Abstract
Hairy/enhancer of split 1 (HES1) is a basic helix–loop–helix transcriptional repressor. Aberrant demethylation has been considered a common mechanism of tumor promoter gene activation. In the current study, we aimed to investigate the methylation status of the HES1 promoter and correlations with clinicopathological parameters and prognosis in colorectal cancer (CRC). The expression of HES1 in 50 paired CRC specimens and adjacent normal tissues was determined by using quantitative real-time polymerase chain reaction and immunohistochemical analysis. Moreover, DNA methylation status was evaluated through methylation-specific polymerase chain reaction and bisulfite sequencing. The correlation of methylation status with HES1 expression level and clinicopathological parameters was statistically analyzed in CRC patients. Our data showed that the methylation level of HES1 was significantly decreased and negatively correlated with HES1 expression in CRC tissues. Moreover, HES1 hypomethylation was associated with a poor histological grade, Dukes’ classification, lymph node metastasis, and clinical stages (P<0.05). Furthermore, survival analyses revealed that a decreased methylation status of HES1 was linked to poor prognosis of CRC patients. In conclusion, promoter hypomethylation upregulates HES1 expression and plays a critical role in the progression and prognosis of CRC patients.
Keywords: colorectal cancer, HES1, hypomethylation, prognosis, progression
Introduction
Colorectal cancer (CRC) is the third most fatal type of cancer and the fourth leading cause of cancer-related deaths worldwide.1 Significant progress has been made to treat the disease, including surgery, simultaneous radio- and chemotherapy, molecular targeted therapy, and biotherapy; however, CRC remains a challenging malignancy with an invariable appearance of tumor recurrence. The 5-year survival rate of CRC is approximately 60%.2 Therefore, investigating the pathogenesis and underlying molecular mechanisms involved in CRC progression is critical for early diagnosis and will provide more effective methods for CRC prevention and therapy.
Hairy/enhancer of split 1 (HES1) is a basic helix-loop-helix (bHLH) transcriptional repressor, regulated by the Notch signaling pathway.3 Previous studies have proven that HES1 is involved in various biological processes, including neurogenesis, inflammation, and cellular development.4–6 Moreover, several studies have reported that HES1 is overexpressed in a variety of human malignancies, including CRC.7–9 This indicates a potential role in the process of tumorigenesis. Recently, it was demonstrated that HES1 supported cell proliferation and migration in human colon cancer, via activating B cell-specific Moloney murine leukemia virus integration site-1 (Bmi-1) and the PTEN/Akt/GSK3β pathway.10 In addition, Weng et al11 reported that HES1 promoted the invasion ability of CRC cells via the STAT3-MMP14 signaling pathway. HES1 was identified as a self-renewal marker in colon carcinoma in nude mice, which improved the tumor-initiating capability of colon cancer cells.12 Moreover, Yuan et al13 showed that HES1 promoted tumor metastasis by inducing epithelial–mesenchymal transition and functioned as a poor prognostic factor for CRC patients. Taken together, these results suggested that upregulation of HES1 may play a role in the occurrence and progression of CRC.
Although HES1 may play an important role in the progression and prognosis of CRC, the underlying mechanism of action that is involved in HES1 upregulation in cancer cells remains to be elucidated. DNA methylation, a process in which a methyl group is added to cytosine in cytosine-phosphate-guanine (CpG) dinucleotides, is a type of epigenetic modification in the human genome that plays a critical role in the regulation of gene expression.14,15 Previous studies have shown that decreased methylation of tumor promoter genes frequently occurs in a variety of human malignancies, including CRC,16–18 which accelerated tumor progression and was related to poor prognosis.16 Until now, HES1 hypomethylation has been found in intestinal stem and progenitor cells, which leads to upregulation of HES1.19,20 So far, several regulatory mechanisms of HES1 expression have been identified;21,22 however, whether the methylation status affects HES1 expression in CRC remains unknown.
In the present study, we compared the methylation status of the HES1 promoter in CRC tissues with adjacent normal specimens and correlated this with clinicopathological parameters and prognosis.
Patients and methods
Patients and tissue specimens
A total of 50 samples of tumor tissues and adjacent normal specimens (3 cm from the tumor edge) were gained from patients undergoing tumor resections between 2012 and 2013 in Zhejiang Provincial People’s Hospital. All participants provided written informed consent. The median age of the patients was 66 years (39–76 years). All patients had a definite histological diagnosis of CRC. None of the patients had received chemotherapy or radiotherapy prior to surgery. Patients were followed up postoperatively via telephone consultations. This study was approved by the institutional ethics committee of Zhejiang Provincial People’s Hospital. Resected tissues were directly frozen in liquid nitrogen after resection from the body and were stored at −80°C until further analysis or were fixed in 4% paraformaldehyde for paraffin embedding.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was removed from tissues using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA), and cDNA was synthesized using a TaqMan Reverse Transcription kit (Thermo Fisher Scientific). For mRNA analysis, the expression of HES1 was detected with SYBR Green PCR Master Mix (Thermo Fisher Scientific). GAPDH mRNA levels were used for normalization. The cycling programmer was conducted in a total volume of 20 µL by using the following procedure: preliminary denaturation at 95°C for 10 min, followed by 39 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 60 s, followed by a final elongation step at 95°C for 10 min. qPCR was operated in triplicate, involving no-template controls, using the Bio-Rad CFX-96 Real-time PCR system and iQ SYBR Green Supermix (Bio-Rad Laboratories Inc., Hercules, CA, USA). Relative gene expression of mRNA was quantified via the 2−ΔΔCt method. The primers (GeneCopoeia, Guangzhou, China) used were as follows: HES1 (forward) 5′-TCAACACGACACCGGATAAAC-3′ and (reverse) 5′-GCCGCGAGCTATCTTTCTTCA-3′; and GAPDH (forward) 5′-CTTTGGTATCGTGGAAGGACTC-3′ and (reverse) 5′-GTAGAGGCAGGGGATGATGTTCT-3′.
Immunohistochemical detection
Immunohistochemical staining was performed with the Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA, USA). For all CRC tissues the following procedures were performed: 4% paraformaldehyde fixation at room temperature, paraffin embedding, section (4 µm thickness), deparaffinization in xylene with series concentration gradients, then rehydration, 3% hydrogen peroxide incubation for 25 min, antigen retrieval at 100°C by citrate buffer solution, 10% goat serum (Thermo Fisher Scientific) blocking, followed by Rabbit anti-HES1 antibody incubation (1:200; Abcam, Shanghai, China) at 4°C overnight, next biotinylated anti-rabbit secondary antibody incubation (1:100; BioWorld, Minneapolis, MN, USA) at 37°C for 20 min, and then incubation of streptavidin–horseradish peroxidase complex. Finally, the slides were visualized in diaminobenzidine solution (Sigma-Aldrich Co., St Louis, MO, USA). All sections were separately assessed by two certified pathologists who were uninformed of patients’ clinical pathology and other information. Quantitative analysis for the percentage of HES1-positive cells was performed.
DNA extraction and bisulfite modification
Genomic DNA was extracted from resected tumor tissues and adjacent normal specimens using the QIAamp DNA mini kit (Qiagen NV, Venlo, the Netherlands) according to the manufacturer’s protocol. The quality and quantity of the isolated DNA were measured by NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). Bisulfite treatment of genomic DNA was performed using an EpiTect Fast DNA Bisulfite Kit (Qiagen NV) according to the manufacturer’s instructions. Bisulfite-treated DNA samples were next kept at −20°C for further use.
Methylation-specific PCR (MSP)
CpG islands in the HES1 promoter region were predicted using MethPrimer tools. The methylation level of CpG islands in HES1 promoter regions was initially determined by MSP. Briefly, PCR was conducted in a total volume of 25 µL containing 80 ng of bisulfite-modified DNA template under the following condition: 95°C for 5 min, followed by 35 cycles each containing 95°C denature for 30 s, 55°C annealing for 30 s, and 72°C elongation for 30 s, followed by a final extension at 72°C for 10 min. Nuclease-free water served as a negative control. Aliquots from PCR products (172 bp for methylation and 175 bp for unmethylation) were visualized on 2% agarose gel containing 5 mg/mL ethidium bromide, analyzed, and photographed using ChemiDoc™ MP Imaging System (Bio-Rad Laboratories Inc.) under ultraviolet (UV) illumination. The methylation-specific PCR (MSP) primers (GeneCopoeia) designed according to specific sequences of CpG islands in the HES1 promoter regions were as follows: methylated (M) (forward) 5′-CGGAGGTAGGAGGTTGATTC-3′ and (reverse) 5′-TTAAAATTTTCACTCGACCG-3′; and unmethylated (U) (forward) 5′-GAGTGGAGGTAGGAGGTTGATTT-3′ and (reverse) 5′-TTAAAATTTTCACTCAACCAAAA-3′.
Sodium bisulfite genomic sequencing (BGS)
The CpG island region of HES1 promoter was amplified with sodium bisulfite-modified DNA template under the following PCR conditions: 95°C for 5 min and then 60°C for 50 s, followed by 35 cycles of 95°C for 30 s, 55°C for 50 s, and 72°C for 50 s and a final elongation step at 72°C for 10 min. The sequences of bisulfite-sequencing PCR (BSP) primers (GeneCopoeia) were as follows: (forward) 5′-GGAATATTGTATTAAAGGGTAGGTAGG-3′ and (reverse) 5′-TCAATAATTCCTAACTCTAAATA ACC-3′. Next, the BSP products including four CpG sites were cloned into the pMD18-T vector (TaKaRa, Dalian, China) and then applied to transform competent Escherichia coli T10 cells (Thermo Fisher Scientific) using standard procedures. For all CRC tissues the plasmid DNA was isolated from eight insert-positive clones per each sample using a Qiagen Miniprep kit (Qiagen NV) and then sequenced using the Pyrosequencing PSQ96 HS System (Qiagen NV) depending on the manufacturer’s instructions. The amount of C relative to the total amount of C and T at each CpG sites was determined as the percentage (ie, 0–100). The average of the relative levels of C in the four CpG sites served as the overall HES1 methylation score for each sample. The experiments were performed in triplicate.
Statistical analysis
Statistical analysis was achieved using IBM SPSS Statistics software version 20 (IBM Corporation, Armonk, NY, USA). Quantitative variables, expressed as mean ± SD, were analyzed with Student’s t-test or one-way analysis of variance (ANOVA). Categorical variables were assessed based on Pearson chi-squared test. Survival analysis was performed using the Kaplan–Meier method as well as the log-rank test. Correlations were determined via Spearman rank correlation analysis. The statistical significance was prescribed at P<0.05.
Results
HES1 expression is significantly elevated in CRC tissues
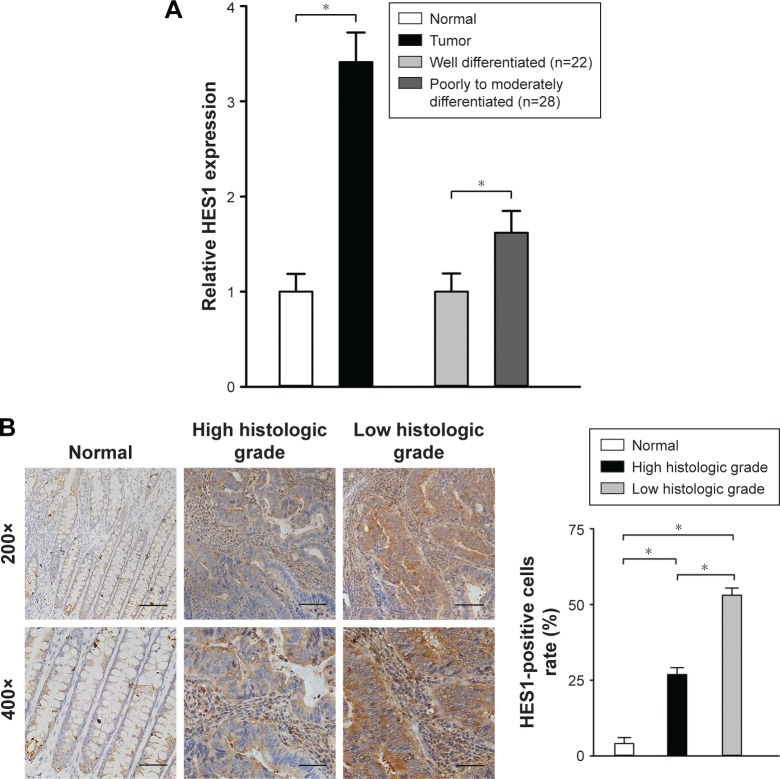
RT-PCR was applied to evaluate HES1 mRNA levels in 50 CRC tissues and paired adjacent normal samples. We observed that, in all 50 CRC tissues, the relative HES1 expression was significantly higher compared to that of adjacent normal samples (Figure 1A, P<0.05). These findings were in line with the results of a previous study.13 In addition, the intensity of HES1 expression was positively associated with the histological grade (Figure 1A, P<0.05). Similar results were obtained when HES1 expression was achieved at the protein level using immunohistochemical staining techniques (Figure 1B). Thus, the aforementioned results suggested that upregulation of HES1 may be involved in the development of CRC.
Figure 1.
HES1 expression in CRC tissues and adjacent normal samples.
Notes: (A) The expression of HES1 in 50 cases of CRC tissues and adjacent normal samples was determined using qRT-PCR. Data are presented as mean ± SD of three independent experiments. GAPDH was used as an internal control. *P<0.05, one-way ANOVA sections. (B) Immunohistochemical staining of HES1 in CRC tissue sections and adjacent normal samples. A quantitative graph for the percentage of HES1-positive cells is presented (five random fields, four sections per sample, *P<0.05, scale bars: 250 µm or 500 µm, two magnifications: 200× or 400×).
Abbreviations: ANOVA, analysis of variance; CRC, colorectal cancer; HES1, hairy/enhancer of split 1; qRT-PCR, quantitative real-time polymerase chain reaction.
In addition, we further correlated the HES1 mRNA expression profile with several clinicopathological characteristics. As shown in Table 1, upregulation of HES1 was significantly correlated with the histological grade, tumor invasion, Dukes’ classification, lymph node metastasis, distant metastasis, and clinical stages (P<0.05). However, no significant correlation was found with age, gender, tumor location, or tumor size (P>0.05). In conclusion, these findings suggest that upregulation of HES1 may be a potential biomarker for CRC malignancy.
Table 1.
Clinicopathological correlation of HES1 mRNA expression and methylation status in 50 primary CRC tissues
| Clinicopathological features | Patients (n) | mRNA expression
|
Methylation status
|
|||
|---|---|---|---|---|---|---|
| Mean ± SD | P-valuea | Hypomethylation (n=38) (%) | Hypermethylation (n=12) (%) | P-valueb | ||
| Age, years | 0.416 | |||||
| ≤60 | 18 | 1.6885±0.0562 | 0.958 | 12 (31.6) | 6 (50.0) | |
| >60 | 32 | 1.6813±0.0127 | 26 (68.4) | 6 (50.0) | ||
| Gender | 0.099 | |||||
| Male | 27 | 1.6535±0.0254 | 0.507 | 23 (60.5) | 4 (33.3) | |
| Female | 23 | 1.6647±0.0338 | 15 (39.5) | 8 (66.7) | ||
| Tumor location | 1.000 | |||||
| Colon | 34 | 1.5924±0.1008 | 0.105 | 26 (68.4) | 8 (66.7) | |
| Rectum | 16 | 1.6296±0.0334 | 12 (31.6) | 4 (33.3) | ||
| Tumor size (cm) | 0.189 | |||||
| ≥5 | 29 | 1.7014±0.0255 | 1.000 | 24 (63.2) | 5 (41.7) | |
| <5 | 21 | 1.6998±0.1824 | 14 (36.8) | 7 (58.3) | ||
| Histological grade | 0.013 | |||||
| Well | 22 | 1.8877±0.0205 | <0.001 | 13 (34.2) | 9 (75.0) | |
| Poor to moderate | 28 | 1.3330±0.0397 | 25 (65.8) | 3 (25.0) | ||
| Invasion | 0.239 | |||||
| T1 + T2 | 16 | 1.6443±0.0451 | 0.019 | 10 (26.3) | 6 (50.0) | |
| T3 + T4 | 34 | 1.5584±0.0189 | 28 (73.7) | 6 (50.0) | ||
| Dukes’ classification | 0.032 | |||||
| A + B | 24 | 1.6214±0.0651 | 0.047 | 15 (39.5) | 9 (75.0) | |
| C + D | 26 | 1.5537±0.1165 | 23 (60.5) | 3 (25.0) | ||
| Lymph node metastasis | <0.001 | |||||
| Negative | 19 | 1.8066±0.2161 | 0.004 | 9 (23.7) | 10 (83.3) | |
| Positive | 31 | 1.5795±0.0142 | 29 (76.3) | 2 (16.7) | ||
| Distant metastasis | 0.400 | |||||
| Negative | 36 | 1.7347±0.0384 | 0.009 | 29 (76.3) | 7 (58.3) | |
| Positive | 14 | 1.6239±0.0655 | 9 (23.7) | 5 (41.7) | ||
| UICC stages | 0.013 | |||||
| I + II | 26 | 1.6584±0.0421 | 0.033 | 16 (42.1) | 10 (83.3) | |
| III + IV | 24 | 1.5812±0.0057 | 22 (57.9) | 2 (16.7) | ||
Notes:
Student’s t-test;
chi-squared test.
Abbreviations: CRC, colorectal cancer; HES1, hairy/enhancer of split 1; UICC, Union for International Cancer Control.
CRC tissues show aberrant HES1 hypomethylation
To investigate whether an epigenetic mechanism contributed to HES1 upregulation in CRC, we examined the methylation status of CpG islands in the HES1 promoter region. Using a bioinformatics approach, we predicted four CpG islands in the HES1 promoter region (Figure 2A). Next, the methylation levels of the CpG islands were determined by MSP. Representative agarose gels of MSP products for each sample are presented in Figure 2B. The data indicated that HES1 was increasingly hypomethylated in cancer tissues compared to matched normal samples. Furthermore, sequences of CpG islands in the HES1 promoter regions after bisulfite modification were analyzed in CRC tissues and normal samples. For this, we adopted a previously described cutoff of ≥8% methylated alleles for hypermethylated colorectal tissues,23 whereas hypomethylation was defined as less than 8%. As shown in Table 2, HES1 hypomethylation was detected in 76.0% (38/50) of CRC tissues, while in non-tumor colorectal tissues hypomethylation was found in 18.0% (9/50) of tissues. Thus, these results suggested that frequent hypomethylation of HES1 may occur as a distinct alteration in CRC tissues.
Figure 2.
Promoter methylation status of HES1 in CRC tissues and paired normal samples.
Notes: (A) A schematic illustration of the CpG islands in the HES1 promoter region. (B) Representative data of MSP analysis in CRC patients. Experiments were performed in triplicate. T, tumor tissue; N, normal tissue; H2O, water control; M, methylated HES1; U, unmethylated HES1; the numbers at the very top of part B represent the case numbers.
Abbreviations: BSP, bisulfite-sequencing PCR; CpG, cytosine-phosphate-guanine; CRC, colorectal cancer; DM, DNA marker; GC, guanine and cytosine; HES1, hairy/enhancer of split 1; MF1, methylated forward primer 1; MR1, methylated reverse primer 1; MSP, methylation-specific PCR; O/E, observed/expected CpG ratio; PCR, polymerase chain reaction; UF1, unmethylated forward primer 1; UR1, unmethylated reverse primer 1.
Table 2.
Methylation status of HES1 in different types of CRC tissues
| Tissue type | Patients (n) | Methylation status
|
|||
|---|---|---|---|---|---|
| Hypomethylation (%) | Hypermethylation (%) | χ2 | P-value* | ||
| Tumor tissue | 50 | 38 (76.0) | 12 (24.0) | 33.762 | <0.001 |
| Normal tissue | 50 | 9 (18.0) | 41 (82.0) | ||
Notes:
Chi-squared test; χ2 means the test value.
Abbreviations: CRC, colorectal cancer; HES1, hairy/enhancer of split 1.
We further analyzed the association between methylation status and clinicopathological characters in the 50 CRC patients. As shown in Table 1, decreased HES1 methylation levels were significantly correlated with a poor histological grade, Dukes’ classification, lymph node metastasis, and clinical stages (P<0.05). However, no significant correlation was found with age, gender, tumor location, tumor size, tumor invasion, or distant metastasis (P>0.05). Overall, these results indicated that HES1 hypomethylation may be a promising predictive indicator for the progression of CRC.
Correlation between HES1 hypomethylation and overexpression in CRC tissues
The relation between HES1 methylation status and the expression of HES1 was evaluated in CRC patients. As shown in Table 3, tumors and adjacent control tissues demonstrated a significantly higher level of hypomethylated HES1 in comparison to hypermethylated HES1. Moreover, the HES1 expression level in hypomethylated tumor tissues was increased compared to that in hypomethylated control tissues. However, no significant differences were found between the two hypermethylated tissue types. Overall, a very strong negative correlation was found between decreased methylation status and increased HES1 mRNA expression.
Table 3.
Association of HES1 promoter hypomethylation with mRNA overexpression in CRC tissues
| Tissue type | Patients (n) | HES1 mRNA expression
|
P-value* | |||
|---|---|---|---|---|---|---|
| Hypomethylation
|
Hypermethylation
|
|||||
| Mean ± SD | P-value‡ | Mean ± SD | P-value‡ | |||
| Tumor tissue | 50 | 1.6882±0.1008 | 0.009 | 1.5534±0.0671 | 0.066 | 0.008 |
| Normal tissue | 50 | 1.5832±0.0624 | 1.5118±0.0338 | 0.031 | ||
Notes: Student’s t-test;
Hypomethylated tumor versus hypomethylated adjacent normal tissues or hypermethylated tumor versus hypermethylated adjacent normal tissues for HES1 mRNA expression.
Hypomethylated HES1 versus hypermethylated HES1 tissue type for HES1 mRNA expression in tumor and adjacent normal tissues.
Abbreviations: CRC, colorectal cancer; HES1, hairy/enhancer of split 1.
Hypomethylation of HES1 is linked to poor prognosis
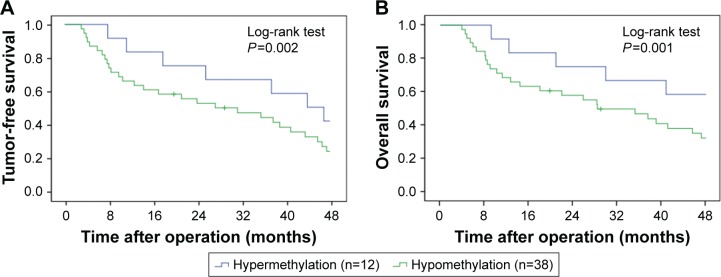
Kaplan–Meier survival analysis was performed to evaluate the survival of CRC patients, who were divided into HES1-hypermethylated and HES1-hypomethylated cases. The data revealed that CRC patients with hypermethylated HES1 promoters showed significantly prolonged median recurrence duration combined with prolonged median survival duration when compared to patients with hypomethylated HES1 promoters (42.0±8.2 months versus 26.2±6.7 months, P=0.002, Figure 3A; 49.1±5.9 months versus 28.4±5.1 months, P=0.001, Figure 3B, log-rank test). In conclusion, these results indicated that HES1 hypomethylation may function as a negative prognostic factor.
Figure 3.
Clinical outcomes by HES1 methylation status.
Notes: Kaplan–Meier survival analysis for tumor-free survival (A) and overall survival (B) was conducted based on the methylation status of HES1 promoter. Results revealed that CRC patients with HES1 promoter hypomethylation showed significantly prolonged median recurrence duration and prolonged median survival duration compared to patients with HES1 promoter hypermethylation (42.0±8.2 months versus 26.2±6.7 months, P=0.002, A; 49.1±5.9 months versus 28.4±5.1 months, P=0.001, B, log-rank test).
Abbreviations: CRC, colorectal cancer; HES1, hairy/enhancer of split 1.
Discussion
Previous studies have demonstrated that an aberrant methylation status (hypermethylation or hypomethylation) is one of the most consistent epigenetic alterations in human cancer.24–26 Indeed, altered DNA methylation patterns have been reported to be significantly correlated with tumor progression and prognosis.27–29 In the current study, we investigated the promoter methylation status of HES1 and correlation with clinicopathological parameters and prognosis in CRC.
HES1 is a novel bHLH transcriptional repressor, which is regulated by the Notch signaling pathway.3 Several reports have demonstrated that HES1 is overexpressed in a variety of human malignancies.7–9 In this study, we found that the expression of HES1 was markedly elevated in CRC tissues, which was consistent with the data presented in a previous study.13 Statistical analysis revealed that HES1 upregulation was significantly correlated with multiple clinicopathological characteristics. The pathological relevance of HES1 has also been described in other malignancies,30–32 suggesting its potential role in tumor development.
It has been proven that aberrant hypomethylation of tumor promoter genes frequently occurs in CRC, such as TCF3 and TIAM1.33,34 In this study, we showed that HES1 methylation was significantly decreased in CRC tissues. The underlying mechanism of HES1 hypomethylation may be mediated by DNA methyltransferase or thymine DNA glycosylase.20,35 Correlation analysis indicated that decreased HES1 methylation in CRC tissues was significantly associated with unfavorable clinicopathological characteristics. However, no correlation was found with invasion or distant metastasis, which was different from the findings regarding the pathological relevance of HES1 expression. This discrepancy may be due to the lower number of cases, or that HES1 expression was affected by factors other than methylation modification. Therefore, in our future studies, the sample size of the studies will be increased.
Kaplan–Meier survival analysis showed that HES1 hypomethylation was associated with poor prognosis in CRC patients. Similar findings were found for many other tumor promoter genes.36,37 A possible explanation for the poor prognosis related to hypomethylation may be the following: 1) decreased promoter methylation results in upregulation of HES1, which is essential for tumor differentiation, migration, invasion, and metastasis and leads to a worse CRC pathological stage; 2) HES1 overexpression contributes to tumor multidrug resistance conceivably by promoting Stat3 phosphorylation and activation.38,39
Although our results are promising, we have not investigated the mechanisms and roles of decreased HES1 methylation in the initiation and development of CRC in vitro and in vivo models. In addition, probable mutations and copy number variations of the HES1 gene in CRC remain to be explored. Thus, additional experiments would be executed in our future studies.
Conclusion
Promoter hypomethylation upregulates HES1 expression and plays a critical role in the progression and prognosis of CRC. Thus, the methylation status of the HES1 promoter may serve as an effective indicator of progression and prognosis of CRC and has great potential for clinical applications and CRC management.
Acknowledgments
This study was supported by the Zhejiang Provincial Natural Science Foundation of China (number LY15H160027).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, DeSantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Duineveld LA, van Asselt KM, Bemelman WA, et al. Symptomatic and asymptomatic colon cancer recurrence: a multicenter cohort study. Ann Fam Med. 2016;14(3):215–220. doi: 10.1370/afm.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134(7):1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 4.Dhanesh SB, Subashini C, James J. Hes1: the maestro in neurogenesis. Cell Mol Life Sci. 2016;73(21):4019–4042. doi: 10.1007/s00018-016-2277-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang Y, Coppo M, He T, et al. The transcriptional repressor Hes1 attenuates inflammation by regulating transcription elongation. Nat Immunol. 2016;17(8):930–937. doi: 10.1038/ni.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Obaldia ME, Bell JJ, Wang X, et al. T cell development requires constraint of the myeloid regulator C/EBP-[alpha] by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14(12):1277–1284. doi: 10.1038/ni.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakahara F, Kitaura J, Uchida T, et al. Hes1 promotes blast crisis in chronic myelogenous leukemia through MMP-9 upregulation in leukemic cells. Blood. 2014;123(25):3932–3942. doi: 10.1182/blood-2013-01-476747. [DOI] [PubMed] [Google Scholar]
- 8.Dailey DD, Anfinsen KP, Pfaff LE, et al. HES1, a target of Notch signaling, is elevated in canine osteosarcoma, but reduced in the most aggressive tumors. BMC Vet Res. 2013;9:130. doi: 10.1186/1746-6148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H-Y, Zhang H-Y, Wang X, Xu J, Ding Y. Expression and clinical significance of Notch signaling genes in colorectal cancer. Tumour Biol. 2012;33(3):817–824. doi: 10.1007/s13277-011-0301-3. [DOI] [PubMed] [Google Scholar]
- 10.Gao F, Huang W, Zhang Y, et al. Hes1 promotes cell proliferation and migration by activating Bmi-1 and PTEN/Akt/GSK3β pathway in human colon cancer. Oncotarget. 2015;6(36):38667–38680. doi: 10.18632/oncotarget.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng MT, Tsao PN, Lin HL, et al. Hes1 increases the invasion ability of colorectal cancer cells via the STAT3-MMP14 pathway. PLoS One. 2015;10(12):e0144322. doi: 10.1371/journal.pone.0144322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao F, Zhang Y, Wang S, et al. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Sci Rep. 2014;4:3963. doi: 10.1038/srep03963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan R, Ke J, Sun L, et al. HES1 promotes metastasis and predicts poor survival in patients with colorectal cancer. Clin Exp Metastasis. 2015;32(2):169–179. doi: 10.1007/s10585-015-9700-y. [DOI] [PubMed] [Google Scholar]
- 14.Jjingo D, Conley AB, Soojin VY, Lunyak VV, Jordan IK. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3(4):462–474. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulis M, Queirós AC, Beekman R, Martín-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829(11):1161–1174. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Kano H, Takayama T, Midorikawa Y, Nagase H. Promoter hypomethylation of RAR-related orphan receptor α1 is correlated with unfavorable clinicopathological features in patients with colorectal cancer. Biosci Trends. 2016;10(3):202–209. doi: 10.5582/bst.2016.01097. [DOI] [PubMed] [Google Scholar]
- 17.Sung HY, Ju W, Ahn J-H. DNA hypomethylation-mediated overexpression of carbonic anhydrase 9 induces an aggressive phenotype in ovarian cancer cells. Yonsei Med J. 2014;55(6):1656–1663. doi: 10.3349/ymj.2014.55.6.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villalba M, Diaz-Lagares A, Redrado M, et al. TMPRSS4 protein overexpression and its promoter hypomethylation predict poor prognosis in squamous lung cancer patients. Eur J Cancer. 2016;61(suppl 1):S14. doi: 10.18632/oncotarget.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueo T, Imayoshi I, Imayoshi T, et al. The role of Hes genes in intestinal development, homeostasis and tumor formation. Development. 2012;139(6):1071–1082. doi: 10.1242/dev.069070. [DOI] [PubMed] [Google Scholar]
- 20.Sheaffer KL, Kim R, Aoki R, et al. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014;28(6):652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller P, Crofts JD, Newman BS, et al. SOX9 mediates the retinoic acid-induced HES-1 gene expression in human breast cancer cells. Breast Cancer Res Treat. 2010;120(2):317–326. doi: 10.1007/s10549-009-0381-6. [DOI] [PubMed] [Google Scholar]
- 22.Pope JL, Ahmad R, Bhat AA, Washington MK, Singh AB, Dhawan P. Claudin-1 overexpression in intestinal epithelial cells enhances susceptibility to adenomatous polyposis coli-mediated colon tumorigenesis. Mol Cancer. 2014;13:167. doi: 10.1186/1476-4598-13-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwagami S, Baba Y, Watanabe M, et al. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg. 2013;257(3):449–455. doi: 10.1097/SLA.0b013e31826d8602. [DOI] [PubMed] [Google Scholar]
- 24.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 25.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 26.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70(10):27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 27.Walker BA, Wardell CP, Chiecchio L, et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. 2011;117(2):553–562. doi: 10.1182/blood-2010-04-279539. [DOI] [PubMed] [Google Scholar]
- 28.Flanagan J, Wilhelm-Benartzi C, Metcalf M, Kaye S, Brown R. Association of somatic DNA methylation variability with progression-free survival and toxicity in ovarian cancer patients. Ann Oncol. 2013;24(11):2813–2818. doi: 10.1093/annonc/mdt370. [DOI] [PubMed] [Google Scholar]
- 29.Hatano Y, Semi K, Hashimoto K, et al. Reducing DNA methylation suppresses colon carcinogenesis by inducing tumor cell differentiation. Carcinogenesis. 2015;36(7):719–729. doi: 10.1093/carcin/bgv060. [DOI] [PubMed] [Google Scholar]
- 30.Rui-xin LI, Ting-ting YA, Yun-yun LI, et al. Effects of Hes1 in metastasis and epithelial-mesenchymal transition in cervical cancer cells. J Int Obstet Gynecol. 2015;42(1):112–115. [Google Scholar]
- 31.Wang SC, Lin XL, Wang HY, et al. Hes1 triggers epithelial-mesenchymal transition (EMT)-like cellular marker alterations and promotes invasion and metastasis of nasopharyngeal carcinoma by activating the PTEN/AKT pathway. Oncotarget. 2015;6(34):36713–36730. doi: 10.18632/oncotarget.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiaschetti G, Abela L, Nonoguchi N, et al. Epigenetic silencing of miRNA-9 is associated with HES1 oncogenic activity and poor prognosis of medulloblastoma. Br J Cancer. 2014;110(3):636–647. doi: 10.1038/bjc.2013.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Cai S, Wang X, Jiang Z. Hypomethylation-associated up-regulation of TCF3 expression and recurrence in stage II and III colorectal cancer. PLoS One. 2014;9(11):e112005. doi: 10.1371/journal.pone.0112005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin H, Li T, Ding Y, et al. Methylation status of T-lymphoma invasion and metastasis 1 promoter and its overexpression in colorectal cancer. Hum Pathol. 2011;42(4):541–551. doi: 10.1016/j.humpath.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Cortellino S, Xu J, Sannai M, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146(1):67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba Y, Nosho K, Shima K, et al. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology. 2010;139(6):1855–1864. doi: 10.1053/j.gastro.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Zhang YN, Lin GL, et al. S100P, a potential novel prognostic marker in colorectal cancer. Oncol Rep. 2012;28(1):303–310. doi: 10.3892/or.2012.1794. [DOI] [PubMed] [Google Scholar]
- 38.Gu F, Ma Y, Zhang Z, et al. Expression of Stat3 and Notch1 is associated with cisplatin resistance in head and neck squamous cell carcinoma. Oncol Rep. 2010;23(3):671–676. doi: 10.3892/or_00000683. [DOI] [PubMed] [Google Scholar]
- 39.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK–STAT signalling. Nat Cell Biol. 2004;6(6):547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]