Abstract
Use and misuse of antibiotics has driven the evolution of serine β-lactamases to better recognize new generations of β-lactam drugs, but the selective pressures driving evolution of metallo-β-lactamases are less clear. Here, we present evidence that New Delhi Metallo-β-lactamase (NDM) is evolving to overcome the selective pressure of zinc(II) scarcity. Studies of NDM-1, NDM-4 (M154L), and NDM-12 (M154L, G222D) demonstrate that the point mutant M154L, contained in 50% of clinical NDM variants, selectively enhances resistance to the penam ampicillin at low zinc(II) concentrations relevant to infection sites. Each of the clinical variants is shown to be progressively more thermostable and to bind zinc(II) more tightly than NDM-1, but a selective enhancement of penam turnover at low zinc(II) concentrations indicates that most of the improvement derives from catalysis rather than stability. X-ray crystallography of NDM-4 and NDM-12, as well as bioinorganic spectroscopy of dizinc(II), zinc(II)/cobalt(II), and dicobalt (II) metalloforms probe the mechanism of enhanced resistance and reveal perturbations of the dinuclear metal cluster that underlie improved catalysis. These studies support the proposal that zinc(II) scarcity, rather than changes in antibiotic structure, is driving the evolution of new NDM variants in clinical settings.
Keywords: Antibiotic resistance, beta-lactamase, zinc, nutritional immunity, evolution
Graphical Abstract
Since the first report of New Delhi Metallo-β-lactamase (NDM), now a worldwide, community-acquired, antibiotic-resistance determinant in Enterobacteriaceae, 18 different variants of this metallo-β-lactamase (MBL) containing altered amino acid sequences have been identified from the clinic or from infected animals.1–19 The emergence of these NDM variants raises a number of questions: How do the variations impact function? What selective pressure drives the changes? Will the variants impact clinical outcomes? Some of these questions have been previously addressed for serine β-lactamases. For instance, extended-spectrum-β-lactamases are an infamous example for which the use and misuse of β-lactam drugs appear to have driven the evolution of variants with altered substrate selectivity that can accommodate new substrate classes and thus extend antibiotic resistance.20–21 However, NDM is already a highly efficient catalyst with broad substrate selectivity that encompasses all of the clinically-used classes of β-lactams, excepting monobactams.18, 22 A need to alter substrate selectivity may not be a significant selective pressure placed on NDM-encoding microorganisms. In support of this idea, it is notable that most of the identified NDM variants confer only limited changes in catalytic activity and substrate selectivity. In contrast, a number of NDM variants have significantly increased thermal stability, which could impact antibiotic resistance by extending the lifetime of the enzyme.22 Clearly, there are different mechanisms by which NDM variants could enhance antibiotic resistance. Herein, we investigate an alternative hypothesis, that some naturally-occurring NDM variants enhance the ability of the enzyme to function at low zinc(II) ion concentrations relevant at typical sites of infection.
Competition between host and pathogen for scarce and essential metal ions at an infection site is well established for iron.23 The Fe(III) ion has limited aqueous solubility (1.4 nM), and pathogens have evolved structurally diverse siderophores to effectively compete with host proteins for binding iron ions.24–25 The proposal that host and pathogen may also compete for zinc(II) is also supported.23 Humans secrete the proteins calprotectin and psoriasin, which can sequester zinc(II) and inhibit microbial infections.26 The total zinc(II) concentration in Escherichia coli is ca. 200 μM, but almost all of these ions are protein-bound, with most binding to the ribosome.27–28 Metalloregulatory proteins in E. coli are triggered at femtomolar concentrations of zinc(II).27 Some bacteria have zinc(II)-selective transporters (e.g. the ZnuABC transporter in E. coli) and the siderophores pyridine-2,6,-dithiocarboxylatic acid and yersiniabactin can also transport zinc(II), essentially acting as zincophores.29 Taken together, these observations suggest that competition for zinc(II) ions might provide a selective pressure driving the evolution of pathogens.
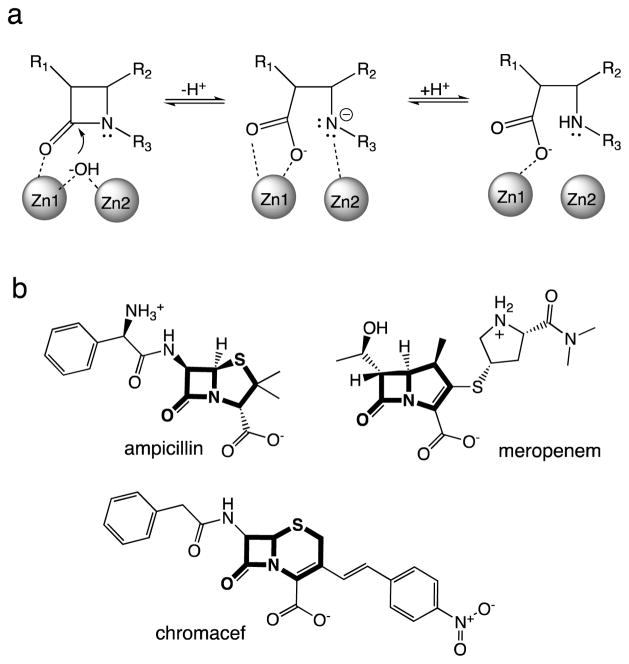
NDM requires metal ions for activity and has been characterized as a dizinc hydrolase. Zinc(II) ions are bound in adjacent Zn1 and Zn2 sites, forming a dinuclear zinc(II) ion cluster.30 In a proposed catalytic mechanism, the metal ions work in tandem to depress the pKa of a bridging hydroxide nucleophile, Zn1 polarizes the β-lactam carbonyl oxygen and stabilizes a tetrahedral adduct, and Zn2 stabilizes the developing negative charge on the leaving group nitrogen (Figure 1A).31 This metalloenzyme may have some advantages when competing for metal ion binding because its periplasmic location could facilitate access to extracellular zinc and because some metallo-β-lactamases can substitute alternative divalent metal ions if zinc(II) is not available.31–34 The Zn1 of NDM-1 is bound tightly, and the Zn2 site has a Kd value estimated, by changes in activity, to be in the low micromolar range (2 μM).35 The concentration of zinc(II) at typical infection sites appears to be in a similar range. Blood plasma has approximately 6 μM labile zinc(II) and 12 μM bound zinc(II) ions.36 Lung sputum from healthy and cystic fibrosis patients contains 0.2 – 28 μM zinc(II), and urine contains 0.6 – 10 μM zinc(II) ions.36–39 Laboratory selection experiments suggest that zinc(II) scarcity may be a selective pressure driving evolution of MBL genes. Artificial variants of a MBL with decreased zinc(II) affinity are less able to confer resistance to cells cultured using low zinc(II) concentrations.40 Specifically, the unusual N-terminal lipidation of NDM has been shown to confer stability that allows NDM to resist degradation at low zinc(II) concentrations.41 Herein, we demonstrate that the M154L substitution found in almost half of all clinical NDM variants decreases antibiotic sensitivity to specific β-lactams at low zinc(II) concentrations. This effect is facilitated, at least in part, by enhanced affinity of these NDM variants for Zn2, suggesting that NDM-bearing bacteria are evolving to overcome the selective pressure of zinc(II) scarcity on antibiotic resistance.
Figure 1.
Structures of selected β-lactams and hydrolysis by NDM-1. A) Abbreviated catalytic mechanism for NDM-1 catalyzed hydrolysis of β-lactams. For a more complete proposed mechanism, see reference (31). B) Structures of selected β-lactam structures with the (core scaffold) shown in bold: ampicillin (penam), meropenem (carbapenem), and chromacef (cephalosporin).
RESULTS AND DISCUSSION
Of the 18 variants of NDM identified from clinical samples or animals, 50% contain a substitution at Met154 (NDM-4: M154L; NDM-5: V88L, M154L; NDM-7: D130N, M154L; NDM-8: D130G, M154L; NDM-11:M154V; NDM-12: M154L, G222D; NDM-13: D95N, M154L; NDM-15: M154L, A233V; NDM-17: V88L, M154L, E170K), with all except NDM-11 incorporating a Leu residue. This change is the most common substitution found in all naturally-occurring NDM variants. Although the variants were identified in geographically distant sources, each of the Leu substitutions are encoded by a CTG codon that presumably arose from A to C point mutation(s) of the Met codon (ATG) found in NDM-1. Since the Leu for Met substitution is quite conservative, the potential for significant structural or functional changes did not, at first, appear high. Inspection of the surrounding protein environment in a crystal structure of NDM-1 revealed that the M154 side chain neighbors the α-carbon of His122, a residue that serves as a Zn1 ligand and is part of a short helical segment that also contains the Zn2 ligand Asp124.30 As previously established in other metalloproteins, changes in second-shell and surrounding residues have the potential to significantly affect active-site metal ion affinity and preference.42 Therefore, due to the prevalence of the M154L substitution in naturally-occurring NDM variants and the proximity of this substitution to the dinuclear zinc cluster, we focused on characterizing differences between NDM-1 and NDM-4 (M154L). NDM-12 (M154L, G222D) was also included in the comparison to see if any differences imparted by M154L might be retained or altered in the presence of additional substitutions.
Using methods previously established with NDM-1, we over-expressed and purified NDM-1, NDM-4 and NDM-12.35, 43 To assay substrate selectivity, we chose substrates to represent three different classes of β-lactam drugs (Figure 1B): ampicillin (penams), meropenem (carbapenems) and cephalothin (cephems). Chromacef (a cephem) was also included as a common β-lactam substrate in laboratory assays.44 For each substrate, we determined steady-state kinetic parameters with NDM-1, -4, and -12, with all assays supplemented by 10 μM ZnSO4 to provide the optimum zinc(II) concentration previously determined for NDM-1 with all three types of substrate (Table 1).35 Although there are differences observed for each parameter (kcat, KM, and kcat/KM) when NDM-1, -4 and -12 are compared, the differences are only of minor magnitude (< 5-fold), a finding that corroborates other published comparisons of substrate hydrolysis kinetics among naturally-occurring NDM variants.22
Table 1.
Steady-state kinetic parameters for NDM variants supplemented with optimum ZnSO4 (10 μM)
| Substrate & Variant | kcat (s−1) | KM (μM) | kcat/KM (M−1s−1) |
|---|---|---|---|
| Ampicillin | |||
| NDM-1 | 810 ± 40 | 240 ± 40 | 3.4 × 106 |
| NDM-4 | 1440 ± 90 | 580 ± 70 | 2.5 × 106 |
| NDM-12 | 450 ± 20 | 260 ± 40 | 1.7 × 106 |
| Meropenem | |||
| NDM-1 | 160 ± 10 | 240 ± 30 | 0.7 × 106 |
| NDM-4 | 150 ± 30 | 360 ± 90 | 0.4 × 106 |
| NDM-12 | 85 ± 5 | 130 ± 20 | 0.6 × 106 |
| Cephalothin | |||
| NDM-1 | 123 ± 3 | 17 ± 2 | 7.2 × 106 |
| NDM-4 | 86 ± 2 | 14 ± 1 | 6.1 × 106 |
| NDM-12 | 54 ± 1 | 10 ± 1 | 5.4 × 106 |
| Chromacef | |||
| NDM-1 | 7.7 ± 0.3 | 1.6 ± 0.2 | 4.8 × 106 |
| NDM-4 | 4.9 ± 0.1 | 0.7 ± 0.1 | 7.0 × 106 |
| NDM-12 | 3.9 ± 0.2 | 3.5 ± 0.6 | 1.1 × 106 |
Previously, we found that the zinc-dependence of NDM-1 activity differs with the structural class of β-lactam used in the assay.35 Under conditions of saturating (or near-saturating) substrate, the rate of NDM-1-catalyzed hydrolysis of penam substrates decreased significantly at low zinc concentrations. Carbapenem substrates showed a similar trend, but the loss of activity was of lesser magnitude. Cephalosporin substrates did not show loss of activity at low zinc concentrations. The effect of substrate structure on zinc-dependence could stem from the possibility of bound substrate enhancing Zn2 affinity, or from the possibility that Zn2 is more essential for catalyzing the hydrolysis of some substrates than others.45–47 For example, the cephalosporins we tested (e.g. chromacef) have a β-lactam nitrogen leaving-group in conjugation with strongly electron withdrawing groups that can facilitate lactam hydrolysis. This type of substrate would be the least reliant on Zn2 to stabilize the leaving group (Figure 1), and show the least dependence on zinc(II) concentrations. The carbapenems we tested have substituents that are less electron withdrawing and show a more moderate dependence on zinc(II) concentrations. The penams do not have an electron withdrawing group in conjugation with the leaving group, and would be most dependent on Zn2 for leaving group stabilization. Accordingly, hydrolysis of these penam substrates exhibit the most dramatic dependence on zinc(II) concentration. Regardless of the exact mechanism, we hypothesized that naturally-occurring NDM variants might have evolved to bind zinc(II) more tightly and to confer stability or enhanced catalysis under conditions of zinc(II) scarcity, and therefore tested the zinc(II) dependence of these variants for turnover of various substrates.
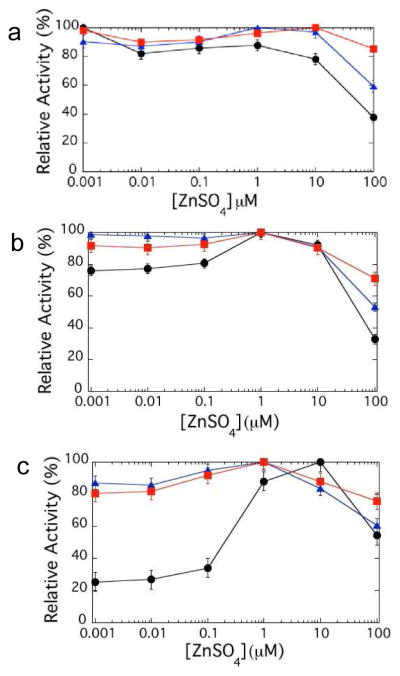
The relative activities of NDM-1, -4, and -12 with saturating substrate concentrations (when possible) were plotted and normalized to the maximum initial rate of each variant to depict the zinc(II) dependence (Figure 2). For cephalothin (a cephem), we observed only minimal changes in activity with decreasing zinc(II) concentration, similar to the behavior of nitrocefin with NDM-1.35 For meropenem (a carbapenem), we observe a moderate decrease of activity at low zinc(II) concentrations for NDM-1, but the magnitude of the decrease is less for NDM-4 and NDM-12. Most dramatically, we observed a significant loss in NDM-1 activity with ampicillin (a penam) at low zinc(II) concentrations, but activity is instead retained by NDM-4 and NDM-12. The loss of ampicillin turnover by NDM-1 under these conditions reflects changes in kcat, since the KM values for each variant decrease under low zinc(II) concentrations (Table 2). In sum, at low zinc(II) concentrations, the retention of NDM activity for each variant is dependent on the type of substrate used. Specifically with the penam ampicillin, the clinical variants NDM-4 and NDM-12 maintain their activity significantly better than NDM-1 when zinc(II) ions are scarce.
Figure 2.
Zinc (II) dependence of NDM variants for hydrolysis of various β-lactams. When possible, measurements were taken with saturating concentrations of substrates using stocks of ‘as-purified’ monozinc NDM variants, with increasing ZnSO4 concentrations in the assay solution, as described in Methods. The variants NDM-1 (●), NDM-4 (M154L,
 ), and NDM-12 (M154L, G22D,
), and NDM-12 (M154L, G22D,
 ), were assayed with the cephalosporin cephalothin (A, 300 μM), the carbapenem meropenem (B, 300 μM), and the penam ampicillin (C, 1 mM). Error bars represent the largest deviation of the mean from two experiments.
), were assayed with the cephalosporin cephalothin (A, 300 μM), the carbapenem meropenem (B, 300 μM), and the penam ampicillin (C, 1 mM). Error bars represent the largest deviation of the mean from two experiments.
Table 2.
Steady-state kinetic parameters for NDM variants supplemented with minimal ZnSO4 (1 nM)
| Substrate & Variant | kcat (s−1) | KM (μM) | kcat/KM (M−1s−1) |
|---|---|---|---|
| Ampicillin | |||
| NDM-1 | 300 ± 10 | 60 ± 8 | 5 × 106 |
| NDM-4 | 1050 ± 50 | 290 ± 40 | 3.6 × 106 |
| NDM-12 | 400 ± 20 | 200 ± 20 | 2.0 × 106 |
We next sought to determine if these NDM variants also showed different zinc(II) dependent behavior when challenged in cell culture, and if this effect differed depending on the class of β-lactam antibiotic used. We determined minimum inhibitory concentrations (MIC) of antibiotics from the three structural classes of β-lactams examined above using ‘standard’ conditions, and using conditions designed to mimic zinc(II) scarcity by including the metal chelator ethylenediaminetetraacetic acid (EDTA). Use of EDTA to limit zinc(II) ion availability for microbial growth has been described previously.40 In general, the MIC values determined using limited zinc(II) concentrations were much lower than those determined using standard conditions for all strains and antibiotics, reflecting an impaired ability of all NDM variants to confer resistance at zinc(II) concentrations similar to those at typical sites of infection (Table 3). Strains of E. coli harboring the same plasmid, but lacking the blaNDM gene have the same MIC values in both conditions, showing the effect is NDM-dependent. A more detailed analysis reveals that the magnitude of the zinc(II) dependence of the NDM variants is different depending on which antibiotic is used (Table 3, structures are shown in Figure S1). Under standard conditions, differences in MIC values for ampicillin were not observed among the variants, but at concentrations where zinc(II) is scarce, the MIC values for NDM-4 and NDM-12 were dramatically higher (≥ 64-fold), indicating that much higher antibiotic concentrations were required to inhibit cell growth. The standard conditions used with carbapenems meropenem and imipenem did not show differences in MIC values between variants, but conditions that mimic zinc(II) scarcity revealed a moderate increase in MIC values for NDM-4 and NDM-12 over NDM-1 (4-fold and 16-fold increase for meropenem (Table 3) and 4-fold and 8-fold increase for imipenem (Table S1), respectively), indicating that these variants can better enhance cell viability at low zinc(II) concentrations. Finally, the cephalosporins cephalothin, ceftazidime, cefotaxime, and cefepime all did not show differences in MIC values between NDM variants under standard conditions (cephalothin MIC values are shown in Table 3 and the other cephalosporins in Tables S2–S5). Conditions mimicking zinc(II) scarcity reveal some increases in MIC values that ranged in magnitude from >1 to 16-fold, depending on the structure of the antibiotic. In sum, the zinc(II) scarcity limits the ability of NDM to provide antibiotic resistance. Notably, the increase in cell viability conferred by NDM-4 and NDM-12 was not uniform for all classes of β-lactams, but instead showed a significantly larger effect on ampicillin, consistent with the substrate-specific changes observed in vitro using enzyme kinetics.
Table 3.
MIC values for NDM variants and various β-lactams in ‘standard’ and zinc (II) limited conditions
| Strain | Antibiotic | MIC (mg/L)Standard Conditionsb | MIC (mg/L)EDTA (50 μM)b |
|---|---|---|---|
| pHSG298a | ampicillin | 2 | 2 |
| NDM-1 | ampicillin | > 8192 | 128 |
| NDM-4 | ampicillin | > 8192 | 8192 |
| NDM-12 | ampicillin | > 8192 | > 8192 |
|
| |||
| pHSG298 | meropenem | ≤ 0.06 | ≤ 0.06 |
| NDM-1 | meropenem | 32 | 0.125 |
| NDM-4 | meropenem | 32 | 0.5 |
| NDM-12 | meropenem | 32 | 2 |
|
| |||
| pHSG298 | cephalothin | 4 | 4 |
| NDM-1 | cephalothin | > 1024 | 128 |
| NDM-4 | cephalothin | > 1024 | 512 |
| NDM-12 | cephalothin | > 1024 | 512 |
Vector-only control
Results are from three biological replicates.
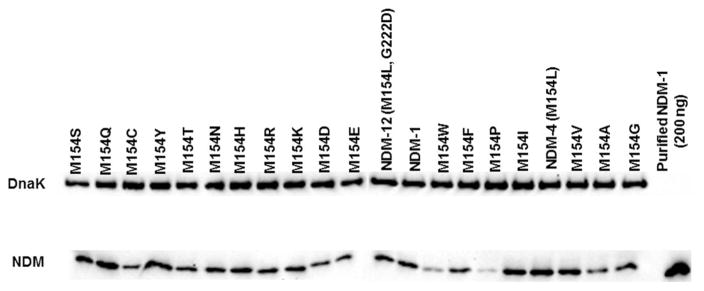
To obtain a better mechanistic understanding of these effects, we constructed a panel of E. coli strains containing plasmids encoding 20 different amino acids at the 154 position of NDM. The MIC values for these strains under standard and scarce zinc(II) conditions were determined for the same panel of antibiotics described above. The results with ampicillin (Table 4) are representative of most of the other antibiotics tested (Tables S1–S6). Under standard conditions, the M154L variant has the same MIC value as NDM-1. Other substitutions at this position generally either result in equivalent MIC values, or MIC values that are substantially decreased. The latter effect indicates increased sensitivity to β-lactams, likely due to lowered NDM concentration (Figure 3), structure, function or a possible combination of effects. The substitutions at position 154 most deleterious for MIC values of each of the antibiotics tested are consistently the non-conservative substitutions Asp or Pro. Low MIC values due to the Pro substitution can likely be attributed to lower expression or stability, but the lowered MIC values of the Asp variant are instead more likely to stem from changes in activity (Figure 3). We note an interesting Trp substitution that retains MIC values even with significantly lowered expression. Next, the panel of mutants were tested in low zinc(II) concentrations, which reveal the unique advantage conferred by the Leu substitution. All other substitutions at this position result in MIC values equal to, or considerably less than, Met (NDM-1). With all the antibiotics tested, the substitutions that are most similar to Leu, M154I and M154V, typically had the second highest MIC values, ranking either equal to Met or resulting in MIC values one or two dilutions below. Accordingly, the only other naturally-occurring variant found to encode a non-Leu substitution at this position contains a Val (NDM-11). In sum, Leu is the only substitution at position 154 that improves cell viability when treated with β-lactams under conditions of zinc(II) scarcity.
Table 4.
MIC values for ampicillin with NDM variants substituted at position 154 in standard and zinc(II) limited conditions
| Strain | MIC (mg/L) Standard Conditionsb | MIC (mg/L) EDTA (50 μM)b |
|---|---|---|
| NDM 154Ma | > 8192 | 128 |
| NDM 154La | > 8192 | 8192 |
| NDM 154I | > 8192 | 128 |
| NDM 154F | 8192 | 64 |
| NDM 154V | > 8192 | 128 |
| NDM 154W | 8192 | 16 |
| NDM 154G | 2048 | 8 |
| NDM 154Q | > 8192 | 32 |
| NDM 154K | 2048 | 8 |
| NDM 154E | 1024 | 4 |
| NDM 154A | 4096 | 8 |
| NDM 154D | 64 | 4 |
| NDM 154N | 8192 | 8 |
| NDM 154P | 64 | 4 |
| NDM 154S | 4096 | 8 |
| NDM 154T | 8192 | 8 |
| NDM 154Y | 8192 | 16 |
| NDM 154C | 4096 | 8 |
| NDM 154H | > 8192 | 16 |
| NDM 154R | 4096 | 8 |
Values are taken from Table 3.
Results from three biological replicates
Figure 3.
Expression levels of NDM in E. coli DH10B cells carrying pHSG-298/blaNDM-1 and M154 variants grown in MH broth to log phase. DNAK was used as a loading control (top band). Purified NDM-1 (200 ng) was used as a control.
Because the selective advantages of the NDM-4 and NDM-12 variants are only revealed at low zinc(II) concentrations, we sought to determine whether these variants have altered affinity for Zn2 (the labile zinc in NDM-135). An established colorimetric assay48 was used to monitor competition for binding zinc(II) between a small molecule complex with the colored zinc chelator, 4-(2-pyridylazo)resorcinol (PAR), when challenged by addition of monozinc NDM variants. Removal of zinc(II) from the PAR complex and subsequent binding to an NDM variant was expected to result in a decrease in absorbance, and these changes can be fitted to determine the Kd of the open metal-binding site in each NDM variant (Figure 4). The Kd determined for NDM-1 (18.6 ± 0.7 μM) is in the low micromolar range, and this protein has the weakest affinity. Notably, affinity for zinc(II) is improved in both of the other variants: NDM-4 (Kd = 10.0 ± 0.8 μM); NDM-12 (Kd = 6.6 ± 0.9 μM). These values are slightly higher than the Zn2 Kd determined earlier for NDM-1 using activity assays, but the difference may be attributable to the presence of substrate, which has been shown to alter zinc(II) Kd values in other MBLs.46–47
Figure 4.
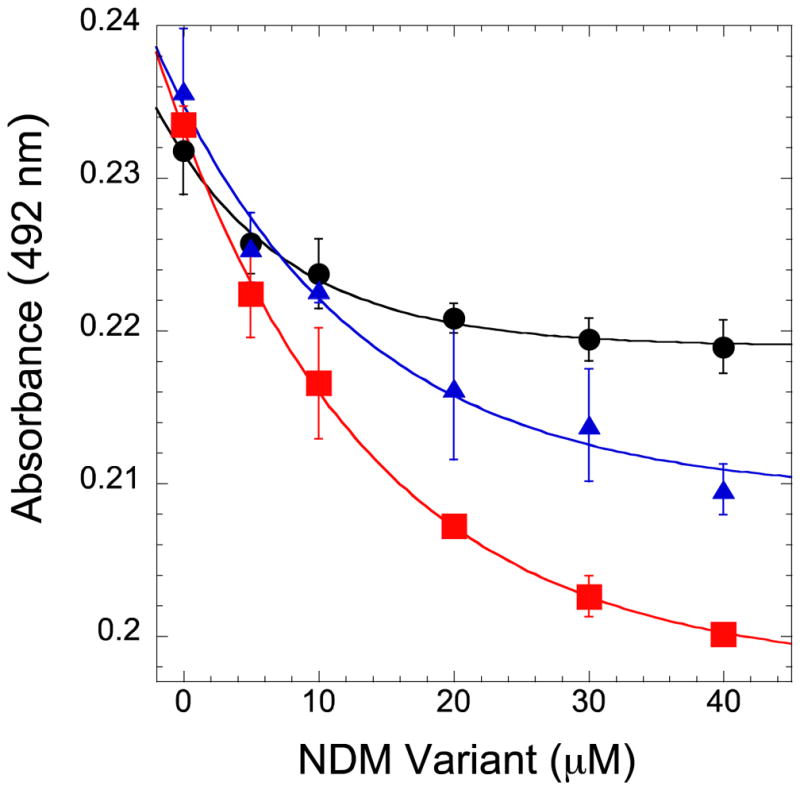
Competition for zinc(II) between NDM variants and PAR. Solutions of PAR (10 μM) and ZnSO4 (4 μM) were titrated with monozinc NDM-1 (●), NDM-4 (M154L,
 ), or NDM-12 (M154L, G222D,
), or NDM-12 (M154L, G222D,
 ). Fitting methods to derive the respective Kd values (18.6 ± 0.7 μM, 10.0 ± 0.8 μM, 6.6 ± 0.9 μM) are referenced in Materials and Methods.
). Fitting methods to derive the respective Kd values (18.6 ± 0.7 μM, 10.0 ± 0.8 μM, 6.6 ± 0.9 μM) are referenced in Materials and Methods.
Since zinc(II) binding might also stabilize enzyme structure, we assayed the zinc(II) dependence on protein stability of NDM-1, -4, and -12. Here, we used differential scanning fluorimetry as a metric of protein stability and determined melting temperatures (TMs) for each NDM variant at different zinc (II) concentrations (Figure 5). NDM-1 showed the lowest TM (57.2 ± 0.3 °C) at high zinc(II) concentrations that fill both Zn1 and Zn2 sites. The TMs for NDM-4 (59.5 ± 0.1 °C) and NDM-12 (61.7 ± 0.2 °C) are successively larger under the same conditions. At lowered zinc(II) concentrations, the TM values are, on average, approximately 20 °C lower than those of the fully-metallated protein, signifying a significant role for stabilization of the global NDM fold through binding zinc(II). The TM values of NDM-1, NDM-4, and NDM-12 are all similar at low zinc(II) concentrations, but the TM of NDM-4 (40.6 ± 0.1 °C) is slightly higher than NDM-1 and NDM-12 (NDM-12 = 36.8 ± 0.6 °C, NDM-1 = 39.1 ± 0.3 °C) with P values of 0.012 and 0.031, respectively as determined by Student’s t-test. When compared with an earlier report22 containing TMs for NDM-1 and NDM-4, the values at high zinc(II) concentrations are quite similar, but TMs at low zinc(II) concentrations differ significantly (approx. 20 °C), likely reflecting differences in the initial metal ion content of the proteins used in each study (the earlier report does not list initial metal ion content of the proteins; the experiments here start with the apo protein). In our current work, we note that the transition between the TMs at high and low zinc(II) concentrations occur at similar values to those determined for Zn2 Kd, and also show the same rank order for the variants (NDM-1 (5.06 ± 0.09 μM) > NDM-4 (4.69 ± 0.04 μM) > NDM-12 (3.7 ± 0.1 μM). Since the DSF system is not at equilibrium and, like the kinetic method, involves additional ligands which might impact zinc(II) affinity, the transitions are not fit as Kd values. However, they do illustrate the importance of metal binding to protein stability, and reveal enhanced stability for the metal-bound variants NDM-4 and NDM-12.
Figure 5.
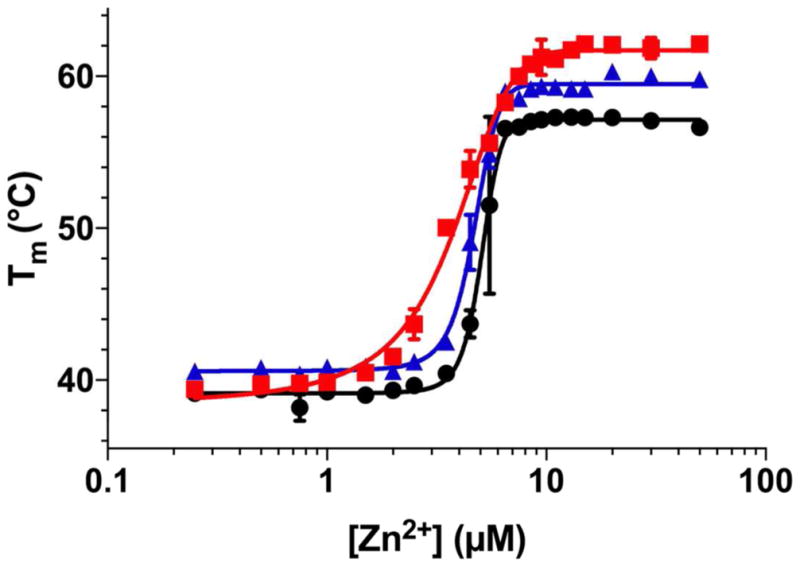
Differential Scanning Fluorimetry of NDM Variants With Varying Zinc (II) Concentrations. Melting points for NDM-1 (●), NDM-4 (M154L,
 ), and NDM-12 (M154L, G222D,
), and NDM-12 (M154L, G222D,
 ) were determined by differential scanning fluorimetry at a range of ZnCl2 concentrations from 0.25 μM to 80 μM. Fitting methods to derive the respective midpoints of the transitions (NDM-1 = 5.06 ± 0.09 μM; NDM-4 = 4.69 ± 0.04 μM; NDM-12 = 3.70 ± 0.1 μM) are described in Materials and Methods.
) were determined by differential scanning fluorimetry at a range of ZnCl2 concentrations from 0.25 μM to 80 μM. Fitting methods to derive the respective midpoints of the transitions (NDM-1 = 5.06 ± 0.09 μM; NDM-4 = 4.69 ± 0.04 μM; NDM-12 = 3.70 ± 0.1 μM) are described in Materials and Methods.
The increase in stability of the metal-bound forms of NDM-4 and NDM-12 over NDM-1 could contribute to increased MIC values of these variants in conditions of zinc(II) scarcity. MIC values for NDM-4 and NDM-12 increased for almost all β-lactams that we tested at low zinc(II) concentrations. However, a global effect on protein stability would predict a similar magnitude of increase for all classes of β-lactams. In contrast, the low zinc(II) concentrations used in both the enzyme activity assays and MIC studies reveal a particularly large improvement for NDM-4 and NDM-12 that is specific for the substrate ampicillin, suggesting instead that improvements in catalysis, rather than overall protein stability, play a more dominant role in enhancing cell survival with this penam when zinc(II) is scarce.
To determine the impact of changes in NDM-4 and NDM-12 on protein structure, we determined the X-ray crystal structures of each. Data collection, refinement, and geometric quality statistics are provided in Table 5. Our structure of NDM-4 was solved in space group P 1211 and contains two chains within the asymmetric unit. While this is the same space group as an existing NDM-4 structure in the PDB (accession ID 4TYF),49 the existing structure (4TYF) contains only one chain per asymmetric unit. Despite this difference in unit cell parameters and asymmetric unit composition, the two chains within our structure of NDM-4 superimpose with 0.16 Å and 0.19 Å RMSD to the existing NDM-4 structure. This is the first report of a NDM-12 structure. Our structure of NDM-12 was solved in space group P 1211, contains a single chain per asymmetric unit, and features unit cell dimensions similar to the existing NDM-4 structure (4TYF). Superimposition of the structural models of NDM-1, NDM-4 and NDM-12 reveal only small changes in the overall structure. Each chain within our structure of NDM-4 superimposes with 0.22 Å and 0.23 Å RMSD to an existing structure of NDM-1 (accession ID 4EXY), and our NDM-12 structure superimposes with 0.26 Å RMSD to NDM-1 (4EXY).
Table 5.
X-Ray crystallography data collection and refinement statistics for NDM-4 and NDM-12.
| NDM-442-270 | NDM-1242-270 | |
|---|---|---|
| Data Collection | ||
|
| ||
| Beam line | ALS 4.2.2 | ALS 4.2.2 |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Space group | P 1 21 1 | P 1 21 1 |
| a, b, c (Å) | 41.4, 59.2, 84.5 | 41.6, 58.7, 41.7 |
| α, β, γ (°) | 90, 98.7,90 | 90, 98.6, 90 |
| Resolution rangea (Å) | 39.18 – 1.40 (1.45 – 1.40) | 33.72 – 1.35 (1.40 – 1.35) |
| Reflectionsa | 280,017 (27,179) | 135,100 (6,384) |
| Unique reflectionsa | 79,231 (7,915) | 41,682 (3,103) |
| Completenessa (%) | 99.6 (99.9) | 95.7 (72.0) |
| Wilson B-factor | 13.3 | 13.2 |
| Redundancy | 3.5 (3.4) | 3.2 (2.1) |
| Mean I/σI | 9.68 (0.81) | 14.71 (0.90) |
| CC1/2a | 0.998 (0.396) | 0.999 (0.442) |
|
| ||
| Refinement | ||
|
| ||
| Number of reflections | 79,193 (7,907) | 41,678 (3,102) |
| PDB ID | 5WIG | 5WIH |
| Rwork / Rfree | 0.200 / 0.225 | 0.188 / 0.222 |
| Number of atoms (protein/water/ligands) | 3,452 / 608 / 4 | 1,926 / 233 / 2 |
| Average B factors (protein/water/ligands) | 19.44 / 30.71 / 18.26 | 20.94 / 34.97 / 22.73 |
|
| ||
| Model Quality | ||
|
| ||
| Bond lengths rmsd (Å) | 0.011 | 0.009 |
| Bond angles rmsd (°) | 1.46 | 1.10 |
| Ramachandran favored (%) | 98.24 | 98.24 |
| Ramachandran outliers (%) | 0.00 | 0.00 |
| Poor rotamersb (%) | 0.86 | 1.19 |
| Cβ deviations >0.25 Åb | 0 | 0 |
| Clash scoreb | 3.09 | 2.42 |
| Clash percentileb | 97th percentile (N=480; 1.40 Å ± 0.25 Å) | 99th percentile (N=466; 1.35 Å ± 0.25 Å) |
| MolProbity scoreb | 1.10 | 1.08 |
| MolProbity score percentileb | 99th percentile (N=3,363; 1.40 Å ± 0.25 Å) | 99th percentile (N=3,057; 1.35 Å ± 0.25 Å) |
Values in parentheses represent data for the highest resolution shell.
Calculated with MolProbity v4.3.1.64
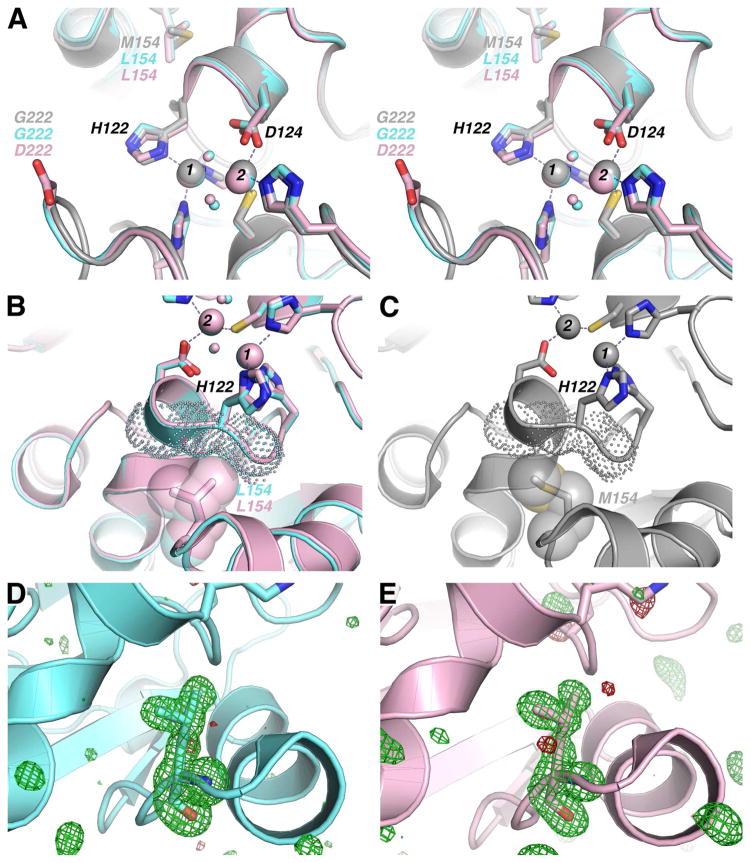
The N- and C-terminal sequences of the proteins have minor differences in conformation, Asn220 is placed as a different rotamer in NDM-4, and the putative substrate-binding β-hairpin loop (residues 63–74) is angled differently among the variants (not shown). These differences likely represent the normal range of conformations available to NDM in solution. A view focused around M154L, G222D and the active-site zinc(II) cluster (Figure 6A) shows the proximity of the Met154 side chain to the α-carbon of the Zn1 ligand His122, as well as to the short helical region that contains the Zn2 ligand Asp124. These structures show that the M154L and G222D mutations do not impart any large structural perturbations in the active-site zinc(II) cluster, or primary and secondary shell ligands in NDM. The mechanism whereby G222D imparts a functional changes is not readily apparent from the structure, although this loop borders the active site and changes in flexibility upon substitution of Gly would not necessarily be visualized in the structure. In contrast, a structural role for the M154L substitution is clear from a close analysis of interactions near the active site. Leu154 more closely buttresses the His122 α-carbon (Figure 6B), as compared to Met154 (Figure 6C), potentially restricting dynamics of the short active-site helical region containing His122 and Asp124 in NDM-4 and NDM-12. We note that only small changes in position or flexibility might be required to achieve the functional differences observed here. For example, changes to hydrophobic residues of carbonic anhydrase that neighbor primary zinc (II) ligands induce only a small difference in the X-ray crystal structure (< 1 Å movement of a β-strand), but induce a large (36-fold) change in zinc (II) ion affinity.50 In the case of NDM variants, the static structures solved crystallographically indicate no large changes in structure. Comparison of the NDM-1 (4EXY) structure with the NDM-4 and NDM-12 structures shows no significant differences in B-factors for residues surrounding the active site, including the 154 and 222 sites or the neighboring β-hairpin loop, although restraints placed on these positions by the crystal lattice may dampen changes to flexibility in solution. The absence of major structural differences suggest that the observed functional changes in the variants may result from small perturbations to the zinc(II) ligands that impact affinity, conformations accessible to zinc(II) ligands not modeled in their lowest energy state, or changes in the array of dynamic conformations available to primary zinc(II) ligands in each variant. NDM-12 was chosen to investigate the additively of other substitutions to M154L, and because of the proximity of G222D to the active site. Comparison of the NDM-4 and NDM-12 structures suggest that there is no synergistic cooperation of the M154L and G222D sites to alter structure, although a more extensive analysis of NDM variants will be required to explore interactions of multiple substitutions to structure and function.
Figure 6.
Superimposition of Structures for NDM Variants. The structural models for NDM-1 (grey; Protein Data Bank Accession Code 4EXY66), NDM-4 (M154L, light blue) and NDM-12 (M154L, G222D, purple) are superimposed using PyMOL with some atoms omitted for clarity. The large spheres in panels A–C represent the Zn1 (1) and Zn2 (2) sites, and the smaller spheres represent ordered water molecules (or bridging hydroxide ion). Atom labels and spheres are colored with respect to the NDM variant to which they belong, or in black if the label refers to all three structures. (A) A stereo image indicates the position of residues that coordinate Zn1 and Zn2, as well as the positions of the M154L and G222D mutations. (B) A close-up view of NDM-4 and NDM-12 L154 (sidechains shown as spheres) demonstrates how the δ-methyl group of the L154 side chain buttresses the backbone atoms of H122 (backbone atoms of residues 121–123 shown as dots). (C) Close-up view of NDM-1 shows that the M154 sidechain (show as spheres and sticks) does not exhibit buttressing of H122 backbone atoms (backbone atoms of residues 121–123 shown as dots). (D) Omit map generated by Polder67 within PHENIX61 by omitting NDM-4 residue L154 (sidechain shown as sticks) with positive density (green) and negative density (red) drawn at 3.5σ. (D) Omit map generated by Polder67 within PHENIX61 by omitting NDM-12 residue L154 (sidechain shown as sticks) with positive density (green) and negative density (red) drawn at 3.5σ.
In order to more sensitively determine structural differences in solution among the metal ion clusters in NDM variants, we prepared two alternative metalloforms of each variant, dicobalt(II) and zinc(II)cobalt(II), the latter of which incorporates cobalt(II) selectively at the metal-2 site coordinated by Cys208, His250 and Asp124. Substitution of diamagnetic zinc(II) with paramagnetic cobalt(II) provides a spectroscopic signal that allows interrogation of changes in structure induced by each mutation. Previously, the dicobalt(II) and zinc(II)cobalt(II) metalloforms of NDM-1 have been characterized and the kinetic parameters (kcat, KM, kcat/KM) differ from the dizinc metalloform only by < 3-fold, indicating that incorporation of these alternative metals does not greatly perturb the enzyme’s function.31
UV-Vis spectroscopy of dicobalt(II) NDM-1 shows the expected four ligand field bands between 500 and 650 nm and a more intense ligand-to-metal charge transfer band near 320 nm that arises from interaction with the Cys208 thiolate, as reported earlier (Supporting Information Figure S2).31 Dicobalt(II) NDM-4 and NDM-12 do now show significant shifts in the position of these absorption bands, although the ligand-field intensities of the two variants are somewhat obscured by a 20 % increase in S-to-Co charge-transfer (CT) intensity. The similarity of the optical spectra suggest there are no large scale changes in coordination number or geometry among the dicobalt(II) NDM variants, consistent with the crystal structures described above.
To probe structural differences in finer detail, the dicobalt(II) metalloforms of the three NDM variants were examined by 1H NMR spectroscopy. The spectrum of dicobalt(II) NDM-1 shows a number of hyperfine shifted resonances, as previously reported (bottom spectrum in Figure 7).31 Resonances at 79, 72, and 64 ppm correspond to solvent-exchangeable protons of the three His ligands that make up the metal-1 binding site (His120, His122, His189), while resonances at 165, 107, and 48 ppm correspond to Cys208, His250 and Asp124 (metal-2 site ligands). As we showed previously, using heterodimetallic forms of NDM-1, a cobalt(II) in the metal-2 site gives rise to a number of additional resonances from +40 to −65 ppm, arising from second sphere interactions with the penta-coordinate metal-2 site cobalt(II) ion.31 The spectrum of dicobalt(II) NDM-1 is strikingly different from that of the dicobalt(II) variants of NDM-4 and NDM-12, though the variant spectra are nearly identical to each other (Figure 7). The spectra show some perturbation of the metal-1 site ligands, with one His proton shifting downfield from 64 ppm, and a significant loss in intensity from the His proton at 79 ppm, which may indicate greater conformational flexibility at the metal-1 site of NDM-4 and -12.
Figure 7.
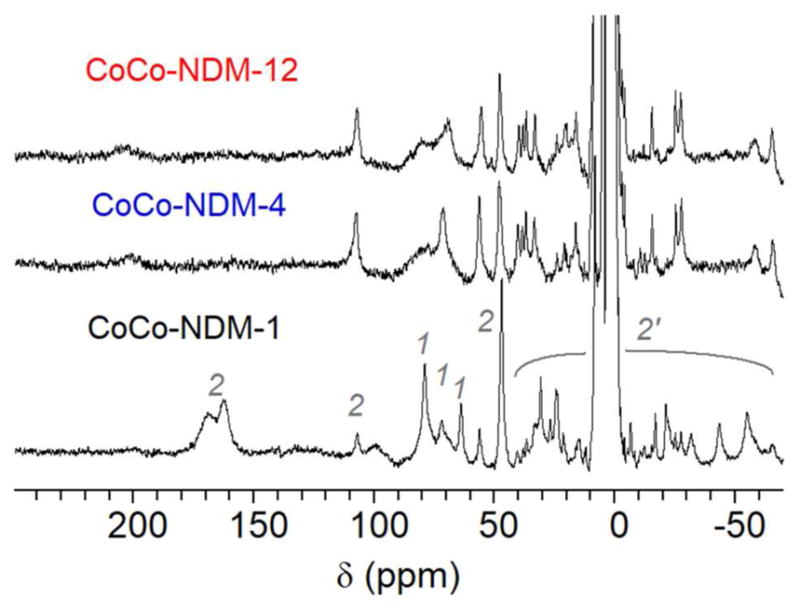
300 MHz 1H NMR spectra of dicobalt(II) NDM variants. Spectra are shown for NDM-1, NDM-4 (M154L), and NDM-12 (M154L, G222D). Labels above the NDM-1 spectrum reflect the origin of the signal: metal-1 (1) or metal-2 ligands (2) and metal-2 secondary interactions (2′).
Meanwhile, perturbation of the metal-2 site is more readily apparent. For example, the α-CH2 protons of Asp124 appear as a magnetically-equivalent geminal pair (to similar Co-O-C-H dihedral angles), leading a peak of two-proton intensity at 48 ppm. Both variants show a similar resonance at 48 ppm, at half the intensity seen for dicobalt(II) NDM-1, along with a resonance at 56 ppm of roughly equal intensity. We suggest this may indicate that the carboxylate ligand has rotated some in the variants, leading to inequivalent dihedral angles, and chemical shifts. Similarly, the resonances near 160 and 165 ppm for dicobalt(II) NDM-1 suggest a nearly symmetric disposition of the cysteine protons about the Co-S-C plane, and this symmetry is clearly lost in NDM-4 and -12, with the appearance of a new resonance near 200 ppm that lacks an obvious germinal partner. The most likely candidate appears to be a broad line that appears in the region where the metal-1 site His ligands are observed (Figure 7). Such a shift, changing the β-CH2 proton chemical shift difference from ~ 5 in NDM-1 to ~ 120 in NDM-4 and NDM-12, would in turn require a relatively large change in dihedral angle,51 and consistent with the increased CT intensity noted above. The spectra for NDM-4 and -12 also show a significant reduction in the number of secondary contacts at the metal-2 site, based on the observation of fewer resonances below 40 ppm. To verify that most of these changes, as indicated in Figure 7, are localized to the metal-2 site, we generated the zinc(II)cobalt(II) form of each NDM variant (zinc(II) in the metal-1 site, and cobalt(II) in the metal-2 site). The 1H-NMR spectra of the zinc(II)cobalt(II) NDM variants (Figure S3) were of insufficient signal-to-noise to reliably identify both Cys proton resonances for NDM-4 and -12, but they support the conclusion above that the mutations in NDM-4 and -12 more clearly affect the metal-2 site.
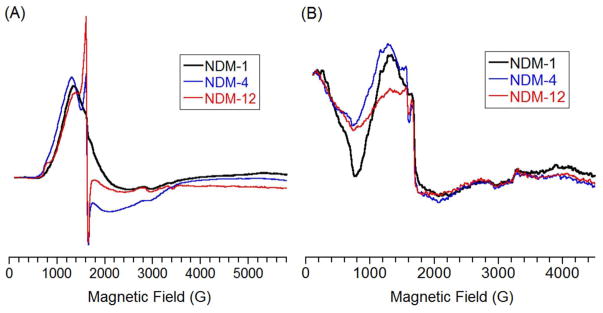
Finally, CW EPR was used to probe the metal ions directly (Figure 8). Standard perpendicular mode (B1⊥B0) spectra (Figure 8A) show commonly broad signals from the high-spin cobalt(II) ions. They show only a small difference between the three variants at very low field (~ 800 G); this small feature has been shown diagnostic for a spin-coupled dicobalt(II) system.52 The signal appears largest for NDM-1, while being difficult to identify for the other two. This perturbation is reflected in the parallel EPR spectra (Figure 8B), which show a reduction in intensity for the corresponding negative feature at ~ 800 G. The (negative) intensity of this signal has been correlated to the coupling strength between the cobalt(II) ions in other MBLs,53 and the data in Figure 8 suggest that the mutations in NDM-4 and -12 serve to further decouple the two metal ions.
Figure 8.
EPR spectra of of dicobalt(II) metalloforms of NDM variants. A) Perpendicular and B) parallel CW-EPR spectra of dicobalt(II) NDM-1, NDM-4 (M154L) and NDM-12 (M154L, G222D), as labeled, all at 1 mM. The sharp spikes at 1600 G are due to minor contamination by iron in NDM-1, -4, and -12 samples (0.05, 0.1, and 0.1 equivalents, respectively).
In conclusion, M154L, the most common point mutation found among clinical variants of NDM, confers enhanced cell viability to ampicillin-treated cultures under conditions where zinc(II) is scarce. The additional mutation in NDM-12 (G222D) can augment this effect in some cases, possibly by a separate additive mechanism. The mutations of NDM-4 and NDM-12 progressively confer additional thermostability at high zinc(II) concentrations and enhanced Zn2 affinity. The selectivity of increased MIC values of these variants for the penam ampicillin is mirrored in kinetic assays using purified enzymes and indicates that improvements in catalysis, rather than increases in overall global stability, are the major factors driving cell survival. Crystal structures of NDM-4 and -12 illustrate the proximity of the mutated sites to the dizinc cluster responsible for catalyzing hydrolysis of β-lactam substrates, and bioinorganic spectroscopy of dicobalt(II) and zinc(II)cobalt(II) metalloforms reveal how the mutations, predominantely the widespread M154L variation, perturb the structure of the catalytic metal cluster. Taken together, these studies support the proposal that clinical NDM variants are evolving to overcome zinc(II) scarcity found in common sites of infection. That zinc(II) scarcity serves as a selective pressure may also help explain the success of some non-selective metal chelators to counter NDM-1 activity in animal infection models.54–55
METHODS
Cloning, Expression, and Purification of NDM Variants for Kinetic Characterization
Recombinant, soluble Δ35 NDM-1 was expressed and purified using a codon-optimized expression plasmid described previously, pET27b-Strep-NDM1, with minor modifications to the protocol.35, 43 The N-terminal truncation removes a sequence that promotes lipidation56 and allows expression of a soluble enzyme. Briefly, cultures of E. coli BL21(DE3) (pET27b-Strep-NDM1) were grown with shaking at 25 °C in LB growth medium supplemented with 50 μM ZnSO4 until reaching mid-log phase. Production of NDM-1 was induced upon addition of isopropyl β-D-1-thiogalctopyranoside (0.5 mM) and continued overnight. Cells were pelleted by centrifugation at 2831 × g for 30 min, the supernatant discarded, and the pellets resuspended in NaH2PO4 buffer (50 mM) with NaCl (300 mM) at pH 8 for subsequent lysis by sonication. After pelleting and discarding the cell debris, soluble proteins were chromatographed using Qiagen Strep-Tactin Superflow Plus affinity medium using the manufacturer’s protocol and desthiobiotin to elute (as described previously).35, 43 The resulting recombinant soluble NDM-1 protein was dialyzed against two changes of 4 L of Storage Buffer (50 mM Tris, 150 mM NaCl, pH 7.5). For the kinetic studies reported here, the Strep-affinity tag at the N-terminus of the Δ35 NDM-1 construct was not removed because kcat and KM values determined for ampicillin and meropenam (described below) averaged within 0.3-fold of those determined after removal of the Strep tag.35 The NDM-4 (M154L) and NDM-12 (M154L, G222D) variants were prepared in the same way, except that the expression plasmids encoding these proteins were first constructed through QuikChange-PCR mutagenesis using the primers described in Supporting Information (Table S7). Before expression, the sequence of the gene inserts in the expression plasmids encoding each variant (pET27b-Strep-NDM4, pET27b-Strep-NDM12) were verified by sequencing to contain only the intended mutations. The purified recombinant NDM-1, NDM-4, and NDM-12 proteins were each judged as homogeneous by Coomassie-stained SDS-PAGE analysis (not shown). Concentrations were determined by either Bradford assay (BioRad), or a calculated extinction coefficient for the tagged variants (ε280 = 34,850 M−1cm−1), The molar zinc (II) content of each was determined using 4-(2-pyridylazo)resorcinol (PAR) in denaturing buffer as described previously,35 with “as purified” NDM-1, NDM-4, and NDM-12 containing 1.5 ± 0.2, 1.3 ± 0.1, and 1.2 ± 0.2 molar equivalents of zinc (II), respectively. Typical yields for each were ≥ 20 mg protein / 1 L culture medium.
Steady-State Kinetic Assays of NDM Variants
Steady state kinetic parameters (kcat, KM, kcat/KM) were determined for NDM-1, NDM-4, and NDM-12 with ampicillin, meropenem (both purchased from Sigma-Aldrich Inc. (St. Louis, MO)) and chromacef (a generous gift from Larry Sutton (Benedictine College, Atchison, KS)) at pH 7.0, in Hepes buffer (50 mM), supplemented by ZnSO4 (10 μM for all substrates and also 1 nM for ampicillin), using disposable polystyrene cuvettes (10 mm path length, Thermo Fisher Scientific Inc. (Fair Lawn, NJ)) and a Cary 50 Bio-UV-Visible Spectrophotometer by following the method described previously.35
To determine the zinc(II) dependence of activity, we followed a previously published procedure, with minor adjustments as noted below.35 Briefly, initial rates were determined for hydrolysis of chromacef (10 μM, monitored at 442 nm44), cephalothin (300 μM, monitored at 255 nm57), ampicillin (1 mM), or meropenem (300 μM) by each purified NDM variant (40 nM) in the presence of varying ZnSO4 concentrations (0.1 nM – 100 μM). When possible, saturating amounts of substrate were used, but lower concentrations were used when the intrinsic absorbance of the substrate was too high. To visualize zinc(II) dependence, observed rates are plotted relative to the maximum observed rate for each variant at any zinc(II) concentration tested.
Cloning of BLANDM Variants For Cell Viability Assays
The blaNDM-1 gene was cloned from K. pneumoniae 246–61A, a clinical isolate from New Delhi, in 2007.58 The upstream primer was designed to include the native promoter and lipidation signal (lipobox), as well as to retain the initial 35 residues that are omitted from constructs used to produce soluble protein for kinetic characterization (above). Both forward and reverse oligomers included a BamH1 restriction site for cloning. The PCR product was first cloned into the PCR XL-TOPO vector (Invitrogen, Carlsbad, CA). After verification by sequencing, the XL-TOPO clone was digested with BamH1 and the NDM-1 sequence inserted into BamH1 digested pHSG-298 vector (Takara, Dalian, China).
Based on the NDM-1/pHSG298 construct, sequences encoding NDM-4 and NDM-12 were generated by Celtek Biosciences (Franklin, TN) and cloned into the pBluescript II SK(+) vector. The sequence was designed with flanking Xba1 and BamH1 restriction sites in order to clone the sequence into the pHSG298 vector in the same orientation as NDM-1/pHSG298. Following restriction digest of each NDM/pBluescript SK(+) construct and pHSG298 with Xba1 and BamH1, the sequences encoding NDM-4 and NDM-12 were ligated into the pHSG298 vector. E. coli DH10B were transformed with the resulting vector and the inserted sequences verified by DNA sequencing.
The QuikChange XL site-directed mutagenesis kit (Agilent, Santa Clara, CA) was used to generate all the remaining variants at the codon encoding M154 in the NDM-1/pHSG298 construct, following the manufacturer’s protocol. For PCR mutagenesis, four degenerate complimentary primer sets and ten specific complimentary primer sets were used to generate the remaining 18 amino acid variants (Thermo Fisher Scientific, Hampton, NH) (Table S8), transformed into E. coli DH10B and the insert sequence verified by DNA sequencing.
Minimum inhibitory Concentration Measurements
MIC measurements were performed in triplicate using the Mueller-Hinton agar dilution method according to the Clinical Laboratory and Standards Institute (CLSI) protocol.59 We performed all cell viability assays in E. Coli DH101B, in order to provide a uniform genetic background in which to evaluate the impact of single amino acid changes. Briefly, bacterial cultures containing blaNDM variants cloned into a uniform vector were grown overnight at 37°C in Mueller-Hinton (M-H) broth. The cultures were diluted, and a Steers replicator was used to deliver 10 μL of a diluted overnight culture containing approximately 104 CFU. MICs were determined for the following: imipenem, meropenem (Fresnius Kabi); cefepime (WG Critical Care); and ampicillin, cephalothin, ceftazidime, cefotaxime (Sigma-Aldrich). MICs were also determined (as above) with the addition of 50 μM EDTA (United States Biochemical Corp.) to the Mueller-Hinton agar in order to limit zinc(II) availability.
Immunoblotting
Immunoblotting was used to evaluate the expression levels of NDM-1 and variants. Five-milliliter cultures of E. coli DH10B cells containing pHSG-298 phagemids harboring the various blaNDM variants in Mueller-Hinton (MH) broth containing 50 μg/mL kanamycin were grown at 37 °C to an optical density at 600 nm (OD600) of 0.8. Fifty-microliter aliquots of whole cells from these cultures were pelleted and frozen overnight. Pellets were resuspended in 20 μL loading dye, separated by SDS-PAGE, and transferred to a polyvinylidene difluoride membrane (Novex, Life Technologies, Carlsbad, CA) by electroblotting. After blocking for 1 h with 5 % nonfat dry milk, blaNDM presence on the blot was detected by incubation in 5 % nonfat dry milk with anti-NDM-1 polyclonal antibody mouse serum (1/200 dilution) and 1/10,000 dilution of mouse anti-DnaK monoclonal Ab (Enzo Life Sciences) overnight at 4°C. The membrane was washed four times, 15 min each, in Tris-buffered saline (pH 7.4) containing 0.1% Tween-20 and subsequently incubated in 5 % nonfat dry milk with 1/10,000 dilution of HRP-goat anti-mouse Ab conjugate (Santa Cruz Biotechnology). After four additional washes, the membrane was processed for exposure using the ECL kit (GE Healthcare) and FOTO/AnalystVR FX (Fotodyne).
PAR Competition Assay
Using an established assay,48 we determined the Kd values for the Zn2 site (the labile zinc ion in NDM-135) for each of the soluble Δ35 NDM variants. Briefly, the colorimetric chelator 4-(2-pyridylazo)resorcinol (PAR, 10 μM, Sigma-Aldrich) is incubated with ZnSO4 (4 μM, diluted from a 0.1M ZnSO4 standard solution, Sigma-Aldrich) in HEPES Buffer (50 mM) at pH 7.4, and then titrated with the ‘as purified’ NDM-1, NDM-4 and NDM-12 (0 – 40 μM). At each condition, concentrations of the ZnHx(PAR)2 complex and total protein were calculated from absorbance values at 492 and 280 nm, respectively, and Kd values were derived from fits as described elsewhere.48
Cloning, Expression, and Purification of NDM Variants for DSF and Crystallography
The NDM-1 coding sequence used for expressing Δ35 NDM-1 was cloned into a pET-24a vector to produce a plasmid encoding residues 42–270 with a stop codon inserted immediately after the codon for residue 270 for use expressing a tagless soluble Δ42 NDM-1 construct that better promotes crystallization. NDM-4 and NDM-12 variants were produced by site directed mutagenesis using the pET24a-NDM-1 construct as a template. NDM variants for DSF and crystallography were expressed in BL21(DE3) E. coli cells grown in Terrific Broth. Cells were harvested and resupsended in 25 mM HEPES, pH 7.5, 150 mM NaCl, 1 mg/mL lysozyme (Sigma), 20 μg/mL DNaseI (MP Biomedicals), and 100 μg/ml 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (Gold Biotechnology). Cells were immediately frozen in liquid nitrogen and thawed at 4°C with rotation the night before purification. Cell lysate was clarified by centrifugation at 17,000 × g for 45 min, followed by filtration of the supernatant through a 0.45μM filter (Fisher). Lysate was dialyzed against 20 mM HEPES, pH 7.6, 100 μM ZnCl2. Dialyzed lysate was loaded onto a HiPrep Q FF 16/10 sepharose column (GE Healthcare) and NDM-1, NDM-4, or NDM-12 were eluted using a linear gradient of dialysis buffer and increasing percentage of 20 mM HEPES, pH 7.6, 500 mM NaCl, 100μM ZnCl2. Protein eluted from the anion exchange column was concentrated using 5 kDa MWCO centrifugal concentrators prior to loading onto a HiLoad 16/600 superdex 75 (S75) size exclusion column (GE Healthcare) equilibrated with 20 mM HEPES, pH 7.6. S75 elution fractions containing NDM-1, NDM-4, or NDM-12 were concentrated and purified by an additional round of size exclusion chromatography using identical parameters. Final S75 elution fractions were concentrated using 5 kDa MWCO centrifugal concentrators prior to use for DSF or crystallization trials.
DSF Studies
Aliquots of 50 μM NDM-1, NDM-4, and NDM12 were treated with EDTA to remove zinc, followed by extensive dialysis against 20 mM HEPES, pH 7.5, 150 mM NaCl. Briefly, NDM-1, NDM-4, and NDM-12 samples were dialyzed twice against 2 mM EDTA, 20 mM HEPES, pH 7.5, 150 mM NaCl, followed by three separate dialysis steps against 20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM TCEP, and a small scoop of Chelex resin. Buffers were exchanged with approximately 12 h intervals. Zinc-depleted NDM-1, NDM-4, and NDM-12 samples were then diluted to a final concentration of 5 μM into 20 mM HEPES, pH 7.5, 150 mM NaCl, 5× SYPRO Orange (Pierce Thermo Inc.), and ZnCl2 at a range of zinc concentrations from 0.25 μM to 50 μM. Protein samples were dispensed into 96-well Frame Star PCR plate sealed with a clear thermal-seal film to prevent evaporation. DSF data was collected on a CFX96 RT-PCR (Bio-Rad) via the preset “HEX” channel for fluorescence excitation and emission. Temperature was increased in 0.5 °C increments with a five second equilibration at each step. Fluorescence intensity data was fit to Boltzmann Sigmoidal curve using Prism (GraphPad, Inc.) to determine melting temperature (Tm) of all proteins. Tm values across the range of zinc concentrations were fit to sigmoidal curves using Prism.
X-Ray Crystallography
Crystallization screening for NDM-4 and NDM-12 was carried out by preparing 96-well sitting drop vapor diffusion screens using a Phoenix crystallization robot (Art Robbins Instruments). The 800 nL sitting drops consisted of a 1:1 ratio of 1.6 mM protein and crystallization condition. Initial screening was carried out using the sparse matrix crystallization screens MCSG 1–4 (Microlytic). Optimization of initial NDM-4 hits produced diffraction quality crystals in 0.1M bis-tris, pH 6.5, and 20 % (w/v) polyethylene glycol monomethyl ether 550. Optimization of NDM-12 hits yielded diffraction quality crystals in 0.2 M ammonium iodide and 20 % (w/v) polyethylene glycol. Crystals were harvested using 50 μm MicroLoops (MiTeGen) and were cryoprotected by transfer through LV CryoOil (MiTeGen) and frozen in liquid nitrogen. Single crystal X-ray diffraction data were collected at 1.0000 Å on beamline 4.2.2 at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory. Phases for NDM-4 were calculated by molecular replacement using the PHASER60 component of PHENIX61 with the zinc-bound structure of NDM-4 (accession code 4TYF). The molecular replacement solution for the NDM-4 dataset resulted in a TFZ-score of 33.6 and a log likelihood gain of 17,847. Phases for NDM-12 were calculated by molecular replacement using PHASER within PHENIX using the final, refined structure of NDM-4 (chain A) reported in this manuscript. The molecular replacement solution for the NDM-12 dataset resulted in a TFZ-score of 29.1 and a log likelihood gain of 7,016. Each molecular replacement solution was subjected to iterative rounds of model building in Coot62 and refinement in PHENIX using individual anisotropic B-factors and TLS refinement63 for all protein atoms and individual isotropic B-factors for the solvent. All molecular structure factors have been deposited in the PDB (NDM-4 accession code 5WIG; NDM-12 accession code 5WIH). Stereochemical and geometric quality analyses of the NDM-4 and NDM-12 structures were performed with MolProbity version 4.3.164 The final NDM-4 structure, in space group P 1211 contains two chains per asymmetric unit that superimpose with 0.116 Å RMSD. Each NDM-4 chain superimposes onto the previously reported zinc-bound structure of NDM-4 (accession code 4TYF) with 0.16 Å and 0.19 Å RMSD. Both chains within the NDM-4 asymmetric unit feature two bound zinc ions and resolved electron density for all amino residues present in the crystallized construct (residues 42–270), including the beta-hairpin above the active site. Bound waters (608) were resolved bound to either or both chains within the NDM-4 asymmetric unit, while no electron density was observed for additional buffer components.
The final NDM-12 structure, also solved in space group P 1211 contains a single protein chain per asymmetric unit. The chain features two bound zinc ions and resolved electron density for all amino residues present in the crystallized construct (residues 42–270), including the beta-hairpin above the active site. Bound waters (233) were resolved within the electron density of the NDM-12 asymmetric unit, while no electron density was observed for additional buffer components.
Preparation of dicobalt(II) and zinc(II)cobalt(II) metalloforms of NDM Variants
The dicobalt(II) metalloforms of purified, tagless, Δ28-NDM-1, -4 and -12 were prepared through a process of chelation, denaturation, refolding, and addition of cobalt(II) ions. Each purified protein was placed within a 10 kDa molecular weight cutoff membrane dialysis bag and allowed to equilibrate against 1 L of dialysis buffer (unless otherwise noted) for 8 h at 4 °C before changing to the subsequent dialysis buffer to include a series of eleven dialysis steps: Four changes of Buffer A (50 mM HEPES, 150 mM NaCl at pH 6.8) with EDTA (2 mM); Urea (6M, 0.5L), Six changes of Buffer A. The resulting apo-enzymes for each NDM variant were quantified with UV-Vis (ε280 = 27,960 M−1cm−1) and concentrated to 1 mM. The dicobalt(II) metalloforms of each NDM-variant were then prepared by addition of 2 molar equivalents of CoCl2 (Strem Chemicals Inc., #27-0405), which in each case resulted in a color change of the solution from colorless to pink.
Preparation of zinc(II)cobalt(II) metalloforms of each NDM variant were prepared as described previously for NDM-1 to generate metalloforms that predominately place zinc(II) in the metal-1 site (coordinated by three His residues), and cobalt(II) in the metal-2 site (coordinated by Cys, His, and Asp residues).31
UV-Vis Spectroscopy
Apo-NDM-1, -4, and -12 were diluted to 300 μM in Buffer A. Two molar equivalents of CoCl2, from a 50 mM stock solution, were added to each protein on ice. Each sample was added to a 500 μL quartz cuvette and UV-Vis spectra were collected with a PerkinElmer Lambda 750 UV/VIS/IR spectrometer measuring absorbance from 300 to 700 nm at room temperature. A blank spectrum of Buffer A was used to generate difference spectra. All data was normalized at 700 nm.
1H-NMR Spectroscopy
NMR samples were buffered with HEPES (50 mM), NaCl (150 mM) at pH 6.8, with 10 % D2O (v/v). Each sample (300 μL) was added to a D2O matched Shigemi Advanced NMR microtube with a 5 mm outer diameter. Spectra were collected at 292 K on a Bruker ASX300 (BBI probe) 1H NMR operating at a frequency of 300.16 MHz. Spectra were collected using a frequency-switching method, applying a long, low power (270 ms) pulse centered at the water frequency, followed by a high power 3 μs pulse centered at 90 ppm.65 This method allows for suppression of the water signal with enhancement of severely hyperfine-shifted resonances. Spectra consisted of 30,000 transients of 16 k data points over a 333 ppm spectral window (taq ≈ 51 ms); signal averaging took approximately 3 h per spectrum.
EPR Spectroscopy
EPR samples containing NDM-1, -4, and -12 included approximately 10 % (v/v) glycerol as glassing agent. Samples were loaded into EPR tubes, degassed by repeated evacuation/purgation with N2 prior to data collection. Spectra were collected on a Bruker EMX EPR spectrometer, equipped with an ER4116-DM dual mode resonator (9.37 GHz, parallel; 9.62 GHz perpendicular). The data in EPR figures were scaled so that the X-axes matched (perpendicular mode field values were scaled by 9.37/9.62). Temperature control was accomplished using an Oxford ESR900 cryostat and temperature controller (4.5 K). Other spectral conditions included: microwave power = 0.2 mW; field modulation = 10 G (100 kHz); receiver gain = 104; time constant/conversion time = 41 ms.
Supplementary Material
Acknowledgments
Funding
This work was supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and National Institute of Allergy and Infectious Disease under award numbers GM111926, R01AI100560 (to RAB), R01AI063517 (to RAB), and R01AI072219 (to RAB), by the National Science Foundation (CHE-1509285 to MWC and DLT), by the Robert A. Welch Foundation (F-1572 to WF), and by Miami University through the Robert H. and Nancy J. Blayney Professorship (to RCP). This study was also supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 (to RAB) from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 (to RAB). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
The Advanced Light Source is supported by the US Department of Energy under contract number DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory. RAB and CRB thank Dr. Brad Spellberg (Professor, USC) for the gift of anti-NDM-1 polyclonal antibody mouse serum.
Footnotes
Accession Codes
Coordinates and structure factors for NDM-4 and NDM-12 have been deposited in the Protein Data Bank with accession codes 5WIG and 5WIH, respectively.
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: Structures of β-lactams used, MIC determinations, UV-Vis and 1H-NMR spectra of NDM metalloforms, and primers used for mutagenesis. (PDF)
References
- 1.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agent Chemother. 2009;53(12):5046–54. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J Antimicrob Chemother. 2011;66(6):1260–2. doi: 10.1093/jac/dkr135. [DOI] [PubMed] [Google Scholar]
- 3.Tada T, Miyoshi-Akiyama T, Shimada K, Kirikae T. Biochemical analysis of metallo-beta-lactamase NDM-3 from a multidrug-resistant Escherichia coli strain isolated in Japan. Antimicrob Agent Chemother. 2014;58(6):3538–40. doi: 10.1128/AAC.02793-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Boulanger AE, Poirel L. NDM-4 metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agent Chemother. 2012;56(4):2184–6. doi: 10.1128/AAC.05961-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agent Chemother. 2011;55(12):5952–4. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson DA, Sidjabat HE, Freeman JT, Roberts SA, Silvey A, Woodhouse R, Mowat E, Dyet K, Paterson DL, Blackmore T, Burns A, Heffernan H. Identification and molecular characterisation of New Delhi metallo-beta-lactamase-1 (NDM-1)- and NDM-6-producing Enterobacteriaceae from New Zealand hospitals. Int J Antimicrob Agent. 2012;39(6):529–33. doi: 10.1016/j.ijantimicag.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J Antimicrob Chemother. 2013;68(8):1737–40. doi: 10.1093/jac/dkt088. [DOI] [PubMed] [Google Scholar]
- 8.Tada T, Miyoshi-Akiyama T, Dahal RK, Sah MK, Ohara H, Kirikae T, Pokhrel BM. NDM-8 metallo-beta-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob Agent Chemother. 2013;57(5):2394–6. doi: 10.1128/AAC.02553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Li H, Zhao C, Chen H, Liu J, Wang Z, Wang Q, Zhang Y, He W, Zhang F, Wang H. Novel NDM-9 metallo-beta-lactamase identified from a ST107 Klebsiella pneumoniae strain isolated in China. Int J Antimicrob Agent. 2014;44(1):90–1. doi: 10.1016/j.ijantimicag.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Khajuria A, Praharaj AK, Kumar M, Grover N. Presence of a novel variant NDM-10, of the New Delhi metallo-beta-lactamase in a Klebsiella pneumoniae isolate. Ind J Med Microbiol. 2016;34(1):121–3. doi: 10.4103/0255-0857.174101. [DOI] [PubMed] [Google Scholar]
- 11.Tada T, Shrestha B, Miyoshi-Akiyama T, Shimada K, Ohara H, Kirikae T, Pokhrel BM. NDM-12, a novel New Delhi metallo-beta-lactamase variant from a carbapenem-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agent Chemother. 2014;58(10):6302–5. doi: 10.1128/AAC.03355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrestha B, Tada T, Miyoshi-Akiyama T, Shimada K, Ohara H, Kirikae T, Pokhrel BM. Identification of a novel NDM variant, NDM-13, from a multidrug-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agent Chemother. 2015;59(9):5847–50. doi: 10.1128/AAC.00332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou D, Huang Y, Zhao X, Liu W, Dong D, Li H, Wang X, Huang S, Wei X, Yan X, Yang Z, Tong Y, Huang L, Yuan J. A novel New Delhi metallo-beta-lactamase variant, NDM-14, isolated in a Chinese Hospital possesses increased enzymatic activity against carbapenems. Antimicrob Agent Chemother. 2015;59(4):2450–3. doi: 10.1128/AAC.05168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazmierczak KM, Rabine S, Hackel M, McLaughlin RE, Biedenbach DJ, Bouchillon SK, Sahm DF, Bradford PA. Multiyear, Multinational Survey of the Incidence and Global Distribution of Metallo-beta-Lactamase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agent Chemother. 2016;60(2):1067–78. doi: 10.1128/AAC.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Wang Y, Walsh TR, Liu D, Shen Z, Zhang R, Yin W, Yao H, Li J, Shen J. Plasmid-Mediated Novel blaNDM-17 Gene Encoding a Carbapenemase with Enhanced Activity in a Sequence Type 48 Escherichia coli Strain. Antimicrob Agents Chemother. 2017;61(5) doi: 10.1128/AAC.02233-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahman M, Prasad KN, Pathak A, Singh A, Mukherjee C, Ahmad S, Zorn BG. Prevalence and molecular characterisation of NDM-4, NDM-5, NDM-7, NDM-8 and a novel variant of the New Delhi Metallo-beta-lactamase NDM-11 in Enterobacteriaceae from South India. GenBank. 2015 KP265939.1. [Google Scholar]
- 17.Basu S, Mitra S. A new metallo-beta-lactamase NDM-15 in an Escherichia coli isolated from the blood of a septicaemic neonate in West Bengal, India. GenBank. 2015 KP735848.1. [Google Scholar]
- 18.Mojica MF, Bonomo RA, Fast W. B1-Metallo-beta-Lactamases: Where Do We Stand? Current drug targets. 2016;17(9):1029–50. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange F, Pfennigwerth N, Schauer J, Gatermann SG. Novel NDM variant found in Escherichia coli. GenBank. 2017 KY503030.1. [Google Scholar]
- 20.Bush K, Fisher JF. Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Ann Rev Microbiol. 2011;65:455–78. doi: 10.1146/annurev-micro-090110-102911. [DOI] [PubMed] [Google Scholar]
- 21.Patel MP, Hu L, Stojanoski V, Sankaran B, Prasad BVV, Palzkill T. The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation. Biochemistry. 2017;56(27):3443–3453. doi: 10.1021/acs.biochem.7b00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makena A, Brem J, Pfeffer I, Geffen RE, Wilkins SE, Tarhonskaya H, Flashman E, Phee LM, Wareham DW, Schofield CJ. Biochemical characterization of New Delhi metallo-beta-lactamase variants reveals differences in protein stability. J Antimicrob Chemother. 2015;70(2):463–9. doi: 10.1093/jac/dku403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer LD, Skaar EP. Transition Metals and Virulence in Bacteria. Ann Rev Genet. 2016;50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chipperfield JR, Ratledge C. Salicylic acid is not a bacterial siderophore: a theoretical study. Biometals. 2000;13(2):165–8. doi: 10.1023/a:1009227206890. [DOI] [PubMed] [Google Scholar]
- 25.Sandy M, Butler A. Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev. 2009;109(10):4580–95. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10(8):525–37. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292(5526):2488–92. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 28.Hensley MP, Tierney DL, Crowder MW. Zn(II) binding to Escherichia coli 70S ribosomes. Biochemistry. 2011;50(46):9937–9. doi: 10.1021/bi200619w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnstone TC, Nolan EM. Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans. 2015;44(14):6320–39. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Hao Q. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J. 2011;25(8):2574–82. doi: 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, Aitha M, Marts AR, Hetrick A, Bennett B, Crowder MW, Tierney DL. Spectroscopic and mechanistic studies of heterodimetallic forms of metallo-beta-lactamase NDM-1. J Am Chem Soc. 2014;136(20):7273–85. doi: 10.1021/ja410376s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schilling O, Wenzel N, Naylor M, Vogel A, Crowder M, Makaroff C, Meyer-Klaucke W. Flexible metal binding of the metallo-beta-lactamase domain: glyoxalase II incorporates iron, manganese, and zinc in vivo. Biochemistry. 2003;42(40):11777–86. doi: 10.1021/bi034672o. [DOI] [PubMed] [Google Scholar]
- 33.Hu Z, Gunasekera TS, Spadafora L, Bennett B, Crowder MW. Metal content of metallo-beta-lactamase L1 is determined by the bioavailability of metal ions. Biochemistry. 2008;47(30):7947–53. doi: 10.1021/bi8004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cahill ST, Tarhonskaya H, Rydzik AM, Flashman E, McDonough MA, Schofield CJ, Brem J. Use of ferrous iron by metallo-beta-lactamases. J Inorg Biochem. 2016;163:185–193. doi: 10.1016/j.jinorgbio.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas PW, Zheng M, Wu S, Guo H, Liu D, Xu D, Fast W. Characterization of purified New Delhi metallo-beta-lactamase-1. Biochemistry. 2011;50(46):10102–13. doi: 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- 36.Jayaram L, Chunilal S, Pickering S, Ruffin RE, Zalewski PD. Sputum zinc concentration and clinical outcome in older asthmatics. Respirology. 2011;16(3):459–66. doi: 10.1111/j.1440-1843.2011.01932.x. [DOI] [PubMed] [Google Scholar]
- 37.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PloS One. 2013;8(9):e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray RD, Duncan A, Noble D, Imrie M, O’Reilly DS, Innes JA, Porteous DJ, Greening AP, Boyd AC. Sputum trace metals are biomarkers of inflammatory and suppurative lung disease. Chest. 2010;137(3):635–41. doi: 10.1378/chest.09-1047. [DOI] [PubMed] [Google Scholar]
- 39.Smith DJ, Anderson GJ, Bell SC, Reid DW. Elevated metal concentrations in the CF airway correlate with cellular injury and disease severity. J Cystic Fibrosis. 2014;13(3):289–95. doi: 10.1016/j.jcf.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez JM, Meini MR, Tomatis PE, Medrano Martin FJ, Cricco JA, Vila AJ. Metallo-beta-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat Chem Biol. 2012;8(8):698–700. doi: 10.1038/nchembio.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat Chem Biol. 2016;12(7):516–22. doi: 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurst TK, Wang D, Thompson RB, Fierke CA. Carbonic anhydrase II-based metal ion sensing: Advances and new perspectives. Biochim Biophys Acta. 2010;1804(2):393–403. doi: 10.1016/j.bbapap.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas PW, Spicer T, Cammarata M, Brodbelt JS, Hodder P, Fast W. An altered zinc-binding site confers resistance to a covalent inactivator of New Delhi metallo-beta-lactamase-1 (NDM-1) discovered by high-throughput screening. Bioorg Med Chem. 2013;21(11):3138–46. doi: 10.1016/j.bmc.2013.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu S, Vosbeek A, Corbella K, Severson J, Schesser J, Sutton LD. A chromogenic cephalosporin for beta-lactamase inhibitor screening assays. Anal Biochem. 2012;428(2):96–8. doi: 10.1016/j.ab.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Yang H, Young H, Yu S, Sutton L, Crowder MW. Targeting metallo-carbapenemases via modulation of electronic properties of cephalosporins. Biochem J. 2014;464(2):271–9. doi: 10.1042/BJ20140364. [DOI] [PubMed] [Google Scholar]
- 46.Wommer S, Rival S, Heinz U, Galleni M, Frere JM, Franceschini N, Amicosante G, Rasmussen B, Bauer R, Adolph HW. Substrate-activated zinc binding of metallo-beta -lactamases: physiological importance of mononuclear enzymes. J Biol Chem. 2002;277(27):24142–7. doi: 10.1074/jbc.M202467200. [DOI] [PubMed] [Google Scholar]
- 47.Llarrull LI, Tioni MF, Vila AJ. Metal content and localization during turnover in B. cereus metallo-beta-lactamase. J Am Chem Soc. 2008;130(47):15842–51. doi: 10.1021/ja801168r. [DOI] [PubMed] [Google Scholar]
- 48.Kocyla A, Pomorski A, Krezel A. Molar absorption coefficients and stability constants of metal complexes of 4-(2-pyridylazo)resorcinol (PAR): Revisiting common chelating probe for the study of metalloproteins. J Inorg Biochem. 2015;152:82–92. doi: 10.1016/j.jinorgbio.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 49.Ferguson JA, Makena A, Brem J, McDonough MA, Schofield CJ. Structure of a Metall-beta-lactamase. Protein Data Bank. 2015:4TYF. [Google Scholar]
- 50.Cox JD, Hunt JA, Compher KM, Fierke CA, Christianson DW. Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II. Biochemistry. 2000;39(45):13687–94. doi: 10.1021/bi001649j. [DOI] [PubMed] [Google Scholar]
- 51.Bertini I, Turano P, Vila AJ. Nuclear Magnetic Resonance of Paramagentic Metalloproteins. Chem Rev. 1993;93:2833–2932. [Google Scholar]
- 52.Bennett B. EPR of Cobalt-Substituted Zinc Enzymes. Biol Mag Res. 2010;29:345–371. [Google Scholar]
- 53.Tioni MF, Llarrull LI, Poeylaut-Palena AA, Marti MA, Saggu M, Periyannan GR, Mata EG, Bennett B, Murgida DH, Vila AJ. Trapping and characterization of a reaction intermediate in carbapenem hydrolysis by B. cereus metallo-beta-lactamase. J Am Chem Soc. 2008;130(47):15852–63. doi: 10.1021/ja801169j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshizumi A, Ishii Y, Livermore DM, Woodford N, Kimura S, Saga T, Harada S, Yamaguchi K, Tateda K. Efficacies of calcium-EDTA in combination with imipenem in a murine model of sepsis caused by Escherichia coli with NDM-1 beta-lactamase. J Infect Chemother. 2013;19(5):992–5. doi: 10.1007/s10156-012-0528-y. [DOI] [PubMed] [Google Scholar]
- 55.King AM, Reid-Yu SA, Wang W, King DT, De Pascale G, Strynadka NC, Walsh TR, Coombes BK, Wright GD. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature. 2014;510(7506):503–6. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.King D, Strynadka N. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Prot Sci. 2011;20(9):1484–91. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cantu C, 3rd, Palzkill T. The role of residue 238 of TEM-1 beta-lactamase in the hydrolysis of extended-spectrum antibiotics. J Biol Chem. 1998;273(41):26603–9. doi: 10.1074/jbc.273.41.26603. [DOI] [PubMed] [Google Scholar]
- 58.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agent Chemother. 2011;55(3):1274–8. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.CLSI. 24th Informational Supplement. Clinical and Laboratory Standards Institute; Wayne, PA: 2015. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 60.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Applied Crystallograph. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallograph D, Biol Crystallograph. 2010;66(Pt 2):213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallograph D, Biol Crystallograph. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallograph D, Biol Crystallograph. 2006;62(Pt 4):439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 64.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallograph D, Biol Crystallograph. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riley EA, Petros AK, Smith KA, Gibney BR, Tierney DL. Frequency-switching inversion-recovery for severely hyperfine-shifted NMR: evidence of asymmetric electron relaxation in high-spin Co(II) Inorg Chem. 2006;45(25):10016–8. doi: 10.1021/ic061207h. [DOI] [PubMed] [Google Scholar]
- 66.King DT, Worrall LJ, Gruninger R, Strynadka NC. New Delhi metallo-beta-lactamase: structural insights into beta-lactam recognition and inhibition. J Am Chem Soc. 2012;134(28):11362–5. doi: 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- 67.Liebschner D, Afonine PV, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallograph D, Struct Biol. 2017;73(Pt 2):148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.