Abstract
This protocol describes procedures for the isolation, cryopreservation, and thawing of HPBMC and analysis of cell surface markers (CSM) for immunophenotyping using polychromatic flow cytometry. This methodology can be used to ensure that cell integrity and phenotype stability is not altered through cryopreservation and extended storage. For this analysis, HPBMC were isolated from seven healthy individuals and 11-color flow cytometry was performed on freshly isolated samples as well as samples cryopreserved for short and long-term periods. There is no significant difference in the percentage of cells expressing the CSM CD3, CD4, CD8, CD45RO, CD16, CD19, or CD56 between freshly isolated and cryopreserved HPBMC. Hence, cryopreservation of HPBMC does not influence the phenotype of distinct cellular subsets in isolated mononuclear cells. This protocol for HPBMC isolation, cryopreservation and thawing of HPBMC is intended for long term-studies of large cohorts requiring sample shipment and subsequent batch analysis.
Keywords: HPBMC, phenotyping, cell surface markers, immune cell subsets, flow cytometry
INTRODUCTION
Assessing potential adverse effects of chemicals, drugs, and environmental agents on the immune system forms the foundation of toxicology research. Currently, human peripheral blood mononuclear cells (HPBMC) are commonly used to detect immune dysfunction following such exposures. Environmental agents including arsenic, wood smoke, and diesel exhaust trigger specific and select responses from HPBMC which may alter immune cells (Burchiel et al., 2014; Burchiel et al., 2016). Lymphocytes (T cells, B cells, and NK cells) and monocytes are the cell subsets found in the mononuclear component of the blood. HPBMC are easily isolated from whole blood using a density gradient separation first employed in the late 1960s (Nobel and Cutts, 1967). These cells can then be used in downstream assays to evaluate the current state and function of the immune system. Many assays utilize fresh blood directly, however due to degradation of whole blood over time, this is not possible when samples are acquired from distant areas, which may not have the tools needed for analysis, or for studies involving large cohorts where batch analysis is required.
For some longitudinal studies it is necessary to acquire blood samples from several donors over an extended period of time or from the same donor at multiple time points. Cryopreservation allows samples to be held in a steady-state so that they can be shipped or held for batch analysis when fresh sample analysis is not possible. Batch analysis reduces experimental variability as well as cost. The quality of the cryopreserved sample and how closely it resembles the fresh sample is extremely important to ensure minimal inter-assay variations exist as a consequence of cryopreservation. Subset analysis or phenotyping of the HPBMC is a commonly used strategy to analyze the integrity of the immune system, and flow cytometry is an optimal method for this analysis (De Rosa et al., 2001; Maecker, 2009; McLaughlin et al., 2008). Heterogeneous matrices such as blood can be characterized by HPBMC subset identification and quantification using specific antibodies conjugated to cell surface markers (CSM) (Maecker et al., 2001). In addition to CSM cell subset identification, intracellular phosphoproteins and cytokines can be stained and used to assess functional and proliferative capabilities of distinct cell types (De Rosa et al., 2001). This will be the topic of a future companion method that will be submitted to Current Protocols in Toxicology using data obtained from the same sample set.
Monitoring the human immune system for shifts in cell subset characteristics, cytokine production, and proliferative capacity allows clinicians and scientists to detect changes which may be indicative of immune system dysfunction or disease progression. In order to assure the accuracy of these readouts, it is essential that the handling of cells is consistent throughout the study and that cryopreservation and duration of cell storage does not alter cell phenotype. Previous studies have addressed the importance of optimizing and standardizing HPBMC handling prior to immunophenotyping (Weinberg et al., 2009; Maecker et al., 2001). Conjointly, assessment of possible short and long-term effects of cryopreservation on phenotype is a critical parameter for longitudinal studies requiring batch analysis. The following protocol outlines procedures for HPBMC isolation, cryopreservation, thawing and subset analysis. A comparative analysis of fresh versus cryopreserved immune cell subsets using cell surface markers in combination with flow cytometry is also provided as a validation of HPBMC integrity despite cryopreservation. Analysis of freshly isolated and directly analyzed HPBMC versus cryopreserved samples stored for either 30–90 days (short-term) or 180–210 days (long-term) showed no difference between the percentages of B cells, NK cells, NKT cells, and T cells including Th cells, T memory cells, and CTL cells (reported as % of live cells as indicated by fixable viability stain). The aim of this procedure is to suggest a standard method for HPBMC isolation, cryopreservation, thawing, and use of polychromatic flow cytometry for immunophenotyping.
Basic Protocol 1. ISOLATION OF HPBMC FROM WHOLE BLOOD
This protocol describes the isolation of human peripheral blood mononuclear cells (HPBMC) from human whole blood using a density (1.077 ± 0.001g/ml) gradient medium. Mononuclear leukocytes which include lymphocytes and monocytes can be isolated from erythrocytes and granular, polymorphonuclear leukocytes which include neutrophils, basophils and eosinophils.
Note:
CAUTION: All waste should be handled as biohazardous material and should be disposed of accordingly. It is recommended to wear appropriate personal protective equipment (PPE) such as gloves, eye protection, and a disposable lab coat while working with human blood, and blood products including cells.
Wash, as used below, refers to the process of centrifuging the sample as directed, aspirating or pouring off the supernatant, leaving the pellet behind, and resuspending the pellet in the next specified solution.
Many of the steps such as layering and collection can be done with transfer, serological or micropipettes and is up to the user to identify which works best and is the most comfortable for them.
Materials
Fico/lite-LymphoH (Atlanta Biologicals Cat #I40150)
DPBS− (Sigma Cat# D8537; does not contain Ca2+ or Mg2+)
Sterile 50 ml conical centrifuge tubes
Sterile 15 ml conical centrifuge tubes
Serological pipettes sterile 5, 10, 25 ml
Sterile transfer pipettes
Refrigerated centrifuge
Micro-pipettes and sterile tips as needed for counting
Automated or manual cell counter (such as Nexcelom Cellometer Auto 2000) and reagents for discriminating live/dead cells.
HPBMC Isolation
-
Decide the appropriate size tube and number of tubes to be used. Blood will be diluted 1:1 with DPBS-. Up to 8 ml blood/DPBS− can be layered over 6 ml Fico/Lite in a 15 ml centrifuge tube and up to 28 ml blood Fico/Lite can be layered over 20 ml Fico/Lite in a 50 ml centrifuge tube.
Example: 84 ml blood diluted 1:1 with DPBS− yields 168 ml total which can be layered over 20 ml of Fico/Lite in each of 6–50 ml centrifuge tubes.
Aliquot 20 ml Fico/Lite into each 50 ml centrifuge tubes (or 6 ml Fico/Lite into a 15 ml centrifuge tube). This can be done the day before use and stored at 4°C.
Place in a dark cabinet and bring to room temperature (RT)
Obtain fresh blood
Dilute blood by aliquoting ≤ 25 ml blood into 50 ml centrifuge tube add an equal amount of DPBS− (at RT)
Mix by inverting tubes or pipetting up and down several times to mix thoroughly.
Following manufacturer’s recommendations, carefully layer up to 28 ml (for a 50 ml tube) of the blood/DPBS− on top of the Fico/Lite without mixing the layers. To layer the blood mixture, hold tube containing Fico/Lite at a 45° angle and use a 10 ml serological pipette to slowly add the blood/DPBS− by allowing it to run down the side of the tube. As the volume of blood/DPBS− is increased the tube can be moved towards an upright position and the rate of addition can be increased.
Centrifuge for 30 min at 400 × g, RT with the brake OFF so as not to disturb the layers as the centrifuge comes to a stop.
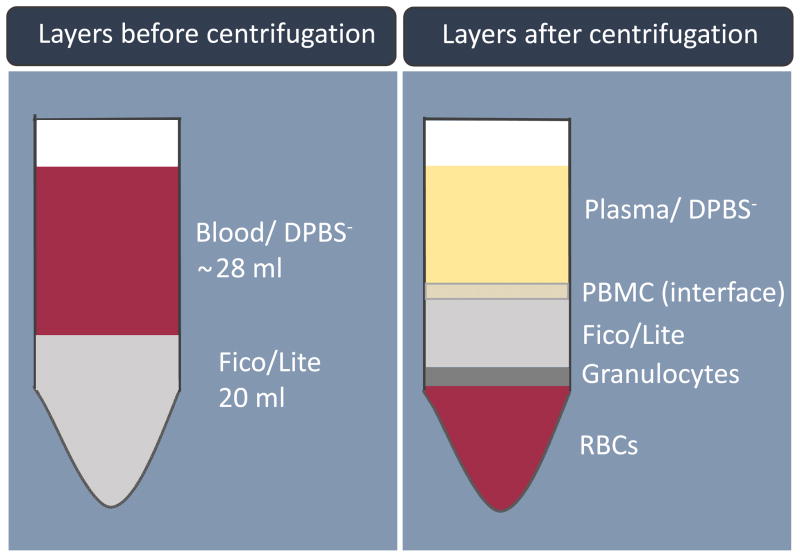
Using a transfer pipette (or serological pipette) carefully collect the mononuclear cell layer at the interface of the Fico/Lite layer (between the plasma and the Fico/Lite layer, see Figure 1) into labeled 15 ml centrifuge tubes. Place up to 7 ml into each tube.
Bring the volume of each tube containing isolated HPBMC up to 15 ml with cold DPBS− and centrifuge at 250 × g for 10 min at 4°C with brake on.
Aspirate (or pour off) supernatant and resuspend cells in 5 ml DPBS− combining pellets into one tube. Wash/centrifuge with 15 ml cold DPBS-. It may be necessary to use more than one 15 ml tube. Tubes can be combined over the next 2 washes.
Repeat wash step two more times.
Following the last wash, aspirate and resuspend the cells in 5 ml cold DPBS-.
-
Count cells (using manual or automated methods); note the number of live cells and the viability.
Cells are now ready to be assayed or cryopreserved for later use.
Figure 1.
Blood components following Fico/Lite gradient separation and centrifugation
Basic Protocol 2. CRYOPRESERVATION OF HPBMC
This protocol describes a procedure for cryopreserving isolated HPBMC which can then be stored for an extended time in liquid nitrogen prior to immunophenotyping by flow cytometry.
Note: Protocols using human peripheral blood cells must first be reviewed and approved by an Institutional Review Board (IRB). All blood samples should be handled with caution as a biohazardous material and should be disposed of accordingly.
Materials
Freshly isolated HPBMC with viabilities greater than 80%
Personal protective equipment (gloves, disposable laboratory coat)
2 ml Cryovials (Nalgene or equivalent for cryopreservation Cat. No. 5000-0020)
Freezing media (pair A+B) Athena Enzyme Systems Cat. No. 0406
Ice Bucket and Ice
Mr. Frosty™ Freezing Container, Nalgene Sigma-Aldrich Cat. No. C1562
Isopropyl alcohol
−80°C freezer
Liquid nitrogen vapor phase storage dewar flask
Automated or manual cell counter and reagent for discriminating live/dead cells
Biological safety cabinet at appropriate containment level
Refrigerated centrifuge
HPBMC Cryopreservation
Once cells have been counted, calculate the number of vials to by frozen at a concentration of 2×107 cells/ml/cryovial. Example: Total cell count is 100× 106 cells, freezing at 20×106 would require 5 vials in 1ml or a total of 5 ml. (See critical parameters: freezing HPBMC)
Label the appropriate number of cryovials for freezing.
Obtain a freezing container (RT) containing the appropriate amount of isopropyl alcohol (as described by manufacturer).
Centrifuge HPBMC at approximately 230 × g for 10 to 15 min @ 4°C with brake.
Aspirate or pour off the supernatant without disturbing the pellet.
Resuspend the cell pellet following manufacturer’s instructions with the appropriate amount of freezing medium ‘A’, which will be half of the total amount needed. Example: Using the example in step 1–5 ml of total freezing media ‘A’+ ‘B’ is needed therefore use 2.5 ml of ‘A’ and 2.5 ml of ‘B’.
Once the cells are resuspended with ‘A’ add the appropriate amount of freezing medium ‘B’, which will be half of the total amount needed (see example in step 6). Aliquot 1 ml per cryovial.
Vials should then be placed into the freezing container and placed at −80°C for at least 24 hr and up to 1 week.
Transfer cryovials to a liquid nitrogen storage container for extended storage. It is best to store cells in the vapor stage of the liquid nitrogen. Most liquid nitrogen storage Dewar flasks are designed for this.
Basic Protocol 3. THAWING OF CRYOPRESERVED HPBMC
This method describes the thawing of cryopreserved samples for use in downstream assays such as multicolored flow cytometry.
Note:
CAUTION: Cryogenic samples are held in liquid nitrogen at very cold temperatures (≤ −196°C). Cryovials may sometimes explode due to liquid nitrogen leaking into the vial and causing an increase in internal pressure due to temperature change and release of gas. Even when they do not explode, they may be under pressure when the cap is opened. To avoid dispersion of mist as a result of the pressure, cover the vial with a paper towel to loosen the vial lid, releasing any pressure while capturing any mist. Furthermore, it is important to wear safety equipment such as eye protection, lab coat, gloves and face shield to prevent exposure or abrasions due to exploding vials.
Wash, as used below, refers to the process of centrifuging the sample as directed, aspirating or pouring off the supernatant, leaving the pellet behind, and resuspending the pellet in the next specified solution.
Materials
15 ml sterile conical centrifuge tube
Water bath at 37°C
Dry ice
Styrofoam container
Cryopreserved samples
Face shield
Sterile Transfer pipette
Thawing cryo-preserved cells
Warm 10 ml of cRPMI medium (see Reagents and Solutions) in a 15 ml conical centrifuge tube for each sample being thawed.
-
Quickly thaw a vial of cryopreserved cells by placing vial into a 37°C water bath and moving the vial in a back and forth motion.
Note: If more there are more than 2 samples to be thawed the additional samples can be held on dry ice in a Styrofoam container. It is recommended that only 2 samples at a time be thawed.
Using a transfer pipette, transfer thawed cells from each cryovial into a labeled 15 ml sterile centrifuge tube containing 10 ml warmed medium (from step 1). After all samples have been thawed and placed in warmed medium, centrifuge 10 min at 230 × g. It is important to thaw and wash the cells as quickly as possible to remove any of the freezing medium.
Wash once more with 5 ml warmed medium, centrifuge 10 min at 230 × g, aspirate the medium, then resuspend the cells in an adequate amount of fresh cRPMI for counting (this amount can be determined by the amount of cells originally cryopreserved in the tube).
-
Count, and adjust cell concentration to 2 ×106 cells/ml in medium accordingly (this concentration can be adjusted depending on the downstream assays).
The cells are now ready to be used for downstream analysis such as immunophenotyping.
Basic Protocol 4. STAINING OF HPBMC FOR FLOW CYTOMETRY
The method described here is for the staining of cell surface markers (CSM) utilizing 11 colors for analysis by flow cytometry. All antibodies used for flow cytometry staining in these procedures were purchased from BD Biosciences and used at the recommended dilutions.
Note:
The antibodies in this procedure are used at the company’s recommend concentration/amount. Upon initial analysis it may be necessary to titrate the antibody.
Tubes for compensation beads + antibody are needed for compensation (one of the unstained samples will be used for compensation of the FVS).
In this procedure samples can be aspirated or emptied/poured onto absorbent paper towel; aspirated = poured.
Fixatives contains formaldehyde. Dispose of waste according to your institutes protocol.
This procedure uses live human cells and formaldehyde. PPE should be worn at all times.
Materials
96- well polypropylene tubes – also called cluster tubes (1.2 ml) in rack (Corning® 4410)
Microcentrifuge tube or other tube adequate for mixing antibody cocktail
12 × 75 mm (5 ml) polystyrene round bottom tubes (Falcon 352008)
Dual position snap caps for 12 × 75 mm tube (Falcon 352032)
Serological pipets sterile 5, 10 and 25 ml
Micropipettes 10, 100, 200 and 1000 μl
Multichannel pipette
Tips sterile to fit micropipettes
Repeat pipette and tips for 50, 200 – 500 μl volumes
Wypall L40 (Kimberly-Clark, Fisher Scientific Cat. No. 19-042-427)
Ice and ice bucket
Aluminum foil
Refrigerated centrifuge with plate carriers for centrifuging 96-well (cluster) tubes.
Staining of cell surface markers
Note:
Two tubes per sample (1 stained and 1 unstained) are required. It is recommended to use approximately 1× 106cell/tube. Prior to sample analysis a pilot assay may be necessary to evaluate non-specific binding using isotype controls and/or a fluorescent minus one (FMO) analysis to determine gating.
Collect 0.5 ml of 2×106cell/ml (1×106) cells (from Basic Protocol 2, step 5) into labeled staining tubes add 200 μl staining buffer.
Wash cells; centrifuge 10 min, 4°C, @ ~250 × g, aspirate, vortex gently and add 500 μl cold stain buffer. (Using the cluster tube rack to hold staining tubes will allow for easier decanting of supernatant during the wash steps. Place several paper towels on top the tubes in the rack. With one hand on the paper towels and one hand on the bottom of the rack, invert the entire rack and gently shake once or twice to remove liquid. Wait several seconds before turning the rack right side up. Remove and discard the paper towels.)
Repeat wash (step 2 above).
Centrifuge 10 min, 4°C, @ ~250 × g, aspirate and resuspend by vortexing gently. Add 50 μl Brilliant Stain Buffer (BSB).
Make a cocktail of the Ab (see Table 1 for recommended volumes) and add 96 μl of the cocktail to each designated flow tube. One set of tubes will contain antibody cocktail and the other set will contain Staining Buffer plus FVS (make this as a cocktail). The unstained tubes will contain stain buffer (95 μl) and FVS780 (1 μl) It is best to calculate for one extra sample so as not to run out of cocktail.
Incubate for 20 min on ice in the dark.
Wash by adding 500 μl cold stain buffer, centrifuge 230 × g for 10 min, aspirate, gently vortex.
Add 250 μl Cytofix, gently vortex.
Incubate 30 min on ice in the dark.
Add 500 μl stain buffer, centrifuge 230 × g for 10 min, aspirate, gently vortex.
Resuspend cell pellet in 500 μl stain buffer.
Hold overnight at 4°C, cover with a lid and aluminum foil to protect from light.
Analyze on LSRFortessa or other cytometer with 4 laser capability within 24 to 34 hours.
Table 1.
Cell surface markers for flow cytometry. Antibody volumes are based on manufacturer’s recommendations to ensure limited inter-assay variability for batch analysis of long-term studies.)
| Laser | Filter | Fluorochrome | Specificity | Clone | Cat. No | Volume (μl) |
|---|---|---|---|---|---|---|
| Blue 488 nm | 530/30 | FITC | CD16 | 3G8 | 555406 | 20 |
| 695/40 | PerCP-Cy™5.5 | HLA-DR | G46-6 | 560652 | 5 | |
|
| ||||||
| Yellow -Green 561 nm | 582/15 | PE | CD56 | B159 | 555516 | 20 |
| 610/20 | PE-CF594 | CD19 | HIB-19 | 562294 | 5 | |
| 780/60 | PE-Cy™7 | CD8 | RPA-T8 | 557746 | 5 | |
|
| ||||||
| Red 640 nm | 670/14 | Alexa Fluor 647 | CD127 | HIL-7R-M21 | 558598 | 20 |
| 730/45 | Alexa Fluor 700 | CD4 | RPA-T4 | 557922 | 5 | |
| 780/60 | APC-Cy™7 | FVS780 | 565388 | 1 | ||
|
| ||||||
| Violet 405 nm | 450/50 | BV421 | CD45RO | UCHL1 | 562641 | 5 |
| 525/50 | BV510 | CD3 | UCHT1 | 563109 | 5 | |
| 605/12 | BV605 | CD14 | M5E2 | 564054 | 5 | |
Stain Anti-mouse Ig,k/Neg control compensation particles the same day as CSM staining. Following manufacturer’s directions, vortex beads before using them.
In flow tubes (12 × 75 mm):
Following manufacturer’s directions: Add 1 drop of Neg and 1 drop of CompBeads to each tube.
Add appropriate antibody (See Table 1). Make 1 tube for each antibody. One tube will contain only compensation beads and no antibody.
Incubate 20 min at RT in the dark.
Add 1ml staining buffer to each tube, centrifuge at 250 × g for 10 min, and aspirate.
Resuspend in 500 μl staining buffer by vortexing gently, cap, and cover with aluminum foil to protect from light.
Run as compensation controls for the samples above.
REAGENTS AND SOLUTIONS
RPMI complete medium (cRPMI)
500 ml RPMI 1640 HEPES modified medium (Sigma Aldrich)
Add 50 ml Fetal Bovine Serum (FBS; Atlanta Biologicals)
Add 5 ml of 200 mM L-glutamine (2 mM final concentration; Sigma Aldrich)
Add 5 ml of 10,000 Units/ml penicillin with 10,000 μg/ml streptomycin sulfate (100 U/ml Penicillin with 100 μg/ml streptomycin sulfate final concentration; Gibco™)
Store up to 1 month at 4°C
Stain Buffer/Wash Solution
500 ml Dulbecco’s Phosphate Buffered Saline (does not contain calcium or magnesium; Sigma Aldrich)
Add 1 ml heat inactivated FBS (0.2% v/v final concentration; Atlanta Biologicals)
Add 0.45 g sodium azide (0.09% w/v final concentration; Sigma Aldrich)
Stable at least 1 month at 4°C
Cytofix Buffer (BD Biosciences Cat. No. 554655) Contains formaldehyde! Dispose of waste as directed by institute protocol.
Brillant Stain Buffer (BD Biosciences Cat. No. 563794)
Anti-mouse Ig, k/Negative Control Compensation Particles Set (BD Biosciences Comp Bead Cat. No. 552843)
COMMENTARY
Background Information
Recent studies have revealed associations between disease states and the numbers of ‘rare’ lymphocyte subsets even if larger cell populations show no significant changes. Detection of minor shifts and surveillance of distinct cell subtypes has been shown to be critically relevant in diseases such as HIV and leukemia (Roederer et al., 1995; Rabin et al., 1995; Migueles et al., 2002; Betts et al., 2006; Duque et al., 1993) These studies highlight the complexity of the immune system and the need for optimization and standardization of current HPBMC protocols. For this reason, previous studies have attempted to standardize methods for isolation as well as cryopreservation (Weinberg et al., 2009; Maecker et al., 2001). Due to the ever changing field of toxicology however, it’s necessary to periodically revisit and optimize these methods. This is of particular importance in longitudinal studies involving large cohorts and with the ongoing efforts of the “Human Immunology Project” (Maecker et al., 2001).
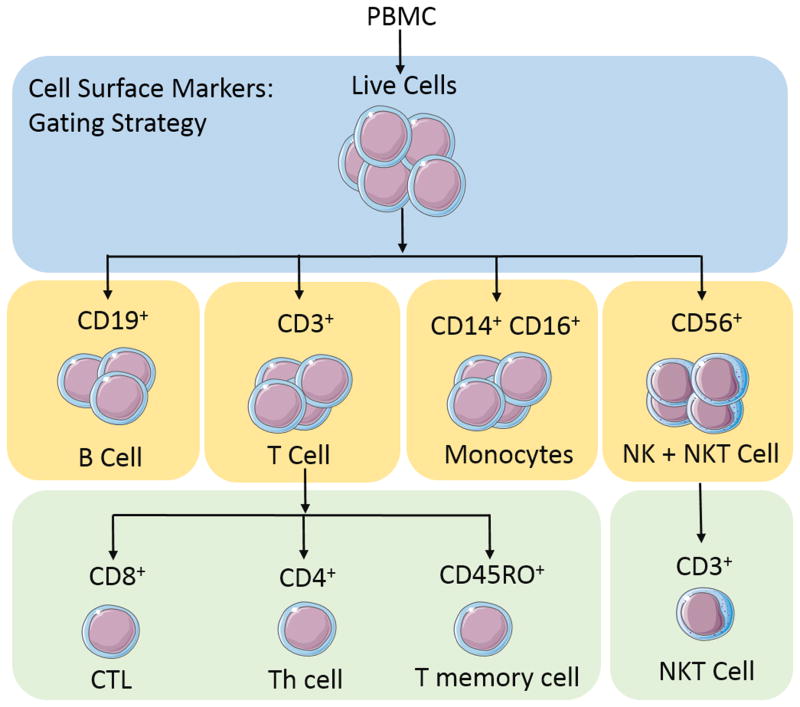
In addition to implementing a standard procedure for HPBMC handling upstream of immunophenotyping, in depth analyses of the effects of cryopreservation with regard to cellular characteristics and function are necessary to distinguish true biological variations from technical artifacts. Therefore, in addition to protocols on HPBMC isolation, cryopreservation, and thawing, this procedure includes a comparative analysis of fresh versus cryopreserved HPBMC phenotypes to ensure that donor phenotypes remain unchanged despite short and long term cryopreservation. Cell surface markers and gating strategies are outlined in Figure 2. The antibodies used in this protocol (purchased from BD) are used according to the manufacturer’s directions to reduce variation between assays and are intended only as suggestions for long-term studies where batch analysis is required.
Figure 2.
Gating strategy used to analyze cell surface markers for immunophenotyping
Implementing standardized methods of cell handling prior to flow cytometry analysis is crucial for the reproducibility of longitudinal assays. A decreased percent viability can be a direct indication of aberrations in cell isolation, cryopreservation, and/or thawing methods as well as the stability of the cells following cryopreservation. It’s also important to note that thorough understanding of equipment and analysis software prior to beginning a study will decrease the likelihood of assay variability. To avoid mistakes and improve reproducibility, CS&T tracking beads and flow cytometry templates are advantageous. Additionally, prior to experimentation, some specific elements of flow cytometry such as color compensation, fluorescence minus one, and antibody titration should be thoroughly explored (see critical parameters, flow cytometry assays for helpful references).
Cell surface markers and flow cytometry
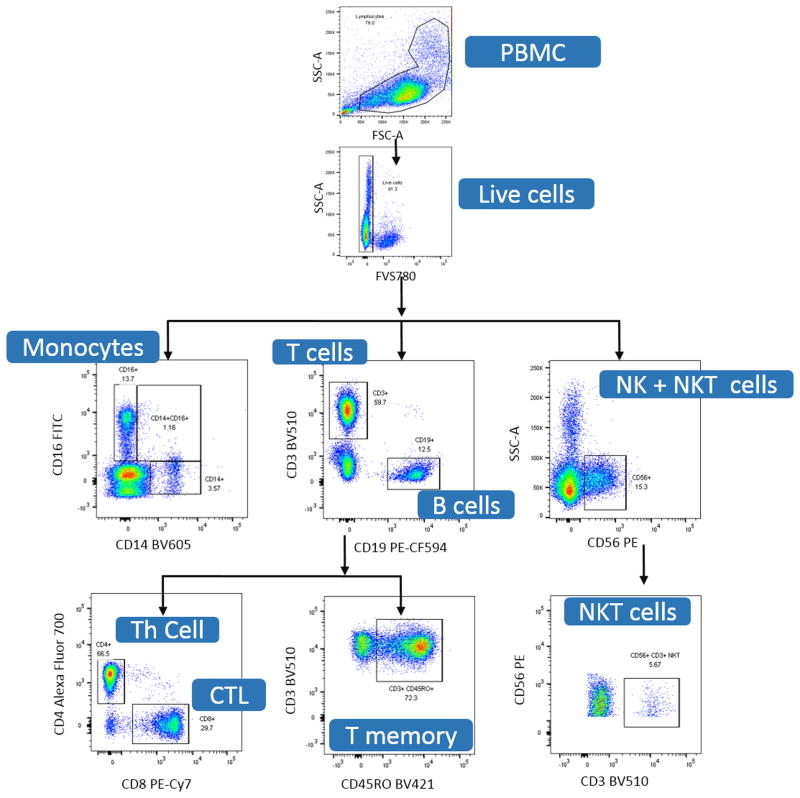
Major T cell subsets can be defined by expression of CD3. From there, subsets can then be split into their relative expression of CD4 for the labeling of T helper (Th) cells, CD8 for the identification of cytotoxic T lymphocytes (CTL) and CD45RO as a marker of T memory cells (Figure 2). The gating strategy used in this protocol not only clearly defines T cell subsets, but also defines additional populations of interest such as B cells (CD19), NK cells (CD56), and monocytes (CD14/CD16) (Figure 2). Cells negative for CD3 as well as CD19 can be delineated into groups of dendritic cells (DC), monocytes, or natural killer (NK) cells (Figure 2). As CD16 phenotyping can include monocytes and NK cells, additional markers must be used if classification and analysis of these subsets is required. In this gating strategy, we further employ the use of a CD14 antibody which is specific for monocyte cell subsets. Classic monocytes are CD16 negative and express high levels of CD14, while non-classical monocytes are CD14 low and CD16 hi. For additional information at specific gating steps, Figure 3 depicts an example of gating using FlowJo single cell analysis software. Table 3 (Valiathan et al., 2014; Chng et al., 2004) summarizes reported ranges for each lineage. These ranges can vary depending on ethnicity, age and gender of donors, or as a result of how authors have chosen to report these ranges. As such, these factors should be considered when comparing results.
Figure 3.
Gating example for immunophenotyping using the suggested cell surface markers from Basic Protocol 4, Table 1.
Table 3.
Lymphoctye subset populations (ranges reported in percentages).
| Marker | Valiathan et al. | Chgn et al. |
|---|---|---|
| CD3 | 65 – 88 | 47 – 79 |
| CD19 | 2 – 27 | 7 – 22 |
| CD4 | 26 – 62 | 23 – 47 |
| CD8 | 14 – 44 | 14 – 40 |
| NK | 2 – 27 | 7 – 37 |
| CD4/CD8 ratio | 0.6 – 4.4 | 0.68 – 2.73 |
Many of the cellular subsets listed above can be identified and analyzed using a wide variety of gating strategies employing distinct and unique cell surface markers. Gating strategies and antibody choice are dependent on experimental design, cellular subsets of interest, and personal preference. Please see critical parameters/troubleshooting for useful resources on antigen selection and use.
The 11 CSMs suggested in this protocol can be used in combination to further identify rare cellular subsets that are not described here. The use of HPBMC and subsequent flow cytometry in toxicology is a growing field. While CSM for major lymphocyte and monocyte lineages will likely remain constant, novel cellular subset identification as well as investigations into new functions for “old” subsets are driving companies to increase antibody variety and availability. These changes require researchers to periodically review current protocols and gating strategies to accommodate this rapidly evolving technology making this both a challenging and exciting field of study.
Critical Parameters
It is important in the isolation, cryopreservation and thawing procedures described here to maintain sterility of the sample. Persons handing blood and blood products should use PPE and should dispose of all waste as biohazard material. The fixative used in this protocol contain formaldehyde and should therefore be handled with care and disposed of appropriately. Collection and analysis should be conducted following a written protocol approved by and Internal Review Board (IRB) and/or a Human Research Review Committee (HRRC).
Isolating HPBMC
It is recommended that blood be processed as soon as possible after being drawn. Blood can be held at room temperature for several hours before isolating HPBMC.
It is not recommended to store blood more than 6 hours before HPBMC isolation. Isolation soon after collection will ensure quality results in the downstream applications.
Collect whole blood into heparinized tubes (other blood anticoagulants maybe desirable depending on the downstream analysis).
It is important that all reagents and blood be at room temperature before using. Place blood on a rocker or rotator to continue mixing while bringing to room temperature. Using reagents or blood warmed to 37° will yield poor separation as well as increase cell death.
Decreasing the amount of time from acquisition to isolation by limiting the number of donors being processed ensures high viability and yield.
It is also important that the initial centrifugation be done in a centrifuge that is at room temperature and that the brake is turned off. Use of the break can disrupt the separation of the reagents/cells leaving a cloudy and hard to distinguish cell layer.
The isolation medium can be toxic to the HPBMC. Therefore, it is important to limit the time the HPBMC are in contact with the isolation media and wash the cells several times to remove isolation medium.
To aide in collection of the HPBMC layer some choose to first remove the Plasma/DPBS− layer, this will not affect the yield as long as the HPBMC layer is not disturbed.
There are a variety of ficoll-based products that may be used for mononuclear cell isolation as well as specifically designed tubes to aide in the separation and collection process. It is up to the user to balance cost and time in order to choose a product that meets their needs. The methods of layering and centrifugation are similar from product to product. Many of the steps such as layering and collection can be done with transfer, serological or micropipettes and is up to the user to identify which works best and is the most comfortable for them.
Freezing HPBMC
The freezing container should allow the temperature to drop at a rate very close to −1°C/min, this is the optimal rate for cell preservation as it limits the amount of ice crystals being formed.
Mr. Frosty Freezing container should be used as directed. There are a limited number of freeze thaws as well as a specified amount of isopropanol to be used.
Cells can be frozen at concentrations of 2×106 cell/ml to 20×106 cells/ml. Optimal concentrations should be determined based on use upon the number of cells needed for downstream analysis. It is not recommended to freeze at 20×106 cells/ml when only a fraction of the cells are needed for downstream analysis as the remainder will be discarded. Also having to thaw multiple aliquots can be time consuming and lead to decreased viability due to time, planning for subsequent use is recommended.
Freezing containers that do not require isopropanol can be purchased or made, most important is that the rate of freezing be optimal. This procedure has not tested the use of alternate freezing containers.
Thawing HPBMC
Thaw cryopreserved HPBMC rapidly by transferring them directly from liquid nitrogen or dry ice to a 37°C water bath. It is imperative that the aliquot be thawed quickly and diluted into medium or DPBS− to reduce the DMSO which can be toxic to cells at concentrations > 0.1%
Addition of FBS can help to pellet the cells during centrifugation and help to reduce the loss of cells during the washing step, but is not required.
Using large amount of wash solution will dilute out the DMSO in the freezing media, which over time is toxic to cells.
Flow cytometry assays
For longitudinal studies it is important to reproduce functionality of the flow cytometer. For studies presented here we initially identified optimum voltage gains for each fluorochrome and then utilized BD Biosciences Cytometer Setup & Tracking (CS&T) to establish a baseline. For subsequent uses the CS&T beads were used to reestablish the cytometer conditions initially used. Once the baseline has been established the CS&T beads can be used daily for measurements of relative fluorescence detection efficiency, relative background, the standard deviation of electronic noise, and various cytometer settings (see specifications sheet supplied with beads) and can be used to reproducibly set up the cytometer from day to day.
It is recommended that a fixable viability stain be used to discriminate live from dead cells. Antibodies can bind non-specifically to dead cells distorting results.
Multi-colored panels require compensation controls. The compensation controls should be stained, stored, and ran under the same conditions and at the same time as the research samples.
Before running multi-colored panels, it is recommended to run fluorescence minus one (FMO) controls to verify identification of subsets of cells.
It is important to collect as many events as possible for analysis, our lab typically collects 30,000 events of the gate lymphocyte population which allows greater than 30,000 cells to be analyzed on the HPBMC population. This number is based on the cell number in the sample and the time required collecting the events.
There are a variety of cytometers which have the capability to run/analyze multicolor assays. Our procedures utilized a 4-laser LSRFortessa with BD FACSDiva software. BD offers many online tutorials to help with the setup of experiments, acquisition of data, and cytometer setup using CS&T beads for standardization and quality assurance as well as in-person training. It is extremely important to understand the cytometer capabilities, such as lasers and filters, as well as the software to design and conduct successful multicolor assays. BD also offers online tools such as the Fluorescence Spectrum Viewer which can be a useful aid in designing multicolor experiments.
Troubleshooting
Isolation of HPBMC
Temperature and time are critical to the viability and stability. Low or cold temperatures can lead to increased red blood cell contamination because erythrocytes aggregate less and it takes longer for the separation to occur. High or warm temperatures can lead to low yields because the erythrocytes aggregate more and trap the mononuclear cells. Additionally, the density of the ficoll is less than 1.077 g/ml allowing mononuclear cells to penetrate the ficoll. A low yield of HPBMC contaminated with granulocytes can be due to brake being used to slow/stop the spin, or vibration from a poorly balanced centrifuge.
Freezing and Thawing HPBMC
Low viability can be due to extended exposure of the cells to the dimethyl sulfoxide (DMSO) contained in the freezing media. It is important to thaw the cells quickly and to dilute and remove the DMSO as quickly as possible. Cells stored long term at −80°C before being transferred to liquid nitrogen will also display lower viability than cells transferred within 1 week following freezing to liquid nitrogen. Low yield of live cells can be associated with loss of cells during the washing steps, but is also dependent on viability. Using care when decanting or respirating the supernatant and gently resuspending the pellet, cell loss can be reduced.
Immunophenotyping by Flow cytometry
If the cell subset numbers do not check out as being accurate (example T cell total CD3≠ CD4+CD8 and Lymphocyte total CD3+CD 56+ CD14+ CD19 ≠ 100%), ensure that the compensation has been set correctly and is being applied to all samples. Verify gating by running and analyzing FMO controls, and ensure that dead cells have been excluded through gating. If the number of dead cells is above the anticipated percentage, this could be an indication of improper HPBMC isolation, cryopreservation or thawing.
Anticipated Results
Isolation of HPBMC
Viabilities (as assessed after isolation) are expected to be greater than 90% and yields should be between 1–2×106 cells per ml of blood.
Thawing HPBMC
Anticipated viabilities are greater or equal to 85%. In our laboratory, and as shown in Table 2, viabilities of thawed cells cryopreserved >120 days remain above 85%.
Immunophenotyping by Flow cytometry
HPBMC populations include lymphocytes (T-cells, B-cells), NK cells, monocytes and dendritic cells (not analyzed in this procedure). While the percentages of each cell type varies between individuals, when performed correctly, phenotypic identification of cell types should yield results close to reported reference ranges (Table 3). Furthermore, T cell totals (T Total) is the combined percentage of CD4+ and CD8+ and should be consistent with the percentage of CD3+ cells. Lymphocyte/leukocyte totals (L Total) is the combined total of CD3+, CD19+, CD14+ and CD56+ cells and should be close to 100% as they account for the T, B, Monocyte, and NK cells found in the peripheral mononuclear cell isolate.
This protocol for HPBMC isolation and cryopreservation is for batch analysis of large cohorts involved in long-term studies. The integrity of each cellular subset following cryopreservation is of utmost importance to ensure a difference in results does not arise due to handling of samples and duration of storage. To analyze whether cryopreservation affects cellular phenotype, 11 color flow cytometry was used on fresh and cryopreserved samples from the same donors. The results found in Table 4 represent the percentages of cellular subsets within the HPBMC population of these donors. Percentages of cell types are represented as percentages mean ± standard deviation (Table 4). This protocol and the gating strategy outlined in it reports percentage of live HPBMC cells including lymphocyte and monocyte populations.
Table 4.
HPBMC cellular subsets analyzed by 11-color flow cytometry
| CD3+ | CD14+ | CD19+ | CD56+ | Total | |
|---|---|---|---|---|---|
| Fresh sample (N=7) | 68.03 ± 7.11 | 3.14 ± 2.55 | 7.75 ± 3.05 | 15.14 ± 4.01 | 94.06 |
| ST cryopreserved (n=6) | 69.67 ± 4.99 | 5.29 ± 1.83 | 6.30 ± 1.75 | 17.00 ± 4.42 | 98.27 |
| LT cryopreserved (n=4) | 67.65 ± 5.32 | 4.32 ± 1.82 | 7.79 ± 3.29 | 17.43 ± 1.70 | 97.18 |
| CD4+ | CD8+ | Total | |
|---|---|---|---|
| Fresh sample (N=7) | 40.4 ± 2.75 | 21.47 ± 4.83 | 61.87 |
| ST cryopreserved (n=6) | 41.47 ± 3.45 | 21.90 ± 4.46 | 63.37 |
| LT cryopreserved (n=4) | 41.63 ± 3.31 | 20.60 ± 4.08 | 62.23 |
| CD3+ CD45RO+ | CD16+ | CD3+ CD56+ | |
|---|---|---|---|
| Fresh sample (N=7) | 28.96 ± 7.66 | 14.22 ± 6.51 | 1.96 ± 0.91 |
| ST cryopreserved (n=6) | 29.70 ± 7.07 | 14.82 ± 4.66 | 2.90 ± 1.41 |
| LT cryopreserved (n=4) | 31.48 ± 8.10 | 16.68 ± 2.25 | 2.38 ± 1.42 |
(For full list of markers, see Table 1: HLA-DR and CD127 not described). ST = Short Term cryopreservation (30–90 days), LT = Long Term cryopreservation (180–210 days). Results shown are in percentages (mean ± standard deviation) of live cells.
Analysis of lymphocyte subsets in a total of 7 donors freshly after isolation revealed an average of 68% CD3+, 3% CD14+, 8% CD19+, and 15% CD56+, for a total of 94%. T-cell markers CD4 and CD8 accounted for approximately 40% and 21% respectively. Additional CSM markers analyzed in these donors showed expression of CD3/CD45RO at 29%, CD16 at 14% and NKT cells at 2% of total live cells. Flow cytometry analysis of these same subsets following cryopreservation and storage of HPBMC aliquots short term (30 – 90 days, n=6 of the 7 original donor samples), or long term (180 – 210 days, 4 of the 7 original samples) revealed no statistical difference in a majority of the cell subsets as compared to freshly isolated samples. In this small subset a significant difference was seen in marker CD14. Taken together, the proposed method of cryopreservation and handling does not affect the percentages of major cell subsets and is therefore an optimal technique for longitudinal studies using HPBMC.
Time Considerations
Isolation of HPBMC
This procedure including warming the reagents and blood to room temperature takes about 2 ½ hours. Time can be dependent on how many samples are being isolated at one time and how fast the blood can be layered onto the Fico/Lite.
Freezing HPBMC
The freezing of HPBMC following isolation will take about 30 minutes. This includes labeling the cryovials, centrifuging the samples, resuspend in freezing media and placing into the −80°C freezer. The frozen samples can be transferred to liquid nitrogen the next day.
Thawing HPBMC
Thawing two samples will take approximately 45 minutes including the warming of the medium and washing and counting of the cells.
Flow cytometry assays
Staining of samples for flow cytometry requires approximately 3 hr, the time can vary depending on the amount of samples being assayed. The first time set up of the experiment, cytometer (CS&T beads for baseline and running) and compensation set up and processing of FMO samples requires approximately 2.5 hr up to about 5 hr. The initial experimental set up and settings can be saved as experimental templates/protocols on the cytometer to reduce the time needed for subsequent experiments. From a template the setup of the cytometer and analyzing the compensation controls can take up to 1 hr. It is recommended to use 96- well polypropylene tubes (cluster tubes) in a rack for staining. This will allow the researcher to handle more samples at one-time. Each tube has the capacity to hold 1.2 ml therefore the researcher should adjust volumes to be less than 1 ml so as to not cross-contaminate samples while handling. By using cluster tubes/rack, reagents can easily be poured off from all tubes at one-time onto a Wypall towel or other absorbent towel. It is important to label all tubes and to use care when inverting or pouring off reagents to avoid dislodging the tube from the rack or spilling reagent. Cells are fixed and can be analyzed the day after staining.
Table 2.
Anticipated percentages of live cells following fresh isolation, short term cryopreservation (ST CP), and long term cryopreservation (LT CP).
| Donor | Fresh | ST CP | LT CP |
|---|---|---|---|
| A | 97 | 88 | |
| B | 98 | 88 | 89 |
| C | 97 | 89 | 89 |
| D | 98 | 88 | 86 |
| E | 98 | 88 | |
| F | 98 | 87 | |
| G | 98 | 89 |
SIGNIFICANCE.
Human peripheral blood mononuclear cells (HPBMC) are critical components of the immune system, the body’s natural defense against infection and foreign invaders. They are widely used in toxicology research to assess potential adverse effects on the immune system by drugs, chemicals, environmental agents, and toxins. Longitudinal toxicology studies of human blood may require samples to be shipped from distant sites or acquired at various time points. Oftentimes direct processing of donor samples is not possible and cryopreservation of HPBMC is necessary for shipping and batch analysis. Therefore, the optimization and standardization of these methods is of utmost importance. It is equally important to ensure that the phenotype of HPBMC cellular subsets is not altered by cryopreservation and extended storage.
Acknowledgments
This work was supported by a grant from the National Institute of Environmental Health Sciences (R01-ES019968 to SWB)
Literature Cited
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Conors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T-cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchiel SW, Lauer FT, Beswick EJ, Gandolfi AJ, Parvez F, Liu KJ, Hudson LG. Differential Susceptibility of Human Peripheral Blood T Cells to Suppression by Environmental Levels of Sodium Arsenite and Monomethylarsonous Acid. PLoS One. 2014;9(10):e109192. doi: 10.1371/journal.pone.0109192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchiel SW, Lauer FT, MacKenzie D, McClain S, Kuehl PJ, McDonald JD, Harrod KS. Changes in HPBMC markers of immmune function following controlled short-term inhalation exposures of humans to hardwood smoke. Inhal Toxicol. 2016;28(2):61–70. doi: 10.3109/08958378.2015.1136714. [DOI] [PubMed] [Google Scholar]
- Chng WJ, Tan GB, Kuperan P. Establishment of Adult Peripheral Blood Lymphocyte Subset Reference Range for an Asian Population by Single-Platform Flow Cytometry: Influence of Age, Sex, Race and Comparison with Other Published Studies. Clinical and Diagnostic Laboratory Immunology. 2004:168–173. doi: 10.1128/CDLI.11.1.168-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa SC, Herzenberg LA, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7(2):245–8. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- Duque RE. Flow cytometric analysis of lymphomas and acute leukemias. Ann NY Acad Sci. 1993;677:309–325. doi: 10.1111/j.1749-6632.1993.tb38786.x. [DOI] [PubMed] [Google Scholar]
- Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2001;12(3):191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker HT. Multiparameter flow cytometry monitoring of T cell responses. Methods Mol Biol. 2009;485:375–91. doi: 10.1007/978-1-59745-170-3_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin BE, Baumgarth N, Bigos M, Roederer M, De Rosa SC, Altman JD, Nixon DF, Ottinger J, Oxford C, Evans TG, Asmuth DM. Nine-color flow cytometry for accurate measurement of T cell subsets and cytokine responses. Part I: Panel design by an empiric approach. Cytometry A. 2008;73(5):400–10. doi: 10.1002/cyto.a.20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- Noble PB, Cutts JH. Separation of blood leukocytes by Ficoll gradient. Can Vet J. 1967;8(5):110–111. [PMC free article] [PubMed] [Google Scholar]
- Rabin RL, Roederer M, Maldonado Y, Pertru A, Herzenberg LA. Altered representation of naïve and memory CD8 T cell subsets in HIV-infected children. J Clin Invest. 1995:2054–2060. doi: 10.1172/JCI117891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M, Dubs JG, Anderson MT, Raju PA, Herzenberg LA. CD8 naïve T cell counts decrease progressively in HIV-infected adults. J Clin Invest. 1995:2061–2066. doi: 10.1172/JCI117892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg A, Song LY, Wilkening C, Sevin A, Blais B, Louzao R, Stein D, Defechereux P, Durand D, Riedel E, Raftery N, Jesser R, Brown B, Keller MF, Dickover R, McFarland E, Fenton T. Optimization and Limitations of Use of Cryopreserved Peripheral Blood Mononuclear Cells for Functional and Phenotypic T-Cell Characterization. Clin Vaccine Immunol. 2009:1176–86. doi: 10.1128/CVI.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiathan R, Deeb K, Daimante M, Ashman M, Sachdeva N, Asthana D. Reference ranges of lymphocyte subsets in healthy adults and adolescence with special mention of T cell maturation subsets in adults of South Florida. Immunology. 2014;219(7):487–96. doi: 10.1016/j.imbio.2014.02.010. [DOI] [PubMed] [Google Scholar]