths) and a low through-plane resolution (large slice thickness). Such images are upsampled to isotropic digital resolution using interpolation. Interpolation of low resolution images causes partial volume artifacts that affect segmentation and need to be accounted for [1]. SR for natural images has been a rich area of research in the computer vision community [3, 14, 11]. There has been a significant amount of research in SR for medical images, especially neuroimaging [7, 8]. Since SR is an ill-posed problem, a popular way to perform SR in MRI is to use example-based super-resolution. Initially proposed by Rousseau [8], example-based methods leverage the high resolution information extracted from a high resolution (HR) image in conjunction with a low resolution (LR) input image to generate an SR version approximating the HR image. A number of approaches have followed up on this idea by using self-similarity [6] and a generative model [5]. However, these methods are reliant on external data that may not be readily available.
Single image SR methods [4] downsample the given LR image to create an even lower resolution image LLR and learn the mapping from the LLR image to the LR image. This mapping is then applied to the LR to generate an SR image. This approach has seen use in MRI [9] where HR and LR dictionaries were trained to learn the LR-HR mapping. However, even these methods depend on learning their dictionaries on external LR-HR training images and are not truly “single image” methods. They assume that the test LR image is a representative sample of the training data, which may not always be the case.
In this paper, we propose a method that only uses information from the available LR image to estimate its SR image. Our approach—called Self Super-resolution (SSR)—takes advantage of the fact that in an anisotropic acquisition, the in-plane resolution is higher than the through-plane resolution. SSR comprises two steps: 1) We generate new additional images, each of which is LR along a certain direction, but is HR in the plane normal to it. Thus, each new image contributes information to a new region in the Fourier space. 2) We combine these images in the Fourier space via Fourier Burst Accumulation [2]. We describe the algorithm in Section 2. In Section 3 we validate SSR results using image quality and tissue classification. We also demonstrate SR on Magnetization Prepared Gradient Echo (MPRAGE) and FLAIR images and show visually improved image appearance and tissue segmentation in the presence of white matter lesions.
2 Method
Background: Fourier Burst Accumulation
The motivation for our approach is from a recent image deblurring method devised by Delbracio et al. [2] called Fourier Burst Accumulation (FBA). Given a series of images of the same scene acquired in the burst mode of a digital camera, FBA was used to recover a single high resolution image with reduced noise. Each of the burst images is blurred due to random motion blur introduced by hand tremors and the blurring directions are independent of each other. Let x be the true high resolution image of the scene. Let yi, i ∈ {1, …, N} be the ith observed image in the burst which is blurred in a random direction with kernel hi. The observation model for yi, is yi = hi * x + σi, where σi is the additive noise in the ith image. For a non-negative parameter p, the FBA estimate x̂p(j) at voxel j is obtained by,
| (1) |
where wi(ω) are weights calculated for each frequency ω, and Yi(ω) = ℱ(yi) are the Fourier transforms of the observed burst images, yi. Simply put, the Fourier transform of the high resolution image x is a weighted average of the Fourier transforms Yi(ω) of the input burst images. The weights are given by,
| (2) |
which determines the contribution of Yi(ω) at the frequency ω. If the magnitude of Yi(ω) is large, it will dominate the summation. Thus, given LR images blurred independently from the same HR image, an estimate of the underlying HR can be calculated using FBA.
Our Approach: SSR
For the SR of MRI, the input is a LR image y0 and the expected output is the SR image x̂. We assume that y0 has low resolution in the through-plane (z) direction and has a higher spatial resolution in the in-plane. Let the spatial resolution of y0 be px × py × pz mm3, and assume that pz > px = py. In the Fourier space, the extent of the Fourier cube is [−Px, Px], where Px = 1/2px mm−1 on the ωx axis, [−Py, Py], Py = 1/2py mm−1 on the ωy axis, and [−Pz, Pz] Pz = 1/2pz mm−1 on the ωz axis. Clearly, Pz < Px = Py. To improve the resolution in the z direction, it is necessary to widen the Fourier limit on the ωz axis by estimating the Fourier coefficients for frequencies that are greater than Pz. We use FBA for estimating the Fourier coefficients at these frequencies.
FBA requires multiple images as input and expects that some of the images have the Fourier coefficients in the desired region and uses those to fill in the missing Fourier information. It is impossible to perform FBA with just a single image y0 as there is no way to estimate the Fourier information outside of [−Pz, Pz] on the ωz axis. Thus, we need additional images that can provide higher frequency information on the ωz axis. The first stage in our proposed SR algorithm is to create these additional images through synthesis given only y0.
Synthesize Intermediate Images
To gain Fourier information from frequencies greater than |Pz|, we need images that have non-zero Fourier magnitudes for frequencies that have the ωz component greater than |Pz|. In other words, we need images that are higher resolution in any direction that has a non-zero z component. We propose to use rotated and filtered versions of the available y0 to generate these images. At the outset, we upsample y0 to an isotropic px×px×px digital resolution using cubic b-spline interpolation. If x is the underlying true high resolution isotropic image, then we have, y0 = h0 * x+σ0, where h0 is the smoothing kernel, which in 2D MRI acquisitions is the inverse Fourier transform of the slice selection pulse. In an ideal case, this is a rect function because the slice selection pulse is assumed to be a sinc function. However, in reality it is usually implemented in the scanner as a truncated sinc.
Given rotation matrices Ri, i ∈ {1, …, N} perform the following steps:
Consider an image Ri(y0), which is y0 rotated by a rotation matrix Ri.
Next, apply the rotated kernel Ri(h0) to y0 to create .
Finally, apply the kernel h0 to Ri(y0) to form .
For each rotation matrix Ri, we have two new images and . From Step 2 and the definition of y0 (ignoring the noise), we know that . We use and y0 as training images by extracting features from and pairing them with corresponding patches in y0 and learn the transformation that essentially deconvolves to get y0. We need to learn this transformation so that we can apply it to and therefore deconvolve it to cancel the effect of convolution by h0. From Step 3 and the definition of y0 (ignoring the noise), we know that . Deconvolving to cancel the effects of Ri(h0) is analogous to deconvolving to cancel the effects of h0, as is also rotated.
With the training pair and y0, and test image , we use a single image SR approach known as Anchored Neighborhood Regression (ANR) [11] to learn the desired transformation. ANR was shown to be effective in 2D super-resolution of natural images with better results than some of the state-of-the-art methods [14, 3]. ANR is computationally very fast as opposed to most other SR methods [11], which is a highly desirable feature in our setting. In brief, ANR creates training data from and y0 by calculating the first and the second gradient images of in all three directions using the Sobel and Laplacian filters respectively. At voxel location j a 3D patch is extracted from each of these gradient images and concatenated to form a feature vector . ANR uses PCA to reduce the dimensionality of the feature to the order of ~ 102. In our case, we do not extract continuous voxels as the LR acquisition means neighboring voxels are highly correlated. Our patch dimensions are linearly proportional to the amount of relative blurring in the x, y, and z directions in y0. This means that patches are cuboids in shape and are longer in the dimension where the blurring is higher. We then calculate the difference image and the extracted patch and pair it with to create the training data. Next, ANR jointly learns paired high resolution and low resolution dictionaries, using the the K-SVD algorithm. The atoms of the learned dictionary are regarded as cluster centers with each feature vector being assigned to one based on the correlation between feature vectors and centers. Cluster centers and their associated feature vectors are used to estimate a projection matrix Pk for every cluster k, by solving a least squares problem such that, . Given an input test image, , feature vectors are computed and a cluster center is assigned based on the arg max of the correlation, following which, the stored Pk is applied to estimate the patch ĝj at voxel j in the newly created image ŷi. Overlapping patches are predicted and the overlapping voxels have their intensities averaged to produce the final output ŷi. This modified ANR is carried out to estimate each ŷi where the rotation matrices can be chosen intelligently to cover the Fourier space. ANR on its own can only add information in a single direction of the Fourier domain. However, when run in multiple directions, we are able to add coefficients for more frequencies in the Fourier space. We rotate ŷi back to their original orientation to get them in the same reference frame. We can now use FBA on the set {y0, } to estimate the SR result x̂ that accumulates Fourier information from all these images to enhance the resolution. Our algorithm is summarized in Algorithm 1.
Algorithm 1.
SSR
| Data: LR image y0 |
| Upsample y0 to isotropic digital resolution |
| Based on the spatial resolution of y0, calculate h0, the slice selection filter |
| for i=1:N do |
| Construct a rotation matrix Ri and apply to y0 to form the rotated image Ri(y0) |
| Apply Ri to h0 to form the rotated filter Ri(h0) |
| Generate |
| Generate |
| Use ANR to synthesize |
| end for |
| Apply FBA to get, |
3 Results
Validation: Super-resolution of T1-weighted Images
Our dataset consists of T1-weighted MPRAGE images from 20 subjects of the Neuromorphometrics dataset. The resolution of images in this dataset is 1 mm3 isotropic and we consider this our HR dataset. We create LR images by modeling a slice selection filter (h0) based on a slice selection pulse modeled as a truncated sinc function. The slice selection filter itself looks like a jagged rect function. We create LR datasets with slice thicknesses of 2 and 3 mm. The in-plane resolution remains 1 ×1 mm2.
Using these LR datasets, we generated SR images and evaluated their quality using peak signal to noise ratio (PSNR) that directly compares them with the original HR images. We also evaluated the sharpness in the SR images by calculating a sharpness metric known as S3 [13]. We compare our algorithm against trilinear and cubic b-spline interpolation, ANR, and non-local means (NLM)-based upsampling [7].
For our algorithm, we use a base patch-size of 4 × 4 × 4, which is multiplied by the blurring factor in each dimension to create a different-sized patch for each direction. We have used N = 9 rotation matrices (⇒ 9 synthetic LR images in a particular direction which are HR in the perpendicular plane). The number of dictionary elements used in ANR was 128. Table 1 details the comparison done using the PSNR metric calculated over 20 subjects. As can be seen, our algorithm outperforms the interpolation methods and ANR statistically significantly (using one-tailed t-test and Wilcoxon rank sum tests) in all the LR datasets. Table 2 shows the S3 values which are the highest for SSR.
Table 1.
Mean PSNR values (dB).
| LR (mm) | Lin. | BSP | ANR | NLM | SSR |
|---|---|---|---|---|---|
| 2 | 35.64 | 35.99 | 36.55 | 36.21 | 37.98* |
| 3 | 31.20 | 31.98 | 26.80 | 32.71 | 33.49* |
Table 2.
Mean S3 values.
| Lin. | BSP | ANR | NLM | SSR | HR |
|---|---|---|---|---|---|
| 0.43 | 0.39 | 0.47 | 0.49 | 0.65* | 0.81 |
| 0.40 | 0.39 | 0.48 | 0.45 | 0.61* | 0.81 |
In the second part of this experiment, we performed segmentation of the HR, LR, and, SR images. In the absence of HR data, we want to show that it is beneficial to apply our algorithm and use the SR images for further processing. We use an in-house implementation of the atlas-based EM algorithm for classification proposed by Van Leemput et al. [12], that we refer to as AtlasEM, which provides white matter (WM), gray matter (GM), sulcal cerebrospinal fluid (CSF), and ventricles (Ven). For each of the 2 and 3 mm LR datasets, we run AtlasEM on (1) cubic b-spline interpolated images, (2) SR images produced by ANR, NLM, and SSR, and (3) available HR images. The HR classifications are used as ground truth against which we compare other results using Dice overlap coefficients. Our observations are recorded in Table 3.
Table 3.
Mean Dice overlap scores with ground truth HR classifications for WM, GM, CSF, and ventricles, over 20 subjects.
| LR (mm) | Method | Mean Tissue Dice Coefficients | |||
|---|---|---|---|---|---|
|
| |||||
| WM | GM | CSF | Ven. | ||
| 2 | BSP | 0.960 | 0.949 | 0.950 | 0.941 |
| ANR | 0.926 | 0.795 | 0.396 | 0.881 | |
| NLM | 0.921 | 0.791 | 0.391 | 0.871 | |
| SSR | 0.973* | 0.960* | 0.957* | 0.959* | |
|
| |||||
| 3 | BSP | 0.891 | 0.773 | 0.405 | 0.832 |
| ANR | 0.877 | 0.782 | 0.490 | 0.846 | |
| NLM | 0.898 | 0.773 | 0.379 | 0.843 | |
| SSR | 0.916* | 0.811* | 0.510* | 0.875* | |
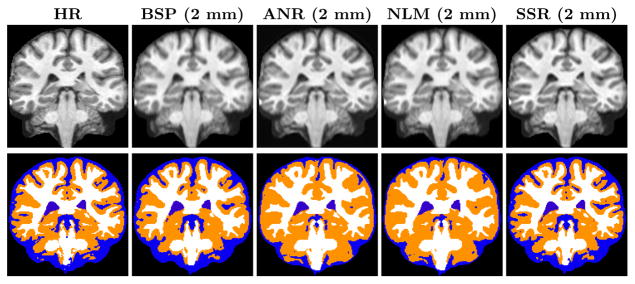
For each of the tissues we demonstrate a significant increase in the Dice, for both LR datasets. The HR, BSP, and different SR results for both the datasets are shown in the top row of Fig. 1. The SSR results are visually sharper as the fine details on the cortex and the tissue boundaries are more apparent. The classifications are shown in the bottom row of Fig. 1. The classification near the cortex and tissue boundaries is better in the SR image than the rest.
Fig. 1.
On the top row from left to right are coronal views of the original HR image, cubic bspline (BSP) interpolated image of the 2 mm LR image, ANR, NLM, and our SSR results. In the bottom row are their respective tissue classifications.
SR of MPRAGE and FLAIR Images
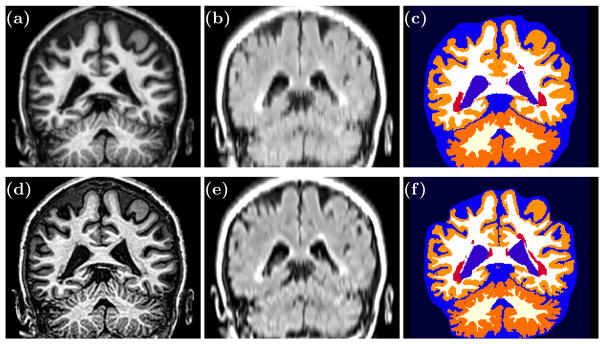
In this section, we describe an application where we use SSR to improve the resolution of FLAIR images that were acquired in a 2D acquisition with a slice thickness of 4.4 mm whereas the axial resolution was 0.828 × 0.828 mm3. The MPRAGE images were also anisotropic with a resolution of 0.828 × 0.828 × 1.17 mm3. These FLAIR images were acquired on multiple sclerosis subjects that present with white matter lesions that appear hyperintense. We generated isotropic SSR MPRAGE (Fig. 2(d)) and SSR FLAIR (Fig. 2(e)). In Figs. 2(a) and 2(b) are their interpolated counterparts. The SSR images are visually sharper than the interpolated LR images. We ran a brain and lesion segmentation algorithm [10] that shows a crisper cortex and lesion segmentation for the SSR images, but we cannot validate these results due to lack of ground truth data.
Fig. 2.
(a) Interpolated MPRAGE, (b) Interpolated FLAIR, (c) segmentation of interpolated images, (d) SSR MPRAGE, (e) SSR FLAIR, and (f) segmentation of SSR images.
4 Discussion and Conclusions
We have described SSR, an MRI super-resolution approach that uses the existing high frequency information in the given LR image to estimate Fourier coefficients of higher frequency ranges where it is absent. SSR is fast (15 mins) and needs minimal preprocessing. We have validated SSR terms of image quality metrics and segmentation accuracy. We have also demonstrated an application of FLAIR and MPRAGE SR that can potentially improve segmentation in the cortex and lesions. It is therefore an ideal replacement for interpolation.
References
- 1.Ballester MAG, et al. Estimation of the partial volume effect in MRI. Medical Image Analysis. 2002;6(4):389–405. doi: 10.1016/s1361-8415(02)00061-0. [DOI] [PubMed] [Google Scholar]
- 2.Delbracio M, Sapiro G. Removing camera shake via weighted fourier burst accumulation. IEEE Trans Image Proc. 2015;24(11):3293–3307. doi: 10.1109/TIP.2015.2442914. [DOI] [PubMed] [Google Scholar]
- 3.Freeman WT, et al. Example-based super-resolution. IEEE Computer Graphics and Applications. 2002;22(2):56–65. [Google Scholar]
- 4.Huang JB, et al. Single image super-resolution from transformed self-exemplars. IEEE Conference on Computer Vision and Pattern Recognition; 2015. [Google Scholar]
- 5.Konukoglu E, et al. Example-Based Restoration of High-Resolution Magnetic Resonance Image Acquisitions. MICCAI; 2013; 2013. pp. 131–138. [DOI] [PubMed] [Google Scholar]
- 6.Manjón JV, et al. MRI superresolution using self-similarity and image priors. International Journal of Biomedical Imaging. 2010;425891:11. doi: 10.1155/2010/425891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manjón JV, et al. Non-local MRI upsampling. Medical Image Analysis. 2010;14(6):784–792. doi: 10.1016/j.media.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Rousseau F. Brain hallucination. 2008 European Conference on Computer Vision (ECCV 2008); 2008. pp. 497–508. [Google Scholar]
- 9.Rueda A, et al. Single-image super-resolution of brain MR images using over-complete dictionaries. Medical Image Analysis. 2013;17(1):113–132. doi: 10.1016/j.media.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Shiee N, et al. A topology-preserving approach to the segmentation of brain images with Multiple Sclerosis lesions. Neuro Image. 2010;49(2):1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timofte R, et al. Anchored Neighborhood Regression for Fast Example-Based Super-Resolution. ICCV; 2013; 2013. pp. 1920–1927. [Google Scholar]
- 12.Van Leemput K, et al. Automated model-based tissue classification of MR images of the brain. IEEE Trans Med Imag. 1999;18(10):897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- 13.Vu CT, et al. S3: A Spectral and Spatial Measure of Local Perceived Sharpness in Natural Images. IEEE Trans Image Proc. 2012;21(3):934–945. doi: 10.1109/TIP.2011.2169974. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, et al. Image Super-resolution via Sparse Representation. IEEE Trans Image Proc. 2010;19(11):2861–2873. doi: 10.1109/TIP.2010.2050625. [DOI] [PubMed] [Google Scholar]