Abstract
Background:
Evaluation of interindividual variability is a challenging step in risk assessment. For most environmental pollutants, including perchloroethylene (PERC), experimental data are lacking, resulting in default assumptions being used to account for variability in toxicokinetics and toxicodynamics.
Objective:
We quantitatively examined the relationship between PERC toxicokinetics and toxicodynamics at the population level to test whether individuals with increased oxidative metabolism are be more sensitive to hepatotoxicity following PERC exposure.
Methods:
Male mice from 45 strains of the Collaborative Cross (CC) were orally administered a single dose of PERC () or vehicle (Alkamuls-EL620) and euthanized at various time points (/strain/time). Concentration–time profiles were generated for PERC and its primary oxidative metabolite trichloroacetate (TCA) in multiple tissues. Toxicodynamic phenotyping was also performed.
Results:
Significant variability among strains was observed in toxicokinetics of PERC and TCA in every tissue examined. Based on area under the curve (AUC), the range of liver TCA levels spanned nearly an order of magnitude (-fold). Expression of liver cytochrome P4502E1 did not correlate with TCA levels. Toxicodynamic phenotyping revealed an effect of PERC on bodyweight loss, induction of peroxisome proliferator activated receptor-alpha (PPAR)-regulated genes, and dysregulation of hepatic lipid homeostasis. Clustering was observed among a) liver levels of PERC, TCA, and triglycerides; b) TCA levels in liver and kidney; and c) TCA levels in serum, brain, fat, and lung.
Conclusions:
Using the CC mouse population model, we have demonstrated a complex and highly variable relationship between PERC and TCA toxicokinetics and toxicodynamics at the population level. https://doi.org/10.1289/EHP788
Introduction
Interindividual variability in susceptibility to toxicity has been observed or assumed to exist for all environmental chemicals and drugs; however, little empirical data are available to quantify such differences (Zeise et al. 2013). Traditional in vivo toxicity testing relies on the use of a single strain of rodent (typically an inbred or hybrid strain), with multiple identical animals being used at each time point or in each dose group; thus, extrapolation from these data to a diverse human population is difficult and necessitates the use of default assumptions. In the past decade, the utility of a mouse population-based approaches to in vivo toxicity testing has been demonstrated for a number of toxic agents and the appreciation of genetics as an important dimension in science and practice of investigative pharmacology and toxicology is increasing (Rusyn et al. 2010). Importantly, genetically heterogeneous mouse models have been combined with the limited data for interindividual differences in chemical toxicity from human studies to characterize the extent of the variability (Chiu et al. 2014), discover genetic determinants (Harrill et al. 2009), and understand the molecular underpinnings of toxicity (French et al. 2015).
Recent advances in mouse genetics have led to the development of highly diverse mouse populations, one of which is known as the Collaborative Cross (CC) (Churchill et al. 2004). The CC is a large panel of recombinant inbred strains that were derived from eight inbred founder strains (Collaborative Cross Consortium 2012; Threadgill et al. 2011). This new resource has been used to make seminal discoveries in biomedical science (Crowley et al. 2015; Phillippi et al. 2014) and enabled creation of mouse models for diseases that were thought not to exist in the mouse (Gralinski et al. 2015; Rasmussen et al. 2014).
One of the most ubiquitous environmental pollutants without solid experimental data on variability in toxicokinetics and toxicodynamics is tetrachloroethylene (perchloroethylene, PERC). PERC is a chlorinated olefin solvent with a variety of industrial applications, a ubiquitous contaminant of groundwater, soil, ambient and urban air, and is one of the most common pollutants present in many hazardous waste sites (National Research Council 2009a). The most well-known application of PERC is in dry-cleaning. Of the approximately 32,000 dry cleaners in the United States operating as of 2006, about 28,000 () were using PERC as a solvent (Goehl and O'Neil 2005). Not only are workers exposed to PERC via dry-cleaning, but consumers are also potentially exposed via “off-gassing” of PERC from treated clothing (Sherlach et al. 2011). PERC is therefore of great concern to public health protection agencies, including the U.S Environmental Protection Agency (EPA) and the International Agency for Research on Cancer (IARC). PERC has been classified by IARC (2013) and the U.S. EPA (2011b) as a possible human carcinogen, mainly based on evidence for carcinogenesis from chronic studies in rodents (National Toxicology Program 1986).
PERC exposure is also associated with noncancer toxicity in a number of tissues (U.S. EPA 2011b). The organ-specific adverse effects associated with exposure to PERC and other structurally similar olefins, such as trichloroethylene (TCE), are consequences of tissue-specific metabolism (Cichocki et al. 2016; Lash and Parker 2001; Lash et al. 2014), which involves both cytochrome P450s (CYPs) and glutathione (GSH) S-transferases (GSTs).
The oxidative metabolism of PERC results in the formation of trichloroacetate (TCA), a chemical with suggestive evidence of carcinogenic potential based on significantly increased incidences of liver tumors in mice (U.S. EPA 2011a). Because TCA is a major metabolite of PERC, it is thought to be critical to establishing an exposure–response relationship for PERC and toxicity (Lash and Parker 2001). Furthermore, it is estimated that the contribution of TCA to PERC-associated liver cancer in rodents can be as little as 12% to as much as 100% (U.S. EPA 2011b). TCA itself provides a useful marker of oxidative metabolism, which has been used as a dose metric for risk assessment extrapolation from mice to humans (U.S. EPA 2011b).
Numerous recent hazard and risk assessments pointed to a critical need for better characterization of toxicokinetics, toxicodynamics, and population variability of PERC in order to improve public health protection (Guha et al. 2012; National Research Council 2010; U.S. EPA 2011b). Although there have been previous attempts at characterizing toxicokinetic variability of PERC (Bois et al. 1996; Covington et al. 2007; Gelman et al. 1996). Chiu and Ginsberg (2011) found that all these efforts were hampered by inadequate experimental data, leading to substantial uncertainty in the ultimate risk assessments. Moreover, variability exists in both toxicokinetics and toxicodynamics, and may be a consequence of multiple factors, including genetics. Therefore, the contribution of such factors to population level variability in both toxicokinetic and toxicodynamic responses to PERC deserves further attention.
We used a genetically diverse mouse population of 45 CC mouse strains to quantify the extent of interstrain variability in response to exposure to a single high dose of PERC. We determined whether individuals with increased hepatic TCA levels are more sensitive to the hepatotoxic effects of PERC. Mice were exposed to a single dose of PERC and were sampled at multiple time points to generate population-level concentration–time profiles for PERC and TCA across multiple tissues. Toxicodynamic phenotyping included study of the expression of CYPs, dysregulation of hepatic lipid metabolism, and histopathological assessment of the liver.
Methods
Chemicals and Reagents
PERC was purchased from Sigma Aldrich (St. Louis, MO; Catalog #270,393-100ML; Lot #SHBD9374V). Alkamuls-EL620 was acquired from Solvay (Deptford, NJ; Lot #SP8J26X07). All reagents used were of the highest purity available.
Animals
Adult (6–8 weeks of age) male mice from 45 strains of the Collaborative Cross were acquired from the Systems Genetics Core at the University of North Carolina (Chapel Hill, NC) or from a colony maintained at Texas A&M University. All animals were acquired between 6 January 2015 and 20 February 2015. Exact dates of birth, dates of acquisition, and original IDs (if applicable) are available online on the Mouse Phenome Database (see “Data Availability” subsection, below). Strain nomenclature is used as suggested by the nomenclature committee (Collaborative Cross Consortium 2012). Of the 45 strains exposed to PERC, one animal from one strain (CC059/TauUnc) died in the 24-hr treatment group as a result of a gavage error. Comparisons between time-matched vehicle- and PERC-exposed individuals within a strain was required for data interpretation, toxicodynamic data are only presented for 44/45 strains. Animals were housed in disposable HEPA-filtered polycarbonate cages with hardword chip bedding (Sanichip, P.J. Murphy Forest Products, Bowling Green, KY) with access to food (Teklad #8,604, Envigo, Indianapolis, IN) and purified water ad libitum. The animal room was maintained on a 12-hr light/dark cycle. Animals were allowed to acclimate to the room for at least 10 days prior to beginning experimentation. All experiments were approved by the Institutional Animal Care and Use Committee of Texas A&M University. For all animal studies, exposures were conducted between 0800 and 1100 hours to limit potential diurnal variation in PERC metabolism. The in-life portion of the study was conducted over a 6-week time frame to limit seasonal variations.
PERC Exposure Protocol and Tissue Collection
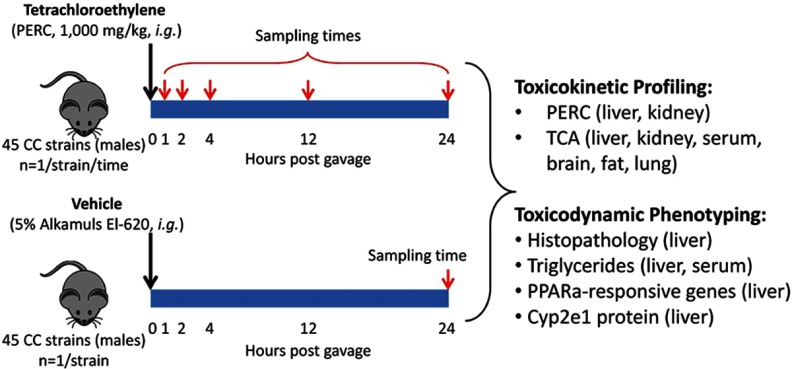
Figure 1 shows a schematic of the overall experimental design. Mice were administered a single dose of PERC () or vehicle (5% Alkamuls EL-620 in saline) by oral gavage (i.g.; ). All dosing solutions were prepared fresh daily (and used within 1.5 hr of preparation) in amber glass vials with foil-lined septa to minimize adsorption of PERC to the container walls and/or septum. For PERC-treated animals, the time points for tissue collection were 1, 2, 4, 12, and 24 hr postgavage (/strain/time point); vehicle-treated mice were euthanized only at the 24-hr time point (/strain). Prior to necropsy, animals were weighed and were deeply anesthetized by intraperitoneal (i.p.) injection of Euthasol™ (; Med-Vet International, Mettawa, IL) and euthanasia was performed via exsanguination through the vena cava. Serum was prepared by centrifugation using Z-gel tubes (Starstedt; Numbrecht, Germany). All other tissues were removed, rinsed in phosphate-buffered saline, and weighed. The left lobe of the liver was separated from the other lobes after weighing. The kidneys, lungs, brain, and epididymal fat pads were collected and snap-frozen in liquid nitrogen. For animals in the 24-hr group, small sections of left and median liver lobes and kidney were fixed in 10% neutral-buffered formalin, embedded in paraffin, sectioned at , and stained with hematoxylin & eosin (H&E).
Figure 1.
Study design consisted of two groups (PERC and vehicle) and five time points (1, 2, 4, 12, and 24 hr postdosing).
Gas Chromatography/Mass Spectrometry (GC/MS) Analysis of PERC and TCA in Tissues
A detailed protocol for the analysis of PERC and TCA in tissues is provided in the Supplemental Material, “Materials and Methods.” Briefly, PERC was analyzed in methanolic tissue extracts via headspace GC/MS. TCA was analyzed via GC/MS after derivatization to its methyl ester based on a modified U.S. EPA method (EPA 815-B-03-002; Domino et al. 2003).
Selection of Strains for TCA Toxicokinetic Profiling
Based upon TCA analysis in liver, kidney, and serum, tissues from nine additional strains were analyzed for TCA levels in brain, gonadal fat pad, and lung tissue. These strains were selected by stratifying strains in quartiles based on TCA AUC levels in liver, kidney, and serum, and then using the randomLHS function of the lhs R package. Unique strains were then selected from each box of the Latin hypercube.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
A detailed protocol for qRT-PCR is provided in the Supplemental Material, “Materials and Methods.” Briefly, qRT-PCR was performed on 50 ng of cDNA from the liver (left lobe) with Taqman® probes targeting Acyl-CoA oxidase 1 (Acox1) and Cyp4a10. -glucuronidase (GusB) was used as the housekeeping gene.
Western Blotting for CYP2E1
A detailed protocol for Western blotting of hepatic CYP2E1 is provided in the Supplemental Material, “Materials and Methods.” Briefly, of hepatic protein was immunoblotted for CYP2E1 and (loading control) using standard sodium dodecyl-sulfate polyacrylamide gel electrophoresis techniques.
Triglyceride Quantitation
Triglycerides were measured in liver (left lobe) and serum using a commercially available colorimetric kit (Dako, Carpinteria, CA) according to the manufacturer’s instructions.
Statistical Analysis
GraphPad Prism (La Jolla, CA) was used to calculate areas under the curves (AUCs) for all analytes and to perform paired and unpaired t-tests between vehicle- and PERC-treated groups. R (v.3.1.2) was used for fitting toxicokinetic data to a nonlinear function (nls), to generate heatmaps (gplots) and boxplots (ggplot2). For all tests, a was required for statistical significance.
Data Availability
All raw data have been made publically available by uploading files into the Mouse Phenome Database (http://phenome.jax.org; MPD: Rusyn7).
Results
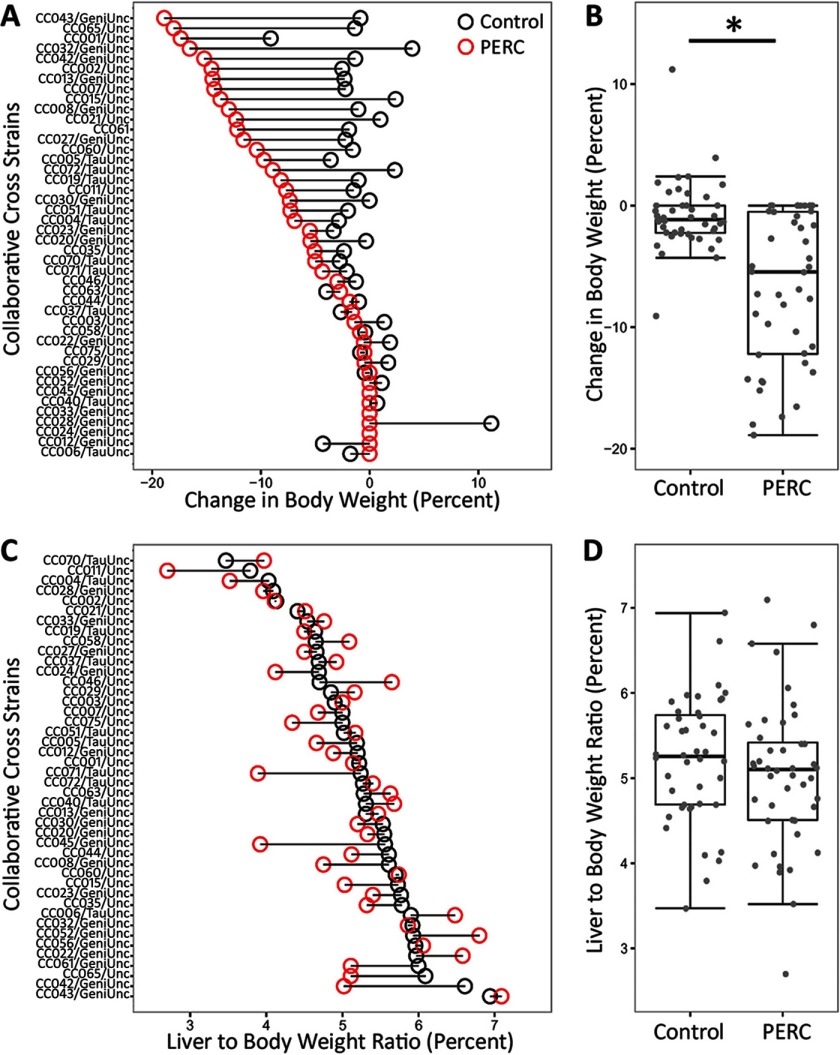
A single dose of PERC resulted in bodyweight loss in of strains, whereas liver-to-bodyweight ratios remained largely unaffected by PERC exposure (Figure 2). On average, a significant loss of body weight by was observed (, t-test), although the response was variable between strains. For instance, strains CC043/GeniUnc, CC065/Unc, and CC001/Unc lost over 15% of their body weight, whereas other strains (e.g., CC012/GeniUnc) maintained their body weight. The liver-to-bodyweight ratio was highly variable in both vehicle- and PERC-treated animals. The median liver-to-bodyweight ratio was lower in PERC-treated animals compared with vehicle-treated animals, although this was not a statistically significant observation (, unpaired t-test).
Figure 2.
Relative change in bodyweight (A,B) and liver to body weight ratio (C,D), as compared to pre-treatment weight, in vehicle- (black dot) and PERC- (, i.g., red dot) exposed Collaborative Cross mice 24 hr following a single intragastric gavage. A and C show change in the individual strains that were ordered according to the change in body weight in PERC-treated mice (A), or liver to body weight ratio in vehicle-treated mice (C). B and D show population averages as box and whisker plots. The box represents the interquartile range (IQR), the black line represents the median, and the whiskers represent and . The asterisk (*) denotes significant difference () between treatment groups (unpaired t-test).
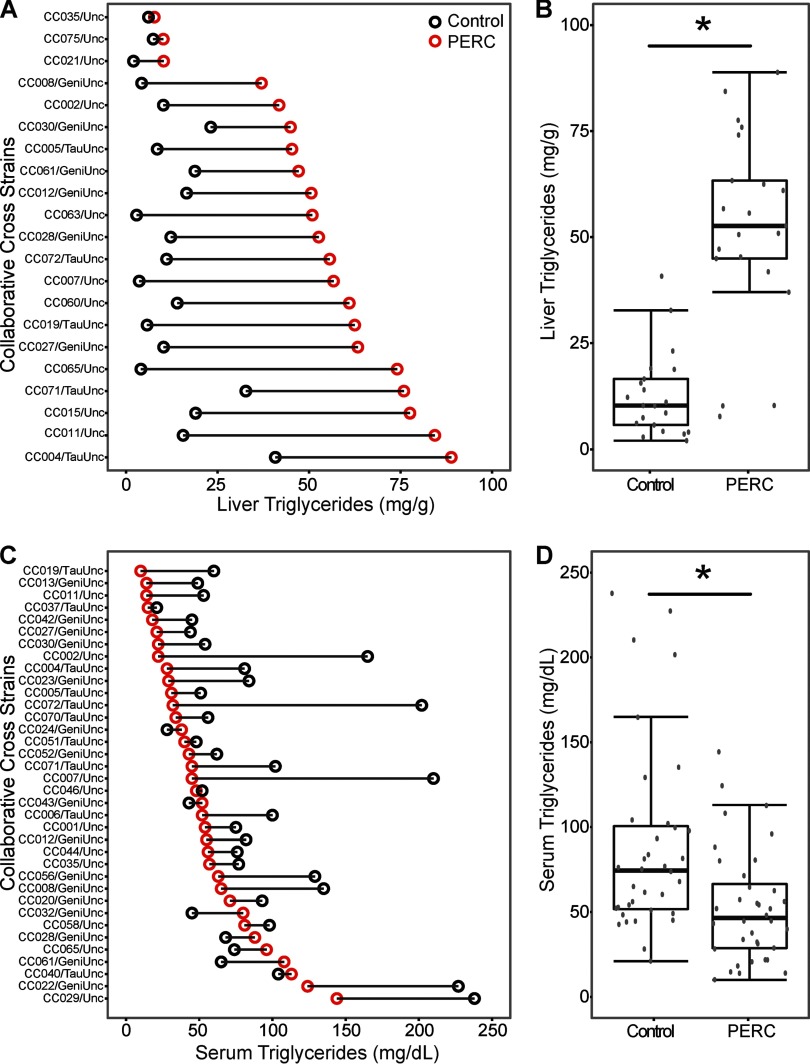
Significant accumulation (, t-test) of triglycerides was observed in the livers of PERC-exposed animals (Figure 3A,B); this finding was confirmed histopathologically (see Figure S1). About one-third of the strains tested exhibited various degrees of hepatosteatosis 24 hr following PERC exposure. Interestingly, the degree and type of steatosis varied among strains; some mice displayed zonal steatosis, others displayed azonal, and both macro- and micro-vesicular (and mixed) steatosis was observed. The accumulation of hepatic lipids was coincident with a decrease (, t-test) in serum triglyceride levels (Figure 3C,D), providing additional evidence that PERC exposure, even at a single dose, is coincident with dysregulation of liver lipid metabolism.
Figure 3.
Differences in liver (A,B) and serum (C,D) triglyceride levels in vehicle- (black dot) and PERC- (, i.g., red dot) exposed Collaborative Cross mice 24 hr following a single intragastric gavage. A and C show change in the individual strains that were ordered according to the values in PERC-treated mice. B and D show population averages as box and whisker plots (see Figure 2 for the legend). The asterisks (*) denote significant differences () between treatment groups (unpaired t-test).
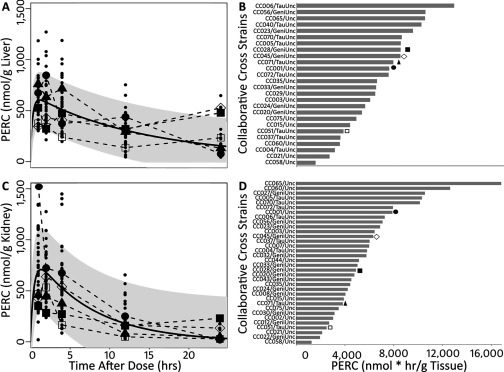
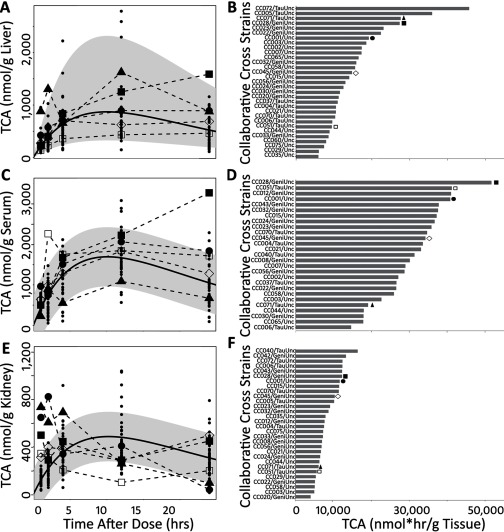
The levels of PERC and its primary oxidative metabolite TCA were assessed in a standard toxicokinetic study design. Levels of PERC in the liver and kidney (Figure 4; see also Table S1) were highly variable across the population (geometric standard deviation and 1.74, respectively), as highlighted by the broad 95% confidence intervals (gray shadow) around the nonlinear least squares regression line (solid black line). It is also evident that in both tissues, the shape of the concentration–time profiles in the individual strains was also variable.
Figure 4.
Concentration–time profiles of PERC in liver (A) and kidney (C) following a single oral dose of PERC (). The black solid line and gray shadowing represent a fitted nonlinear least square regression line and 95% confidence interval (respectively) for the entire data set. Dots represent data from the individual animals. Some strains are highlighted by using enlarged symbols. B and D represent calculated AUCs for strains in which a full concentration–time profile was available (i.e., mouse/timepoint). The symbols in A and C are coordinated (by strain) with the symbols above the bars in B and D.
Similar to PERC, tissue levels of TCA and shapes of the concentration–time profiles were strain-dependent (Figure 5; see also Table S2). On average, TCA levels were about 3-fold higher than PERC on a molar basis. Levels of TCA were highest in serum, followed by the liver and kidney. In the liver, the geometric mean (GM) was liver (). Although most of the strains either exhibit a plateau or clearance from all tissue compartments by the 24 hr time point, some strains appear to accumulate TCA with time. This type of toxicokinetic behavior yields under-prediction of the maximum TCA concentration. Due to this limitation, there is greater uncertainty in peak TCA concentrations for these strains and therefore the actual GM and GSD statistics are likely greater than those reported here.
Figure 5.
Concentration–time profiles of TCA in liver (A), serum (C), and kidney (E) following a single oral dose of PERC (). The black solid line and gray shadowing represent a fitted nonlinear least square regression line and 95% confidence interval (respectively) for the entire data set. Dots represent data from the individual animals. Some strains are highlighted by using enlarged symbols. B, D, and F represent calculated AUCs for strains in which a full concentration–time profile was available (i.e., mouse/timepoint). The symbols in A, C, and E are coordinated (by strain) with the symbols above the bars in B, D, and F.
PERC is structurally similar to trichloroethylene (TCE) and these compounds share qualitatively similar metabolism pathways. Thus, we compared the interstrain differences in serum TCA levels of this study (after exposure to PERC) and the serum TCA levels from (Bradford et al. 2011), in which a single oral dose of TCE in corn oil was administered to 16 strains of male inbred mice. Interstrain variability in TCA levels was similar between two studies (Bradford et al. 2011: ; ; this study: ; ); however, TCA AUCs in the serum of PERC-exposed mice was much greater as compared with TCE-exposed mice even though the dose of TCE was over 2.5-fold higher on a molar basis. Thus, in spite of qualitatively similar metabolism, the conversion of PERC and TCE to TCA in vivo is quantitatively different.
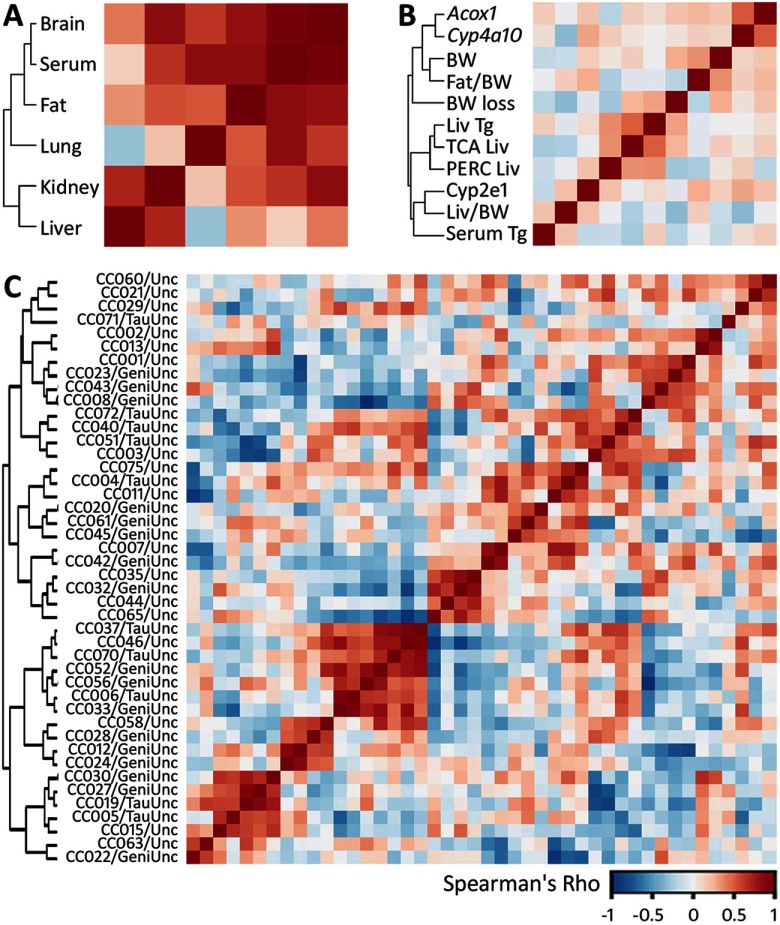
A subset of nine strains was selected for an in-depth profiling of TCA levels across additional tissues, as described in “Methods.” In addition to liver, kidney, and serum, TCA was analyzed in gonadal (epididymal) fat pads, lung, and brain (see Figure S2). Although a large degree of variability existed among individual strains, the general trend for TCA levels was serum<liver<lung<kidney<brain<fat. To better characterize the relationship between tissue compartments, unsupervised hierarchical clustering of correlations of TCA AUCs between tissues was performed (Figure 6A). Levels of TCA in serum, brain, fat, and lung correlate with one another and form a major cluster (as indicated by the dendrogram). Liver and kidney also have a high level of correlation with each other, but not to other tissue sites.
Figure 6.
Correlation matrices (colored according to the value of pairwise Spearman’s correlation for the phenotypes included in each analysis; see color bar scale) for toxicokinetic and toxicodynamic phenotypes across the population of Collaborative Cross strains. Unsupervised hierarchical clustering among parameters is shown by the dendrogram. (A) Heatmap of TCA partial AUCs (0–24 hr) in multiple tissues from a subset of nine strains of the Collaborative Cross treated with a single dose of PERC (see Figure S2 for concentration–time profiles). (B) Heatmap of toxicodynamic and toxicokinetic responses to PERC across the population. Abbreviations: Acox1 and Cyp4a10 = induction of Acox1 and Cyp4a10 after PERC exposure; BW = basal bodyweight; Fat/BW = fat-to-bodyweight ratio; BW loss = loss of bodyweight after PERC exposure, percent; Liv Tg = liver triglycerides following PERC exposure; TCA Liv= TCA AUC in liver; PERC Liv = PERC AUC in liver; CYP2E1 = liver CYP2E1 levels in vehicle-treated mice; Liv/BW = liver-to-bodyweight ratio after PERC treatment; Serum TG = serum triglyceride levels after PERC treatment. For all phenotypic parameters except for metabolite data, values were normalized to vehicle-treated strain-matched controls. (C) Heatmap of correlations among strains based on the phenotypes shown in B.
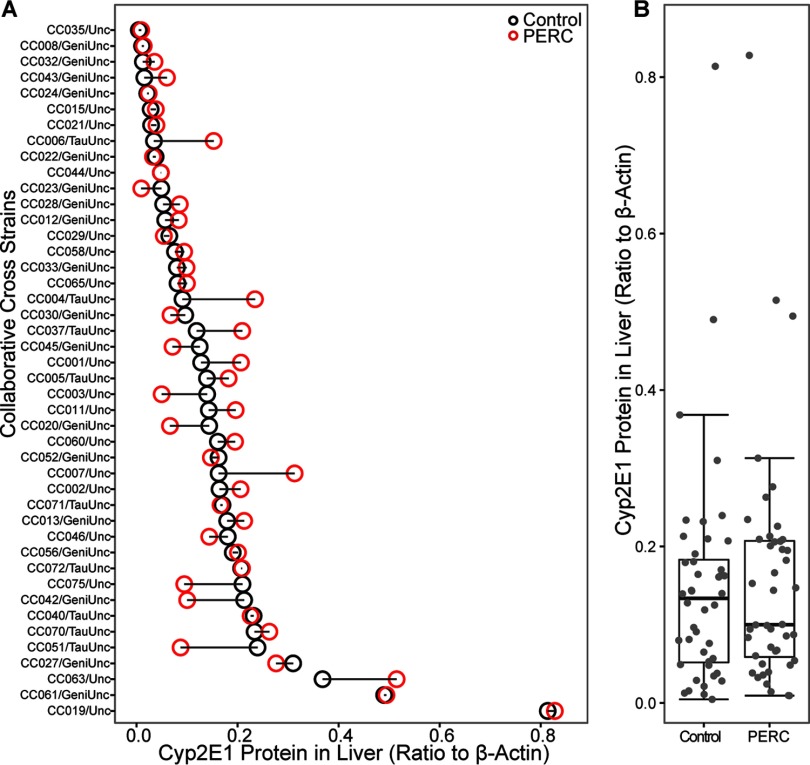
CYP2E1 metabolizes small lipophilic chemicals, such as TCE (Kim and Ghanayem 2006; Lash et al. 2014; Ramdhan et al. 2008); however, little is known about specific enzymes involved in PERC oxidation. Hepatic CYP2E1 protein was measured across CC strains (Figure 7). Levels of CYP2E1 were highly variable among vehicle-treated mice. After exposure to PERC, some strains exhibited marginal increase, whereas others a decrease in hepatic CYP2E1. At the population level, there was no statistically significant effect of PERC exposure on hepatic CYP2E1 levels.
Figure 7.
Liver levels of CYP2E1 protein in vehicle- (black dot) and PERC- (, i.g., red dot) exposed Collaborative Cross mice 24 hr following a single intragastric gavage. A shows change in the individual strains that were ordered according to the values in vehicle-treated mice. B shows population average as box and whisker plots (see Figure 2 for the legend).
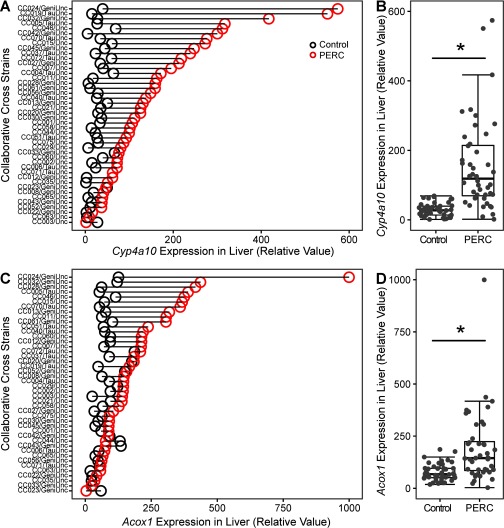
Liver effects of PERC and TCE have been attributed to formation of TCA, which is an activator of PPAR (Maloney and Waxman 1999). One study reported that PERC induces PPAR after acute and subacute (less than 14 days) exposure, but not after 30 days of exposure (Philip et al. 2007); however, the overall database on the role of PPAR in the effects of PERC is limited. Thus, we examined transcript levels for two PPAR target genes, Cyp4a10 and Acox1, in liver tissue 24 hr following exposure (Figure 8). In vehicle-treated mice, levels of individual transcripts were fairly consistent among strains. Almost every strain exhibited induction of both transcripts upon PERC exposure; however, the extent of induction was highly variable for both Acox1 (-fold; ) and Cyp4a10 (-fold; ). At the population level, statistically significant induction of both transcripts was observed.
Figure 8.
Differences in expression of Cyp4a10 (A,B) and Acox1 (C,D) in the liver of vehicle- (black dot) and PERC- (, i.g., red dot) exposed Collaborative Cross mice 24 hr following a single intragastric gavage. A and C show change in the individual strains that were ordered according to the values in PERC-treated mice. B and D show population averages as box and whisker plots (see Figure 2 for the legend). The asterisks (*) denote significant differences ()between treatment groups (unpaired t-test).
To further characterize the relationship among toxicokinetic and toxicodynamic phenotypes investigated in the population of CC mice, we performed unsupervised hierarchical clustering (Figure 6B). The induction of Acox1 and Cyp4a10 was highly correlated, suggesting that PERC exposure is associated with activation of the PPAR pathway in all exposed mouse strains. However, at the population level, no correlation was observed between liver PERC or TCA AUC and induction of these genes. Importantly, liver triglyceride level and liver PERC and TCA AUCs were clustered together, suggesting that either parent chemical or metabolite may be responsible for the observed dysregulation of hepatic lipids. Coincident with this finding was the observation of a negative, albeit weak, correlation between serum triglycerides and PERC and TCA AUCs in the liver. When all toxicodynamic phenotypes were parameterized for strain-to-strain correlations, we observed several clustered subpopulations, as displayed in the heatmap (Figure 6C); however, overall there was no obvious clustering of strains based on hepatic TCA levels.
Discussion
In this study, PERC was used as a model toxicant to investigate the contribution of genetic background to interindividual variability in toxicokinetics and subsequent toxicity associated with environmental chemical exposure. PERC is an ideal case-study chemical as its metabolism is proposed to play a critical role in its mode of toxicity. Therefore, it was hypothesized that individuals with an increased capacity to oxidize PERC to the hepatotoxic metabolite TCA would be at an increased risk for acute hepatotoxicity following a single exposure of PERC (Guyton et al. 2014). Moreover, TCA is a widely accepted principal marker of oxidative metabolism of PERC, which is implicated in the hepatocarcinogenic effects of PERC in mice and used as the basis for extrapolating cancer risk from mice to humans (U.S. EPA 2011b).
A similar approach has been taken before using a panel of inbred mice for the structurally similar chlorinated olefin solvent TCE (Bradford et al. 2011). In that study, however, the levels of parent TCE and/or metabolites were only measured in serum as a surrogate for target tissue. In the current study, levels of PERC and TCA were measured in a large number of target and nontarget tissues, including liver, kidney, serum, brain, lung, and fat across a much larger panel of inbred strains of the CC. We found that serum TCA AUCs were highly correlated with those in fat, lung, and brain, but were uncorrelated to those in liver and kidney. Thus, for PERC, sampling serum TCA levels may not be predictive of hepatic TCA levels. These data do suggest, however, that analysis of both liver and serum may provide sufficient toxicokinetic data to model systemic TCA exposure.
The toxicokinetics of TCA in the livers of CC mice was highly strain dependent, with an approximate 8-fold difference in TCA levels between strains with the highest (CC072/TauUnc) and least (CC035/Unc) amounts of this metabolite in the target tissue. Interestingly, in a human volunteer study with seven individuals exposed to 1 ppm of PERC by inhalation, the AUC of TCA in blood varied about 7-fold (Chiu et al. 2007), suggesting a similar order of magnitude variation despite the differences in sample size and the relative homogeneity of the human subjects.
TCA toxicokinetics following oral exposure to PERC in a similar vehicle has previously been investigated in male Swiss Webster mice (Philip et al. 2007). In the present study, the average level for maximum hepatic TCA concentration () was similar to the reported value of from Philip et al. (2007). Similarities were also observed between the levels of PERC measured in liver and kidney in Philip et al. (2007) and the population means in the current study. It is noteworthy that the concentration–time profile for Swiss Webster mice in Philip et al. (2007) does not fit the profile for all of the strains examined in our study.
It is commonplace for risk assessors of environmental contaminants to apply a so-called “uncertainty factor” of three for interindividual variability in toxicokinetics, in the absence of chemical-specific data. Based on the empirical experimental evidence presented here (see Tables S1 and S2), assuming a goal of protecting 99% of the population, a factor of 3.16 may be appropriate for estimating interindividual variability in PERC and TCA AUCs in liver, and TCA AUCs in kidney, lung, brain, and serum. However, a factor of 3.16 does not cover the 1% most sensitive individual for TCA toxicokinetics in adipose tissue or PERC toxicokinetics in kidney. The latter may be of particular importance considering that genotoxic conjugative metabolites of PERC are formed in renal tissue (Lash and Parker 2001).
Both national (U.S. EPA) and international (WHO) agencies have encouraged replacing these fixed “uncertainty factors” with values derived from chemical-specific data, so-called “chemical-specific adjustment factors” or “data-derived extrapolation factors” (IPCS 2005; U.S. EPA 2014). For example, factors derived from population PBPK modeling protective of 99% population were used to replace the uncertainty factor for toxicokinetic variability in several U.S. EPA assessments, including TCE (U.S. EPA 2011c).
The interstrain toxicokinetic data from Bradford et al. (2011) were used to generate a population-based physiologically based pharmacokinetic (PBPK) model for TCE (Chiu et al. 2014). Importantly, the extent of variability observed among inbred strains of mice intragastrically exposed to a single high dose of TCE closely approximated that predicted from PBPK models calibrated to population toxicokinetic data in humans that included both occupational (e.g., Monster et al. 1976) as well as lower, more environmentally relevant exposures (Chiu et al. 2007). Therefore, population-based PBPK modeling of rodent-derived toxicokinetic data is useful for understanding interindividual variability in toxicokinetics among the human population, and potentially applicable to risk assessment by replacing the “uncertainty factors” with values based on chemical-specific data. Specifically, data-derived estimates for toxicokinetic variability could conceivably be made by incorporating the current population-level data into a population PERC PBPK model. Because of the single high dose level in this study, such a PBPK model would likely to be incorporate previously collected data (Chiu and Ginsberg 2011) in a Bayesian manner. The anticipated result would be additional confidence in decision-making when considering uncertainties associated with interindividual differences within a population.
The focus of this study was on the oxidative metabolism of PERC to TCA. Relatively little is known about the pathways of PERC oxidation to TCA. However, for TCE, metabolism to TCA is proposed to occur mainly through CYP2E1 (Kim and Ghanayem 2006; Lash et al. 2014; Ramdhan et al. 2008), although other enzymes are likely involved (Lash et al. 2014). In this study, we measured hepatic levels of CYP2E1 in vehicle- and PERC-treated mice 24 hr after dosing. Hepatic protein levels of CYP2E1 in vehicle-treated mice were variable across strains, and overall there was no effect of PERC treatment on modulation of hepatic CYP2E1 levels. Similarly, Philip et al. (2007) reported no effect of PERC treatment on hepatic CYP2E1 levels. When hepatic CYP2E1 levels in vehicle-treated mice were correlated by strain to hepatic TCA AUCs, there was no correlation at the population level (), suggesting that CYP2E1 may not be a major contributor to PERC metabolism to TCA. Indeed, in human lymphoblastoma-derived MCL-5 cells transfected with various human CYPs (hCYP), hCYP2B6 (and, to a lesser extent, hCYP2C8 and hCYP1A2) was able to catalyze the reaction of PERC to acetylated neoantigens, which were recognized by immunoreactivity to anti-trifluoroacetylated antibodies (White et al. 2001). In isolated male F344 rat hepatocytes, incubation with PERC in the presence of CYP inhibitors had little effect on cytotoxicity as assessed by LDH leakage into the cell medium (Lash et al. 2007). In the same study, isolated hepatocytes from rats pretreated with the CYP2E1 inducer pyridine did not increase cytotoxicity of PERC, but did increase the cytotoxicity of TCE. However, it has been shown that pretreatment of male B6C3F1 mice with the suicide CYP2E1 inhibitor carbon tetrachloride did decrease TCA levels after PERC exposure (Fisher et al. 2004). These historic data in conjunction with the current study highlight the complexity of PERC metabolism as a consequence of interspecies and interindividual variability and warrant further mechanistic investigation into the enzymes involved in PERC metabolism.
A variety of acute toxicity end points were evaluated in this study, with dysregulation of hepatic lipid homeostasis being the most consistent finding. Hepatic lipid accumulation was observed histopathologically in about 33% of all PERC-treated mice 24 hr after dosing, and there was a statistically significant increase in liver triglyceride levels in PERC-exposed animals relative to vehicle-treated controls. Coincident with this finding, there was a decrease in serum triglycerides, and an induction of the PPAR-responsive genes Cyp4a10 and Acox1 in the liver, suggesting that PERC exposure results in dysregulation of liver lipid metabolism and that this phenotype is largely independent of genetic background. Lipid accumulation in the liver has been observed following a single dose of PERC in mice (Philip et al. 2007) and also following 6 weeks of oral administration of in corn oil to Male Swiss-Cox (outbred) mice (Buben and O'Flaherty 1985).
The availability of a large number of phenotypes from a large number of individual strains permitted the establishment of a relationship between interindividual variability in toxicokinetics and toxicodynamics of PERC at the population level. Although there was a significant effect of PERC exposure on a number of toxicodynamic phenotypes, not a single one was correlated with hepatic TCA levels. This demonstrates a complex and highly variable relationship between PERC and TCA toxicokinetics and toxicodynamics at the population level, specifically that correlation between metabolism and toxicity of PERC is not to be presumed.
Interestingly, TCA levels were correlated to induction of PPAR-regulated genes in TCE-exposed mice (Yoo et al. 2015), which further highlights the differences between these two highly structurally related chlorinated solvents. Using unsupervised hierarchical clustering, there was a cluster formed among PERC and TCA AUCs and liver triglyceride accumulation. However, all of the correlation coefficients between any two of these individual parameters suggest only a weak correlation. It was previously reported that neither administered PERC dose nor total urinary PERC metabolite levels were correlated with liver triglyceride accumulation in mice (Buben and O'Flaherty 1985). Nonetheless, at the population level, there appears to be some relationship among PERC, TCA, and hepatic triglyceride accumulation. Thus, we posit that interindividual differences in PERC metabolism may be only partially responsible for the variability in the toxicity of this compound, and that additional research is needed to determine whether genetic modifiers may account for these differences.
This study was not without limitations. We focused on the toxicokinetic study design and have examined the effects of a single high dose exposure only. Future studies examining the sub-chronic or chronic effects, and exposures to lower doses of PERC are warranted. This may be particularly true for PERC, which displays saturable enzyme kinetics at high exposure concentrations in male Sprague-Dawley rats (Pegg et al. 1979; Schumann et al. 1980) and male B6C3F1 (Schumann et al. 1980), outbred Swiss-Cox (Buben and O'Flaherty 1985) and Swiss-Webster mice (Philip et al. 2007). This study probed interindividual variation in metabolic “capacity” (i.e., related to Vmax) and is less informative to variation in “affinity” (i.e., related to KM). Because metabolism is proportional to Vmax/KM at lower exposure, this study still likely addresses a portion of the overall variability, but is less sensitive to additional variability at low doses due to interindividual differences in KM among CC mice. The high dose level also limits interpretation of the lack of correlation between metabolism and CYP2E1, because a stronger correlation may exist at lower exposure levels.
In this work, we did not evaluate intrastrain variability and future studies may benefit from concomitant evaluation of intra- and interstrain variability. Furthermore, we only investigated the role of genetics in variability in toxicokinetics and toxicodynamics of PERC. Other factors, including sex, diet, and underlying disease may contribute to interindividual variability in PERC toxicokinetics or toxicodynamics (National Research Council 2009b, 2010).
Despite its limitations, this study has important implications for conducting animal experiments for the purpose of addressing interindividual variability and informing human health assessments. Specifically, this is the first study to use CC mice to perform parallel and tissue-specific toxicokinetic and toxicodynamic phenotyping at the population level. Using this approach, the variability in toxicokinetics of PERC in multiple tissues across a population was quantified. Utilizing an inbred mouse population model permits genetically identical individuals to be tested at multiple time points, with different doses, and with different chemicals, thereby increasing statistical power and confidence and allowing chemical-to-chemical comparisons. Even though such a study design does not yield toxicity phenotypes that are more traditionally used for human health assessments, it serves as a prerequisite step to subsequent sub-chronic and chronic studies. By assaying a large number of genetically diverse but genetically defined individuals through an acute study design one can select a smaller set of strains that are representative of the diversity in toxicokinetics for the follow up studies. For instance, subpopulations of strains based on a particular phenotype (e.g., hepatic TCA levels) can be selected for a 90-day study where the use of a large number of strains would be challenging because of ethical and financial considerations.
In summary, the CC is an invaluable resource for toxicologists studying interindividual variability in toxicokinetics and toxicodynamics. By taking advantage of this mouse population model, intervariability in toxicokinetics of PERC and TCA was quantified across multiple tissues. These data will be useful for updating the current PBPK model for PERC (Chiu and Ginsberg 2011) to a population-based model and will likely be of use in future risk management decisions for PERC. Unsupervised hierarchical clustering of toxicodynamic phenotypes highlighted the complexity of the relationship between toxicokinetics and toxicodynamics of tetrachloroethylene at the population level and suggested a potential relationship between hepatic PERC and TCA levels and liver triglyceride accumulation. Our hypothesis that individuals with increased hepatic TCA levels would be more susceptible to PERC-associated hepatotoxicity was largely unsupported, suggesting that TCA is not the only active toxicant and/or that other genetic determinants contribute to interindividual susceptibility to PERC-associated hepatotoxicity.
Supplemental Material
References
- Bois FY, Gelman A, Jiang J, Maszle DR, Zeise L, Alexeef G. 1996. Population toxicokinetics of tetrachloroethylene. Arch Toxicol 70:347–355, PMID: 8975633, 10.1007/s002040050284. [DOI] [PubMed] [Google Scholar]
- Bradford BU, Lock EF, Kosyk O, Kim S, Uehara T, Harbourt D, et al. 2011. Interstrain differences in the liver effects of trichloroethylene in a multistrain panel of inbred mice. Toxicol Sci 120:206–217, PMID: 21135412, 10.1093/toxsci/kfq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buben JA, O'Flaherty EJ. 1985. Delineation of the role of metabolism in the hepatotoxicity of trichloroethylene and perchloroethylene: a dose-effect study. Toxicol Appl Pharmacol 78:105–122, PMID: 2994252. [DOI] [PubMed] [Google Scholar]
- Chiu WA, Campbell JL Jr, Clewell HJ III, Zhou YH, Wright FA, Guyton KZ, et al. 2014. Physiologically based pharmacokinetic (PBPK) modeling of interstrain variability in trichloroethylene metabolism in the mouse. Environ Health Perspect 122:456–463, PMID: 24518055, 10.1289/ehp.1307623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WA, Ginsberg GL. 2011. Development and evaluation of a harmonized physiologically based pharmacokinetic (PBPK) model for perchloroethylene toxicokinetics in mice, rats, and humans. Toxicol Appl Pharmacol 253:203–234, PMID: 21466818, 10.1016/j.taap.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Chiu WA, Micallef S, Monster AC, Bois FY. 2007. Toxicokinetics of inhaled trichloroethylene and tetrachloroethylene in humans at 1 ppm: empirical results and comparisons with previous studies. Toxicol Sci 95:23–36, PMID: 17032701, 10.1093/toxsci/kfl129. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, et al. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36:1133–1137, PMID: 15514660, 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Cichocki JA, Guyton KZ, Guha N, Chiu WA, Rusyn I, Lash LH. 2016. Target organ metabolism, toxicity, and mechanisms of trichloroethylene and perchloroethylene: key similarities, differences, and data gaps. J Pharmacol Exp Ther 359:110–123, PMID: 27511820, 10.1124/jpet.116.232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Cross Consortium. 2012. The genome architecture of the collaborative cross mouse genetic reference population. Genetics 190:389–401, 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington TR, Robinan GP, Van Landingham CB, Andersen ME, Kester JE, Clewell HJ. 2007. The use of Markov chain Monte Carlo uncertainty analysis to support a Public Health Goal for perchloroethylene. Regul Toxicol Pharmacol 47:1–18, PMID: 16901594, 10.1016/j.yrtph.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Crowley JJ, Zhabotynsky V, Sun W, Huang S, Pakatci IK, Kim Y, et al. 2015. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat Genet 47:353–360, PMID: 25730764, 10.1038/ng.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domino MM, Pepich BV, Munch DJ, Fair PS, Xie Y. 2003. Determination of haloacetic acids and dalapon in drinking water by liquid-liquid microextraction, derivatization, and gas chromatography with electron capture detection. Cincinnati, OH:U.S. Environmental Protection Agency, Office of Ground Water and Drinking Water; http://nepis.epa.gov/Exe/ZyPDF.cgi?Dockey=901V0400.PDF [accessed 19 May 2017]. [Google Scholar]
- Fisher J, Lumpkin M, Boyd J, Mahle D, Bruckner JV, El-Masri HA. 2004. PBPK modeling of the metabolic interactions of carbon tetrachloride and tetrachloroethylene in B6C3F1 mice. Environ Toxicol Pharmacol 16:93–105, PMID: 21782696, 10.1016/j.etap.2003.10.006. [DOI] [PubMed] [Google Scholar]
- French JE, Gatti DM, Morgan DL, Kissling GE, Shockley KR, Knudsen GA, et al. 2015. Diversity outbred mice identify population-based exposure thresholds and genetic factors that influence benzene-induced genotoxicity. Environ Health Perspect 123:237–245, PMID: 25376053, 10.1289/ehp.1408202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Bois F, Jiang J. 1996. Physiological pharmacokinetic analysis using population and informative prior distributions. J Am Stat Assoc 91:1400–1412, 10.2307/2291566. [DOI] [Google Scholar]
- Goehl E, O'Neil J. 2005. Dry Cleaning Docket: Background Information Document, http://www3.epa.gov/airtoxics/dryperc/11-14-05background.pdf [Online 1 June 2016].
- Gralinski LE, Ferris MT, Aylor DL, Whitmore AC, Green R, Frieman MB, et al. 2015. Genome wide identification of SARS-CoV susceptibility loci using the Collaborative Cross. PLoS Genet 11:e1005504, PMID: 26452100, 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha N, Loomis D, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, et al. 2012. Carcinogenicity of trichloroethylene, tetrachloroethylene, some other chlorinated solvents, and their metabolites. Lancet Oncol 13:1192–1193, PMID: 23323277. [DOI] [PubMed] [Google Scholar]
- Guyton KZ, Hogan KA, Scott CS, Cooper GS, Bale AS, Kopylev L, et al. 2014. Human health effects of tetrachloroethylene: key findings and scientific issues. Environ Health Perspect 122:325–334, PMID: 24531164, 10.1289/ehp.1307359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, et al. 2009. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res 19:1507–1515, 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer). 2013. Trichloroethylene, tetrachloroethylene and some other chlorinated agents. IARC Monogr Eval Carcinog Risk Hum 106. [PMC free article] [PubMed] [Google Scholar]
- IPCS (International Programme on Chemical Safety). 2005. Chemical-Specific Adjustment Factors For Interspecies Differences And Human Variability: Guidance Document For Use Of Data In Dose/Concentration–Response Assessment. Geneva:World Health Organization. [Google Scholar]
- Kim D, Ghanayem BI. 2006. Comparative metabolism and disposition of trichloroethylene in Cyp2e1–/– and wild-type mice. Drug Metab Dispos 34:2020–2027, PMID: 16959879, 10.1124/dmd.106.010538. [DOI] [PubMed] [Google Scholar]
- Lash LH, Chiu WA, Guyton KZ, Rusyn I. 2014. Trichloroethylene biotransformation and its role in mutagenicity, carcinogenicity and target organ toxicity. Mutat Res Rev Mutat Res 762:22–36, PMID: 25484616, 10.1016/j.mrrev.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash LH, Parker JC. 2001. Hepatic and renal toxicities associated with perchloroethylene. Pharmacol Rev 53:177–208, PMID: 11356983. [PubMed] [Google Scholar]
- Lash LH, Putt DA, Huang P, Hueni SE, Parker JC. 2007. Modulation of hepatic and renal metabolism and toxicity of trichloroethylene and perchloroethylene by alterations in status of cytochrome p450 and glutathione. Toxicology 235:11–26, PMID: 17433522, 10.1016/j.tox.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney EK, Waxman DJ. 1999. trans-Activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol Appl Pharmacol 161:209–218, PMID: 10581215, 10.1006/taap.1999.8809. [DOI] [PubMed] [Google Scholar]
- Monster AC, Boersma G, Duba WC. 1976. Pharmacokinetics of trichloroethylene in volunteers, influence of workload and exposure concentration. Int Arch Occup Environ Health 38:87–102, PMID: 1002309, 10.1007/BF00378619. [DOI] [PubMed] [Google Scholar]
- National Research Council. 2009a. Contaminated Water Supplies at Camp Lejeune: Assessing Potential Health Effects. Washington, DC:National Academies Press. [PubMed] [Google Scholar]
- National Research Council. 2009b. Science and Decisions: Advancing Risk Assessment. Washington, DC:National Academies Press. [PubMed] [Google Scholar]
- National Research Council. 2010. Review of the Environmental Protection Agency’s Draft IRIS Assessment of Tetrachloroethylene. Washington, DC:National Academies Press. [PubMed] [Google Scholar]
- National Toxicology Program. 1986. NTP Toxicology and Carcinogenesis Studies of Tetrachloroethylene (Perchloroethylene) (CAS no. 127-18-4) in F344/N rats and B6C3F1 Mice (Inhalation Studies). Natl Toxicol Program Tech Rep Ser 311:1–197. [PubMed] [Google Scholar]
- Pegg DG, Zempel JA, Braun WH, Watanabe PG. 1979. Disposition of tetrachloro(14C)ethylene following oral and inhalation exposure in rats. Toxicol Appl Pharmacol 51:465–474, PMID: 538758. [DOI] [PubMed] [Google Scholar]
- Philip BK, Mumtaz MM, Latendresse JR, Mehendale HM. 2007. Impact of repeated exposure on toxicity of perchloroethylene in Swiss Webster mice. Toxicology 232:1–14, PMID: 17267091, 10.1016/j.tox.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Phillippi J, Xie Y, Miller DR, Bell TA, Zhang Z, Lenarcic AB, et al. 2014. Using the emerging Collaborative Cross to probe the immune system. Genes Immun 15:38–46, PMID: 24195963, 10.1038/gene.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdhan DH, Kamijima M, Yamada N, Ito Y, Yanagiba Y, Nakamura D, et al. 2008. Molecular mechanism of trichloroethylene-induced hepatotoxicity mediated by CYP2E1. Toxicol Appl Pharmacol 231:300–307, PMID: 18565563, 10.1016/j.taap.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AL, Okumura A, Ferris MT, Green R, Feldmann F, Kelly SM, et al. 2014. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 346:987–991, PMID: 25359852, 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusyn I, Gatti DM, Wiltshire T, Kleeberger SR, Threadgill DW. 2010. Toxicogenetics: population-based testing of drug and chemical safety in mouse models. Pharmacogenomics 11:1127–1136, PMID: 20704464, 10.2217/pgs.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann AM, Quast JF, Watanabe PG. 1980. The pharmacokinetics and macromolecular interactions of perchloroethylene in mice and rats as related to oncogenicity. Toxicol Appl Pharmacol 55:207–219, PMID: 7423514. [DOI] [PubMed] [Google Scholar]
- Sherlach KS, Gorka AP, Dantzler A, Roepe PD. 2011. Quantification of perchloroethylene residues in dry-cleaned fabrics. Environ Toxicol Chem 30:2481–2487, PMID: 21898565, 10.1002/etc.665. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Miller DR, Churchill GA, de Villena FPM. 2011. The Collaborative Cross: a recombinant inbred mouse population for the systems genetic era. ILAR J 52:24–31, PMID: 21411855. [DOI] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2011a. Toxicological Review of Trichloroacetic Acid. Washington, DC:U.S. EPA. [Google Scholar]
- U.S. EPA. 2011b. Toxicological Review of Tetrachloroethylene (CAS no. 127-18-4): In Support of Summary Information on the Integrated Risk Information System (IRIS). EPA/635/R-08/011A. Washington, DC:U.S. EPA. [Google Scholar]
- U.S. EPA. 2011c. Toxicological Review of Trichloroethylene (CAS no. 79-01-6): In Support of Summary Information on the Integrated Risk Information System (IRIS). EPA/635/R-09/011F. Washington, DC:U.S. EPA. [Google Scholar]
- U.S. EPA. 2014. Next Generation Risk Assessment: Incorporation of Recent Advances in Molecular, Computational, and Systems Biology. EPA/600/R-14/004. Washington, DC:U.S. EPA. [Google Scholar]
- White IN, Razvi N, Gibbs AH, Davies AM, Manno M, Zaccaro C, et al. 2001. Neoantigen formation and clastogenic action of hydrochlorofluorocarbons-123 and perchloroethylene in human MCL-5 cells. Toxicol Lett 124:129–138, PMID: 11684365. [DOI] [PubMed] [Google Scholar]
- Yoo HS, Bradford BU, Kosyk O, Shymonyak S, Uehara T, Collins LB, et al. 2015. Comparative analysis of the relationship between trichloroethylene metabolism and tissue-specific toxicity among inbred mouse strains: liver effects. J Toxicol Environ Health Part A 78:15–31, PMID: 25424544, 10.1080/15287394.2015.958417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeise L, Bois FY, Chiu WA, Hattis D, Rusyn I, Guyton KZ. 2013. Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ Health Perspect 121:23–31, PMID: 23086705, 10.1289/ehp.1205687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data have been made publically available by uploading files into the Mouse Phenome Database (http://phenome.jax.org; MPD: Rusyn7).