Abstract
The family Cystoviridae includes enveloped viruses with a tri-segmented dsRNA genome and a double-layered protein capsid. The innermost protein shell is a polymerase complex responsible for genome packaging, replication and transcription. Cystoviruses infect Gram-negative bacteria, primarily plant-pathogenic Pseudomonas syringae strains. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the taxonomy of the Cystoviridae, which is available at http://www.ictv.global/report/cystoviridae.
Keywords: Cystoviridae, ICTV, taxonomy, Pseudomonas phage phi6
Abbreviation
PC, polymerase complex.
Virion
The spherical virion of a cystovirus has three structural layers (Fig. 1 and Table 1). The outermost layer is the lipid bilayer envelope, consisting of host-derived phospholipids [1] and four virally encoded integral membrane proteins (P6, P9, P10, P13). Host attachment spikes (formed by P3) are anchored to the envelope via fusogenic protein P6 (2). The envelope encloses the nucleocapsid, consisting of two concentric protein layers: the nucleocapsid surface shell and the polymerase complex (PC) core [2]. The nucleocapsid surface shell contains 200 copies of protein P8 trimers arranged into a T=13 icosahedral lattice [3]. The internal PC core consists of four protein species: the major structural protein P1, the RNA-dependent RNA polymerase P2 [4], the hexameric packaging NTPase P4 [5] and the assembly cofactor P7 [6]. The structural framework of the PC core is formed by 120 copies of protein P1, arranged as asymmetric dimers on a T=1 icosahedral lattice.
Fig. 1.
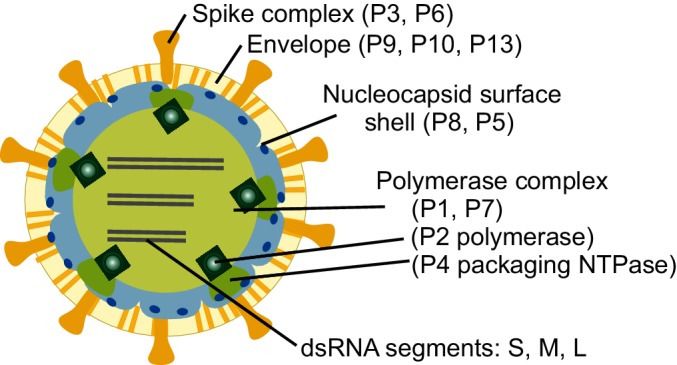
Schematic presentation of cystovirus particle (Pseudomonas phage phi6) with location of virion proteins.
Table 1. Characteristics of the family Cystoviridae.
| Typical member: | Pseudomonas phage phi6 (Segment S, M12921; Segment M, M17462; Segment L, M17461), species Pseudomonas virus phi6, genus Cystovirus |
|---|---|
| Virion | Enveloped virions (~85 nm) with two concentric, icosahedrally symmetric protein layers: the nucleocapsid surface shell (T=13) and the polymerase complex core (T=1). Spikes protrude from the virion surface |
| Genome | Three segments of linear, double-stranded RNA, totaling 13.4 kbp, encoding 13 genes |
| Replication | Single-stranded genomic precursor molecules are packaged into the viral polymerase complex. The packaged RNA molecules are replicated and transcribed within the particle |
| Translation | Viral proteins are translated from polycistronic messenger RNA molecules |
| Host range | Gram-negative bacteria, mostly Pseudomonas species |
| Taxonomy | One genus (Cystovirus) and one species (Pseudomonas virus phi6) |
Genome
Cystoviruses have three segments of linear, double-stranded RNA, named according to their size as L (large, 6.4 kbp), M (medium, 4.1 kbp) and S (small, 2.9 kbp) (Fig. 2). One copy of each genome segment is encapsidated in a virion. In each segment, genes are clustered into functional groups. The coding regions are flanked by terminal non-coding regions containing signals for genome packaging and replication [2].
Fig. 2.
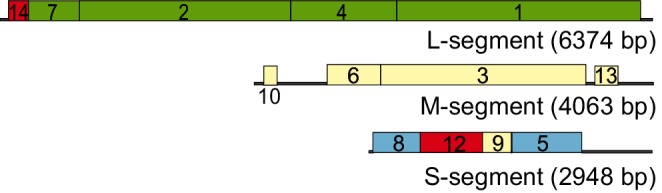
Genome organization of Pseudomonas phage phi6. The gene and protein numbers are the same. Colourings indicate genes encoding constituents of the polymerase complex (green), nucleocapsid (blue), envelope-associated proteins (cream) and non-structural proteins (red).
Replication
Infection is initiated when the virion adsorbs to host pili [7]. As the pilus retracts, the virus particle reaches the bacterial outer membrane. Subsequently, envelope protein P6 induces fusion between the viral envelope and the host outer membrane, resulting in the release of the nucleocapsid into the periplasmic space [8]. The peptidoglycan layer is digested by virion-associated lytic enzyme P5 and the nucleocapsid is exposed to the host cytoplasmic membrane. Via an endocytic-like process, the nucleocapsid enters the cytoplasm [9]. The virion-associated RNA polymerase [4] is activated and viral transcription commences. Transcription is semi-conservative [10] and produces full-length, polycistronic copies of the genome segments (Table 1). Early in the infection approximately equal amounts of messenger RNA molecules are produced from each genome segment. The early proteins translated from the L transcript assemble to form empty PC cores [6]. One copy of each type of transcript is packaged into an empty PC core, ultimately triggering the negative-strand synthesis within the core [6]. After replication, a second round of transcription initiates, resulting in the predominant production of S and M transcripts that direct the production of late proteins needed in virion assembly [2]. The nucleocapsid surface shell assembles on the genome-containing polymerase complex [6]. Finally, the envelope derived from the host plasma membrane [1] encloses the nucleocapsid and spikes attach on the virion surface. Ultimately, mature virions are released upon virus-induced host cell lysis [7].
Taxonomy
A single genus, Cystovirus, with one species, Pseudomonas virus phi6.
Resources
Full ICTV Online (10th) Report: http://www.ictv.global/report/cystoviridae.
Funding information
The authors acknowledge funding from the Academy of Finland FinSynBio programme (grant 272507) and Finnish Centre of Excellence Program (the CoE in Biological Interactions, grant 252411). Production of this summary, the online chapter and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
The authors acknowledge Professor Dennis Bamford, who has contributed to the description of the Cystoviridae family in the previous ICTV Reports. Members of the ICTV Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Michael J. Adams, Donald B. Smith, Richard J. Orton and Andrew Kropinski.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Laurinavičius S, Bamford DH, Somerharju P. Transbilayer distribution of phospholipids in bacteriophage membranes. Biochim Biophys Acta. 2007;1768:2568–2577. doi: 10.1016/j.bbamem.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Poranen MM, Tuma R, Bamford DH. Assembly of double-stranded RNA bacteriophages. Adv Virus Res. 2005;64:15–43. doi: 10.1016/S0065-3527(05)64002-X. [DOI] [PubMed] [Google Scholar]
- 3.Sun Z, El Omari K, Sun X, Ilca SL, Kotecha A, et al. Double-stranded RNA virus outer shell assembly by bona fide domain-swapping. Nat Commun. 2017;8:14814. doi: 10.1038/ncomms14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 5.Pirttimaa MJ, Paatero AO, Frilander MJ, Bamford DH. Nonspecific nucleoside triphosphatase P4 of double-stranded RNA bacteriophage φ6 is required for single-stranded RNA packaging and transcription. J Virol. 2002;76:10122–10127. doi: 10.1128/JVI.76.20.10122-10127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poranen MM, Paatero AO, Tuma R, Bamford DH. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol Cell. 2001;7:845–854. doi: 10.1016/S1097-2765(01)00228-3. [DOI] [PubMed] [Google Scholar]
- 7.Bamford DH, Palva ET, Lounatmaa K. Ultrastructure and life cycle of the lipid-containing bacteriophage φ6. J Gen Virol. 1976;32:249–259. doi: 10.1099/0022-1317-32-2-249. [DOI] [PubMed] [Google Scholar]
- 8.Bamford DH, Romantschuk M, Somerharju PJ. Membrane fusion in prokaryotes: bacteriophage φ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 1987;6:1467–1473. doi: 10.1002/j.1460-2075.1987.tb02388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poranen MM, Daugelavičius R, Ojala PM, Hess MW, Bamford DH. A novel virus-host cell membrane interaction. Membrane voltage-dependent endocytic-like entry of bacteriophage φ6 nucleocapsid. J Cell Biol. 1999;147:671–682. doi: 10.1083/jcb.147.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Usala SJ, Brownstein BH, Haselkorn R. Displacement of parental RNA strands during in vitro transcription by bacteriophage φ6 nucleocapsids. Cell. 1980;19:855–862. doi: 10.1016/0092-8674(80)90076-8. [DOI] [PubMed] [Google Scholar]


