Abstract
Phosphoinositide 3-kinase (PI3K) activity is stimulated by diverse oncogenes and growth factor receptors, and elevated PI3K signaling is considered a hallmark of cancer. Many PI3K pathway-targeted therapies have been tested in oncology trials, resulting in regulatory approval of one isoform-selective inhibitor (idelalisib) for treatment of certain blood cancers, and a variety of other agents at different stages of development. In parallel to PI3K research by cancer biologists, investigations in other fields have uncovered exciting and often unpredicted roles for PI3K catalytic and regulatory subunits in normal cell function and in disease. Many of these functions impinge upon oncology by influencing the efficacy and toxicity of PI3K-targeted therapies. Here we provide a perspective on the roles of class I PI3Ks in the regulation of cellular metabolism and in immune system functions, two topics closely intertwined with cancer biology. We also discuss recent progress developing PI3K-targeted therapies for treatment of cancer and other diseases.
Introduction and Historical Context
Reversible phosphorylation of inositol lipids controls diverse functions in cells. The head group of phosphatidylinositol can be phosphorylated on three of the free hydroxyls to form seven different phosphoinositide species with distinct roles in vesicle trafficking and signal transduction. Studies from several laboratories in the 1980s established that activated growth factor receptors and oncoproteins associate with an enzyme that phosphorylates PtdIns (Sugimoto et al., 1984; Whitman et al., 1985). At that time, only two phosphoinositides were known to exist: phosphatidylinositol-4-phosphate (PtdIns-4-P) and phosphatidylinositol-4,5-bisphosphate (PtdIns-4,5-P2). In 1988 the enzymatic activity that associated with oncoproteins (specifically polyoma middle T antigen) was shown to phosphorylate the 3′-hydroxyl substituent of the inositol ring to produce phosphatidylinositol-3-phosphate (PtdIns-3-P) (Whitman et al., 1988) and a follow up paper (Auger et al., 1989) revealed that platelet-derived growth factor (PDGF) stimulates this enzyme to produce phosphatidylinositol-3,4-bisphosphate (PtdIns-3,4-P2) and phosphatidylinositol-3,4,5-trisphosphate (PtdIns-3,4,5-P3) in smooth muscle cells. These findings led to the proposal that the bioactive product of phosphoinositide 3-kinase (PI3K) activity is important for cellular responses to growth factors and for malignant transformation. This prediction has been confirmed by thirty years of research showing that elevated PI3K signaling can contribute to tumorigenesis and is a hallmark of human cancer. Driven by this discovery, medicinal chemistry efforts have yielded a large toolbox of PI3K pathway inhibitors with varied selectivity profiles, many of which are being tested in clinical trials for cancer (Table S1). Along the way, we have learned that PI3K transmits important signals that regulate a variety of physiological processes in virtually all tissue types studied to date. Consequently, it comes as no surprise that the development of PI3K inhibitors to treat cancer has been challenged by the emergence of dose-limiting, on-target adverse effects. Inhibitors specific to mutated forms of PI3K that are commonly found in a wide variety of cancers could circumvent the on-target toxicities and lead to far better efficacy/toxicity profiles. Furthermore, the increasingly refined view of how various PI3K enzymes function in different cell types continues to unveil new opportunities for therapeutic intervention in cancer and in other diseases.
The PI3K field provides a prime example of the importance of basic research to understanding a family of proteins with relevance to human disease. Indeed, studies of PI3K genetics in model organisms have provided some of the most fundamental insights into the function of PI3K enzymes and their lipid products. The first PI3K gene to be cloned was S. cerevisiae Vps34, which is required for vacuolar protein sorting in yeast and is the only PI3K gene in that organism (Herman and Emr, 1990). Similarly, the human ortholog, hVPS34 (encoded by PIK3C3), is required for vesicle trafficking and for autophagy (Backer, 2016). These findings highlight that the most evolutionarily conserved function of 3′-phosphoinositides is to direct traffic of cargo between cellular organelles. An elegant chemogenomic strategy in budding yeast identified the target of rapamycin (TOR) and established its role in nutrient sensing (Heitman et al., 1991) before the mammalian target of rapamycin (mTOR; also known as the mechanistic target of rapamycin) was discovered and shown to integrate signals from nutrients and PI3K. In another example, a genetic screen in C. elegans provided the first clue that PI3K controls metabolism and aging (Dorman et al., 1995; Morris et al., 1996), conclusions that were supported by later studies of the PI3K/mTOR pathway in mice (Foukas et al., 2013; Selman et al., 2009; Wu et al., 2013). Studies in D. melanogaster also revealed critical roles for this pathway in growth control of cells and organs and reinforced the connection of PI3K with FOXO transcription factors first identified in worms (Hay, 2011). The first direct demonstration that PI3K genes have transforming potential was provided by a study of chicken cells infected with an avian retrovirus encoding an activated PI3K catalytic subunit (Chang et al., 1997), although much earlier mutational studies of polyoma middle T antigen had shown that binding and activation of PI3K was critical for the transforming function of this oncoprotein (Whitman et al., 1985). Later cancer genomic analyses revealed that activating mutations in PI3K genes (most commonly the PIK3CA gene encoding p110α) occur frequently in human tumors (Samuels et al., 2004).
Generation of mice with deletion or mutation of PI3K genes has been instrumental in delineating the unique and redundant functions of PI3K isoforms in mammalian cells and tissues (Okkenhaug, 2013; Vanhaesebroeck et al., 2010). The complexity of PI3K signaling is well illustrated by studies of the immune system. Indeed, one of the most important themes arising from mouse genetic models has been that the signaling outputs from the various PI3K isoforms must be carefully balanced for proper immune cell development and to optimize responses to pathogens. In accordance with these preclinical observations, it is now appreciated that human immunodeficiencies can result from either loss- or gain-of-function mutations in certain PI3K-encoding genes (Lucas et al., 2016). Additionally, knowledge gained from mouse genetics has led to the concept that drug-mediated inhibition of PI3K isoforms expressed in immune cells (p110γ and p110δ) can reprogram the immune system to combat solid tumor cells more effectively (Okkenhaug et al., 2016).
The knowledge accumulated during the past three decades of lipid kinase research indicates that the PI3K family members participate in an extraordinarily broad range of cellular regulatory processes, including cell growth and proliferation, metabolism, migration, and secretion. Moreover, aberrations in PI3K signaling contribute to an equally broad spectrum of human diseases, such as cancer, immunological disorders, neurological disorders, diabetes, localized tissue overgrowth and cardiovascular disease. This review will highlight recent advances in our understanding of the molecular mechanisms that underpin the PI3K signaling network, with particular emphasis on the contributions of this network to cellular metabolism and immune regulation – two complex processes that offer both challenges and opportunities for the development of PI3K pathway targeted agents.
The PI3K signaling network
Class I PI3K enzyme structure and activation
Human cells express three classes of PI3K enzymes. This review focuses on the class I PI3Ks, their mechanisms of activation, and the signaling networks in which they participate. There are three class II PI3Ks (PI3K-C2α, β, γ) and a single class III PI3K (hVPS34). The reader is referred to other recent reviews concerning class II and III PI3K function (Backer, 2016; Falasca and Maffucci, 2012; Hawkins and Stephens, 2016; Okkenhaug, 2013).
Mammals express four class I catalytic isoforms (p110α, β, γ, and δ encoded by PIK3CA, PIK3CB, PIK3CG, and PIK3CD) that catalyze the phosphorylation of PtdIns-4,5-P2 to generate PtdIns-3,4,5-P3 (Figure 1). This phospholipid acts as a second messenger to recruit cytoplasmic proteins to specific plasma membrane or endomembrane locations. The p110α and p110β proteins are expressed ubiquitously, whereas expression of p110γ and p110δ is enriched in immune cells. Each catalytic isoform forms a dimer with a regulatory subunit that modulates the activity and subcellular localization of the complex (Figure 1). In normal cells, PtdIns-3,4,5-P3 is induced transiently by growth factor stimulation and is rapidly metabolized by lipid phosphatases, including the tumor suppressor PTEN, which terminates PI3K signaling via removal of the 3′-phosphate from PtdIns-3,4,5-P3. Cancer cells frequently contain elevated amounts of PtdIns-3,4,5-P3 due to increased activity of oncogenic signaling proteins residing upstream of PI3K, or to mutational activation of PI3K itself. Many cancers also exhibit loss of PTEN function, which elevates basal and stimulated PtdIns-3,4,5-P3 abundance by reducing the turnover rate of this second messenger. In a meta-analysis of cancer genome sequencing studies, PIK3CA and PTEN were found to be the second and third most highly mutated genes in human cancers (Lawrence et al., 2014).
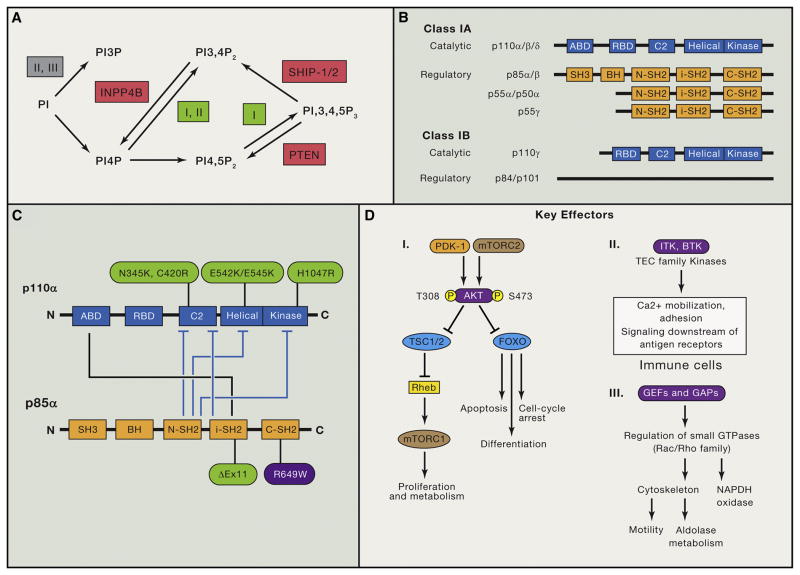
Figure 1. Overview of Phosphoinositides, Class I PI3K Protein Isoforms, p110α Activity Regulation, and PI3K Downstream Effectors.
(A) Schematic overview of the major synthesis and degradation pathways for PtdIns-3-P (PI3P), PtdIns-3,4-P2 and PtdIns-3,4,5-P3. The classes of PI3K (I, II, or III) that mediate reactions are indicated. Lipid phosphatases are in red. INPP4B, inositol polyphosphate-4-phosphatase, type II.
(B) Domain structure of class I PI3K catalytic and regulatory subunits. ABD, adaptor-binding domain; RBD, Ras-binding domain; BH, breakpoint cluster region homology.
(C) Diagram of the intramolecular interactions between class IA catalytic and regulatory subunits (p110α and p85α are displayed as well studied examples). Tight binding of the ABD to iSH2 confers stability to p110α. The other contacts shown in blue block arrows diminish basal activity and are relieved upon regulatory subunits binding to pTyr. Cancer-associated activating mutations are shown in green. SHORT syndrome mutation in p85α (R649W) is in purple.
(D) Brief summary of key PI3K effectors: PDK-1, AKT, TEC family kinases, and GEFs/GAPs for small GTPases. AKT has many other important substrates not shown here (Manning and Toker, 2017). The specific GEFs that mediate PI3K-dependent Rac activation to promote motility and aldolase release are not known.
Activation of Class I PI3Ks occurs through multiple upstream pathways that couple a broad range of cell surface receptors to specific PI3K isoforms. Generally, PI3Ks are capable of being activated by receptor-coupled tyrosine kinase activities, small Ras-related GTPases, and heterotrimeric G proteins. Each class I isoform has a domain that interacts with members of the Ras GTPase superfamily (Figure 1). For p110α, p110γ and p110δ, this domain binds to Ras or R-ras subfamily members whereas p110β interacts with the Rac/cdc42 subfamily. Three of the class I catalytic isoforms (For p110α, β and δ; collectively known as the class IA subgroup) associate with regulatory subunits whose SH2 domains bind to phosphotyrosyl residues on growth factor receptors or adaptor proteins such as IRS1. The other catalytic isoform (p110γ; known as class IB) associates with regulatory subunits (p101, p87) that mediate binding to βγ subunits of heterotrimeric G proteins following activation of G protein-coupled receptors (GPCRs). Adding to this complexity, the p110β isoform contains a Gβγ binding site that enables this isoform to be a coincidence detector for GPCR and tyrosine kinase signaling (Houslay et al., 2016). Through an unknown mechanism, p110δ in B lymphocytes is activated by chemokine receptors, which are members of the GPCR family. In murine macrophages, p110γ can be activated downstream of tyrosine kinases as well as GPCRs (Schmid et al., 2011). In summary, GPCRs and RTKs exhibit considerable plasticity in terms of coupling to the various Class I PI3Ks, determined in part by the cellular context.
Structural and biophysical studies have clarified the mechanisms of activation of different class I isoforms (Backer, 2010; Burke and Williams, 2015). As an example, we will discuss the p110α isoform and its activation by physiological signals as well as by cancer-associated PIK3CA mutations. p110α associates with one of five different regulatory subunits (p85α, p55α, p50α, encoded by PIK3R1; p85β, PIK3R2; p55γ, PIK3R3). Each of these subunits contains two SH2 domains (N-SH2, C-SH2) flanking a coiled-coil region known as the inter-SH2 (iSH2) domain (Figure 1). The catalytic and regulatory subunits make additional contacts that maintain the enzyme in a low activity state under basal conditions. The helical, kinase and C2 domains of the catalytic subunit contact the p85-N-SH2 domain; the C2 domain also contacts the p85-iSH2 domain. Binding of the regulatory subunit’s SH2 domains to phosphotyrosines relieves these inhibitory contacts and positions the dimer near the membrane where it can access substrate and receive further inputs from Ras and other signaling components.
Activating PI3K mutations in cancer, immune deficiency and tissue overgrowth
Many distinct PIK3CA activating mutations have been identified in human tumors. The two most common mutation “hotspots” are H1047R and E542K/E545K (Figure 1). The H1047R mutation enhances interaction of the kinase domain with membranes and bypasses the requirement for association with Ras (Burke and Williams, 2015). In contrast, E542K and E545K mutations disrupt the inhibitory interface with the N-SH2 domains of the regulatory subunits (Burke et al., 2012; Miled et al., 2007). Other less common PIK3CA mutations (e.g. N345K, C420R) disrupt the interface of the C2 domain with iSH2. Tumor-associated mutations in other class I PI3K genes are very rare. However, C-terminal truncation and deletion mutants that disrupt part of the iSH2 domains of regulatory subunits (encoded by PIK3R1 and PIK3R2) are oncogenic and frequently occur in brain and endometrial cancers (Figure 1). Transformation by these regulatory subunit variants requires activation of the p110α catalytic isoform (Sun et al., 2010).
Some cancer-associated PIK3CA mutations can also occur during development, and result in mosaic tissue overgrowth syndromes, venous malformations and brain malformations associated with severe epilepsy (Kurek et al., 2012). Analogous activating mutations in PIK3CD encoding p110δ (E1021K, E525K, N334K) have been identified in approximately 100 patients worldwide (Lucas et al., 2016), with distinct mutations affecting other domains discovered recently (Heurtier et al., 2017). Affected individuals suffer from a dominant immunodeficiency disorder termed Activated PI3K-Delta Syndrome (APDS). Disease-causing PIK3CD mutations elevate basal activity and membrane binding of p110δ, the dominant class I isoform in lymphocytes. Another subgroup of patients harbor PIK3R1 germline deletions and develop an immunodeficiency termed APDS2, based on its clinical similarity to the syndrome in patients with activating PIK3CD mutations. Interestingly, the most common PI3KR1 deletion in APDS2 selectively enhances basal activity of p85α/p110δ complexes, relative to p85α/p110α complexes (Dornan et al., 2017). This finding helps to explain why APDS2 patients do not have elevated risk of solid tumors associated with expression of mutationally-activated p110α.
PI3K effectors
The most proximal outcome of PtdIns-3,4,5-P3 production by class I PI3Ks is the recruitment of specific proteins to membrane signaling complexes. The shared property of these PI3K effectors is a pleckstrin homology (PH) domain selective for PtdIns-3,4,5-P3 and/or PtdIns-3,4-P2. Within the family of PI3K effectors are subsets with distinct enzymatic or signaling functions. These include serine/threonine kinases of the AGC kinase family, tyrosine kinases of the TEC (tyrosine kinase expressed in hepatocellular carcinoma) family, and modulators of small GTPase activities, termed guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (Figure 1). In this way, multiple, diverging downstream pathways can be simultaneously triggered by PI3K activation. A few canonical examples are discussed below.
Compared to other effectors, members of the AKT sub-family of AGC serine/threonine kinases (AKT1, AKT2, AKT3) seem to be activated more universally downstream of receptor-mediated PI3K activation. In fact, AKT phosphorylation often serves as a surrogate readout of class I PI3K activation. This tight coupling of PI3K to AKT is likely the result of two factors. First, phosphorylation of the AKT activation loop (Thr 308 on AKT1) occurs through a relatively straightforward mechanism, involving dual recruitment to the plasma membrane of AKT and its upstream activating kinase, phosphoinositide-dependent kinase-1 (PDK-1) activation (Manning and Toker, 2017). Membrane colocalization of the constitutively active PDK-1 with AKT facilitates PDK1-mediated phosphorylation of AKT. Second, the PH domains of both AKT and PDK-1 have affinity for both PtdIns-3,4,5-P3 and PtdIns-3,4-P2. The latter lipid can be produced from PtdIns-3,4,5-P3 by SHIP-1 and SHIP-2 (Figure 1), is often sustained after a transient peak of PtdIns-3,4,5-P3, and may promote AKT activation at endomembranes (Manning and Toker, 2017).
Although AKT phosphorylation on Thr 308 is both necessary and sufficient to mediate many downstream events, additional phosphorylation sites control substrate selectivity, stability and possibly subcellular localization (Manning and Toker, 2017). mTOR complex-2 (mTORC2) phosphorylates Ser 473 of the AKT hydrophobic motif (Sarbassov et al., 2005); this modification promotes maximal AKT activity and seems particularly important for a subset of substrates including Forkhead Box, subgroup O (FOXO) transcription factors (Jacinto et al., 2006). The mechanisms by which mTORC2 is activated to phosphorylate AKT have not been fully resolved (Ebner et al., 2017; Liu et al., 2015).
AKT phosphorylates many substrates involved in cell proliferation, metabolism, survival and motility (Manning and Toker, 2017). Mutations in the PH domain that promote membrane localization occur frequently in cancer (e.g. AKT1-E17K in 4–8% of breast cancer patients), supporting the idea that AKT is an important PI3K effector in oncogenic signaling. Notably, AKT plays an evolutionarily conserved role in growth factor signaling downstream of PI3K. In C. elegans, two AKT orthologs act downstream of an insulin receptor homolog (DAF2) and PI3K (AGE1) to suppress activity of DAF-16, a transcription factor homologous to human FOXO proteins. Likewise, the response to insulin in mammalian cells involves AKT-mediated inactivation of FOXO-dependent transcription.
TEC family tyrosine kinases are key PI3K effectors in lymphocytes. BTK, ITK and TEC all possess PH domains with exquisite selectivity for PtdIns-3,4,5-P3. A key function is to phosphorylate phospholipase C to promote hydrolysis of PtdIns-4,5-P2. Although a TEC homolog exists in flies (Tec29), the expansion of this kinase family in vertebrates and prominent expression in lymphoid cells is consistent with crucial roles in adaptive immunity. Indeed, humans lacking the BTK gene or with mutations affecting the BTK PH domain have a profound block in B cell development and fail to produce antibodies, a genetic immunodeficiency known as X-linked agammaglobulinemia. There is a strong link between PI3K and BTK function in B cells, first shown by knockout studies in mice where deletion of Pik3r1 or Pik3cd caused defects in B cell development and survival similar to those in mice lacking BTK (Deane and Fruman, 2004). More recently, pharmacological inhibitors of BTK (ibrutinib; Imbruvica® and ACP-196) and p110δ (idelalisib; Zydelig®) have shown a strong convergence of clinical activity in cancer, with best responses in malignancies of mature B cells (Fruman and Cantley, 2014).
TEC family kinases including BTK are expressed in various leukocyte subsets (mast cells, macrophages) where they function downstream of Fc receptors. Elucidation of these functions has led to an appreciation that BTK inhibitors have potential utility in solid tumors by disrupting the supportive roles of macrophages (Gunderson et al., 2016).
GEFs for Rho/Rac/cdc42 family GTPases are less widely appreciated, but nonetheless critical effectors of class I PI3K signaling. These small GTPases are regulated by many GEFs, of which only a subset bear PH domains with selectivity for PtdIns-3,4,5-P3. In neutrophils, P-Rex1 is a signal integrator activated by βγ subunits of heterotrimeric G proteins together with PtdIns-3,4,5-P3 produced by p110γ. P-Rex1 stimulates a GTPase cascade involving RhoG and Rac, leading to activation of the NADPH oxidase as well as neutrophil migration (Damoulakis et al., 2014; Welch et al., 2002) (Figure 1). In cancer cells and growth factor-stimulated fibroblasts, PI3K activation drives Rac-mediated actin re-organization. In addition to modulating cell morphology and motility, this PI3K/Rac signaling axis drives increases in glycolytic flux through the release of aldolase from actin filaments (Hu et al., 2016) (Figure 1).
The last PI3K effector we will discuss in this section is mTOR. This serine-threonine kinase forms two cellular complexes known as mTORC1 and mTORC2, with distinct subunit composition and substrate selectivity (Figure 2) (Saxton and Sabatini, 2017). Apart from AKT-Ser473, established substrates of mTORC2 include analogous sites in serum- and glucocorticoid regulated kinases (SGKs) and protein kinase C (PKC) isoforms. mTORC1 phosphorylates numerous substrates that promote anabolic metabolism to support cell growth and proliferation. mTORC1 activity can be increased by mitogenic signals through PI3K/AKT, RAS/ERK and other pathways (Dibble and Cantley, 2015), but also requires coordinate signals delivered through nutrient sensing pathways (Saxton and Sabatini, 2017). Thus, mTORC1 represents a key signaling node that coordinates anabolic metabolism and cell mass accumulation with growth factor receptor stimulation and nutrient availability.
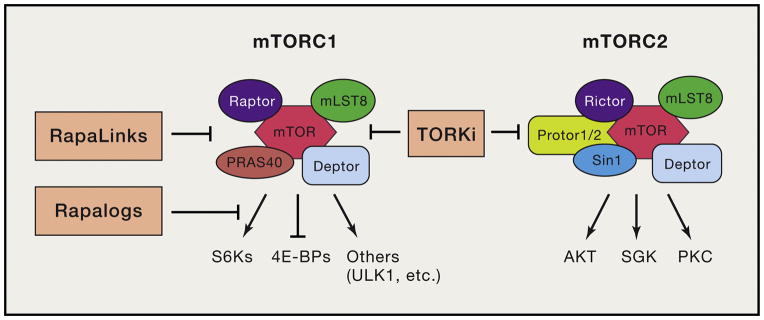
Figure 2. Overview of mTORC1 and mTORC2 complexes, key substrates, and inhibitors.
The processes inhibited by different classes of mTOR inhibitor are shown. First generation rapalogs are partial inhibitors of mTORC1 that inhibit phosphorylation of S6Ks more than 4E-BP1. Second generation TORKi fully inhibit mTORC1 and mTORC2. Third generation RapaLinks fully but selectively inhibit mTORC1, and also overcome single resistance mutations to rapalogs and TORKi.
Rapamycin is a bacterially-derived product that binds to intracellular FKBP12, thereby generating a complex that binds to mTORC1 at an allosteric site, termed the FKBP12-rapamycin binding (FRB) domain. Importantly, although rapamycin is exquisitely selective for mTORC1, this drug has differential effects on the phosphorylation of distinct mTORC1 substrates (Figure 2). ATP-competitive mTOR kinase inhibitors (TORKi) fully suppress kinase activity of both mTORC1 and mTORC2 without affecting integrity of the complexes (Figure 2). Comparisons of rapamycin and TORKi have provided valuable insights into the function of mTOR complexes and their substrates.
The direct mTORC1 substrate S6 kinase-1 (S6K1) contributes to metabolic reprogramming by increasing glycolysis and protein, lipid, and nucleotide biosynthesis (Figure 2) (Magnuson et al., 2012). mTORC1 also initiates powerful negative feedback regulation of growth factor receptor signaling, such that inhibition of mTORC1 or S6K1 leads to elevated activation of PI3K, AKT and the ERK pathway (Carracedo et al., 2008; Saxton and Sabatini, 2017). S6K1 is highly sensitive to inhibition by rapamycin, and the disruption of S6K1-mediated negative feedback might contribute to limited efficacy of rapamycin and its derivatives (termed rapalogs) in cancer.
The eukaryotic initiation factor-4E (eIF4E)-binding proteins (4E-BPs) are key mTORC1 substrates that control cell proliferation and survival. Phosphorylation of 4E-BPs by mTORC1 inhibits their binding to eIF4E, enabling assembly of the latter with eIF4G and eIF4A to form an active, cap-binding translation initiation complex known as eIF4F. Among cap-dependent mRNA transcripts, those that are more sensitive to decreased eIF4F activity are enriched in cell cycle and survival factors. Pharmacological and genetic studies have validated eIF4F as an oncogenic node and targetable vulnerability in cancer cells (Malka-Mahieu et al., 2017). Importantly, 4E-BP phosphorylation is inhibited to a greater extent by TORKi than by rapamycin (Figure 2), and the more penetrating inhibition of translation initiation by TORKi contributes to the more profound inhibition of cell growth and proliferation by these agents.
The PI3K pathway in cellular and organismal metabolism
PI3K signaling is evolutionarily conserved among multicellular organisms as a mechanism to respond to external growth cues. In mammals PI3K signaling is activated downstream of a myriad of growth factor receptors, including PDGF receptor (PDGFR) and epidermal growth factor receptor (EGFR), which drive proliferation and migration, insulin-like growth factor receptor (IGFR) which stimulates growth and survival, and insulin receptor (INSR) which regulates metabolic homeostasis. To coordinate responses to extracellular queues, the effectors of PI3K need to alter multiple facets of the cell, e.g. signaling that drives cell cycle progression also generates increased demand for metabolic programs to produce the energy and macromolecular synthesis to support cell growth and mitotic cell division. In order to meet these biosynthetic requirements, the PI3K/AKT/mTOR network must orchestrate a complex set of metabolic responses in the host cell.
Upon growth factor stimulation, receptor tyrosine kinases undergo conformational changes allowing them to autophosphorylate and become active. INSR autophosphorylation recruits the insulin receptor substrate (IRS) proteins, which the INSR phosphorylates on several sites to generate an optimal binding motif for the SH2 domains of p85 (Cantley and Songyang, 1994).
The immediate effect of insulin-driven PI3K signaling in muscle and fat cells is an increase in glucose uptake, attributable to enhanced glucose transporter translocation to the membrane (Huang and Czech, 2007) as well as increases in transcription and translation of the genes encoding these transporters (Lien et al., 2016). In muscle and adipose tissue, AKT2 is the primary isoform that phosphorylates and inhibits the function of the RabGAP, AS160, which allows intracellular vesicles containing the glucose transporter GLUT4 to migrate to the plasma membrane (Yuasa et al., 2009). This sequence of events supports enhanced glucose uptake into these tissues within minutes of serum insulin elevation. Most other tissues rely on increased insulin or IGF1-dependent transcription and translation of GLUT1 or other glucose transporters to increase glucose uptake, a much slower process. PI3K signaling has also been implicated in GLUT1 translocation to the cell membrane; however the mechanism for this translocation has not been fully elucidated (Bentley et al., 2003; Rathmell et al., 2003). Since GLUT1 is the major glucose transporter in many cancers, understanding how PI3K contributes to GLUT1 translocation may have clinical significance as oncologists attempt to modulate cancer cell metabolism as a therapeutic strategy. PI3K signaling controls transcription of GLUT1 through multiple mechanisms, including activation of mTORC1 that indirectly elevates the expression of HIF1α (Thomas et al., 2006; Wieman et al., 2007), and c-Myc (Osthus et al., 2000), transcription factors that drive the expression of genes involved in glucose metabolism, including SLC2A1 encoding GLUT1. AKT can also acutely stimulate glucose uptake by phosphorylating the adaptor protein TXNIP, reducing endocytosis of glucose transporters GLUT1 and GLUT4 (Waldhart et al., 2017).
AKT also enhances glucose metabolism by phosphorylating hexokinase 2 to facilitate its association with voltage-dependent anion channels at the mitochondrial membrane (Roberts et al., 2013) and indirectly by activating PFKFB2 which generates fructose 2,6-bisphosphate, an allosteric activator of PFK1 (Deprez et al., 1997). PI3K and AKT regulate other aspects of cellular metabolism through the activation of mTORC1. The activation of S6K1/2 and inhibition of 4EBP1 by mTORC1 (Figure 2) drives anabolic processes including protein and nucleotide synthesis as well as transcriptional activation of genes encoding enzymes of glycolysis and the pentose phosphate pathway (Dibble and Cantley, 2015). Both PI3K/AKT and mTORC1 promote lipid synthesis through activation of SREBP1 and SREBP2 transcription factors (Düvel et al., 2010; Porstmann et al., 2008).
A variety of negative feedback loops have evolved to maintain homeostasis of the PI3K/mTOR signaling pathway, to ensure that cells do not attempt to grow under conditions of energy stress or nutrient starvation and to protect multicellular organisms from localized tissue overgrowth. For example, when mTORC1 is highly active, it phosphorylates and stabilizes the adaptor protein GRB10, which binds and down-regulates the insulin receptor (Hsu et al., 2011; Yu et al., 2011). Variants of the GRB10 gene were implicated in type 2 diabetes (Prokopenko et al., 2014).
PI3K activity also initiates AKT-independent signaling cascades to impact cellular metabolism. In a study examining the mechanism by which PI3K inhibitors impact the progression of BRCA1/TP53 mutant tumors, Juvekar et al. demonstrated that synthesis of ribose and a set of glycolytic intermediates were impaired in response to PI3K inhibition, with lesser effects of AKT inhibitors (Juvekar et al., 2012). This result was dependent on the release of aldolase from F-Actin filaments mediated by PI3K-dependent Rac activation and consequent remodeling of the cytoskeleton. The aldolase-dependent increase in glyceraldehyde-3-phosphate provides a mechanism for increased ribose synthesis via the non-oxidative pentose phosphate pathway, allowing cells to generate the RNA and DNA needed for cell growth and proliferation (Hu et al., 2016). Thus, cytoskeletal remodeling driven by the PI3K-Rac axis drives not only cell motility but also metabolic reprogramming downstream of growth factor receptors. The role of PI3K signaling in effecting cytoskeletal remodeling is further highlighted by the recent identification of NT5C as a novel AKT substrate with a role in cytoskeletal remodeling that is mediated through interaction with ARP2/3 (Moniz et al., 2017).
p110α mediates most tissue responses to insulin and IGF1, driving tissue growth and maintaining glucose homeostasis throughout development (Engelman et al., 2006; Foukas et al., 2006). Early studies in mice demonstrated that the genes encoding class IA regulatory subunits (Pik3r1 and Pik3r2, encoding p85α and p85β) or the p110 catalytic subunits (Pik3ca, encoding p110α) play complex roles in insulin response in muscle and liver (Engelman et al., 2006). These studies reveal that insulin-dependent growth of heart and skeletal muscle is mediated by p110α (Engelman et al., 2006). Together the data show that the p85 regulatory subunit has both positive and negative regulatory functions in insulin signaling, as p85 is required for p110 stability and function, but suppresses insulin signaling in some tissues when in excess over p110 (Luo et al., 2005a). Suppression of insulin signaling by p85 subunits appears to occur by multiple mechanisms, including through the sequestration of IRS-1 and via activation of PTEN and JNK (Luo et al., 2005b, 2006; Taniguchi et al., 2006).
Keeping the complex PI3K-AKT-mTOR network homeostatically balanced is critical to prevent aberrant cellular proliferation and to maintain glucose homeostasis. This is highlighted by the deleterious impact of sporadic activating mutations in PIK3CA, as well as other genes in the PI3K-AKT-mTOR pathway, which arise during early embryonic development and give rise to mosaic tissue overgrowth syndromes including CLOVES (congenital lipomatous overgrowth, vascular malformations, and epidermal nevi) (Kurek et al., 2012), as discussed below. Reciprocally, inhibitory point mutations affecting the C-SH2 domain of p85α (primarily R649W; Figure 1) result in a dominant growth defect known as SHORT (short stature, hyperextensibility of joints, ocular depression, Rieger anomaly, and teething delay) Syndrome. In addition to developmental abnormalities and short stature, these patients have characteristics of Type 1 diabetes, without defects in insulin production or the insulin receptor, but as the result of impaired ability of insulin to activate PI3K (Chudasama et al., 2013). The underlying mechanism for this has not yet been established; however, the contrast of the severe phenotypes of these patients with the earlier mouse models of complete p85α deletion that demonstrated increased insulin sensitivity (Fruman et al., 2000a; Terauchi et al., 1999) highlights our incomplete understanding of the intricacies of this signaling network. The existence of patients with these phenotypes confirms the critical role of PI3K signaling downstream of IR/IGFR1 both for regulating metabolism as well as controlling cellular growth and proliferation.
Alterations leading to dysregulated insulin/PI3K signaling result in highly complex pathologies at the organismal level. In a normal setting, increases in blood glucose (typically from eating) will induce the pancreas to release insulin, thereby signaling to muscle and fat to take up more glucose, and to the liver to suppress glucose release until the system is brought back into homeostasis (Hopkins et al., 2016). If this pathway is perturbed, as is the case in patients with insulin resistance, the resulting, persistent hyperglycemia can lead serious, multi-organ pathology and even death. Obesity, which is usually caused by excessive food intake, frequently results in insulin resistance and is associated with an array of other metabolic changes correlated with increased cancer risk. However, it is difficult to draw direct connections between any one of the multiple concurrent changes that occur with obesity as many of them - increased inflammation, hyper-insulinemia, and changes in hormone signaling - have been shown to promote cancer. Although excessive PI3K signaling is a hallmark of cancer cells, too little PI3K signaling in the liver and muscle can lead to insulin resistance and type 2 diabetes. Understanding at the molecular level how PI3K signaling is maintained in tumors at the same time that PI3K signaling in muscle and liver is suppressed (insulin resistance) could facilitate the development of new therapies that target the dysregulation of PI3K/insulin signaling in tumors without disrupting normal tissues, such as the development of drugs that specifically target mutant isoforms of oncogenic proteins (e.g. p110α with mutant H1047R) thus sparing endogenous signaling molecules which are critical for the maintenance of normal homeostasis.
One key consideration in targeting PI3K or AKT for cancer treatment is how to manage the on-target toxicity to systemic metabolism. The PI3K gene that is most commonly mutated in human cancers, PIK3CA, is also the gene that encodes the isoform of PI3K (p110α) that mediates insulin responses in muscle, liver and fat. Because most p110α inhibitors that have entered clinical trials for solid tumors inhibit both the mutant and wild type p110α at therapeutic doses, these drugs induce acute insulin resistance, resulting in severe hyperglycemia, which, in turn, leads to severe hyperinsulinemia. In tumors that express IR, this systemic feedback may play a significant role to limit the therapeutic efficacy of these compounds (Figure 3), particularly in the setting of patients who are already insulin resistant (Gallagher et al., 2012). In the clinic, prolonged hyperglycemia caused by long term treatment with PI3K inhibitors is typically managed with biguanides (Bendell et al., 2012) that increase systemic insulin sensitivity and reduce basal blood glucose and insulin levels, though systemic insulin is typically still elevated, and is likely to compromise therapeutic responses to PI3K inhibitors. Moving forward, the clinical success of PI3K targeted therapies may be dependent on either identifying patient populations for whom the systemic metabolic impact of these compounds will not inhibit the therapeutic efficacy, perhaps patients whose tumors do not express IR/IGFR, or implementing new ways to limit the hyperinsulinemia through diet and drug combinations. Alternatively, drugs that have higher selectivity for mutant versus wild type PI3K could circumvent this systemic feedback. Both of these approaches would be expected to reduce the hyperglycemia and systemic metabolic disruptions that occur due to the on-target effects of compounds that inhibit the PI3K signaling cascade.
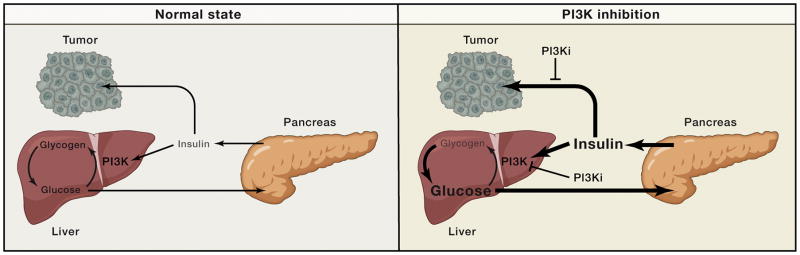
Figure 3. Cartoon of systemic glucose homeostasis in the normal state (left) and upon PI3K inhibitor treatment (right).
In the normal state blood glucose levels are maintained in homeostasis through the actions of insulin, which stimulates glucose uptake and glycogen storage thereby keeping the system balanced. Changes in blood glucose levels (such as increases upon eating) stimulate commensurate changes in insulin release which drive either increased glucose uptake (when insulin levels are high) or gluconeogenesis (when insulin levels are low). When PI3K inhibitors are used they perturb insulin signaling in cells, thereby pushing the systemic balance to favor glucose release. This causes blood glucose levels to acutely increase, which in turn signals to the pancreas to release a bolus of insulin. As indicated by the cartoon these high insulin levels have the potential to reactivate insulin signaling both in metabolic tissues, which is critical in order for the system to come back to homeostasis, as well as in tumors, where insulin has the potential to reactivate PI3K signaling thereby undercutting the efficacy of the PI3K inhibitors.
PI3K in innate and adaptive immunity
General concepts
Host defense in vertebrates is mediated by secreted proteins (including antibodies, complement, anti-microbial peptides) and by a diverse array of leukocytes with distinct functions. Each cell type of the immune system expresses receptors that elicit cellular responses in part through activation of class I PI3Ks. Notably, the study of PI3K signaling in leukocytes has revealed a number of important distinctions from other cellular systems. A central difference is that p110γ and p110δ are the dominant class I isoforms that produce PtdIns-3,4,5-P3 following receptor engagement (Okkenhaug, 2013). p110α and p110β are expressed in immune cells but their roles are restricted (Kulkarni et al., 2011; Ramadani et al., 2010). A second distinct feature is that, unlike growth factor receptor signaling, PI3K activation in leukocytes is not always an “on switch” that promotes a more powerful immune response. Depending on the receptor, the cell type, and the degree of PI3K activation, this pathway can activate or dampen responses, or skew cellular differentiation fates. A third key difference is the wiring of signaling cascades downstream of PtdIns-3,4,5-P3 production. For example, for some lymphocyte responses TEC family kinases seem to play a greater role than AKT in PI3K signaling output, and mTORC1 is more dependent on nutrient inputs than on PI3K/AKT activity. Each of these distinctions has important implications when considering the action of PI3K-mTOR pathway inhibitors on the immune system. Before elaborating further on the complexity of PI3K signaling in leukocytes, we will summarize some general concepts in host defense and tumor immunity.
Immune responses are generally categorized in terms of two broad sets of responses, denoted as innate immunity or adaptive immunity. Cells of the innate immune system provide a first line of defense that acts quickly via receptors with invariant ligand specificity, but lacks specificity or memory for pathogens. The adaptive immune system involves a diverse repertoire of lymphocyte clones (T and B cells) each with a unique antigen receptor; clonal expansion and differentiation provides delayed but powerful antigen-specific immunity that can last a lifetime. Adaptive immunity requires prior activation of innate immune cells, usually dendritic cells (DCs), by pathogen-associated molecular patterns that induce the DCs to migrate to lymph nodes where they present antigen and provide costimulation to T cells. In turn, lymphocytes differentiate and produce factors (cytokines and antibodies) that enhance pathogen destruction by innate immune cells and complement proteins. In addition to clonally diverse T and B cells, there exist multiple lymphocyte subsets with innate immune cell-like properties, including natural killer (NK) cells, γδ T cells, B-1 B cells, and innate lymphoid cells (ILCs). Conversely, mast cells are usually categorized as an innate immune cell yet these cells are activated to degranulate by diverse antigens, recognized by cell surface-bound IgE antibodies.
Although the primary role of vertebrate immune systems is to detect and destroy pathogens, both innate and adaptive arms also protect from cancer. Cytotoxic T cells can recognize and kill tumor cells presenting neo-antigen peptides on class I major histocompatibility complex (MHC) molecules. NK cells can kill tumor cells that downregulate class I MHC and/or upregulate stress ligands, or tumor cells coated with antibodies bound to cell surface tumor antigens. There is ample evidence from both mouse and humans that intrinsic or drug-induced immunosuppression increases cancer incidence. Furthermore, anti-cancer immune responses exert selective pressure on heterogeneous tumor cells, a process known as immunoediting, such that cells capable of evading innate and/or adaptive immune responses are selected during tumor evolution. Escape mechanisms include loss of tumor antigens, expression of checkpoint receptor ligands, secretion of suppressive cytokines, sequestration of nutrients, recruitment of various immunoregulatory cell types, and creation of physical barriers. In recent years multiple strategies have emerged to boost the immune system’s ability to detect and destroy tumor cells. Several immunotherapies have been approved for clinical use in a broad range of cancers, with many more in clinical trials. To broaden and deepen clinical responses, there is considerable interest in combining immunotherapies with small molecule targeted agents. Thus, defining the impact of PI3K-mTOR pathway inhibitors on immune cell subsets is essential to designing effective combinations for cancer treatment, and for expanding the application of these agents to immune disorders.
PI3K in innate immunity
Inflammation is an immediate response to pathogen detection by the innate immune system. Production of chemokines by macrophages in the infected tissue, together with complement fragments C3 and C5, increase local vascular permeability and attract neutrophils. Both resident macrophages and infiltrating neutrophils phagocytose bacteria via a variety of cell surface receptors. Neutrophils activate an intracellular NADPH oxidase that produces reactive oxygen species (ROS) to kill engulfed bacteria. Each of these processes requires class I PI3K activation (Hawkins and Stephens, 2015).
Most chemoattractants bind G protein-coupled receptors (GPCRs) that activate p110γ (Hawkins and Stephens, 2015). PtdIns-3,4,5-P3 recruits the guanine nucleotide exchange factor p-Rex-1 to activate a G protein cascade of Rho and Rac GTPases leading to cytoskeletal remodeling and ROS production (Damoulakis et al., 2014). Evidence that p110γ inactivation suppresses these responses led to initial excitement about developing p110γ inhibitors for inflammatory disease (Rückle et al., 2006). However, initial challenges for discovery of highly selective compounds and concerns about maintaining host defense have slowed progress for p110γ inhibitors in this therapeutic area. Instead, exciting evidence that p110γ inhibitors can reprogram the immune milieu in tumors (discussed below) has led to clinical trials in cancer of a selective p110γ inhibitor (IPI-549) combined with immunotherapy (De Henau et al., 2016). Given the key functions of p110γ in neutrophils, it will be important to carefully monitor frequencies of bacterial infections in cancer patients enrolled in p110γ inhibitor trials.
The p110δ and p110β catalytic isoforms also contribute to cellular responses promoting inflammation. After GPCR triggering of p110γ in human neutrophils, the subsequent activation of p110δ is needed to sustain NADPH oxidase activity (Condliffe et al., 2005). Both p110δ and p110β regulate the spreading of neutrophils and macrophages on extracellular matrix and the engulfment of IgG-opsonized particles. p110β plays a dominant role in ROS production by neutrophils recognizing immobilized immune complexes, and p110β-deficient mice were resistant to immune complex-mediated inflammation in vivo (Kulkarni et al., 2011). In microglia, p110δ is expressed and required for efficient release of TNFα in response to glucose deprivation and restoration, an in vitro correlate of ischemic stroke. In an in vivo mouse model of ischemia and reperfusion, selective inhibition of p110δ reduced cerebral damage and improved neurological outcome (Low et al., 2014). There is also evidence that p110δ inhibition is effective in mouse models of chronic obstructive pulmonary disease (Marwick et al., 2010), and clinical trials of a p110δ inhibitor (GSK2269557) are underway in this disease.
An evolutionarily conserved mechanism by which innate immune cell recognize pathogen-associated molecular patterns (PAMPs) is via Toll-like receptors (TLRs). TLR engagement activates NFκB and interferon-regulatory factors to induce transcriptional changes needed for immune responses. TLRs also augment PI3K-mTOR pathway activity, with either positive or negative regulatory consequences in different TLR signaling contexts. During innate antiviral responses triggered by TLR7 and TLR9 in plasmacytoid dendritic cells, the PI3K-mTOR pathway has a primarily positive role in type I interferon (IFNα and IFNβ) production (Costa-Mattioli and Sonenberg, 2008). Selective inhibitors of p110δ or mTORC1 suppress type I IFN production, and both S6Ks and 4E-BPs have been implicated in this process. Despite these findings, it is not yet apparent whether human patients treated with PI3K or mTOR inhibitors have specific impairments in virus-induced interferon production.
Innate receptors for bacterial cell wall components include TLR2 and TLR4; TLR5 recognizes the conserved flagellin protein complex. Engagement of these TLRs on myeloid cells (macrophages, monocytes and conventional DCs) promotes production of pro-inflammatory cytokines including IL-1, TNFα, and IL-12 that are balanced by production of anti-inflammatory products including IL-10. Interestingly, in many contexts PI3K-mTOR pathway activation by TLRs serves to attenuate the inflammatory response (Weichhart et al., 2015). Consequently, PI3K-mTOR pathway inhibitor treatment of mouse and human myeloid cells increases transcription of genes encoding inflammatory cytokines, decreases IL-10 production, and enhances their capacity to prime T cells. The PI3K-mTOR pathway antagonizes TLR signaling, in part by promoting STAT3 activity while suppressing the pro-inflammatory NFκB-mediated transcriptional program. In addition, p110δ has an isoform-specific role in LPS responses by promoting internalization of TLR4 and dissociation of the adaptor protein TIRAP (Aksoy et al., 2012). Interestingly, hyperactivation of mTORC1 signaling in macrophages lacking TSC2 suppresses NFκB function and drives metabolic reprogramming, loss of quiescence, and macrophage proliferation leading to formation of granulomas (Linke et al., 2017).
The pro-inflammatory effect of PI3K-mTOR pathway inhibitors has several important implications. First, it likely contributes to common long-term side effects of rapamycin or rapalog treatment, namely mucositis and pneumonitis. Mucositis is also one of the main dose-limiting toxicities of the TORKi compounds (Table S1) AZD2014 (Basu et al., 2015) and CC-223 (Bendell et al., 2015a), and occurred frequently in a phase I trial of TAK-228 (Ghobrial et al., 2016). Mucositis was reported in phase I studies of pan-PI3K inhibitor buparlisib (BKM120) (Bendell et al., 2012; Ragon et al., 2017) and the dual PI3K/mTOR inhibitor BEZ235 (Bendell et al., 2015b; Carlo et al., 2016). The p110δ inhibitor idelalisib is associated with autoimmune colitis as discussed below, but also with lung and liver inflammation (Coutré et al., 2015) that might arise from increased innate immune stimulation. Thus, a significant impediment to clinical application of PI3K-mTOR pathway inhibitors is the development of inflammatory conditions.
On the other hand, the pro-inflammatory potential of PI3K-mTOR pathway inhibitors is potentially advantageous for the immunotherapy of cancer. Many solid tumors have a resident population of tumor-associated macrophages (TAMs), and a pro-inflammatory gene expression profile (high IL-12, interferon-γ) in several tumor types correlates with extended patient survival (Kaneda et al., 2016a). The PI3K catalytic isoform p110γ is abundantly expressed in TAMs and promotes a more immune suppressive phenotype characterized by expression of IL-10 and TGFβ (Kaneda et al., 2016a). Gene expression signatures associated with high p110γ expression in TAMs are associated with reduced patient survival (Kaneda et al., 2016a). Notably, genetic or pharmacological inhibition of p110γ delayed tumor growth in several mouse models, stimulated anti-tumor T cell responses and enhanced the efficacy of immune checkpoint blockade (De Henau et al., 2016; Kaneda et al., 2016a). In a separate study of pancreatic cancer, p110γ inhibition opposed tumor progression by augmenting CD8 T cell responses and reducing tumor cell invasion and the protective fibrosis (known as desmoplasia) characteristic of this disease (Kaneda et al., 2016b). It should be stressed that the p110γ isoform is not expressed in most solid tumors, and adoptive transfer studies confirmed the TAM-intrinsic role of p110γ in tumor immunosuppression (Kaneda et al., 2016a, 2016b). Thus, the anti-tumor effect of p110γ inhibition occurs entirely via reprogramming the immune microenvironment. Another point of emphasis is that the anti-tumor effect of p110γ inhibition is relatively modest, but more dramatic when combined with immune checkpoint blockade.
Another scenario where the pro-inflammatory action of PI3K-mTOR pathway inhibitors has potential utility for cancer control is in the development of DC-based tumor vaccines. In murine DCs stimulated with flagellin in the presence of killed tumor cells, PI3K inhibitors suppressed production of IL-10 and TGFβ while preserving or enhancing IL-12 release (Marshall et al., 2012). In a tumor vaccine model, adoptive transfer of PI3K inhibitor-treated DCs enhanced anti-tumor efficacy and fostered the expansion of effector T cells secreting inflammatory cytokines IFNγ and IL-17 (Marshall et al., 2012). This type of approach holds promise for improving efficacy of DC-based vaccines while avoiding systemic administration of PI3K-mTOR pathway inhibitors. mTORC2 might play a key role downstream of PI3K in the programming of DC function, as injection of rictor-deficient DCs into B16 melanoma tumors stimulated T cell responses that slowed tumor growth (Raïch-Regué et al., 2016).
PI3K in adaptive immunity
The adaptive immune system is important for antigen-specific immune responses and for immunological memory to pathogens and vaccines. In addition, the balance of anti-tumor versus immunosuppressive lymphocyte subsets is a key factor in tumor progression and response to immunotherapies. Class I PI3K signaling is activated by antigen receptors expressed by T and B cells, and by other inputs including costimulatory molecules and cytokine receptors. Several comprehensive reviews have detailed how PI3K signaling is engaged by different receptors to regulate a variety of lymphocyte responses, and how genetic deficiency or hyperactivity of PI3K isoforms can both lead to immunodeficiency (Hawkins and Stephens, 2015; Lucas et al., 2016; Okkenhaug and Vanhaesebroeck, 2003). The roles of mTORC1 and mTORC2 in the metabolic programming and differentiation of effector lymphocytes is another topic that has been thoroughly reviewed (Chi, 2012; Jellusova and Rickert, 2016; Waickman and Powell, 2012). Here we will emphasize and discuss the distinct wiring of PI3K signaling networks in lymphocytes. Some of these differences help to explain the remarkable efficacy of p110δ-targeted inhibitors in B cell tumors, as well as the immunosuppressive mechanism of rapamycin.
The TEC family proteins BTK, ITK and TEC have key roles in PI3K signaling responses downstream of antigen receptors in T and B cells (Figure 4). These tyrosine kinases possess a PH domain for PtdIns-3,4,5-P3-dependent membrane recruitment, along with SH2 and SH3 domains for protein-protein interactions. In B cells, engagement of the antigen receptor (B cell receptor; BCR) and co-receptor (CD19/CD21/CD81) triggers formation of signaling microclusters, also known as signalosomes, that drive activation of phospholipase Cγ, leading to production of diacylglycerol and inositol trisphosphate. These second messengers ultimately trigger Ca2+ mobilization and the activation of NFκB as well as the Ras-Raf-Mek-Erk pathway. Within the BCR signalosome, BTK and the PI3K p85α/p110δ dimer are required for maximal signaling output as measured by Ca2+ flux, IκB degradation and Erk phosphorylation. Notably, BTK inhibition can reduce PI3K signaling output in B cells (Bojarczuk et al., 2016; Compagno et al., 2017; Saito et al., 2003), supporting the concept that PI3K and BTK cooperate in the signalosome rather than operating in a simple linear fashion. The functional link between BTK and p85α/p110δ is strongly supported by genetic evidence. Humans lacking BTK have X-linked agammaglobulinemia, an immunodeficiency syndrome associated with few mature B cells and a profound defect in antibody production. Similarly, rare autosomal recessive loss-of-function mutations in PIK3R1 (p85α) or PIK3CD (p110δ) result in agammaglobulinemia (Conley et al., 2012; Zhang et al., 2013). In mice, loss of p85α or p110δ causes similar defects in B cell development and activation as those observed in B cells lacking BTK and TEC (which is partially redundant with BTK in mouse B cells). Consistent with the “signalosome” model of activation, inactivation of other BCR signaling components causes similar B cell phenotypes in mice and germline mutations cause agammaglobulinemia in humans (Fruman et al., 2000b).
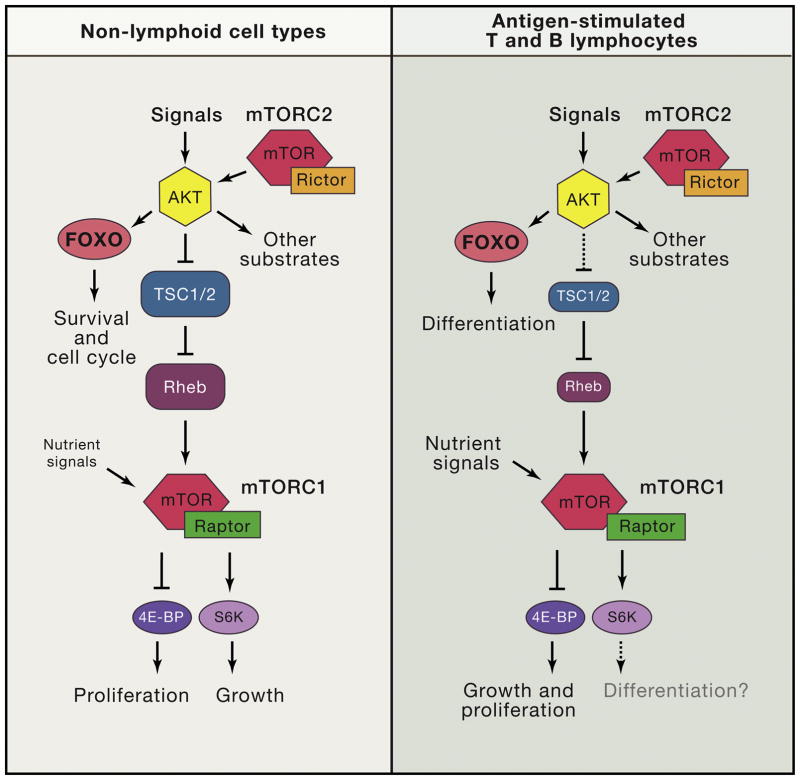
Figure 4. Distinct wiring of PI3K/mTOR network in lymphocytes (T and B cells) compared to other commonly studied non-lymphoid cell types such as fibroblasts.
In lymphocytes, FOXO transcription factors have prominent roles in differentiation. mTORC1 is tightly coupled to nutrient access and often uncoupled from PI3K/AKT activity. Downstream of mTORC1, the 4E-BP/eIF4E axis controls both growth and proliferation whereas in other cell types, S6Ks are crucial for cell growth. S6Ks might contribute to lymphocyte differentiation but this is unproven.
p110δ and BTK also function in shared signaling pathways in human B cell tumors. This is supported by convergent clinical responses to selective inhibitors of p110δ (idelalisib, duvelisib) and BTK (ibrutinib, acalabrutinib) in chronic lymphocytic leukemia (CLL), as well as by in vitro studies of B cell leukemia and lymphoma cells. In CLL and activated B cell-type diffuse large B cell lymphoma, chronic BCR signaling drives cell survival that is reduced by p110δ or BTK inhibitors. PI3K and BTK also function downstream of other functionally important receptors including CD40 and receptors for chemokines. Chemokine signaling via PI3K and BTK drives migration towards stromal cells secreting pro-survival factors such as BAFF, and increased adhesion to these supportive cells and to extracellular matrix. As a consequence, most CLL patients treated with p110δ or BTK inhibitors experience rapid lymph node shrinkage that is mainly due to impaired chemokine-dependent homing and reduced retention of leukemia cells in lymph node niches.
In T cells, antigen receptor (T cell receptor; TCR) signaling also involves formation of signalosomes containing p110δ and TEC family kinases ITK and TEC. In mice, loss of ITK or p110δ causes some similarities in T cell phenotypes including reduced TCR-mediated Ca2+ flux and adhesion. CD4 T cells lacking p110δ or ITK display impaired Th2 responses (Miller et al., 2004; Nashed et al., 2007; Okkenhaug et al., 2006; Soond et al., 2012) and are resistant to Th2-driven asthma. However, an inhibitor of ITK kinase activity failed to protect in an asthma model and actually enhanced Th2-mediated inflammation (Sun et al., 2015). In addition, it has not been firmly established that PI3K activation by the TCR or other receptors on T cells is required for ITK/TEC activation, or vice versa. Notably, p110δ and TEC kinases also serve important functions downstream of Fc receptors in innate leukocytes including mast cells and macrophages. These findings suggested possible applications of p110δ inhibitors to ameliorate symptoms of allergy (Ali et al., 2004) and BTK inhibitors to reprogram the myeloid compartment of pancreatic tumors (Gunderson et al., 2016).
Another prominent feature of class I PI3K signaling in B and T cells is the importance of FOXO regulation by AKT (Figure 4). Combined loss of Akt1 and Akt2 recapitulates some B cell development phenotypes associated with loss of p85α or p110δ, particularly reduced numbers of marginal zone (MZ) and B-1 cells (Calamito et al., 2010). Conversely, deletion of Foxo1 increased MZ B cell numbers and corrected the MZ B cell deficiency in mice lacking CD19 (Chen et al., 2010). It is also important for AKT activity to be attenuated at various decision points in B cell development to allow FOXO-dependent transcriptional programming. For example, Foxo1 is required for expression of the IL-7 receptor in pro-B cells and Rag genes at the pre-B cell stage (Amin and Schlissel, 2008; Dengler et al., 2008). On the other hand, some level of PI3K activation by the pre-BCR is required for extinction of Rag gene expression and further developmental progression (Ramadani et al., 2010). Intermediate levels of PI3K/AKT signaling output also seem to be required for survival of B-cell acute lymphoblastic leukemia cells, as deletion of PTEN caused hyperactivation of AKT leading to p53-mediated cell death, in a process analogous to the physiological deletion of autoreactive immature B cells (Shojaee et al., 2016).
When B cells are activated by antigen, they undergo clonal expansion and differentiate to secrete antigen-specific antibodies of various classes. Some B cells quickly differentiate into plasmablasts, which mainly produce low-affinity IgM, while others adopt a germinal center fate to undergo class switch recombination (CSR) and somatic hypermutation (SHM), resulting in secretion of higher affinity class-switched antibodies. This differentiation decision depends in part on the level of PI3K/AKT signaling versus the activity of FOXO transcription factors (Limon and Fruman, 2012), which control expression of the Aicda gene encoding activation-induced cytidine deaminase (AID) (Dengler et al., 2008). Elevated PI3K signaling through the loss of PTEN strongly suppresses class switching while increasing the plasmablast fate (Omori et al., 2006). Class switching can be restored in vitro by expression of constitutively active Foxo1 or AID (Omori et al., 2006). Conversely, PI3Kδ inhibition increases AID expression and CSR while reducing plasmablast differentiation (Omori et al., 2006). Similarly, inactivation of mTORC2 or AKT promotes class switching in a FOXO-dependent manner (Limon et al., 2014).
Within the germinal center B cell compartment, cyclical changes in the activity of PI3K/AKT versus FOXO are essential for proper trafficking and differentiation. The architecture of germinal centers (GCs) includes dark and light zones defined by histological staining. GCB cells undergo cycles of movement between the dark zone where they proliferate rapidly and undergo CSR and SHM, and the light zone where they are selected for antigen binding affinity. In mouse GCs, PI3K activity is restricted to the light zone while nuclear Foxo1 is largely absent. A fraction of cells in the light zone that do express Foxo1 are destined for dark zone re-entry as Foxo1 is needed to instruct the dark zone gene program including the chemokine receptor Cxcr4 (Dominguez-Sola et al., 2015; Sander et al., 2015). Foxo1 deletion or increased PI3K activity lead to loss of architectural polarity and lack of dark zones while impairing SHM and class switching.
Dynamic changes in the balance between AKT and FOXO function are also important for the fate of CD4+ and CD8+ T cells. In resting T cells, FOXO transcription factors maintain expression of homing receptors that allow recirculation between blood and lymphoid tissue. Engagement of the TCR and costimulatory molecules activates PI3K/AKT to cause FOXO nuclear exit, thereby reprogramming homing receptor expression to favor lymph node exit and trafficking to infected tissue (Fabre et al., 2008; Kerdiles et al., 2009). In the context of T helper differentiation, the T follicular helper (TFH) subset requires engagement of the costimulatory receptor, ICOS, which activates PI3K/AKT to suppress Foxo1-dependent gene expression (Rolf et al., 2010; Stone et al., 2015). Foxo1 also plays a role in the differentiation of murine and human regulatory T cells (Tregs) (Hsu et al., 2015; Kerdiles et al., 2010); notably, Foxo1 activity must be finely tuned to preserve Treg trafficking and function (Luo et al., 2016). The decision of CD8+ T cells to adopt effector or memory gene expression programs also depends on the balance of AKT and FOXO activity (Hess Michelini et al., 2013; Macintyre et al., 2011).
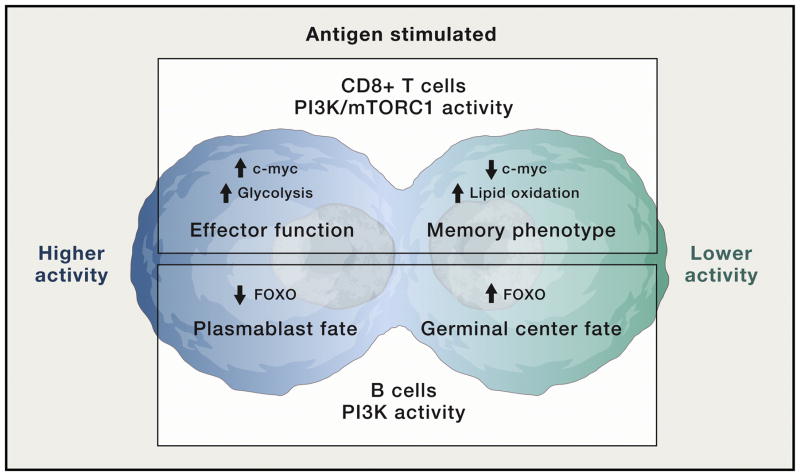
Accumulating evidence suggest that distinct differentiation fates are programmed at the first lymphocyte cell division via asymmetric partitioning of signaling proteins between daughter cells (Reiner and Adams, 2014). This was shown first in CD8+ T cells, where the first division results in one CD8-high daughter cell with high potential to proliferate and differentiate into cytotoxic effectors, and one CD8-low daughter cell destined to seed the memory CD8 T cell pool (Chang et al., 2007). A later study showed that the CD8-high daughter cells retain higher mTORC1 activity (Figure 5), which drives c-Myc expression to promote glycolytic metabolism required for the effector CD8 T cell fate (Verbist et al., 2016). Bifurcation of PI3K/mTORC1 activity also contributes to differentiation fates in B cells and CD4 T cells (Lin et al., 2015; Nish et al., 2017; Pollizzi et al., 2016) (Figure 5).
Figure 5. Asymmetric partitioning of PI3K/mTOR signaling during initial division of activated T and B cells results in distinct cell fates of daughter cells.
This figure illustrates that the first division of activated lymphocytes produces two cells with differential levels of PI3K/mTOR signaling, which in turn drive distinct metabolic and differentiation programs. Top: CD8 T cells. Bottom: B cells.
The wiring of mTORC1 signaling in lymphocytes has important differences from other frequently studied cell types. For example, activated lymphocytes frequently sustain mTORC1 signaling that is disconnected from PI3K/AKT activity (Figure 4). This was first observed in B-lymphoid tumor cells (Kharas et al., 2008; Wlodarski et al., 2005) and in mouse splenic B cell subsets (Donahue and Fruman, 2007), and later in CD8+ T cells (Salmond et al., 2009). mTORC1 activity in B cells is highly dependent on nutrients in vitro (Donahue and Fruman, 2007; Wlodarski et al., 2005) and suppressed under hypoxic conditions in germinal centers (Cho et al., 2016; Jellusova et al., 2017). Similarly, in activated CD8+ T cells, leucine uptake via the system L amino acid transporter Slc7a5 is required for mTORC1 activity, c-Myc translation and metabolic reprogramming (Sinclair et al., 2013) whereas PI3K/AKT signaling is dispensable for these outcomes (Macintyre et al., 2011). In cancer cells, ERK can phosphorylate TSC2 to promote mTORC1 activity but it is not clear whether this pathway is active in lymphocytes. However, ERK and RSK kinases provide a PI3K/AKT-independent input to S6 phosphorylation in TCR-activated CD8+ cells (Salmond et al., 2009). In established IL-2-dependent CD8+ effector T cells, mTORC1 activity is sustained by both JAK and SRC family tyrosine kinases, whereas PI3K/AKT activity is primarily dependent on Src family members (Ross et al., 2016).
Another indication of altered wiring of the mTOR network in lymphocytes is the role of mTORC1 effectors in cell growth and proliferation (Figure 4). In fibroblasts, mTORC1 promotes cell size increase mainly through S6Ks while driving cell cycle progression mainly through the 4E-BP/eIF4E axis (Dowling et al., 2010). In contrast, lymphocyte growth and proliferation are coupled through the 4E-BP/eIF4E axis while S6Ks are dispensable (So et al., 2016). The function of S6Ks in lymphocytes remains unclear but these kinases may regulate T helper cell differentiation (Kurebayashi et al., 2012; Pai et al., 2016; Sasaki et al., 2016). This convergence of signaling via 4E-BP/eIF4E might allow lymphocytes to more tightly couple cell mass accumulation to cell proliferation, to accommodate the extraordinarily rapid cell-doubling times of antigen-stimulated lymphocytes. Surprisingly, eIF4E function is more rapamycin-sensitive in lymphocytes than in fibroblasts and many cancer cell types. This difference correlates with predominant expression of 4E-BP2, whose phosphorylation is more rapamycin-sensitive than 4E-BP1 on key mTORC1 phospho-sites (So et al., 2016). This 4E-BP isoform switch likely contributes to the stronger anti-proliferative effect of rapamycin in lymphocytes compared to other cell types. The central role of eIF4E in lymphocyte activation also is important to consider when developing eIF4E-targeted agents for cancer therapy.
While mTORC1 activity is essential for lymphocyte proliferation and effector subset differentiation, hyperactivation of mTORC1 in T or B cells lacking TSC1 impairs development, homeostasis and function (Jellusova and Rickert, 2016; Pollizzi et al., 2015). These observations support a unifying theme that PI3K/AKT/mTOR activation in lymphocytes is not an on/off switch for adaptive immunity. Instead, the degree of signaling output determines the outcome of B and T cell differentiation. A related concept is that the overall effect of PI3K inhibition in vivo is determined by opposing actions in different lymphocyte subsets. An informative example is provided by studies of the p110δ catalytic isoform. Inactivation of p110δ in T cells impairs differentiation of effector CD4+ (Okkenhaug et al., 2006; Soond et al., 2010) and CD8+ (Macintyre et al., 2011) T cells, yet also impairs Treg function (Ali et al., 2014; Patton et al., 2006). The Treg defect is likely responsible for autoimmune colitis that develops in p110δ-deficient mice (Okkenhaug et al., 2002; Patton et al., 2006). Likewise, diarrhea and colitis is a frequent side effect of idelalisib in human patients (Coutré et al., 2015). On the other hand, impaired Treg function has a beneficial outcome in the context of tumor immunity, where genetic or chemical inhibition of p110δ promoted tumor regression in several mouse models (Ahmad et al., 2017; Ali et al., 2014). The latter observation has raised interest in testing p110δ inhibitors to enhance immunotherapy response in solid tumors.
The phenotype of APDS patients also helps illustrate the opposing roles of p110δ in adaptive immune cells. Mutations in these patients elevate p110δ activity and promote lymphoproliferation. However, these persistently activated lymphocytes are prone to activation-induced cell death or senescence, and most patients have low IgG titers and poor vaccine responses consistent with impaired CSR and SHM. Thus, the overall outcome of hyperactivation of p110δ in lymphoid cells is a life-threatening immunodeficiency. Some APDS patients have been treated with rapalogs; there is hope that patients will benefit more from treatment with selective p110δ inhibitors, and clinical trials have been initiated (Table 1). Dosage and scheduling will need to be adjusted to minimize the inflammatory side effects of idelalisib described above. Additional concerns arose following regulatory approval of idelalisib, when some patients developed fatal infections. A phase 3 study of idelalisib with bendamustine and rituximab also reported an increased risk of infection including some deaths (Zelenetz et al., 2017). It is reasonable to propose that the infection risk is due to impaired CD4+ and CD8+ effector T cell differentiation.
Table 1.
Non-malignant diseases associated with hyperactive PI3K/mTOR signaling
| Disease | Genetic defect | Targeted treatment (approved or tested) |
|---|---|---|
| Cowden Syndrome | PTEN haploinsufficiency | Rapalogs tested |
| CLOVES and other tissue overgrowth syndromes | Somatic PIK3CA mutation | Rapalogs tested; p110α inhibitors planned |
| APDS | Germline PIK3CD mutation | Rapalogs tested; p110δ inhibitors in trials |
| Tuberous sclerosis | TSC haploinsufficiency | Rapalogs approved |
| Lymphangioleiomyomatosis | Somatic TSC mutations | Rapalogs approved |
| Parkinson’s Disease | PARK2 mutations | Rapalogs * |
In patients with early-onset Parkinson’s linked to PARK2 or alpha synuclein mutations, mTORC1 inhibition might be beneficial if treatment were started early in disease. Two potentially beneficial actions in this setting are: (1) tempering inappropriate protein synthesis and growth signaling, and (2) increasing autophagy and the clearance of neurotoxic protein aggregates.
Therapeutic targeting of the PI3K pathway in cancer
The recognition that PI3K signaling was aberrantly activated in the majority of human cancers, together with the presence of actionable target proteins in the PI3K/mTOR network, spurred expectations that PI3K/mTOR pathway inhibitors would spawn a major paradigm shift in cancer therapy. It is now widely appreciated that the actual clinical results have fallen considerably short of this extremely high expectation. Three major factors have contributed to the underwhelming performance of the PI3K/mTOR pathway inhibitors. First, this pathway is activated via a myriad of cell surface receptors, and cancer cells have shown remarkable plasticity when it comes to amplifying upstream mechanisms to maintain signal flow through the PI3K/mTOR pathway and other compensatory pathways in the presence of pharmacological inhibitors. For example, exposure to the inhibitors themselves causes the disruption of negative feedback mechanisms that limit the activity of the pathway to a range compatible with normal cell physiology. Drug-induced interference with negative feedback regulation can reduce PI3K/mTOR pathway inhibitor therapeutic activities in the absence of genetic mutations, a phenomenon termed adaptive resistance. Second, intrinsic or acquired resistance to PI3K/mTOR pathway inhibitors is commonly associated with mutations or copy number alterations of regulatory genes within the pathway or parallel oncogenic pathways, or activation of growth factor receptors that stimulate both PI3K and MAPK signaling. Third, systemic administration of PI3K/mTOR pathway inhibitors is associated with dose-limiting toxicities that prevent sufficient target engagement in tumor tissues to maintain pathway suppression. To some extent, these challenges to effective therapy with PI3K/mTOR pathway inhibitors were anticipated based on the numerous roles that the PI3K/mTOR pathway plays in tissue growth, metabolism, and physiological functions.
Mechanisms of resistance to PI3K/mTOR pathway inhibitors
Adaptive resistance
Pharmacological inhibition of the PI3K pathway in cancer cell lines in culture is frequently followed, within hours to days, by the induction of adaptive (non-genetic) resistance mechanisms (Thorpe et al., 2015) (Figure 6). A well-established adaptive response that restores PI3K/mTOR pathway signaling in the presence of mTORC1-selective inhibitors involves the disruption of a S6K1-mediated feedback loop that destabilizes IRS-1. In the absence of mTORC2 inhibition, loss of this negative feedback leads to AKT hyperactivation (O’Reilly et al., 2006). Identification of this feedback loop and another involving adaptor protein Grb10 (Hsu et al., 2011; Yu et al., 2011) prompted support for the development of TORKi that target both mTORC1 and mTORC2. Cancer cells in culture also respond to PI3K/mTOR pathway inhibitors by “rebound” signaling driven in part by FOXO activity. One frequent mechanism involves increased transcription of genes encoding RTKs, most notably HER3, EGFR, and INSR/IGFR1 (Chakrabarty et al., 2012; Chandarlapaty et al., 2011; Muranen et al., 2012). These responses can, in principle, be addressed by combining PI3K pathway inhibitors with RTK inhibitors or blocking antibodies (García-García et al., 2012; Garrett et al., 2013); however, it will be challenging to predict and address the specific RTKs conferring PI3K inhibitor resistance in individual patients. RTK upregulation may be an especially challenging problem when using isoform-selective PI3K inhibitors, because one or more of the remaining class I PI3Ks may assume the signaling functions of the drug-inhibited PI3K isoform, thereby augmenting the resistance conferred by RTK upregulation. Another mechanism of FOXO-mediated rebound signaling is via upregulation of Rictor, leading to increased AKT phosphorylation in renal cancer cells (Lin et al., 2014).
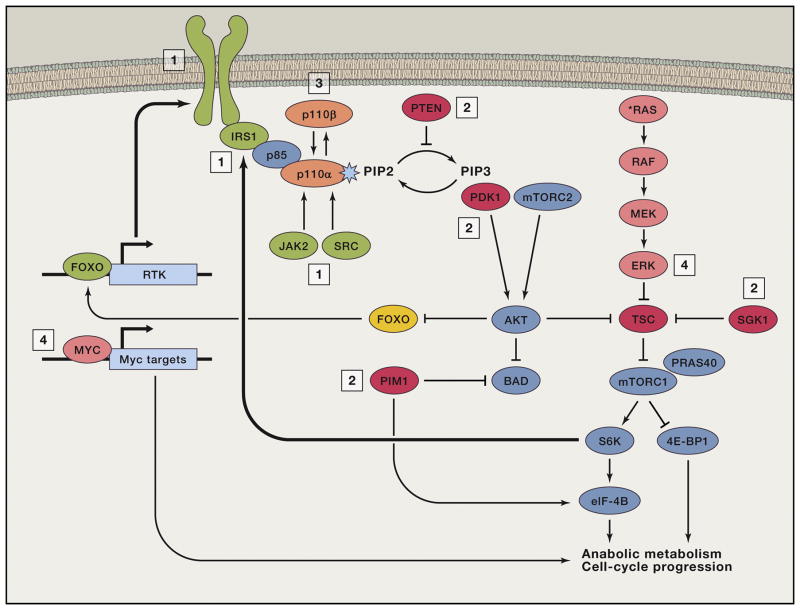
Figure 6. Mechanisms of Adaptive, Primary, and Acquired Resistance to PI3K/mTOR Pathway Inhibitors.
Adaptive resistance (in green, also labeled “1”) often involves upregulation of upstream regulators, including RTKs (HER3, INSR, IGF-1R), IRS-1, JAK2, or SRC by disruption of negative-feedback loops. Primary or acquired resistance (in red, “2”) can arise by expression or activation of kinases with downstream targets in common with AKT or mTORC1 (SGK1, PDK1, PIM1), constitutive activation of mTORC1 signaling (e.g., due to loss of TSC function), or loss of PTEN expression. Primary or acquired resistance (in orange, “3”) can also arise by PI3K isoform switching; selective inhibition of p110α can lead to substitution by p110β or vice versa. Finally, primary or acquired resistance can also arise by activation of heterologous pathways leading to common endpoints; for instance, MYC-dependent transcriptional activation or ERK activity (in pink, “4”).
A distinct mechanism of adaptive resistance was reported in triple negative breast cancer cells, which rapidly activated JAK2/STAT5 signaling during exposure to PI3K/mTOR pathway inhibitors (Figure 6). This adaptive response was addressed by co-treatment with a JAK inhibitor (Britschgi et al., 2012). A different nonreceptor tyrosine kinase, Src, which normally functions downstream of the EGFR and other RTKs, mediated adaptive resistance to a TORKi in a preclinical model of glioblastoma multiforme (GBM) (Wei et al., 2016). Src-mediated adaptive resistance is potentially targetable with the pan-Src family kinase inhibitor, dasatinib.
Bypass activation of mTOR and downstream targets
The mechanisms underlying primary and acquired resistance (Figure 6) are at least as diverse as those described above for adaptive resistance. Persistent mTORC1 activity in PIK3CA mutant breast cancer cell lines was responsible for resistance to the p110α-selective inhibitor alpelisib (BYL719; Table S1), and was reversed by combination with the mTORC1 inhibitor everolimus (Elkabets et al., 2013). An inverse correlation was also observed between the efficacy of alpelisib and phospho-S6 levels (a downstream target of mTORC1-S6K) after 28 days of treatment, indicating that inhibition of mTORC1 activity is pivotal for the antitumor activities of p110α inhibitors. Interestingly, persistent mTORC1 activity also predicts resistance to RAF or MEK inhibitors in BRAF-mutant melanoma models (Corcoran et al., 2013), indicating that drug-refractory mTORC1 activation confers broad-based resistance to both PI3K/mTOR pathway- and Ras pathway-targeted agents (Ilagan and Manning, 2016).
Although incomplete PI3K/mTOR pathway inhibition undoubtedly contributes to persistent mTORC1 activation, we now recognize that mTORC1 is also stimulated through PI3K-independent signaling mechanisms. Using a library screening approach, the protein serine-threonine kinase, Pim1, was shown to confer resistance to several PI3K/mTOR pathway inhibitors in breast cancer cells derived from the luminal A/B and HER2+ lineages (Le et al., 2016). Pim1 overexpression supported the activities of both AKT and mTORC1 in the setting of PI3K pathway inhibition. Elevated Pim1 expression correlated with alpelisib primary resistance in a panel of breast cancer cell lines, and a small molecule Pim1 kinase inhibitor conferred increased alpelisib sensitivity to three of the four breast cancer designated as high Pim1 expressors. Importantly, two of four breast tumor biopsies taken at the time of progression on alpelisib displayed upregulation of Pim1 expression relative to the pretreatment samples. Pim kinases may be significant contributors to resistance to PI3K/mTOR pathway inhibitors in breast cancer, as increased expression of mRNAs encoding Pim1, Pim2, or Pim3 were observed in more than ten percent of the nearly 1000 tumor samples analyzed in this study.
Aberrant activation of PDK1 or SGK1 has also been identified as mediators of resistance to p110α inhibitors in PIK3CA mutant breast cancer cells (Castel et al., 2016). Treatment of a PIK3CA-mutant, alpelisib-resistant breast cancer xenograft model with a combination of alpelisib and either a PDK1 or a SGK1 inhibitor caused significant tumor regression or stasis, respectively. Like AKT, SGK1 is activated by sequential phosphorylation by mTORC2 and PDK1, and shares with AKT the ability to phosphorylate TSC2, leading to mTORC1 activation. In the alpelisib-resistant cells, maximal mTORC1 inhibition was only achievable when alpelisib was combined with a PDK1 or SGK1 inhibitor. Analysis of 18 biopsies from patients treated with alpelisib showed that samples with high SGK1 mRNA and phospho-N-Myc downstream regulated 1 (NDRG1; a substrate for and biomarker of SGK1 activation) levels correlated with intrinsic resistance to alpelisib, whereas most phospho-NDRG1-negative tumors were from patients who attained partial responses or stable disease. Similarly, SGK3 overexpression and/or activation has been repeatedly linked to PI3K/mTOR inhibitor resistance in breast cancer cells (Bago et al., 2016; Gasser et al., 2014). These reports offer a common mechanism for intrinsic or acquired resistance to PI3K or AKT inhibitors.
Activation of parallel pathways
Amplification of MYC drives resistance to PI3K/mTOR pathway inhibitors in breast cancer cell lines (Figure 6), and MYC copy number and/or c-Myc expression is often elevated in breast cancer (Ilic et al., 2011; Liu et al., 2012) and lymphoid malignancies (Dang, 2012). Although direct targeting of c-Myc remains an elusive clinical goal, preclinical studies suggest that inhibition of the bromodomain and extra-terminal (BET) family protein, BRD4, antagonizes c-Myc-dependent PI3K pathway inhibitor resistance. BRD4 functions as an epigenetic reader that recognizes histones bearing acetylated lysine residues, and plays a key role in the regulation of transcriptionally-active chromatin. Inhibitors of BRD4 suppress the expression of super-enhancer-associated genes such as MYC (Delmore et al., 2011; Mertz et al., 2011; Zuber et al., 2011), and small molecule BET inhibitors are known to interfere with c-MYC-dependent transcriptional responses (Venkataraman et al., 2014). In ER+ breast cancer cells, BET inhibitors also overcame resistance to everolimus caused by c-Myc overexpression (Bihani et al., 2015), and exerted synergistic antitumor activity with PI3K inhibitors in mice with mammary tumors initiated by oncogenic PI3K-H1047R plus MYC transgene expression (Stratikopoulos et al., 2015). Interestingly, in breast cancer cell lines derived from either mutant PIK3CA-Myc double transgenic or Pten-null mice, the BET inhibitor, JQ1, attenuated PI3K pathway reactivation after treatment with the p110α/δ-selective inhibitor, pictilisib (GDC-0941; Table S1), by blocking upregulation of IGFR1 and insulin receptors. Moreover, rebound PI3K pathway activation by RTK expression was blocked by JQ1 in PIK3CA or PTEN mutant cell lines derived from breast, colorectal, prostate, brain and ovarian tumors (Stratikopoulos et al., 2015). In luminal breast cancer cell lines resistance to JQ1 was closely correlated with PIK3CA mutation, and JQ1 enhanced the anti-tumor efficacy of everolimus in a xenograft model, whereas PTEN loss was associated with increased sensitivity to BET inhibitors in basal breast cancer cells, suggesting that interplay between PI3K and BET signaling will be tumor cell context-dependent (Marcotte et al., 2016). Synergistic induction of cell death has also been observed in patient-derived ovarian cells treated with pictilisib and BET inhibitors (Kurimchak et al., 2016). The adverse event profile of the BET – PI3K inhibitor combination remains to be determined, but the evidence that BET inhibitors address multiple modalities of PI3K resistance supports further progression of this combination into clinical testing.
Tumor heterogeneity
Intratumoral heterogeneity is increasingly appreciated as an important contributor to tumor evolution and relapse following either chemotherapy or targeted therapy (McGranahan and Swanton, 2017). Increasingly sophisticated gene sequencing methodologies now support a branching model of tumor subclonal evolution, indicating that solid tumor tissues comprise an intertwined ecosystem of sub-dominant and dominant clones that communicate with one another and normal host elements in the tumor microenvironment. Clonal dominance is determined in part by extrinsic selection pressures imposed on the tumor, by alterations in tumor location (in metastatic disease), host metabolism and immune responsiveness, and cancer therapy. It is now appreciated that the PI3K pathway can be oncogenically activated in a variable fashion at the subclonal level in primary lesions and metastatic lesions from the same patient. Parallel genomic analyses of 86 diverse, primary tumors and brain metastases from individual patients revealed that more than half of brain metastases contained actionable mutations not seen in the corresponding primary tumor (Brastianos et al., 2015). Mutations in PTEN and PIK3CA were featured prominently as heterogeneous drivers of PI3K/mTOR pathway activation in these brain lesions. These same two genes appear to be particularly prone to heterogeneous mutation patterns in primary tumor tissues. Subclonal mutations in PIK3CA were reported in several cancer subtypes, including NSCLC, colorectal cancer, breast cancer, and others (Harbst et al., 2016; de Bruin et al., 2014; McGranahan and Swanton, 2017; Uchi et al., 2016; Yates et al., 2015). Consistent with these findings, a study spanning nine cancer subtypes demonstrated that subclonal mutations in the PI3K-AKT-mTOR pathway were considerably more frequent than subclonal mutations in the RAS-MAPK pathway, which tended to be ubiquitously expressed in all subclones in the tumor (McGranahan et al., 2015). Convergent evolution of distinct subclonal mutations leading to loss of PTEN function have been documented in recent publications. Following acquired resistance to the p110α-selective inhibitor alpelisib in an ER+ breast cancer patient, Juric and colleagues uncovered six distinct mutational events in PTEN across ten metastatic lesions, all on a common background of a clonal, mono-allelic PTEN deletion (Juric et al., 2015a). The PTEN deletion event occurred prior to PI3K inhibitor therapy, and it was striking that the majority of metastatic lesions impacted the remaining PTEN allele, albeit in different fashions, to achieve full resistance to alpelisib. Heterogeneous genetic alterations leading to PI3K pathway activation represent a potentially major contributor to the limited efficacy of PI3K/mTOR pathway inhibitors, as distinct subclonal alterations might translate into variable levels of sensitivity to these drugs, and in turn, shifts in clonal dominance during PI3K/mTOR pathway inhibitor therapy.
Systemic toxicity as a barrier to PI3K/mTOR pathway inhibitor development
A major hindrance to the broad development of PI3K pathway inhibitors has been the challenge of achieving sufficient depth of target inhibition in tumor tissue, while avoiding dose-limiting toxicities in the patient. Previous studies have established that deep (>90%) inhibition of hepatic PI3K/AKT signaling is required to generate a hyperglycemic response in mice (Taniguchi et al., 2006), and it is reasonable to expect that a similarly profound reduction in tumor tissue-associated p110α activity will be needed to achieve optimal therapeutic responses in most patients. PI3K/mTOR pathway inhibitors with the highest potentials for broad therapeutic activity are the pan-specific PI3K/mTOR and pan-PI3K inhibitors, but these also carry highest toxicity burdens in the PI3K/mTOR pathway inhibitor class. As is the case with the majority of targeted agents, deep, durable clinical responses will require combinations of PI3K/mTOR pathway inhibitors with chemotherapy, immunotherapy, and other targeted therapies. Many of these potential therapeutic partners present their own set of adverse effects, which often overlap with those of PI3K pathway inhibitors.
The results of breast cancer clinical trials provide clear evidence that the toxicity of broad PI3K pathway inhibition is a significant barrier to optimal therapeutic effectiveness. The BELLE series of clinical trials assessed the efficacy and safety of various drug combinations with the pan-PI3K inhibitor buparlisib (BKM120; Table S1) in patients with ER+, HER2- breast cancer. Patients who had progressed on aromatase inhibitor monotherapy received fulvestrant plus either buparlisib or placebo in the Belle-2 phase 3 trial (Baselga et al., 2017). Although the primary endpoint of extended progression-free survival (PFS) was met, the overall response rate (ORR, equals PFS plus stable disease) was disappointingly low. However, in patients with PIK3CA hotspot mutations in circulating tumor cell DNA (ctDNA), PFS and ORR were significantly improved. Similar results were seen in BELLE-3, a phase 3 trial in patients who had progressed on everolimus receiving fulvestrant with buparlisib or placebo (Di Leo et al., 2017). Enthusiasm for the efficacy data was unfortunately dampened by the frequency of serious adverse events leading to frequent discontinuations in both BELLE trials, with elevation of liver transaminases as a dose-limiting toxicity. Buparlisib has off-target tubulin-binding activity, which prompted the identification of a close analog, PQR309, that is more selective for PI3K (Bohnacker et al., 2017) and is now in early clinical trials.
With respect to dual PI3K/mTOR inhibitors, a phase 1 trial of BEZ235 in advanced renal cell carcinoma was terminated early due to frequent dose-limiting toxicities without objective responses (Carlo et al., 2016). In a phase 2 trial of BEZ235 for pancreatic neuroendocrine tumors, 29 of 31 patients discontinued treatment due to adverse events and the primary PFS endpoint was not met, resulting in trial termination (Fazio et al., 2016). Apitolisib (GDC-0980, a pan-PI3K-mTOR inhibitor; Table S1) proved inferior to everolimus in a phase 2 trial in patients with renal cell carcinoma, and serious adverse events and trial discontinuations were more frequent in apitolisib-treated patients (Powles et al., 2016). The extent to which toxicities limited the dose intensity achievable in tumor tissues, but it is plausible that sub-optimal dosing has contributed to the generally disappointing results obtained with these agents.
Opportunities and challenges with isoform-selective PI3K inhibitors
Isoform-selective and isoform-sparing PI3K inhibitors that preferentially inhibit the activity of one or more PI3K isoforms might circumvent the intrinsic toxicity associated with pan-PI3K inhibition, and might be more permissive for exploration of combination therapies. Preclinical studies have shown that cancer cells bearing mutant PIK3CA or HER2 amplification are frequently sensitive to p110α inhibition (Fritsch et al., 2014), whereas PTEN-mutant or -null tumors are more sensitive to p110β inhibition (Edgar et al., 2010; Ni et al., 2012). However, increased dependency on p110β is not an obligate outcome of loss of PTEN function. In a mouse model of ovarian endometrioid adenocarcinoma driven by Pten deletion and expression of oncogenic Kras, inhibition of p110α, but not p110β, was sufficient to prevent tumor growth (Schmit et al., 2014). Similarly, ovarian epithelial cells deficient for PTEN and p53 were either p110β or p110α-dependent, depending on the absence or presence of an oncogenic KRAS allele (Schmit et al., 2014). Moreover, endometrial tumor cells deficient for PTEN, either with or without KRAS mutation, required dual p110α and p110β inhibition to decrease phospho-AKT levels and cell viability (Weigelt et al., 2013). NSCLC cell lines bearing either PIK3CA or PTEN mutation also exhibit resistance to isoform-selective inhibitors, but retain responsiveness to pan-PI3K inhibitors (Stamatkin et al., 2015). Therefore, both tissue of origin and genomic context influence PI3K isoform dependence in cancer cells, making the elucidation of robust, predictive biomarkers for PI3K/mTOR pathway inhibitor responsiveness a daunting challenge for translational oncologists.
PI3K isoform switching represents a well-documented mechanism of resistance to isoform-selective PI3K inhibitors. The Engelman group (Costa et al., 2015) found that inhibition of PI3K signaling in HER2-amplified or PIK3CA-mutant breast cancer lines by the p110α-selective inhibitor, alpelisib, was followed by a rebound in PtdIns-3,4,5-P3 levels after 24 hours. Reversal of the drug effect was attributable to activation of p110β and was associated with increased p110β recruitment to HER3. Interestingly, in mutant PIK3CA-expressing cells, the rebound in PtdIns-3,4,5-P3 level was not associated with a corresponding increase in phospho-AKT, suggesting that changes in PtdIns-3,4,5-P3 levels are not obligatorily linked to downstream pathway activation. In breast cancer xenografts bearing overexpressed and mutationally activated PIK3CA, tumor regression was only observed after treatment with a combination of p110α and p110β-selective inhibitors. Similar results were reported by Baselga and colleagues (Schwartz et al., 2015), who found that treatment with a selective p110β inhibitor led to transient suppression of, followed by a significant rebound in phospho-AKT levels, which were attributed to the upregulation of the IGFR1-IRS1-p110α signaling cascade. Similarly, combined treatment with p110α- and p110β-selective inhibitors caused greater tumor growth inhibition in both a PTEN-null prostate cancer and ER+ breast cancer models (Hosford et al., 2016). In PTEN-deficient, ER+ breast cancer xenografts, treatment with the triple combination of fulvestrant (a selective estrogen receptor degrader) together with inhibitors of p110α and p110β was required to trigger maximal, sustained tumor regressions. In contrast, treatment with the fulvestrant and p110β inhibitor doublet resulted in transient inhibition followed by a striking rebound in phospho-AKT, cyclin D1/3, and phospho-pRb levels. Collectively, these studies and others provide compelling support for the conclusion that the clinical activity of p110β-selective inhibitors is limited by the development of resistance due to isoform switching to p110α-mediated signaling. Parenthetically, it is noteworthy that an activating mutation in the human PIK3CB gene (encoding p110β D1067Y) leads to broad resistance to PI3K inhibitors; however, in contrast to PIK3CA mutations, this mutation is rarely observed in cancer patients (Nakanishi et al., 2016).
Despite the evidence for p110α/p110β redundancy in preclinical models, growing clinical evidence points to the potential of p110α-selective inhibitors in defined patient populations. Taselisib (GDC-0032; Table S1) inhibits p110α, p110γ and p110δ but not p110β, and furthermore has some selectivity for PIK3CA mutant proteins. A phase I dose-escalation trial of taselisib reported a 36% response rate for patients with PIK3CA mutant tumors, versus 0% in patients whose tumors lacked a PIK3CA hotspot mutation (Juric et al., 2017). In a phase 2 trial of fulvestrant plus taselisib, patients with ER+, mutant PIK3CA-expressing breast cancer had substantially better overall responses than patients with wild-type PIK3CA tumors (Dickler et al., 2016). Although treatment discontinuation remained an issue in this study, the safety profile was considered sufficiently acceptable to warrant a phase 3 trial. In an open-label trial of the p110α-selective inhibitor alpelisib with fulvestrant, ER+ breast cancer patients with PIK3CA mutations displayed a significantly improved clinical benefit rate compared to patients with wild type PIK3CA (Juric et al, 2016 abstract), and a similar selectivity for mutant PIK3CA tumors was observed in a phase 1b trial of alpelisib with letrozole (Mayer et al., 2017). Importantly, the safety profile of alpelisib plus letrozole was superior to that of buparlisib plus fulvestrant in the BELLE-2 and 3 trials, and there were fewer discontinuations due to adverse events. Although more than half of patients manifested mild to moderate hyperglycemia, this issue was well managed with metformin. The phase 3 SOLAR-1 trial will study the effects of taselisib or placebo plus fulvestrant in greater that five hundred ER+ breast cancer patients with PI3KCA mutations and, in a separate cohort, patients with wild-type PIK3CA. This large study will go a long way toward establishing whether the PI3K isoform-selective inhibitors will deliver both efficacy and acceptable safety in patients with tumors expressing mutationally activated p110α.
Combination approaches
In some cases, resistance to PI3K inhibitors can be addressed with combination therapy involving other clinically established agents. In ER+ positive breast cancer, PI3K and ER signaling mediate mutual antagonism, which results in pharmacological inhibition of one pathway driving enhanced signaling through the other. Inhibition of PI3K was previously reported to stimulate ER-dependent transcription (Bosch et al., 2015). A recent paper provided mechanistic insights into this phenomenon by demonstrating that inhibition of PI3K triggers activation of the lysine methyltransferase, KMT2D, which functions as an enhancer of ER-dependent transcription (Toska et al., 2017). The concept of mutual antagonism involving the PI3K and ER signaling pathways has led to a series of clinical trials testing PI3K inhibitors combined with ER degraders or aromatase inhibitors, as described in other sections of this review. It is hoped that these strategies will provide additional therapeutic options after the successful combination of mTORC1 inhibitor everolimus with exemestane (Baselga et al., 2012).
Activation of the Ras-Raf-Mek-Erk axis, which occurs in many preclinical tumor models following PI3K pathway blockade, has led to the clinical testing of several PI3K plus MEK inhibitor combinations (Britten, 2013; Juric et al., 2015b; LoRusso et al., 2012; Shimizu et al., 2012). However, clinical signs of efficacy were associated with serious adverse events, raising significant questions about the future development of this combination. The sensitivity of breast cancer models to PI3K inhibition often correlates with retinoblastoma protein (pRb) phosphorylation, with high phospho-pRb levels correlated with resistance (Vora et al., 2014). Key mediators of pRb phosphorylation are the G1-phase-associated cyclin D-CDK4/6 complexes (Sherr and Bartek, 2017). The CDK4/6 inhibitor, ribociclib, sensitized resistant breast cancer cells to alpelisib, prevented the development of adaptive resistance, and provoked tumor regression in breast cancer xenograft models (Herrera-Abreu et al., 2016; Jansen et al., 2017). Combinations of PI3K pathway and CDK4/6 inhibitors have also shown promising activities in pancreatic, HNSCC and NSCLC models (Franco et al., 2014; Gopalan et al., 2013; Ku et al., 2016). The therapeutic pairing of CDK4/6 inhibitors with PI3K inhibitors is particularly appealing in ER+ breast cancer, a cancer subtype with well-documented sensitivity to anti-estrogen plus CDK4/6 inhibitor therapy. Given that thirty percent of ER+ breast cancers bear activating PIK3CA mutations, triplet combinations of PI3K and CDK4/6 inhibitors with estrogen antagonists have attracted considerable interest, and clinical studies are now underway.
An intriguing combination opportunity for PI3K inhibitors has been uncovered in triple negative breast cancer (TNBC). Preclinical studies demonstrated that exposure of TNBC cell lines or patient-derived xenografts (PDXs) to the pan-PI3K inhibitor, buparlisib, resulted in reduced BRCA1 expression, and increased DNA damage (Ibrahim et al., 2012). Tumor growth inhibition in PTEN-null or PIK3CA-mutant PDX models was enhanced by the combination of buparlisib with the poly-ADP ribose polymerase (PARP) inhibitor, olaparib. PARP inhibitors have shown clinically significant activity in BRCA1- or BRCA2-mutated ovarian cancers (later-stage studies in BRCA-deficient TNBC patients are ongoing), due to a synthetic lethal mechanism in cells deficient in homologous combination-mediated DNA repair (Lord and Ashworth, 2016). Combinations of PI3K/mTOR pathway inhibitors and PARP inhibitors have also shown promising activity in prostate, ovarian and SCLC models (Cardnell et al., 2013; González-Billalabeitia et al., 2014; Rehman et al., 2012; Wang et al., 2016). Once again, results from an early clinical trial with buparlisib and olaparib suggest that toxicity will be a formidable obstacle to attaining the drug exposures needed to achieve an optimal therapeutic effect with this drug combination (Matulonis et al., 2016). The possibility that toxicity would be mitigated by combining a PARP inhibitor with a p110α-selective, rather than the pan-PI3K inhibitor, buparlisib, will undoubtedly be addressed in future clinical studies.
PI3K pathway activation and resistance to therapy
Activation of the PI3K/mTOR is a common mediator of resistance to numerous anticancer agents, including conventional chemotherapy and agents targeting other oncogenic nodes (Brown and Toker, 2015; Ilagan and Manning, 2016). Emerging evidences points to PI3K/mTOR signaling as a mechanism of resistance to cancer immunotherapy. Immuno-oncology is currently the most explosive area of cancer therapeutic development, due mainly to the unprecedented durability of the responses seen in patients with normally intractable metastatic cancers (Farkona et al., 2016; Pardoll, 2012). Despite the impressive responses seen in a subset of patients receiving immune PD1/L1- and CTLA4-targeted therapies, objective response rates are commonly in the 25–40% range, indicating that the majority of tumors are intrinsically resistant to these immune checkpoint inhibitors. Immunotherapy resistance was recently linked to tumor-cell autonomous PI3K pathway activation (Peng et al., 2016). PTEN silencing in BRAF-mutant melanoma cells led to impaired T cell tumor recruitment and anti-tumor immunity. In melanoma patients, reduced expression of PTEN was correlated with resistance to anti-PD1 therapy. Moreover, in melanoma patients with regional heterogeneity of PTEN expression in tumor tissues, T cell infiltration was consistently lower in sub-regions that lacked PTEN protein. In a spontaneous Braf-mutant, Pten-null mouse melanoma model, the combination of a selective PI3Kβ inhibitor with anti-PD1 significantly improved tumor growth inhibition and survival compared to each agent alone. Although a pan-PI3K inhibitor impaired the vaccine-induced proliferation of gp100-specific T cells in a preclinical melanoma model, a p110α-selective inhibitor had no effect on this T-cell response, suggesting that isoform-selective inhibitors will be less likely to compromise anti-tumor T cell activity. PI3K/mTOR pathway activation in lung cancer models was associated with increased expression of the PD-1 ligand, PD-L1, and combination therapy with rapamycin plus anti-PD1 antibody significantly reduced tumor burden in a KRAS-driven mouse NSCLC model (Lastwika et al., 2016). However, in a separate study, the investigators observed no consistent effect of PTEN mutation or AKT amplification on the expression of PD-L1 by melanoma cell lines (Atefi et al., 2014). Additional studies are clearly needed to fully understand the mechanisms that regulate PD-L1 expression on tumor cells, and the complex interplay between tumor cell-intrinsic alterations in PI3K/mTOR pathway activity and antitumor immunity, before and during therapy with immunostimulatory agents, and in the absence or presence of traditional tumor cell-targeted therapeutics.
Progress with mTOR-selective agents as anticancer drugs
Activation of mTOR, specifically mTORC1, is a key consequence of tumor-associated alterations that drive PI3K pathway activation. This observation, together with the early availability of rapamycin and its analogs (rapalogs) as clinically established mTORC1 inhibitors in organ transplantation, led to clinical oncology trials that resulted in the approval of two rapalogs for the treatment of ER+ breast, renal and pancreatic neuroendocrine tumors (Table S1). However, despite the positive PFS outcomes in these trials, rapalogs have shown limited efficacy in other trials, likely due to the incomplete inhibition of mTORC1, and compensatory upregulation of the PI3K signaling network (Figure 6). Subsequent medicinal chemistry efforts led to the discovery of a series of mTOR kinase inhibitors (TORKi) that, in contrast to the allosteric mechanism of inhibition employed by rapalogs, blocked mTOR signaling by interacting with the mTOR catalytic domain in an ATP-competitive fashion. As commonly seen with other similarly targeted protein kinase inhibitors, cancer cells acquired resistance to the TORKi through the outgrowth of clones bearing mTOR mutations that reduced the antiproliferative effects of these drugs. Genomic sequencing of ER+ breast cancer cells that had acquired resistance to mTOR inhibitors identified two mutations in the FKBP12-rapamycin binding (FRB) domain that induced high-level resistance to rapalogs, and a second mutation in the catalytic domain of mTOR that decreased sensitivity to a TORKi (Rodrik-Outmezguine et al., 2016). Interestingly, these mutations conferred drug resistance through hyper-activation of mTORC1, as opposed to interference with inhibitor binding to the target protein.
To overcome these mTOR-intrinsic mechanisms of resistance, Shokat and coworkers designed a bivalent inhibitor, termed RapaLink-1, containing a TORKi substituent that targets the catalytic domain, connected with a precisely designed linker to the second pharmacophore (a rapalog) to simultaneously engage the FRB domain of mTOR (Rodrik-Outmezguine et al., 2016). RapaLink-1 effectively blocked mTORC1 signaling by both FRB- and kinase domain-mutated mTOR proteins, and overcame resistance to single-agent rapamycin- or TORKi in these cells. Importantly, RapaLink-1 also effectively inhibited wild-type mTOR signaling. Thus, the RapaLink design may more effectively delay the acquisition of mTOR resistance mechanisms. A follow up study highlighted two additional advantages of RapaLinks compared to TORKi (Fan et al., 2017). First, RapaLinks are somewhat selective for mTORC1 versus mTORC2 (Figure 2), possibly reducing toxicities associated with mTORC2 inhibition. Second, RapaLinks achieve a much longer duration of mTORC1 inhibition, correlating with greater anti-tumor activity in glioma models. Whether these third generation mTORC1 inhibitors will improve the efficacy-toxicity profile of mTOR-targeted therapies in cancer patients remains to be determined.
Optimizing therapeutic index through alternative dosing regimens
The vast majority of clinical studies with PI3K/mTOR pathway inhibitors have employed standard daily dosing regimens, with dose level and frequency based on maximal-tolerated dose (MTD) determinations. Although this approach ensures an acceptable safety profile, it does carry the risk that a safe dose may not be an effective dose due to insufficient target engagement in the tumor tissue. Strategies aimed at achieving optimal target modulation in the tumor, while sparing normal tissue, may be necessary for the PI3K/mTOR pathway inhibitors to achieve their full potential as anticancer agents. To this end, several groups are exploring alternative dosing strategies, particularly those involving high-dose, intermittent therapy as potential paths forward for the clinical applications of PI3K/mTOR pathway inhibitors. For example, the p110α/δ inhibitor copanlisib (BAY80-6946; Table S1), which has a half-life of only 0.7 hr after intravenous (IV) administration in mice, was far more effective in mice bearing BT474 breast cancer xenografts when administered intermittently (3 times per week) than when administered by continuous dosing (Liu et al., 2013). Similarly, intermittent dosing of the p110α/δ-selective inhibitor, AZD8835, was clearly superior to daily dosing of this drug in a preclinical breast cancer model (Hudson et al., 2016). A pharmacokinetic-pharmacodynamic model, based on measurements of caspase-3 cleavage, was used to derive simulations of apoptotic rates. This simulation revealed that intermittent dosing elicited repeated waves of intratumoral apoptosis with no loss of the peak apoptosis, whereas continuous dosing produced a lower level of apoptosis that waned with continued dosing. Higher levels of caspase 3 cleavage were correlated with more penetrating PI3K inhibition, as measured by phospho-AKT levels, during the intermittent, high-dose scheduling. The mTOR inhibitor, vistusertib (AZD2014; Table S1), caused similar growth inhibition of MCF7 ER+ breast cancer xenografts when dosed either continuously or intermittently, but higher rates of tumor cell apoptosis were observed during intermittent dosing (Guichard et al., 2015). These preclinical studies suggest that intermittent dosing schedules might allow higher dosing, and lead to more complete PI3K pathway inhibition and higher levels of apoptosis, even in the monotherapy setting. The clinical success of this proposal is contingent on the notion that normal tissues recover more adeptly during the inter-dose interval than does the tumor tissue.
Early clinical results with intermittent dosing of PI3K/mTOR pathway inhibitors are also yielding encouraging data. In a phase I trial of the pan-PI3K inhibitor, buparlisib, an intermittent five days on, two days off schedule produced fewer early onset adverse events, at the same daily dose of buparlisib (Ma et al., 2016). Intermittent dosing may also permit more pronounced pathway inhibition with equivalent toxicity for less toxic inhibitors such as the isoform-selective PI3K inhibitors. A phase 1 dose escalation trial of the p110α-selective inhibitor, MLN1117 (now TAK-117; Table S1), compared two different, three day per week schedules at higher dose (900 mg maximum dose) with daily dosing (150 mg maximum) (Juric et al., 2015b). The intermittent dosing schedules allowed the investigators to achieve approximately 4-fold higher exposure than was attainable with daily dosing. Analysis of skin biopsies revealed enhanced inhibition of PI3K pathway, as measured by the pharmacodynamic biomarkers, phospho-4EBP1 and phospho-S6. Finally, gedatolisib (PKI-587; PF-05212384; Table S1), a potent inhibitor of both Class I PI3Ks and mTOR, is in early trials in combination with palbociclib and anti-estrogen therapy in women with metastatic ER+ breast cancer, and with cis-platinum in TNBC patients. The dosing protocol for gedatolisib in these study involves intravenous administration on a once-weekly basis (Mallon et al., 2011). The results of studies exploring intermittent dosing with PI3K/mTOR pathway inhibitors are eagerly anticipated, and the hope that these novel regimens will at least partially address the opposing requirements deep inhibition of the PI3K/mTOR pathway in tumor tissue, while minimizing toxic outcomes in normal tissues. Results of a phase 2 trial of copanlisib, presented at the 2017 AACR annual meeting (Martin Dreyling, personal communication), showed an approximate 70% response rate in patients with indolent lymphomas given once-weekly i.v. doses of copanlisib. Importantly, common toxicities associated with continuous inhibition of p110α (hyperglycemia) or p110δ (colitis) were transient or less frequent in the intermittently dosed cohorts.
Mitigation of toxicity is likely to be especially important in the development of combinations of PI3K pathway inhibitors with other targeted agents, particularly those with overlapping toxicity profiles. For instance, clinical trials conducted so far suggest that combinations of pan-PI3K inhibitors with MEK inhibitors will face considerable tolerability challenges (Shimizu et al, 2012; LoRusso et al, 2012). While the use of isoform-selective or isoform-sparing PI3K inhibitors will likely reduce toxicity burden of rational combinations, in many cases the design of an effective dose ratio and schedule may be as critical as the design of the combination itself. Intermittent dosing of either one or both combination partners may ameliorate toxicity and enhance therapeutic index. This concept was tested in a mouse breast xenograft model with combinations of the p110α/γ selective inhibitor, AZD8835, dosed intermittently, and partnered with either intermittently dosed fulvestrant, with continuously administered palbociclib (Hudson et al, 2016). These combinations produced robust tumor regressions; indeed, a triplet combination (AZD8835 plus fulvestrant plus palbociclib), with each drug delivered according to the above protocol, induced profound tumor regressions. Importantly, the MTD of each agent in the triplet combination was the same as that for monotherapy, suggesting that there is no combinatorial toxicity when two of three agents are administered intermittently.
Pulsatile dosing is also being investigated in an ongoing trial of the AKT inhibitor AZD5363 combined with the PARP inhibitor olaparib, in which AZD5363 is dosed according to three intermittent schedules (Michalarea et al., 2016). Although the rationale underlying the choice of an AKT inhibitor (versus a PI3K inhibitor) for this combination treatment protocol is not completely clear, published evidence suggests that PARP inhibition triggers AKT activation and tumor progression in certain settings (Cardnell et al., 2016). It may also be possible to reduce systemic toxicity by sequentially dosing combination partners, as in some chemotherapy combinations. The effectiveness of such a schedule would presumably be optimized by partnering a PI3K-mTOR pathway inhibitor with an agent that left a sustained “mark” of the tumor; for example, a DNA-damaging cytotoxic agent, or a drug targeting the epigenome, such as a histone deacetylase inhibitor.
The encapsulation of PI3K pathway inhibitors in tumor-targeted nanoparticles represents an appealing strategy to promote asymmetric distribution of PI3K/mTOR pathway inhibitors in tumor versus other host tissues. In a preclinical study, P-selectin-binding, fucoidan-based nanoparticles served as delivery vehicles for alpelisib (Mizrachi et al., 2017). Once weekly dosing of the alpelisib-loaded nanoparticles in animals bearing head and neck cancer xenografts caused substantially improved survival, and sensitized the tumors to radiotherapy to a greater extent than daily free alpelisib dosing. In addition, nanoparticle-mediated drug delivery attenuated drug-induced hyperglycemia and hyperinsulinemia, and diminished the attrition of pancreatic islet beta cells seen after alpelisib treatment. These results support further exploration of tumor-targeted nanoparticles as promising tumor-selective delivery vehicles for PI3K/mTOR pathway inhibitors.
Biomarkers: Predicting response and resistance to PI3K pathway inhibitors
As with other targeted therapies, the success of PI3K pathway inhibitors in the clinical setting will be predicated on the accurate identification of those patients who are most likely to respond to treatment. As noted above, a phase 3 study (the BELLE-2 trial) of fulvestrant and buparlisib in letrozole-resistant patients revealed a clear difference in response to buparlisib depending on PIK3CA mutation status, as determined by circulating tumor (ct) DNA analysis (Baselga et al., 2017). In patients with PIK3CA mutations, response rates were clearly increased (18.4% versus 3.5%), as was the median PFS (7.0 v. 3.2 months) after buparlisib treatment relative to placebo. In contrast, patients lacking PIK3CA mutations realized no additional benefit from the fulvestrant plus buparlisib combination relative to fulvestrant alone (11.6% v. 10.6%), and experienced no PFS benefit (6.8 versus 6.8 months). Interestingly, no significant improvements in response rate or PFS were seen in patients with PIK3CA mutations or loss of PTEN expression when these parameters were assessed in archival tumor tissue. Similar increases in PFS benefit were seen in mutant PIK3CA-bearing patients in the BELLE-3 trial of fulvestrant plus buparlisib, which enrolled patients who were resistant to prior rapalog therapy (Di Leo et al., 2017). The BELLE trials confirmed that PIK3CA mutations predict sensitivity to PI3K inhibitors, and argue that analysis of real-time PIK3CA mutation status in ctDNA provides a more predictive biomarker of drug responsiveness than does analysis of archival tumor tissue. In contrast to archival samples from primary tumors, ctDNA analysis takes into account the dynamic changes in cancer genomes (and hence biomarker status) that occur during tumor evolution and in response to earlier therapies. The presence of PIK3CA mutations in ctDNA at the time of buparlisib treatment will logically identify drug-sensitive cell populations more accurately than a historical sample, in which potentially drug-responsive clones may have undergone attrition during tumor progression (McGranahan et al., 2015).
Recent evidence suggests that different PIK3CA mutations results have distinct consequences with regard to PI3K/mTOR pathway inhibitor therapy. Early clinical trials of the isoform-selective inhibitors, alpelisib and taselisib, in breast cancer have produced results consistent with a previous meta-analysis of clinical trials of PI3K pathway inhibitors in patients with diverse cancers, revealing that patients with H1047R mutations had higher response rates than those with other PIK3CA mutations, or no mutations in this gene (Janku et al., 2013; Mayer et al., 2017). The p110β-sparing inhibitor, taselisib, (Table S1) was reported to induce degradation of mutant p110α (Friedman et al., 2017). It is conceivable that distinct activating mutations in p110α might confer differential susceptibilities drug-induced degradation; if this proves to be the case, perhaps drug-induced turnover of the p110α H1047R mutant underlies the increased taselisib sensitivity of tumors expressing this particular p110α oncoprotein. Nonetheless, activating mutations in PIK3CA are not sufficient to define the PI3K/mTOR pathway inhibitor-responsive population. Other positive or negative (e.g., Ras pathway alterations) biomarkers, preferably amenable to longitudinal measurements, are needed for optimal development of drug combinations containing the PI3K/mTOR pathway inhibitors.
The BOLERO-2 trial established the efficacy of everolimus plus exemestane in ER+/HER2- breast cancer (Baselga et al., 2012). In this trial, amplification of cyclin D1 had no impact on everolimus efficacy, whereas mutation or amplification of FGFR1 or FGFR2 resulted in less benefit from everolimus treatment, though the trial was not sufficiently powered to deliver definitive conclusions in this regard. A meta-analysis of the BOLERO-1 and BOLERO-3 phase 3 trials of everolimus or placebo in combination with trastuzumab and chemotherapy (paclitaxel or vinorelbine, respectively) in patients with HER2+ advanced breast cancer revealed a significant PFS benefit from everolimus in patients with either PIK3CA mutation or low PTEN expression (by IHC), whereas patients with neither of these alterations experienced no PFS benefit from the addition of everolimus (André et al., 2016).
The AKT E17K mutation causes constitutive membrane localization and pathway activation, and is found at a frequency of 1–4 percent across multiple tumor types. In early phase trials, the competitive AKT inhibitor AZD5363 showed promising activity in patients with tumors carrying the E17K mutation, as determined by tumor tissue and ctDNA analysis (Hyman et al., 2015; Tamura et al., 2016). In a trial of the AKT inhibitor ipatasertib (GDC-0068; Table S1) with abiraterone in metastatic castration-resistant prostate cancer (CRPC), an improved PFS response to ipatasertib in patients with PTEN loss (determined by FISH and NGS) but not in patients with intact PTEN (Hyman et al., 2015). Hence, PTEN loss and the AKT E17K mutation appear to be promising biomarkers of therapeutic sensitivity to AKT inhibitors.
Lessons learned from clinical experience with idelalisib
Idelalisib, a selective PI3Kδ inhibitor, was approved in 2014 for relapsed chronic lymphocytic leukemia (CLL) and two subtypes of non-Hodgkins lymphoma (NHL) in combination with rituximab. Clinical trials provided important insights into the mechanisms underlying the efficacy of idelalisib, as well as its toxicities (Furman et al., 2014; Gopal et al., 2014). Treatment of CLL patients with idelalisib leads to redistribution of transformed B cells from lymphoid compartments to the peripheral circulation, resulting in lymphocytosis. These responses are shared with those elicited by the BTK inhibitor ibrutinib (Byrd et al., 2013; Herman et al., 2014), consistent with the convergent roles of BTK and p110δ in the BCR signalosome (Fruman and Cantley, 2014). The strong dependence on PI3Kδ for signaling through the BCR in CLL and indolent NHL explains the sensitivity of these transformed B cells to p110δ inhibition. Idelalisib-induced lymphocytosis deprives the malignant B cells of their protective niches in the peripheral tissues, and renders B cells susceptible to apoptotic signals provoked by rituximab immunotherapy or bendamustine chemotherapy. Thus, the predominant mechanism of action of idelalisib differs substantially from the presumptive direct actions of p110α on cancer cells; rather, p110δ inhibition suppresses host responses that nurture the survival and expansion of transformed B cell clones in CLL.
Adverse events associated with idelalisib treatment included elevation of liver enzymes, diarrhea, pneumonitis and pneumonia (Furman et al., 2014; Gopal et al., 2014). Subsequent trials of idelalisib, which included trials of other combinations and trials in younger and treatment-naïve patients, encountered more severe adverse events, such as colitis, pneumonitis and high-grade liver enzyme elevations (Barr et al., 2016; Cheah et al., 2015; Lampson et al., 2016). Unfortunately, several drug-related deaths occurred as a result of Pneumocystis jirovecii or cytomegalovirus infection, leading to trial discontinuation. Currently idelalisib plus rituximab is used only in a salvage setting, with systemic immunosuppression and/or inflammatory complications managed by treatment interruption or corticosteroid treatment. Toxicities associated with idelalisib are considered mechanism-based, given the similarity to the pathologies seen in mice genetically engineered to lack p110δ activity. Patients with grade 3 or higher colitis or transaminitis after idelalisib treatment displayed higher plasma levels of chemokines and lower numbers of Tregs (Lampson et al., 2016), suggesting that the toxic effects of p110δ inhibition may stem in part from Treg depletion that relieves restraints on cytotoxic T cells. Interestingly Pneumocystis jirovecii infections have also been reported in cancer patients receiving the mTOR inhibitor, everolimus (Loron et al., 2015). The apparent suppression of Treg function in idelalisib-treated patients suggested that p110δ inhibition might enhance anti-tumor activity by inhibiting the functions of an important cellular contributor to the immunosuppressive tumor microenvironment. Inhibition of p110δ may interfere with Treg functions, in part, by suppressing the activation of AKT in antigen-responsive Treg cells. Monoallelic expression of an AKT-insensitive FOXO1 mutant in Treg cells resulted in decreased tumor infiltration of activated Tregs accompanied by enhanced activation of tumor-infiltrating cytotoxic T lymphocytes, and in turn increased inhibition of tumor growth in three different preclinical models (Luo et al., 2016). The potential of p110δ inhibitors to enhance antitumor immunity in mice via Treg inhibition was discussed above (Ahmad et al., 2017; Ali et al., 2014). The potential immunostimulatory effects of p110δ inhibitors are being explored in combination with anti-PD-1 antibody in ongoing clinical studies.
Tissue Overgrowth and Hamartoma Syndromes
Syndromes Caused by Deregulated mTORC1 Activation
In spite of the challenges experienced during the clinical development of PI3K/mTOR pathway inhibitors, some noteworthy success stories have emerged from the identification of patient populations bearing mutations, downstream of PI3K, which drive hyper-activation of mTORC1 signaling (Table 1). A canonical example of mTORC1-driven disease pathology is tuberous sclerosis, an autosomal, dominantly inherited genetic disorder with a frequency of 1 in 6000 live births (Nathan et al., 2017; Switon et al., 2016). Consistent with the ubiquitous roles of mTORC1 in the stimulation of cell growth and proliferation, tuberous sclerosis is a multi-organ disease characterized by the outgrowth of benign tumors, termed hamartomas, in the brain, kidneys, heart, and other organs. Surprisingly, these patients have a relatively normal lifespan, but the propensity of tumors to develop in the central nervous system leads to cognitive defects and seizures that can be debilitating and even life threatening. Commonly observed brain lesions are cerebral hamartomas and subependymal giant cell tumors (SEGAs) (Franz et al., 2014; JóŸwiak et al., 2016).
Encouraging results obtained in TSC-deficient mouse models prompted clinical trials with rapalogs in tuberous sclerosis patients. Everolimus is now an established therapeutic agent for adults with high-risk renal angiomyolipomas associated with a diagnosis of tuberous sclerosis, and for the both young and adult patients with surgically non-resectable SEGAs (Bissler et al., 2008, 2013). Both rapamycin and everolimus have shown benefit in reducing the severity of epilepsy in tuberous sclerosis patients (Krueger et al., 2016; Switon et al., 2016). However, rapalog monotherapy is not curative, likely due in part to failure to completely block inappropriate mTORC1 signaling at clinically tolerable doses of these agents. In addition, chronic exposure to rapalogs often leads to serious complications, such as mucositis, pneumonitis, and increased risks of infection, which force discontinuation of therapy. In the absence of chronic exposure to the rapalog, the patient experiences tumor regrowth and symptomatic progression.
Lymphangioleiomyomatosis (LAM) is a syndrome that occurs almost exclusively in females, and is characterized by the emergence of smooth muscle-like cysts in the lung, and a gradual decline in pulmonary function (Henske and McCormack, 2012; Lam et al., 2017). Most women with LAM are diagnosed with a sporadic form of the disease, in which the parental, disease-causing clone has incurred biallelic loss of function mutations in TSC1 or, in the majority of cases, TSC2. LAM is termed a “benign neoplasm”, because accumulating evidence suggests that LAM cells resemble, in phenotype and behavior, low-grade sarcoma cells. Indeed, the primary LAM cells that give rise to the cardinal symptom of cystic lung disease appear to be emigrants from distant organs, with the uterus suspected as a prominent source of pre-metastatic LAM cells. The overwhelming gender difference with regard to LAM incidence suggests that hormonal factors, specifically estrogen, collaborate with loss of TSC function during disease development and progression. Until recently, no effective treatment for LAM was available to the patients, who were left to suffer an inexorable decline in lung function, ultimately leading to the need for a life-saving lung transplant. However, the strong genetic linkage between loss of TSC function and LAM suggested that therapeutic targeting of mTORC1 might prove effective in halting or delaying LAM progression (Yates, 2016). A subsequent clinical study (the MILES trial) demonstrated that rapamycin (Sirolimus™) treatment significantly reduced the rate of decline in several parameters of pulmonary function (McCormack et al., 2011), leading to the drug’s approval by the FDA as the first therapy for LAM (Table 1 and Table S1). Unfortunately, as is the case for tuberous sclerosis patients, rapamycin only delays disease progression and does not reverse damage already done to the lungs. Recently, an important new sirolimus trial (the MILED trial) was announced and is predicted to further increase the impact of rapalog therapy on the disease course of LAM (NCT03150914). This trial will focus on patients at a much earlier stage of the disease than those admitted to the MILES trial. Importantly, the dose of sirolimus selected for MILED (1 mg per day) will be half of the dose tested in the MILES trial. The intention is to reduce the drop-out rate due to adverse drug effects, and to allow these healthier, early-stage patients to derive greater benefit from the mTORC1 inhibitor, administered on a more chronic basis. Given the similarity of LAM lesions to overt cancerous tumors, it is anticipated that combinations involving rapalogs and/or other targeted agents will ultimately be required for optimal therapeutic benefit in this disease.
The efficacy of rapalog therapy in the setting of somatic loss of mutations involving TSC has also been observed in frank malignant disease. Solit and coworkers used deep genomic sequencing to identify somatic mutations in TSC1 in patients with metastatic bladder cancer, and observed that these patients fared significantly better on everolimus therapy than did patients whose bladder tumors contained the wild-type TSC1 gene (Iyer et al., 2012). A subsequent publication identified an advanced, anaplastic thyroid cancer patient bearing a nonsense mutation in TSC1, who also gave an extraordinary response to everolimus (Lim et al., 2016; Wagle et al., 2014). The patient’s disease progressed after 18 months on drug, and re-sequencing of the tumor genome revealed a mutation in MTOR that encoded a single amino acid substitution in the FKBP12-binding domain. Although this alteration confers resistance to the rapalogs, the tumor should, in principle, retain sensitivity to ATP-competitive mTOR kinase inhibitors or the bidentate RapaLinks discussed above. Finally, a recent study identified a cohort of renal cancer patients with loss of function in TSC1 and/or activating mutations in mTOR who were exceptional responders to rapalog therapy (Voss et al., 2014). Interestingly, the cancer-associated TSC mutations described above involve the TSC1 gene, whereas the majority of alterations that underpin tuberous sclerosis and LAM target the TSC2 subunit. It is tempting to speculate that in addition to activating mTORC1, these mutations disrupt an mTORC1-independent function(s) of TSC1, such as the regulation of transforming growth factor-β signaling (Thien et al., 2015).
Syndromes Caused by Deregulated PI3K Signaling
Heritable germline loss of function mutations in PTEN, a key negative regulator of the PI3K/mTOR pathway signaling network, give rise to a cluster of tissue overgrowth syndromes collectively termed PTEN hamartoma tumor syndrome (PHTS) (Gammon et al., 2016; Worby and Dixon, 2014). These PTEN-linked diseases are transmitted generationally in an autosomal dominant fashion. The archetypal member of this disease group is Cowden’s syndrome (CS). Like the tuberous sclerosis complex syndrome described above, CS is a multi-organ system disorder characterized by anomalous tissue overgrowths. However, CS confers a considerably more significant risk of cancer development, with breast, kidneys, colon, thyroid, and endometrium being prime targets for tumorigenesis. Management of CS patients entails mostly supportive care, with particular attention paid to screening for various cancers. Sirolimus has been tested in CS patients (Table 1), and appears to ameliorate disease symptoms, but the response is only partial and escape from the drug has been observed after a few months of therapy (Munoz and Kurzrock, 2012). The less than optimal outcomes to date with rapalog therapy likely reflect the challenge associated with treating the mTORC1 component of a disease stemming from inappropriate activation of the entire PI3K/mTOR pathway network.
CLOVES syndrome is a rare, non-malignant disease resulting from somatically-acquired, mosaic activating mutations in the PIK3CA gene (Kurek et al., 2012). This syndrome is characterized by regional tissue overgrowths and malformations affecting the epidermis, internal organs, skeleton, and central nervous system. CLOVES syndrome was recognized as a separate disease entity only ten years ago, and the main disease-modifying treatment is surgery. Both the disease itself and the surgery can be disfiguring and potentially life-threatening. In patients with vascular anomalies associated with elevated PI3K/AKT/mTOR signaling, rapamycin produced partial responses in 47 of 57 patients enrolled in a phase 2 study (Adams et al., 2016) (Table 1). Patients with PIK3CA-dependent vascular anomalies and other tissue overgrowths may achieve even greater benefit from treatment with a selective p110α inhibitor, which would block mTORC1-independent functions of p110α not covered by rapamycin. As expected, PI3K inhibitors strongly suppress the proliferation of CLOVES syndrome cells in culture (Loconte et al., 2015). We expect that p110α inhibitors will be tested in CLOVES syndrome patients in the near future, and, toxicity issues notwithstanding, it is hoped that these drugs will offer the possibility of complete remissions not achievable with rapalog therapy.
A recent report offered evidence for an alternative pathway of PTEN inactivation driven by somatic disruption of PARK2, which encodes Parkin, an E3 ligase involved in the elimination of dysfunctional mitochondria by mitophagy (Gupta et al., 2017; Pickrell and Youle, 2015). PARK2-deficient cancer cells exhibit elevated reactive oxygen species (ROS) and nitrogen oxide synthase activity, presumably due to defective mitochondrial function. These abnormalities result in loss of PTEN lipid phosphatase activity and reduced expression of the PTEN protein, attributable to the nitrosylation of a critical cysteine residue in the catalytic domain, and subsequent protein ubiquitination and degradation. The results of this study suggest that PARK2 is a haploinsufficient tumor suppressor, and that as many as to two-thirds of human cancers exhibit reduced expression of the PARK2 gene product. Extrapolation of these results to PTEN function argues that at least an equal proportion of human cancers are prone to PI3K/mTOR pathway hyper-activity due to impaired PtdIns-3,4,5-P3 degradation. Moreover, these findings also offer a fascinating link to the mechanism of neurodegeneration in Parkinson’s disease. Loss of function mutations in PARK2 were first identified in individuals with early onset Parkinson’s disease and might be an indication for testing PI3K/mTOR pathway inhibitors (Table 1). The beneficial effects of mTORC1 inhibition in Parkinson’s disease may not be restricted to those cases associated with PARK2 mutations. A study in D. melanogaster revealed that increases in 4E-BP1-dependent translational repression induced by rapamycin or genetic manipulation increased resistance of dopaminergic neurons to cell death in flies bearing disease-associated mutations in the homologs of PARK, PINK1, and LRRK2 (Tain et al., 2009). Previous studies uncovered a link between Parkinson’s disease and an increased risk of certain cancers (Feng et al., 2015). It would be interesting to learn whether reduced Parkin activity in the dopaminergic neurons of the substantia nigra also leads to loss of PTEN activity, and, in turn, whether an inappropriate increase in PI3K/mTOR signaling is causally related to the death of these neurons on Parkinson’s patients.
Perspectives
Over three decades of intensive basic research and drug development have resulted in slow yet undeniable progress in targeting the PI3K/mTOR pathway for the treatment of cancer, along with renewed optimism for future success. The rapalogs have generally failed to deliver on expectations that these drugs would be broadly impactful anti-cancer agents, yet the combination of everolimus with aromatase inhibitors is now an approved second-line therapy in hormone-dependent breast cancer. Although early trials with PI3K/mTOR pathway inhibitors were not as successful as hoped, the next wave of clinical trial approaches is likely to improve the clinical outcome in many oncology settings. For example, the new generation of isoform-selective PI3K inhibitors shows great promise in combination with fulvestrant and/or CDK4/6 inhibitors in breast cancer. The long-sought goal of developing compounds that selectively inhibit mutationally activated forms of p110α has nearly been achieved, with taselisib in phase 3 trials for patients whose tumors harbor PIK3CA mutations. The p110δ inhibitor idelalisib is a clinical option in various blood cancers, and other p110δ-selective compounds with apparently improved safety are currently in development. Intermittent dosing schedules and nanoparticle-mediated drug delivery may broaden the therapeutic window for pan-PI3K inhibitors and TORKi. Development of treatment schemes that minimize systemic feedback through elevated glucose and insulin may further increase the efficacy-safety profiles of these compounds. Refinement of biomarkers for patient selection and prediction of relapse will undoubtedly improve the success rate of PI3K/mTOR pathway inhibitors. To produce even greater gains in application of PI3K/mTOR inhibitors, basic research is necessary to uncover synthetic lethality in different cancer types and leverage this knowledge for effective combinations.
A greater appreciation for the complex roles of PI3K/mTOR signaling in the immune system has uncovered unexpected strategies to modulate the tumor microenvironment for therapeutic benefit. Inhibition of p110δ or p110γ, neither of which is commonly expressed in solid tumor cells, can overcome immune tolerance and/or the immunosuppressive milieu to promote T cell anti-tumor responses and potentiate the efficacy of immunotherapies in preclinical models. Inhibition of PI3K and/or mTOR in dendritic cells also holds promise to enhance cancer vaccine approaches.
Another unexpected outcome of PI3K/mTOR research has been the identification of several disease indications outside of oncology that are likely to benefit substantially from PI3K/mTOR-targeted inhibitors (Table 1). Although relatively rare in incidence, these diseases share a common etiology, based on dysregulated PI3K/mTOR signaling, and cause immense suffering to patients and their families. The evolutionarily conserved role of PI3K and mTOR in organismal aging also provides new opportunities to extend lifespan and, more importantly, healthspan through partial pathway inhibition (Johnson et al., 2013; Pan and Finkel, 2017; Selman et al., 2009; Wu et al., 2013). In the coming years, studies of PI3K/mTOR biology will surely uncover additional surprises and new approaches to ameliorate human disease.
Supplementary Material
Acknowledgments
We thank Ji Luo for reading the manuscript and providing helpful comments. PI3K/mTOR-related research in our laboratories was supported by NIH grants R01-CA158383 and R21-AI124146 (to D.A.F.), and GM041890 and R35 CA197588 (to L.C.C.). D.A.F. receives research support from Revolution Medicines. L.C.C. is a founder and member of the Board of Directors of Agios Pharmaceuticals and is a founder and receives research support from Petra Pharmaceuticals. R.T.A. and S.B. are employees and shareholders at Pfizer. These companies are developing novel therapies for cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams DM, Trenor CC, Hammill AM, Vinks AA, Patel MN, Chaudry G, Wentzel MS, Mobberley-Schuman PS, Campbell LM, Brookbank C, et al. Efficacy and safety of sirolimus in the treatment of complicated vascular anomalies. Pediatrics. 2016;137:e20153257. doi: 10.1542/peds.2015-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Abu-Eid R, Shrimali R, Webb M, Verma V, Doroodchi A, Berrong Z, Samara R, Rodriguez PC, Mkrtichyan M, et al. Differential PI3Kδ Signaling in CD4(+) T-cell Subsets Enables Selective Targeting of T Regulatory Cells to Enhance Cancer Immunotherapy. Cancer Res. 2017;77:1892–1904. doi: 10.1158/0008-5472.CAN-16-1839. [DOI] [PubMed] [Google Scholar]
- Aksoy E, Taboubi S, Torres D, Delbauve S, Hachani A, Whitehead MA, Pearce WP, Berenjeno IM, Nock G, Filloux A, et al. The p110δ isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nat Immunol. 2012;13:1045–1054. doi: 10.1038/ni.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali K, Bilancio A, Thomas M, Pearce W, Gilfillan AM, Tkaczyk C, Kuehn N, Gray A, Giddings J, Peskett E, et al. Essential role for the p110delta phosphoinositide 3-kinase in the allergic response. Nature. 2004;431:1007–1011. doi: 10.1038/nature02991. [DOI] [PubMed] [Google Scholar]
- Ali K, Soond DR, Piñeiro R, Hagemann T, Pearce W, Lim EL, Bouabe H, Scudamore CL, Hancox T, Maecker H, et al. Inactivation of PI(3)K p110δ breaks regulatory T-cell-mediated immune tolerance to cancer. Nature. 2014;510:407–411. doi: 10.1038/nature13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André F, Hurvitz S, Fasolo A, Tseng LM, Jerusalem G, Wilks S, O’Regan R, Isaacs C, Toi M, Burris H, et al. Molecular Alterations and Everolimus Efficacy in Human Epidermal Growth Factor Receptor 2-Overexpressing Metastatic Breast Cancers: Combined Exploratory Biomarker Analysis From BOLERO-1 and BOLERO-3. J Clin Oncol. 2016;34:2115–2124. doi: 10.1200/JCO.2015.63.9161. [DOI] [PubMed] [Google Scholar]
- Atefi M, Avramis E, Lassen A, Wong DJ, Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, et al. Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin Cancer Res. 2014;20:3446–3457. doi: 10.1158/1078-0432.CCR-13-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- Backer JM. The regulation of class IA PI 3-kinases by inter-subunit interactions. Curr Top Microbiol Immunol. 2010;346:87–114. doi: 10.1007/82_2010_52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem J. 2016;473:2251–2271. doi: 10.1042/BCJ20160170. [DOI] [PubMed] [Google Scholar]
- Bago R, Sommer E, Castel P, Crafter C, Bailey FP, Shpiro N, Baselga J, Cross D, Eyers PA, Alessi DR. The hVps34-SGK3 pathway alleviates sustained PI3K/Akt inhibition by stimulating mTORC1 and tumour growth. EMBO J. 2016;35:1902–1922. doi: 10.15252/embj.201693929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr PM, Saylors GB, Spurgeon SE, Cheson BD, Greenwald DR, O’Brien SM, Liem AKD, Mclntyre RE, Joshi A, Abella-Dominicis E, et al. Phase 2 study of idelalisib and entospletinib: pneumonitis limits combination therapy in relapsed refractory CLL and NHL. Blood. 2016;127:2411–2415. doi: 10.1182/blood-2015-12-683516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Campone M, Piccart M, Burris HA, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Im SA, Iwata H, Cortés J, De Laurentiis M, Jiang Z, Arteaga CL, Jonat W, Clemons M, Ito Y, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:904–916. doi: 10.1016/S1470-2045(17)30376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu B, Dean E, Puglisi M, Greystoke A, Ong M, Burke W, Cavallin M, Bigley G, Womack C, Harrington EA, et al. First-in-Human Pharmacokinetic and Pharmacodynamic Study of the Dual m-TORC 1/2 Inhibitor AZD2014. Clin Cancer Res. 2015;21:3412–3419. doi: 10.1158/1078-0432.CCR-14-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, Demanse D, De Buck SS, Ru QC, Peters M, et al. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2012;30:282–290. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- Bendell JC, Kelley RK, Shih KC, Grabowsky JA, Bergsland E, Jones S, Martin T, Infante JR, Mischel PS, Matsutani T, et al. A phase I dose-escalation study to assess safety, tolerability, pharmacokinetics, and preliminary efficacy of the dual mTORC1/mTORC2 kinase inhibitor CC-223 in patients with advanced solid tumors or multiple myeloma. Cancer. 2015a;121:3481–3490. doi: 10.1002/cncr.29422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell JC, Kurkjian C, Infante JR, Bauer TM, Burris HA, Greco FA, Shih KC, Thompson DS, Lane CM, Finney LH, et al. A phase 1 study of the sachet formulation of the oral dual PI3K/mTOR inhibitor BEZ235 given twice daily (BID) in patients with advanced solid tumors. Invest New Drugs. 2015b;33:463–471. doi: 10.1007/s10637-015-0218-6. [DOI] [PubMed] [Google Scholar]
- Bentley J, Itchayanan D, Barnes K, McIntosh E, Tang X, Downes CP, Holman GD, Whetton AD, Owen-Lynch PJ, Baldwin SA. Interleukin-3-mediated cell survival signals include phosphatidylinositol 3-kinase-dependent translocation of the glucose transporter GLUT1 to the cell surface. J Biol Chem. 2003;278:39337–39348. doi: 10.1074/jbc.M305689200. [DOI] [PubMed] [Google Scholar]
- Bihani T, Ezell SA, Ladd B, Grosskurth SE, Mazzola AM, Pietras M, Reimer C, Zinda M, Fawell S, D’Cruz CM. Resistance to everolimus driven by epigenetic regulation of MYC in ER+ breast cancers. Oncotarget. 2015;6:2407–2420. doi: 10.18632/oncotarget.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, Schmithorst VJ, Laor T, Brody AS, Bean J, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358:140–151. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissler JJ, Kingswood JC, Radzikowska E, Zonnenberg BA, Frost M, Belousova E, Sauter M, Nonomura N, Brakemeier S, de Vries PJ, et al. Everolimus for angiomyolipoma associated with tuberous sclerosis complex or sporadic lymphangioleiomyomatosis (EXIST-2): a multicentre, randomised, double-blind, placebo-controlled trial. The Lancet. 2013;381:817–824. doi: 10.1016/S0140-6736(12)61767-X. [DOI] [PubMed] [Google Scholar]
- Bohnacker T, Prota AE, Beaufils F, Burke JE, Melone A, Inglis AJ, Rageot D, Sele AM, Cmiljanovic V, Cmiljanovic N, et al. Deconvolution of Buparlisib’s mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention. Nat Commun. 2017;8:14683. doi: 10.1038/ncomms14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojarczuk K, Sasi BK, Gobessi S, Innocenti I, Pozzato G, Laurenti L, Efremov DG. BCR signaling inhibitors differ in their ability to overcome Mcl-1-mediated resistance of CLL B cells to ABT-199. Blood. 2016;127:3192–3201. doi: 10.1182/blood-2015-10-675009. [DOI] [PubMed] [Google Scholar]
- Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, Tao JJ, Spratt DE, Viola-Villegas NT, Castel P, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med. 2015;7:283ra51. doi: 10.1126/scitranslmed.aaa4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Müller U, Murakami M, Radimerski T, Bentires-Alj M. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell. 2012;22:796–811. doi: 10.1016/j.ccr.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Britten CD. PI3K and MEK inhibitor combinations: examining the evidence in selected tumor types. Cancer Chemother Pharmacol. 2013;71:1395–1409. doi: 10.1007/s00280-013-2121-1. [DOI] [PubMed] [Google Scholar]
- Brown KK, Toker A. The phosphoinositide 3-kinase pathway and therapy resistance in cancer. F1000Prime Rep. 2015;7:13. doi: 10.12703/P7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JE, Williams RL. Synergy in activating class I PI3Ks. Trends Biochem Sci. 2015;40:88–100. doi: 10.1016/j.tibs.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc Natl Acad Sci U S A. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamito M, Juntilla MM, Thomas M, Northrup DL, Rathmell J, Birnbaum MJ, Koretzky G, Allman D. Akt1 and Akt2 promote peripheral B-cell maturation and survival. Blood. 2010;115:4043–4050. doi: 10.1182/blood-2009-09-241638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC, Songyang Z. Specificity in recognition of phosphopeptides by src-homology 2 domains. J Cell Sci Suppl. 1994;18:121–126. doi: 10.1242/jcs.1994.supplement_18.18. [DOI] [PubMed] [Google Scholar]
- Cardnell RJ, Feng Y, Diao L, Fan YH, Masrorpour F, Wang J, Shen Y, Mills GB, Minna JD, Heymach JV, et al. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin Cancer Res. 2013;19:6322–6328. doi: 10.1158/1078-0432.CCR-13-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardnell RJ, Feng Y, Mukherjee S, Diao L, Tong P, Stewart CA, Masrorpour F, Fan Y, Nilsson M, Shen Y, et al. Activation of the PI3K/mTOR Pathway following PARP Inhibition in Small Cell Lung Cancer. PLoS ONE. 2016;11:e0152584. doi: 10.1371/journal.pone.0152584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlo MI, Molina AM, Lakhman Y, Patil S, Woo K, DeLuca J, Lee CH, Hsieh JJ, Feldman DR, Motzer RJ, et al. A Phase Ib Study of BEZ235, a Dual Inhibitor of Phosphatidylinositol 3-Kinase (PI3K) and Mammalian Target of Rapamycin (mTOR), in Patients With Advanced Renal Cell Carcinoma. Oncologist. 2016;21:787–788. doi: 10.1634/theoncologist.2016-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel P, Ellis H, Bago R, Toska E, Razavi P, Carmona FJ, Kannan S, Verma CS, Dickler M, Chandarlapaty S, et al. PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kα Inhibition. Cancer Cell. 2016;30:229–242. doi: 10.1016/j.ccell.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A, Sánchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci U S A. 2012;109:2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Aoki M, Fruman D, Auger KR, Bellacosa A, Tsichlis PN, Cantley LC, Roberts TM, Vogt PK. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- Cheah CY, Nastoupil LJ, Neelapu SS, Forbes SG, Oki Y, Fowler NH. Lenalidomide, idelalisib, and rituximab are unacceptably toxic in patients with relapsed/refractory indolent lymphoma. Blood. 2015;125:3357–3359. doi: 10.1182/blood-2015-03-633156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Limon JJ, Blanc C, Peng SL, Fruman DA. Foxo1 regulates marginal zone B-cell development. Eur J Immunol. 2010;40:1890–1896. doi: 10.1002/eji.200939817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Raybuck AL, Stengel K, Wei M, Beck TC, Volanakis E, Thomas JW, Hiebert S, Haase VH, Boothby MR. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature. 2016;537:234–238. doi: 10.1038/nature19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudasama KK, Winnay J, Johansson S, Claudi T, König R, Haldorsen I, Johansson B, Woo JR, Aarskog D, Sagen JV, et al. SHORT syndrome with partial lipodystrophy due to impaired phosphatidylinositol 3 kinase signaling. Am J Hum Genet. 2013;93:150–157. doi: 10.1016/j.ajhg.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M, Wang Q, Pighi C, Cheong TC, Meng FL, Poggio T, Yeap LS, Karaca E, Blasco RB, Langellotto F, et al. Phosphatidylinositol 3-kinase δ blockade increases genomic instability in B cells. Nature. 2017;542:489–493. doi: 10.1038/nature21406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condliffe AM, Davidson K, Anderson KE, Ellson CD, Crabbe T, Okkenhaug K, Vanhaesebroeck B, Turner M, Webb L, Wymann MP, et al. Sequential activation of class IB and class IA PI3K is important for the primed respiratory burst of human but not murine neutrophils. Blood. 2005;106:1432–1440. doi: 10.1182/blood-2005-03-0944. [DOI] [PubMed] [Google Scholar]
- Conley ME, Dobbs AK, Quintana AM, Bosompem A, Wang YD, Coustan-Smith E, Smith AM, Perez EE, Murray PJ. Agammaglobulinemia and absent B lineage cells in a patient lacking the p85α subunit of PI3K. J Exp Med. 2012;209:463–470. doi: 10.1084/jem.20112533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Rothenberg SM, Hata AN, Faber AC, Piris A, Nazarian RM, Brown RD, Godfrey JT, Winokur D, Walsh J, et al. TORC1 suppression predicts responsiveness to RAF and MEK inhibition in BRAF-mutant melanoma. Sci Transl Med. 2013;5:196ra98. doi: 10.1126/scitranslmed.3005753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa C, Ebi H, Martini M, Beausoleil SA, Faber AC, Jakubik CT, Huang A, Wang Y, Nishtala M, Hall B, et al. Measurement of PIP3 levels reveals an unexpected role for p110β in early adaptive responses to p110α-specific inhibitors in luminal breast cancer. Cancer Cell. 2015;27:97–108. doi: 10.1016/j.ccell.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sonenberg N. RAPping production of type I interferon in pDCs through mTOR. Nat Immunol. 2008;9:1097–1099. doi: 10.1038/ni1008-1097. [DOI] [PubMed] [Google Scholar]
- Coutré SE, Barrientos JC, Brown JR, de Vos S, Furman RR, Keating MJ, Li D, O’Brien SM, Pagel JM, Poleski MH, et al. Management of adverse events associated with idelalisib treatment: expert panel opinion. Leuk Lymphoma. 2015;56:2779–2786. doi: 10.3109/10428194.2015.1022770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoulakis G, Gambardella L, Rossman KL, Lawson CD, Anderson KE, Fukui Y, Welch HC, Der CJ, Stephens LR, Hawkins PT. P-Rex1 directly activates RhoG to regulate GPCR-driven Rac signalling and actin polarity in neutrophils. J Cell Sci. 2014;127:2589–2600. doi: 10.1242/jcs.153049. [DOI] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JA, Fruman DA. Phosphoinositide 3-kinase: diverse roles in immune cell activation. Annu Rev Immunol. 2004;22:563–598. doi: 10.1146/annurev.immunol.22.012703.104721. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickler MN, Saura C, Richards DA, Krop IE, Cervantes A, Bedard PL, Patel MR, Pusztai L, Oliveira M, Ware JA, et al. A phase II study of the PI3K inhibitor taselisib(GDC-0032) combined with fulvestrant (F) in patients (pts) with HER2-negative(HER2-), hormone receptor-positive (HR+) advanced breast cancer (BC) J Clin Oncol. 2016;34(suppl) doi: 10.1158/1078-0432.CCR-18-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Kung J, Holmes AB, Wells VA, Mo T, Basso K, Dalla-Favera R. The FOXO1 transcription factor instructs the germinal center dark zone program. Immunity. 2015;43:1064–1074. doi: 10.1016/j.immuni.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Donahue AC, Fruman DA. Distinct signaling mechanisms activate the target of rapamycin in response to different B-cell stimuli. Eur J Immunol. 2007;37:2923–2936. doi: 10.1002/eji.200737281. [DOI] [PubMed] [Google Scholar]
- Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan GL, Siempelkamp BD, Jenkins ML, Vadas O, Lucas CL, Burke JE. Conformational disruption of PI3Kδ regulation by immunodeficiency mutations in PIK3CD and PIK3R1. Proc Natl Acad Sci U S A. 2017;114:1982–1987. doi: 10.1073/pnas.1617244114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner M, Lučić I, Leonard TA, Yudushkin I. PI(3,4,5)P3 engagement restricts akt activity to cellular membranes. Mol Cell. 2017;65:416–431. e6. doi: 10.1016/j.molcel.2016.12.028. [DOI] [PubMed] [Google Scholar]
- Edgar KA, Wallin JJ, Berry M, Lee LB, Prior WW, Sampath D, Friedman LS, Belvin M. Isoform-specific phosphoinositide 3-kinase inhibitors exert distinct effects in solid tumors. Cancer Res. 2010;70:1164–1172. doi: 10.1158/0008-5472.CAN-09-2525. [DOI] [PubMed] [Google Scholar]
- Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, Tao J, Campos AB, Rodon J, Ibrahim YH, et al. mTORC1 inhibition is required for sensitivity to PI3K p110α inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med. 2013;5:196ra99. doi: 10.1126/scitranslmed.3005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Fabre S, Carrette F, Chen J, Lang V, Semichon M, Denoyelle C, Lazar V, Cagnard N, Dubart-Kupperschmitt A, Mangeney M, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181:2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- Falasca M, Maffucci T. Regulation and cellular functions of class II phosphoinositide 3-kinases. Biochem J. 2012;443:587–601. doi: 10.1042/BJ20120008. [DOI] [PubMed] [Google Scholar]
- Fan Q, Aksoy O, Wong RA, Ilkhanizadeh S, Novotny CJ, Gustafson WC, Truong AYQ, Cayanan G, Simonds EF, Haas-Kogan D, et al. A Kinase Inhibitor Targeted to mTORC1 Drives Regression in Glioblastoma. Cancer Cell. 2017;31:424–435. doi: 10.1016/j.ccell.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkona S, Diamandis EP, Blasutig IM. Cancer immunotherapy: the beginning of the end of cancer? BMC Med. 2016;14:73. doi: 10.1186/s12916-016-0623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazio N, Buzzoni R, Baudin E, Antonuzzo L, Hubner RA, Lahner H, DE Herder WW, Raderer M, Teulé A, Capdevila J, et al. A Phase II Study of BEZ235 in Patients with Everolimus-resistant, Advanced Pancreatic Neuroendocrine Tumours. Anticancer Res. 2016;36:713–719. [PMC free article] [PubMed] [Google Scholar]
- Feng DD, Cai W, Chen X. The associations between Parkinson’s disease and cancer: the plot thickens. Transl Neurodegener. 2015;4:20. doi: 10.1186/s40035-015-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- Foukas LC, Bilanges B, Bettedi L, Pearce W, Ali K, Sancho S, Withers DJ, Vanhaesebroeck B. Long-term p110α PI3K inactivation exerts a beneficial effect on metabolism. EMBO Mol Med. 2013;5:563–571. doi: 10.1002/emmm.201201953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco J, Witkiewicz AK, Knudsen ES. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget. 2014;5:6512–6525. doi: 10.18632/oncotarget.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz DN, Belousova E, Sparagana S, Bebin EM, Frost M, Kuperman R, Witt O, Kohrman MH, Flamini JR, Wu JY, et al. Everolimus for subependymal giant cell astrocytoma in patients with tuberous sclerosis complex: 2-year open-label extension of the randomised EXIST-1 study. Lancet Oncol. 2014;15:1513–1520. doi: 10.1016/S1470-2045(14)70489-9. [DOI] [PubMed] [Google Scholar]
- Friedman LS, Edgar KA, Song K, Schmidt S, Kirkpatrick DS, Phu L, Nannini MA, Hong R, Cheng E, Crocker L, et al. Abstract S6-04: The PI3K inhibitor, taselisib, has enhanced potency in PIK3CA mutant models through a unique mechanism of action. Cancer Res. 2017;77:S6-04–S6–04. [Google Scholar]
- Fritsch C, Huang A, Chatenay-Rivauday C, Schnell C, Reddy A, Liu M, Kauffmann A, Guthy D, Erdmann D, De Pover A, et al. Characterization of the novel and specific PI3Kα inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol Cancer Ther. 2014;13:1117–1129. doi: 10.1158/1535-7163.MCT-13-0865. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Cantley LC. Idelalisib — A PI3Kδ Inhibitor for B-Cell Cancers. N Engl J Med. 2014;370:1061–1062. doi: 10.1056/NEJMe1400055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman DA, Mauvais-Jarvis F, Pollard DA, Yballe CM, Brazil D, Bronson RT, Kahn CR, Cantley LC. Hypoglycaemia, liver necrosis and perinatal death in mice lacking all isoforms of phosphoinositide 3-kinase p85 alpha. Nat Genet. 2000a;26:379–382. doi: 10.1038/81715. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Satterthwaite AB, Witte ON. Xid-like phenotypes: a B cell signalosome takes shape. Immunity. 2000b;13:1–3. doi: 10.1016/s1074-7613(00)00002-9. [DOI] [PubMed] [Google Scholar]
- Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, Zelenetz AD, Kipps TJ, Flinn I, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370:997–1007. doi: 10.1056/NEJMoa1315226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher EJ, Fierz Y, Vijayakumar A, Haddad N, Yakar S, LeRoith D. Inhibiting PI3K reduces mammary tumor growth and induces hyperglycemia in a mouse model of insulin resistance and hyperinsulinemia. Oncogene. 2012;31:3213–3222. doi: 10.1038/onc.2011.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon A, Jasperson K, Champine M. Genetic basis of Cowden syndrome and its implications for clinical practice and risk management. Appl Clin Genet. 2016;9:83–92. doi: 10.2147/TACG.S41947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García C, Ibrahim YH, Serra V, Calvo MT, Guzmán M, Grueso J, Aura C, Pérez J, Jessen K, Liu Y, et al. Dual mTORC1/2 and HER2 blockade results in antitumor activity in preclinical models of breast cancer resistant to anti-HER2 therapy. Clin Cancer Res. 2012;18:2603–2612. doi: 10.1158/1078-0432.CCR-11-2750. [DOI] [PubMed] [Google Scholar]
- Garrett JT, Sutton CR, Kurupi R, Bialucha CU, Ettenberg SA, Collins SD, Sheng Q, Wallweber J, Defazio-Eli L, Arteaga CL. Combination of antibody that inhibits ligand-independent HER3 dimerization and a p110α inhibitor potently blocks PI3K signaling and growth of HER2+ breast cancers. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser JA, Inuzuka H, Lau AW, Wei W, Beroukhim R, Toker A. SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Mol Cell. 2014;56:595–607. doi: 10.1016/j.molcel.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghobrial IM, Siegel DS, Vij R, Berdeja JG, Richardson PG, Neuwirth R, Patel CG, Zohren F, Wolf JL. TAK-228 (formerly MLN0128), an investigational oral dual TORC1/2 inhibitor: A phase I dose escalation study in patients with relapsed or refractory multiple myeloma, non-Hodgkin lymphoma, or Waldenström’s macroglobulinemia. Am J Hematol. 2016;91:400–405. doi: 10.1002/ajh.24300. [DOI] [PubMed] [Google Scholar]
- González-Billalabeitia E, Seitzer N, Song SJ, Song MS, Patnaik A, Liu XS, Epping MT, Papa A, Hobbs RM, Chen M, et al. Vulnerabilities of PTEN-TP53-deficient prostate cancers to compound PARP-PI3K inhibition. Cancer Discov. 2014;4:896–904. doi: 10.1158/2159-8290.CD-13-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal AK, Kahl BS, de Vos S, Wagner-Johnston ND, Schuster SJ, Jurczak WJ, Flinn IW, Flowers CR, Martin P, Viardot A, et al. PI3Kδ inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med. 2014;370:1008–1018. doi: 10.1056/NEJMoa1314583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan PK, Gordillo-Villegas A, Zajac-Kaye M, Kaye FJ. Abstract 693: Inhibitory effect of the CDK4/6 inhibitor, PD 0332991, is enhanced by mTOR inhibition in Non-Small Cell Lung Cancer (NSCLC) Cancer Res. 2013;73:693–693. [Google Scholar]
- Guichard SM, Curwen J, Bihani T, D’Cruz CM, Yates JWT, Grondine M, Howard Z, Davies BR, Bigley G, Klinowska T, et al. AZD2014, an Inhibitor of mTORC1 and mTORC2, Is Highly Effective in ER+ Breast Cancer When Administered Using Intermittent or Continuous Schedules. Mol Cancer Ther. 2015;14:2508–2518. doi: 10.1158/1535-7163.MCT-15-0365. [DOI] [PubMed] [Google Scholar]
- Gunderson AJ, Kaneda MM, Tsujikawa T, Nguyen AV, Affara NI, Ruffell B, Gorjestani S, Liudahl SM, Truitt M, Olson P, et al. Bruton Tyrosine Kinase-Dependent Immune Cell Cross-talk Drives Pancreas Cancer. Cancer Discov. 2016;6:270–285. doi: 10.1158/2159-8290.CD-15-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Anjomani-Virmouni S, Koundouros N, Dimitriadi M, Choo-Wing R, Valle A, Zheng Y, Chiu YH, Agnihotri S, Zadeh G, et al. PARK2 Depletion Connects Energy and Oxidative Stress to PI3K/Akt Activation via PTEN S-Nitrosylation. Mol Cell. 2017;65:999–1013. e7. doi: 10.1016/j.molcel.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbst K, Lauss M, Cirenajwis H, Isaksson K, Rosengren F, Törngren T, Kvist A, Johansson MC, Vallon-Christersson J, Baldetorp B, et al. Multiregion Whole-Exome Sequencing Uncovers the Genetic Evolution and Mutational Heterogeneity of Early-Stage Metastatic Melanoma. Cancer Res. 2016;76:4765–4774. doi: 10.1158/0008-5472.CAN-15-3476. [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Stephens LR. PI3K signalling in inflammation. Biochim Biophys Acta. 2015;1851:882–897. doi: 10.1016/j.bbalip.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Stephens LR. Emerging evidence of signalling roles for PI(3,4)P2 in Class I and II PI3K-regulated pathways. Biochem Soc Trans. 2016;44:307–314. doi: 10.1042/BST20150248. [DOI] [PubMed] [Google Scholar]
- Hay N. Interplay between FOXO, TOR, and Akt. Biochim Biophys Acta. 2011;1813:1965–1970. doi: 10.1016/j.bbamcr.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH, Budhu S, Ghosh A, Pink M, Tchaicha J, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature. 2016;539:443–447. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henske EP, McCormack FX. Lymphangioleiomyomatosis - a wolf in sheep’s clothing. J Clin Invest. 2012;122:3807–3816. doi: 10.1172/JCI58709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman SEM, Niemann CU, Farooqui M, Jones J, Mustafa RZ, Lipsky A, Saba N, Martyr S, Soto S, Valdez J, et al. Ibrutinib-induced lymphocytosis in patients with chronic lymphocytic leukemia: correlative analyses from a phase II study. Leukemia. 2014;28:2188–2196. doi: 10.1038/leu.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Abreu MT, Palafox M, Asghar U, Rivas MA, Cutts RJ, Garcia-Murillas I, Pearson A, Guzman M, Rodriguez O, Grueso J, et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res. 2016;76:2301–2313. doi: 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM. Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med. 2013;210:1189–1200. doi: 10.1084/jem.20130392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurtier L, Lamrini H, Chentout L, Deau M-C, Bouafia A, Rosain J, Plaza J-M, Parisot M, Dumont B, Turpin D, et al. Mutations in the adaptor-binding domain and associated linker region of p110δ cause Activated PI3K-δ Syndrome 1 (APDS1) Haematologica. 2017 doi: 10.3324/haematol.2017.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BD, Goncalves MD, Cantley LC. Obesity and cancer mechanisms: cancer metabolism. J Clin Oncol. 2016;34:4277–4283. doi: 10.1200/JCO.2016.67.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosford SR, Dillon LM, Bouley SJ, Rosati R, Yang W, Chen VS, Demidenko E, Morra RP, Miller TW. Combined Inhibition of Both p110α and p110β Isoforms of Phosphatidylinositol 3-Kinase Is Required for Sustained Therapeutic Effect in PTEN-Deficient, ER(+) Breast Cancer. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houslay DM, Anderson KE, Chessa T, Kulkarni S, Fritsch R, Downward J, Backer JM, Stephens LR, Hawkins PT. Coincident signals from GPCRs and receptor tyrosine kinases are uniquely transduced by PI3Kβ in myeloid cells. Sci Signal. 2016;9:ra82. doi: 10.1126/scisignal.aae0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P, Santner-Nanan B, Hu M, Skarratt K, Lee CH, Stormon M, Wong M, Fuller SJ, Nanan R. IL-10 Potentiates Differentiation of Human Induced Regulatory T Cells via STAT3 and Foxo1. J Immunol. 2015;195:3665–3674. doi: 10.4049/jimmunol.1402898. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Juvekar A, Lyssiotis CA, Lien EC, Albeck JG, Oh D, Varma G, Hung YP, Ullas S, Lauring J, et al. Phosphoinositide 3-Kinase Regulates Glycolysis through Mobilization of Aldolase from the Actin Cytoskeleton. Cell. 2016;164:433–446. doi: 10.1016/j.cell.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Czech MP. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Hudson K, Hancox UJ, Trigwell C, McEwen R, Polanska UM, Nikolaou M, Morentin Gutierrez P, Avivar-Valderas A, Delpuech O, Dudley P, et al. Intermittent High-Dose Scheduling of AZD8835, a Novel Selective Inhibitor of PI3Kα and PI3Kδ, Demonstrates Treatment Strategies for PIK3CA-Dependent Breast Cancers. Mol Cancer Ther. 2016;15:877–889. doi: 10.1158/1535-7163.MCT-15-0687. [DOI] [PubMed] [Google Scholar]
- Hyman DM, Smyth L, Bedard PL, Oza A, Dean E, Armstrong A, Lima J, Bando H, Kabos P, Perez-Fidalgo JA, et al. Abstract B109: AZD5363, a catalytic pan-Akt inhibitor, inAkt1 E17K mutation positive advanced solid tumors. Mol Cancer Ther. 2015;14:B109–B109. [Google Scholar]
- Ibrahim YH, García-García C, Serra V, He L, Torres-Lockhart K, Prat A, Anton P, Cozar P, Guzmán M, Grueso J, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2:1036–1047. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan E, Manning BD. Emerging role of mTOR in the response to cancer therapeutics. Trends in Cancer. 2016;2:241–251. doi: 10.1016/j.trecan.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A. 2011;108:E699–E708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, Pirun M, Sander C, Socci ND, Ostrovnaya I, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, Fu S, Piha-Paul SA, Lee JJ, Luthra R, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73:276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen VM, Bhola NE, Bauer JA, Formisano L, Lee K-M, Hutchinson KE, Witkiewicz AK, Moore PD, Estrada MV, Sánchez V, et al. Kinome-Wide RNA Interference Screen Reveals a Role for PDK1 in Acquired Resistance to CDK4/6 Inhibition in ER-Positive Breast Cancer. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-16-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellusova J, Rickert RC. The PI3K pathway in B cell metabolism. Crit Rev Biochem Mol Biol. 2016;51:359–378. doi: 10.1080/10409238.2016.1215288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellusova J, Cato MH, Apgar JR, Ramezani-Rad P, Leung CR, Chen C, Richardson AD, Conner EM, Benschop RJ, Woodgett JR, et al. Gsk3 is a metabolic checkpoint regulator in B cells. Nat Immunol. 2017;18:303–312. doi: 10.1038/ni.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JóŸwiak S, Sadowski K, Kotulska K, Schwartz RA. Topical Use of Mammalian Target of Rapamycin (mTOR) Inhibitors in Tuberous Sclerosis Complex-A Comprehensive Review of the Literature. Pediatr Neurol. 2016;61:21–27. doi: 10.1016/j.pediatrneurol.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature. 2015a;518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D, De Bono JS, LoRusso P, Nemunaitis JJ, Heath EI, Kwak EL, Macarulla T, Geuna E, de Miguel Luken MJ, Patel C, et al. First-inhuman, phase I, dose-escalation study of selective PI3Kα isoform inhibitor MLN1117 in patients (pts) with advanced solid malignancies. J Clin Oncol. 2015b;33(suppl) doi: 10.1158/1078-0432.CCR-16-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D, Krop I, Ramanathan RK, Wilson TR, Ware JA, Sanabria Bohorquez S, Savage H, Sampath D, Salphati L, Lin R, et al. Phase I Dose Escalation Study of Taselisib (GDC-0032), an Oral PI3K Inhibitor, in Patients with Advanced Solid Tumors. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-16-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmañà J, Rajendran A, Papa A, Spencer K, Lyssiotis CA, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2:1048–1063. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, Woo G, Nguyen AV, Figueiredo CC, Foubert P, et al. PI3Kγ is a molecular switch that controls immune suppression. Nature. 2016a;539:437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda MM, Cappello P, Nguyen AV, Ralainirina N, Hardamon CR, Foubert P, Schmid MC, Sun P, Mose E, Bouvet M, et al. Macrophage pi3kγ drives pancreatic ductal adenocarcinoma progression. Cancer Discov. 2016b;6:870–885. doi: 10.1158/2159-8290.CD-15-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch’en IL, Stockmann C, Katayama CD, Hedrick SM. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharas MG, Janes MR, Scarfone VM, Lilly MB, Knight ZA, Shokat KM, Fruman DA. Ablation of PI3K blocks BCR-ABL leukemogenesis in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells. J Clin Invest. 2008;118:3038–3050. doi: 10.1172/JCI33337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger DA, Wilfong AA, Mays M, Talley CM, Agricola K, Tudor C, Capal J, Holland-Bouley K, Franz DN. Long-term treatment of epilepsy with everolimus in tuberous sclerosis. Neurology. 2016;87:2408–2415. doi: 10.1212/WNL.0000000000003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku BM, Yi SY, Koh J, Bae YH, Sun JM, Lee SH, Ahn JS, Park K, Ahn MJ. The CDK4/6 inhibitor LY2835219 has potent activity in combination with mTOR inhibitor in head and neck squamous cell carcinoma. Oncotarget. 2016;7:14803–14813. doi: 10.18632/oncotarget.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Sitaru C, Jakus Z, Anderson KE, Damoulakis G, Davidson K, Hirose M, Juss J, Oxley D, Chessa TA, et al. PI3Kβ plays a critical role in neutrophil activation by immune complexes. Sci Signal. 2011;4:ra23. doi: 10.1126/scisignal.2001617. [DOI] [PubMed] [Google Scholar]
- Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, et al. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ. Cell Rep. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Kurek KC, Luks VL, Ayturk UM, Alomari AI, Fishman SJ, Spencer SA, Mulliken JB, Bowen ME, Yamamoto GL, Kozakewich HPW, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O’Brien S, Gabbasov R, Fink LS, Li Y, Lounsbury N, et al. Resistance to BET bromodomain inhibitors is mediated by kinome reprogramming in ovarian cancer. Cell Rep. 2016;16:1273–1286. doi: 10.1016/j.celrep.2016.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HC, Nijmeh J, Henske EP. New developments in the genetics and pathogenesis of tumours in tuberous sclerosis complex. J Pathol. 2017;241:219–225. doi: 10.1002/path.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson BL, Kasar SN, Matos TR, Morgan EA, Rassenti L, Davids MS, Fisher DC, Freedman AS, Jacobson CA, Armand P, et al. Idelalisib given front-line for treatment of chronic lymphocytic leukemia causes frequent immune-mediated hepatotoxicity. Blood. 2016;128:195–203. doi: 10.1182/blood-2016-03-707133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastwika KJ, Wilson W, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le X, Antony R, Razavi P, Treacy DJ, Luo F, Ghandi M, Castel P, Scaltriti M, Baselga J, Garraway LA. Systematic functional characterization of resistance to PI3K inhibition in breast cancer. Cancer Discov. 2016;6:1134–1147. doi: 10.1158/2159-8290.CD-16-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leo A, Seok Lee K, Ciruelos E, Lønning P, Janni W, O’Regan R, Mouret Reynier MA, Kalev D, Egle D, Csoszi T, et al. Abstract S4-07: BELLE-3: A phase III study of buparlisib + fulvestrant in postmenopausal women with HR+, HER2–, aromatase inhibitor-treated, locally advanced or metastatic breast cancer, who progressed on or after mTOR inhibitor-based treatment. Cancer Res. 2017;77:S4-S07–S4–S07. [Google Scholar]
- Lien EC, Lyssiotis CA, Cantley LC. Metabolic Reprogramming by the PI3K-Akt-mTOR Pathway in Cancer. Recent Results Cancer Res. 2016;207:39–72. doi: 10.1007/978-3-319-42118-6_3. [DOI] [PubMed] [Google Scholar]
- Lim SM, Park HS, Kim S, Kim S, Ali SM, Greenbowe JR, Yang IS, Kwon NJ, Lee JL, Ryu MH, et al. Next-generation sequencing reveals somatic mutations that confer exceptional response to everolimus. Oncotarget. 2016;7:10547–10556. doi: 10.18632/oncotarget.7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, Fruman DA. Akt and mTOR in B Cell Activation and Differentiation. Front Immunol. 2012;3:228. doi: 10.3389/fimmu.2012.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, So L, Jellbauer S, Chiu H, Corado J, Sykes SM, Raffatellu M, Fruman DA. mTOR kinase inhibitors promote antibody class switching via mTORC2 inhibition. Proc Natl Acad Sci U S A. 2014;111:E5076–E5085. doi: 10.1073/pnas.1407104111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Piao HL, Zhuang L, Sarbassov DD, Ma L, Gan B. FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacologic inhibition of the PI3K-AKT pathway. Cancer Res. 2014;74:1682–1693. doi: 10.1158/0008-5472.CAN-13-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Adams WC, Nish SA, Chen YH, Yen B, Rothman NJ, Kratchmarov R, Okada T, Klein U, Reiner SL. Asymmetric PI3K signaling driving developmental and regenerative cell fate bifurcation. Cell Rep. 2015;13:2203–2218. doi: 10.1016/j.celrep.2015.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke M, Pham HT, Katholnig K, Schnöller T, Miller A, Demel F, Schütz B, Rosner M, Kovacic B, Sukhbaatar N, et al. Chronic signaling via the metabolic checkpoint kinase mTORC1 induces macrophage granuloma formation and marks sarcoidosis progression. Nat Immunol. 2017;18:293–302. doi: 10.1038/ni.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Radisky DC, Yang D, Xu R, Radisky ES, Bissell MJ, Bishop JM. MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat Cell Biol. 2012;14:567–574. doi: 10.1038/ncb2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Rowley BR, Bull CO, Schneider C, Haegebarth A, Schatz CA, Fracasso PR, Wilkie DP, Hentemann M, Wilhelm SM, et al. BAY 80-6946 is a highly selective intravenous PI3K inhibitor with potent p110α and p110δ activities in tumor cell lines and xenograft models. Mol Cancer Ther. 2013;12:2319–2330. doi: 10.1158/1535-7163.MCT-12-0993-T. [DOI] [PubMed] [Google Scholar]
- Liu P, Gan W, Chin YR, Ogura K, Guo J, Zhang J, Wang B, Blenis J, Cantley LC, Toker A, et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer Discov. 2015;5:1194–1209. doi: 10.1158/2159-8290.CD-15-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loconte DC, Grossi V, Bozzao C, Forte G, Bagnulo R, Stella A, Lastella P, Cutrone M, Benedicenti F, Susca FC, et al. Molecular and Functional Characterization of Three Different Postzygotic Mutations in PIK3CA-Related Overgrowth Spectrum (PROS) Patients: Effects on PI3K/AKT/mTOR Signaling and Sensitivity to PIK3 Inhibitors. PLoS ONE. 2015;10:e0123092. doi: 10.1371/journal.pone.0123092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- Loron MC, Grange S, Guerrot D, Di Fiore F, Freguin C, Hanoy M, Le Roy F, Poussard G, Etienne I, Legallicier B, et al. Pneumocystis jirovecii pneumonia in everolimus-treated renal cell carcinoma. J Clin Oncol. 2015;33:e45–e47. doi: 10.1200/JCO.2013.49.9277. [DOI] [PubMed] [Google Scholar]
- LoRusso P, Shapiro G, Pandya SS, Kwak EL, Jones C, Belvin M, Musib LC, de Crespigny A, McKenzie M, Gates MR, et al. A first-in-human phase Ib study to evaluate the MEK inhibitor GDC-0973, combined with the pan-PI3K inhibitor GDC-0941, in patients with advanced solid tumors. J Clin Oncol. 2012;30(suppl) [Google Scholar]
- Low PC, Manzanero S, Mohannak N, Narayana VK, Nguyen TH, Kvaskoff D, Brennan FH, Ruitenberg MJ, Gelderblom M, Magnus T, et al. PI3Kδ inhibition reduces TNF secretion and neuroinflammation in a mouse cerebral stroke model. Nat Commun. 2014;5:3450. doi: 10.1038/ncomms4450. [DOI] [PubMed] [Google Scholar]
- Lucas CL, Chandra A, Nejentsev S, Condliffe AM, Okkenhaug K. PI3Kδ and primary immunodeficiencies. Nat Rev Immunol. 2016;16:702–714. doi: 10.1038/nri.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo CT, Liao W, Dadi S, Toure A, Li MO. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. 2016;529:532–536. doi: 10.1038/nature16486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Field SJ, Lee JY, Engelman JA, Cantley LC. The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex. J Cell Biol. 2005a;170:455–464. doi: 10.1083/jcb.200503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sobkiw CL, Logsdon NM, Watt JM, Signoretti S, O’Connell F, Shin E, Shim Y, Pao L, Neel BG, et al. Modulation of epithelial neoplasia and lymphoid hyperplasia in PTEN+/- mice by the p85 regulatory subunits of phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2005b;102:10238–10243. doi: 10.1073/pnas.0504378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sobkiw CL, Hirshman MF, Logsdon MN, Li TQ, Goodyear LJ, Cantley LC. Loss of class IA PI3K signaling in muscle leads to impaired muscle growth, insulin response, and hyperlipidemia. Cell Metab. 2006;3:355–366. doi: 10.1016/j.cmet.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Ma CX, Luo J, Naughton M, Ademuyiwa F, Suresh R, Griffith M, Griffith OL, Skidmore ZL, Spies NC, Ramu A, et al. A Phase I Trial of BKM120 (Buparlisib) in Combination with Fulvestrant in Postmenopausal Women with Estrogen Receptor-Positive Metastatic Breast Cancer. Clin Cancer Res. 2016;22:1583–1591. doi: 10.1158/1078-0432.CCR-15-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, Tamas P, Feijoo C, Okkenhaug K, Cantrell DA. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- Malka-Mahieu H, Newman M, Désaubry L, Robert C, Vagner S. Molecular Pathways: The eIF4F Translation Initiation Complex-New Opportunities for Cancer Treatment. Clin Cancer Res. 2017;23:21–25. doi: 10.1158/1078-0432.CCR-14-2362. [DOI] [PubMed] [Google Scholar]
- Mallon R, Feldberg LR, Lucas J, Chaudhary I, Dehnhardt C, Santos ED, Chen Z, dos Santos O, Ayral-Kaloustian S, Venkatesan A, et al. Antitumor efficacy of PKI-587, a highly potent dual PI3K/mTOR kinase inhibitor. Clin Cancer Res. 2011;17:3193–3203. doi: 10.1158/1078-0432.CCR-10-1694. [DOI] [PubMed] [Google Scholar]
- Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017 doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, Virtanen C, Bradner JE, Bader GD, Mills GB, et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NA, Galvin KC, Corcoran AM, Boon L, Higgs R, Mills KH. Immunotherapy with PI3K inhibitor and Toll-like receptor agonist induces IFN-γ+IL-17+ polyfunctional T cells that mediate rejection of murine tumors. Cancer Res. 2012;72:581–591. doi: 10.1158/0008-5472.CAN-11-0307. [DOI] [PubMed] [Google Scholar]
- Marwick JA, Caramori G, Casolari P, Mazzoni F, Kirkham PA, Adcock IM, Chung KF, Papi A. A role for phosphoinositol 3-kinase delta in the impairment of glucocorticoid responsiveness in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2010;125:1146–1153. doi: 10.1016/j.jaci.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Matulonis UA, Wulf GM, Barry WT, Birrer M, Westin SN, Farooq S, Bell-McGuinn KM, Obermayer E, Whalen C, Spagnoletti T, et al. Phase I dose escalation study of the PI3kinase pathway inhibitor BKM120 and the oral poly (ADP ribose) polymerase (PARP) inhibitor olaparib for the treatment of high grade serous ovarian and breast cancer. Ann Oncol. 2016 doi: 10.1093/annonc/mdw672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer IA, Abramson VG, Formisano L, Balko JM, Estrada MV, Sanders ME, Juric D, Solit D, Berger MF, Won HH, et al. A Phase Ib Study of Alpelisib (BYL719), a PI3Kα-Specific Inhibitor, with Letrozole in ER+/HER2- Metastatic Breast Cancer. Clin Cancer Res. 2017;23:26–34. doi: 10.1158/1078-0432.CCR-16-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack FX, Inoue Y, Moss J, Singer LG, Strange C, Nakata K, Barker AF, Chapman JT, Brantly ML, Stocks JM, et al. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N Engl J Med. 2011;364:1595–1606. doi: 10.1056/NEJMoa1100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7:283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalarea V, Roda D, Drew Y, Carreira S, O’Carrigan BS, Shaw H, Roux R, Kumar S, Ward S, Parmar M, et al. Abstract CT010: Phase I trial combining the PARP inhibitor olaparib (Ola) and AKT inhibitor AZD5363 (AZD) in germline (g)BRCA and non-BRCA mutant (m) advanced cancer patients (pts) incorporating noninvasive monitoring of cancer mutations. Cancer Res. 2016;76:CT010–CT010. [Google Scholar]
- Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- Miller AT, Wilcox HM, Lai Z, Berg LJ. Signaling through Itk promotes T helper 2 differentiation via negative regulation of T-bet. Immunity. 2004;21:67–80. doi: 10.1016/j.immuni.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Mizrachi A, Shamay Y, Shah J, Brook S, Soong J, Rajasekhar VK, Humm JL, Healey JH, Powell SN, Baselga J, et al. Tumour-specific PI3K inhibition via nanoparticle-targeted delivery in head and neck squamous cell carcinoma. Nat Commun. 2017;8:14292. doi: 10.1038/ncomms14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz LS, Surinova S, Ghazaly E, Velasco LG, Haider S, Rodríguez-Prados JC, Berenjeno IM, Chelala C, Vanhaesebroeck B. Phosphoproteomic comparison of Pik3ca and Pten signalling identifies the nucleotidase NT5C as a novel AKT substrate. Sci Rep. 2017;7:39985. doi: 10.1038/srep39985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Munoz J, Kurzrock R. Targeted therapy in rare cancers--adopting the orphans. Nat Rev Clin Oncol. 2012;9:631–642. doi: 10.1038/nrclinonc.2012.160. [DOI] [PubMed] [Google Scholar]
- Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y, Walter K, Spoerke JM, O’Brien C, Huw LY, Hampton GM, Lackner MR. Activating Mutations in PIK3CB Confer Resistance to PI3K Inhibition and Define a Novel Oncogenic Role for p110β. Cancer Res. 2016;76:1193–1203. doi: 10.1158/0008-5472.CAN-15-2201. [DOI] [PubMed] [Google Scholar]
- Nashed BF, Zhang T, Al-Alwan M, Srinivasan G, Halayko AJ, Okkenhaug K, Vanhaesebroeck B, Hayglass KT, Marshall AJ. Role of the phosphoinositide 3-kinase p110delta in generation of type 2 cytokine responses and allergic airway inflammation. Eur J Immunol. 2007;37:416–424. doi: 10.1002/eji.200636401. [DOI] [PubMed] [Google Scholar]
- Nathan N, Keppler-Noreuil KM, Biesecker LG, Moss J, Darling TN. Mosaic disorders of the pi3k/pten/akt/tsc/mtorc1 signaling pathway. Dermatol Clin. 2017;35:51–60. doi: 10.1016/j.det.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Liu Q, Xie S, Carlson C, Von T, Vogel K, Riddle S, Benes C, Eck M, Roberts T, et al. Functional characterization of an isoform-selective inhibitor of PI3K-p110β as a potential anticancer agent. Cancer Discov. 2012;2:425–433. doi: 10.1158/2159-8290.CD-12-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nish SA, Zens KD, Kratchmarov R, Lin WW, Adams WC, Chen YH, Yen B, Rothman NJ, Bhandoola A, Xue HH, et al. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J Exp Med. 2017;214:39–47. doi: 10.1084/jem.20161046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003;3:317–330. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- Okkenhaug K, Patton DT, Bilancio A, Garçon F, Rowan WC, Vanhaesebroeck B. The p110delta isoform of phosphoinositide 3-kinase controls clonal expansion and differentiation of Th cells. J Immunol. 2006;177:5122–5128. doi: 10.4049/jimmunol.177.8.5122. [DOI] [PubMed] [Google Scholar]
- Okkenhaug K, Graupera M, Vanhaesebroeck B. Targeting PI3K in cancer: impact on tumor cells, their protective stroma, angiogenesis, and immunotherapy. Cancer Discov. 2016;6:1090–1105. doi: 10.1158/2159-8290.CD-16-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori SA, Cato MH, Anzelon-Mills A, Puri KD, Shapiro-Shelef M, Calame K, Rickert RC. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 2006;25:545–557. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- Pai C, Walsh CM, Fruman DA. Context-Specific Function of S6K2 in Th Cell Differentiation. J Immunol. 2016;197:3049–3058. doi: 10.4049/jimmunol.1600167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Finkel T. Key Proteins and Pathways that Regulate Lifespan. J Biol Chem. 2017 doi: 10.1074/jbc.R116.771915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DT, Garden OA, Pearce WP, Clough LE, Monk CR, Leung E, Rowan WC, Sancho S, Walker LS, Vanhaesebroeck B, et al. Cutting edge: the phosphoinositide 3-kinase p110 delta is critical for the function of CD4+CD25+Foxp3+ regulatory T cells. J Immunol. 2006;177:6598–6602. doi: 10.4049/jimmunol.177.10.6598. [DOI] [PubMed] [Google Scholar]
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, Delgoffe GM, Powell JD. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J Clin Invest. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi KN, Sun IH, Patel CH, Lo YC, Oh MH, Waickman AT, Tam AJ, Blosser RL, Wen J, Delgoffe GM, et al. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8(+) T cell differentiation. Nat Immunol. 2016;17:704–711. doi: 10.1038/ni.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles T, Lackner MR, Oudard S, Escudier B, Ralph C, Brown JE, Hawkins RE, Castellano D, Rini BI, Staehler MD, et al. Randomized Open-Label Phase II Trial of Apitolisib (GDC-0980), a Novel Inhibitor of the PI3K/Mammalian Target of Rapamycin Pathway, Versus Everolimus in Patients With Metastatic Renal Cell Carcinoma. J Clin Oncol. 2016;34:1660–1668. doi: 10.1200/JCO.2015.64.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko I, Poon W, Mägi R, Prasad BR, Salehi SA, Almgren P, Osmark P, Bouatia-Naji N, Wierup N, Fall T, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10:e1004235. doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragon BK, Kantarjian H, Jabbour E, Ravandi F, Cortes J, Borthakur G, DeBose L, Zeng Z, Schneider H, Pemmaraju N, et al. Buparlisib, a PI3K inhibitor, demonstrates acceptable tolerability and preliminary activity in a phase I trial of patients with advanced leukemias. Am J Hematol. 2017;92:7–11. doi: 10.1002/ajh.24568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raïch-Regué D, Fabian KP, Watson AR, Fecek RJ, Storkus WJ, Thomson AW. Intratumoral delivery of mTORC2-deficient dendritic cells inhibits B16 melanoma growth by promoting CD8(+) effector T cell responses. Oncoimmunology. 2016;5:e1146841. doi: 10.1080/2162402X.2016.1146841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadani F, Bolland DJ, Garcon F, Emery JL, Vanhaesebroeck B, Corcoran AE, Okkenhaug K. The PI3K isoforms p110alpha and p110delta are essential for pre-B cell receptor signaling and B cell development. Sci Signal. 2010;3:ra60. doi: 10.1126/scisignal.2001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol. 2003;23:7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman FL, Lord CJ, Ashworth A. The promise of combining inhibition of PI3K and PARP as cancer therapy. Cancer Discov. 2012;2:982–984. doi: 10.1158/2159-8290.CD-12-0433. [DOI] [PubMed] [Google Scholar]
- Reiner SL, Adams WC. Lymphocyte fate specification as a deterministic but highly plastic process. Nat Rev Immunol. 2014;14:699–704. doi: 10.1038/nri3734. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Tan-Sah VP, Smith JM, Miyamoto S. Akt phosphorylates HK-II at Thr-473 and increases mitochondrial HK-II association to protect cardiomyocytes. J Biol Chem. 2013;288:23798–23806. doi: 10.1074/jbc.M113.482026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de Stanchina E, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534:272–276. doi: 10.1038/nature17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolf J, Bell SE, Kovesdi D, Janas ML, Soond DR, Webb LM, Santinelli S, Saunders T, Hebeis B, Killeen N, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185:4042–4052. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- Ross SH, Rollings C, Anderson KE, Hawkins PT, Stephens LR, Cantrell DA. Phosphoproteomic Analyses of Interleukin 2 Signaling Reveal Integrated JAK Kinase-Dependent and -Independent Networks in CD8(+) T Cells. Immunity. 2016;45:685–700. doi: 10.1016/j.immuni.2016.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückle T, Schwarz MK, Rommel C. PI3Kgamma inhibition: towards an “aspirin of the 21st century”? Nat Rev Drug Discov. 2006;5:903–918. doi: 10.1038/nrd2145. [DOI] [PubMed] [Google Scholar]
- Saito K, Tolias KF, Saci A, Koon HB, Humphries LA, Scharenberg A, Rawlings DJ, Kinet JP, Carpenter CL. BTK regulates PtdIns-4,5-P2 synthesis: importance for calcium signaling and PI3K activity. Immunity. 2003;19:669–678. doi: 10.1016/s1074-7613(03)00297-8. [DOI] [PubMed] [Google Scholar]
- Salmond RJ, Emery J, Okkenhaug K, Zamoyska R. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8 T cells. J Immunol. 2009;183:7388–7397. doi: 10.4049/jimmunol.0902294. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sander S, Chu VT, Yasuda T, Franklin A, Graf R, Calado DP, Li S, Imami K, Selbach M, Di Virgilio M, et al. PI3 kinase and FOXO1 transcription factor activity differentially control B cells in the germinal center light and dark zones. Immunity. 2015;43:1075–1086. doi: 10.1016/j.immuni.2015.10.021. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sasaki CY, Chen G, Munk R, Eitan E, Martindale J, Longo DL, Ghosh P. p(70S6K1) in the TORC1 pathway is essential for the differentiation of Th17 Cells, but not Th1, Th2, or Treg cells in mice. Eur J Immunol. 2016;46:212–222. doi: 10.1002/eji.201445422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MC, Avraamides CJ, Dippold HC, Franco I, Foubert P, Ellies LG, Acevedo LM, Manglicmot JR, Song X, Wrasidlo W, et al. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kγ, a single convergent point promoting tumor inflammation and progression. Cancer Cell. 2011;19:715–727. doi: 10.1016/j.ccr.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit F, Utermark T, Zhang S, Wang Q, Von T, Roberts TM, Zhao JJ. PI3K isoform dependence of PTEN-deficient tumors can be altered by the genetic context. Proc Natl Acad Sci U S A. 2014;111:6395–6400. doi: 10.1073/pnas.1323004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V, Will M, Yellen P, de Stanchina E, Baselga J, et al. Feedback suppression of PI3Kα signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kβ. Cancer Cell. 2015;27:109–122. doi: 10.1016/j.ccell.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Tullet JMA, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Bartek J. Cell Cycle–Targeted Cancer Therapies. Annual Review of Cancer Biology. 2017;1:41–57. [Google Scholar]
- Shimizu T, Tolcher AW, Papadopoulos KP, Beeram M, Rasco DW, Smith LS, Gunn S, Smetzer L, Mays TA, Kaiser B, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin Cancer Res. 2012;18:2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- Shojaee S, Chan LN, Buchner M, Cazzaniga V, Cosgun KN, Geng H, Qiu YH, von Minden MD, Ernst T, Hochhaus A, et al. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat Med. 2016;22:379–387. doi: 10.1038/nm.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So L, Lee J, Palafox M, Mallya S, Woxland CG, Arguello M, Truitt ML, Sonenberg N, Ruggero D, Fruman DA. The 4E-BP-eIF4E axis promotes rapamycin-sensitive growth and proliferation in lymphocytes. Sci Signal. 2016;9:ra57. doi: 10.1126/scisignal.aad8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soond DR, Bjørgo E, Moltu K, Dale VQ, Patton DT, Torgersen KM, Galleway F, Twomey B, Clark J, Gaston JS, et al. PI3K p110delta regulates T-cell cytokine production during primary and secondary immune responses in mice and humans. Blood. 2010;115:2203–2213. doi: 10.1182/blood-2009-07-232330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soond DR, Slack EC, Garden OA, Patton DT, Okkenhaug K. Does the PI3K pathway promote or antagonize regulatory T cell development and function? Front Immunol. 2012;3:244. doi: 10.3389/fimmu.2012.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatkin C, Ratermann KL, Overley CW, Black EP. Inhibition of class IA PI3K enzymes in non-small cell lung cancer cells uncovers functional compensation among isoforms. Cancer Biol Ther. 2015;16:1341–1352. doi: 10.1080/15384047.2015.1070986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EL, Pepper M, Katayama CD, Kerdiles YM, Lai CY, Emslie E, Lin YC, Yang E, Goldrath AW, Li MO, et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. 2015;42:239–251. doi: 10.1016/j.immuni.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratikopoulos EE, Dendy M, Szabolcs M, Khaykin AJ, Lefebvre C, Zhou MM, Parsons R. Kinase and BET inhibitors together clamp inhibition of PI3K signaling and overcome resistance to therapy. Cancer Cell. 2015;27:837–851. doi: 10.1016/j.ccell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Whitman M, Cantley LC, Erikson RL. Evidence that the Rous sarcoma virus transforming gene product phosphorylates phosphatidylinositol and diacylglycerol. Proc Natl Acad Sci U S A. 1984;81:2117–2121. doi: 10.1073/pnas.81.7.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Hillmann P, Hofmann BT, Hart JR, Vogt PK. Cancer-derived mutations in the regulatory subunit p85alpha of phosphoinositide 3-kinase function through the catalytic subunit p110alpha. Proc Natl Acad Sci U S A. 2010;107:15547–15552. doi: 10.1073/pnas.1009652107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Peng I, Webster JD, Suto E, Lesch J, Wu X, Senger K, Francis G, Barrett K, Collier JL, et al. Inhibition of the kinase ITK in a mouse model of asthma reduces cell death and fails to inhibit the inflammatory response. Sci Signal. 2015;8:ra122. doi: 10.1126/scisignal.aab0949. [DOI] [PubMed] [Google Scholar]
- Switon K, Kotulska K, Janusz-Kaminska A, Zmorzynska J, Jaworski J. Tuberous sclerosis complex: From molecular biology to novel therapeutic approaches. IUBMB Life. 2016;68:955–962. doi: 10.1002/iub.1579. [DOI] [PubMed] [Google Scholar]
- Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Hashimoto J, Tanabe Y, Kodaira M, Yonemori K, Seto T, Hirai F, Arita S, Toyokawa G, Chen L, et al. Safety and tolerability of AZD5363 in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol. 2016;77:787–795. doi: 10.1007/s00280-016-2987-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Kondo T, Sajan M, Luo J, Bronson R, Asano T, Farese R, Cantley LC, Kahn CR. Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKClambda/zeta. Cell Metab. 2006;3:343–353. doi: 10.1016/j.cmet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Terauchi Y, Tsuji Y, Satoh S, Minoura H, Murakami K, Okuno A, Inukai K, Asano T, Kaburagi Y, Ueki K, et al. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat Genet. 1999;21:230–235. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- Thien A, Prentzell MT, Holzwarth B, Kläsener K, Kuper I, Boehlke C, Sonntag AG, Ruf S, Maerz L, Nitschke R, et al. TSC1 activates TGF-β-Smad2/3 signaling in growth arrest and epithelial-to-mesenchymal transition. Dev Cell. 2015;32:617–630. doi: 10.1016/j.devcel.2015.01.026. [DOI] [PubMed] [Google Scholar]
- Thomas GV, Tran C, Mellinghoff IK, Welsbie DS, Chan E, Fueger B, Czernin J, Sawyers CL. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, Dickler MN, Scaltriti M, Leslie CS, Armstrong SA, et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science. 2017;355:1324–1330. doi: 10.1126/science.aah6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchi R, Takahashi Y, Niida A, Shimamura T, Hirata H, Sugimachi K, Sawada G, Iwaya T, Kurashige J, Shinden Y, et al. Integrated multiregional analysis proposing a new model of colorectal cancer evolution. PLoS Genet. 2016;12:e1005778. doi: 10.1371/journal.pgen.1005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- Venkataraman S, Alimova I, Balakrishnan I, Harris P, Birks DK, Griesinger A, Amani V, Cristiano B, Remke M, Taylor MD, et al. Inhibition of BRD4 attenuates tumor cell self-renewal and suppresses stem cell signaling in MYC driven medulloblastoma. Oncotarget. 2014;5:2355–2371. doi: 10.18632/oncotarget.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbist KC, Guy CS, Milasta S, Liedmann S, Kamiński MM, Wang R, Green DR. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature. 2016;532:389–393. doi: 10.1038/nature17442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, Lockerman EL, Pollack SF, Liu M, Li X, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss MH, Hakimi AA, Pham CG, Brannon AR, Chen YB, Cunha LF, Akin O, Liu H, Takeda S, Scott SN, et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res. 2014;20:1955–1964. doi: 10.1158/1078-0432.CCR-13-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Grabiner BC, Van Allen EM, Amin-Mansour A, Taylor-Weiner A, Rosenberg M, Gray N, Barletta JA, Guo Y, Swanson SJ, et al. Response and acquired resistance to everolimus in anaplastic thyroid cancer. N Engl J Med. 2014;371:1426–1433. doi: 10.1056/NEJMoa1403352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldhart AN, Dykstra H, Peck AS, Boguslawski EA, Madaj ZB, Wen J, Veldkamp K, Hollowell M, Zheng B, Cantley LC, et al. Phosphorylation of TXNIP by AKT mediates acute influx of glucose in response to insulin. Cell Rep. 2017;19:2005–2013. doi: 10.1016/j.celrep.2017.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Wang M, Jiang N, Zhang Y, Bian X, Wang X, Roberts TM, Zhao JJ, Liu P, Cheng H. Effective use of PI3K inhibitor BKM120 and PARP inhibitor Olaparib to treat PIK3CA mutant ovarian cancer. Oncotarget. 2016;7:13153–13166. doi: 10.18632/oncotarget.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Shin YS, Xue M, Matsutani T, Masui K, Yang H, Ikegami S, Gu Y, Herrmann K, Johnson D, et al. Single-Cell Phosphoproteomics Resolves Adaptive Signaling Dynamics and Informs Targeted Combination Therapy in Glioblastoma. Cancer Cell. 2016;29:563–573. doi: 10.1016/j.ccell.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Hengstschläger M, Linke M. Regulation of innate immune cell function by mTOR. Nat Rev Immunol. 2015;15:599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Warne PH, Lambros MB, Reis-Filho JS, Downward J. PI3K pathway dependencies in endometrioid endometrial cancer cell lines. Clin Cancer Res. 2013;19:3533–3544. doi: 10.1158/1078-0432.CCR-12-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch HCE, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, Tempst P, Hawkins PT, Stephens LR. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108:809–821. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell. 2007;18:1437–1446. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarski P, Kasprzycka M, Liu X, Marzec M, Robertson ES, Slupianek A, Wasik MA. Activation of mammalian target of rapamycin in transformed B lymphocytes is nutrient dependent but independent of Akt, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase, insulin growth factor-I, and serum. Cancer Res. 2005;65:7800–7808. doi: 10.1158/0008-5472.CAN-04-4180. [DOI] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013;4:913–920. doi: 10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates DH. mTOR treatment in lymphangioleiomyomatosis: the role of everolimus. Expert Rev Respir Med. 2016;10:249–260. doi: 10.1586/17476348.2016.1148603. [DOI] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21:751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa T, Uchiyama K, Ogura Y, Kimura M, Teshigawara K, Hosaka T, Tanaka Y, Obata T, Sano H, Kishi K, et al. The Rab GTPase-activating protein AS160 as a common regulator of insulin- and Galphaq-mediated intracellular GLUT4 vesicle distribution. Endocr J. 2009;56:345–359. doi: 10.1507/endocrj.k08e-216. [DOI] [PubMed] [Google Scholar]
- Zelenetz AD, Barrientos JC, Brown JR, Coiffier B, Delgado J, Egyed M, Ghia P, Illés Á, Jurczak W, Marlton P, et al. Idelalisib or placebo in combination with bendamustine and rituximab in patients with relapsed or refractory chronic lymphocytic leukaemia: interim results from a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2017;18:297–311. doi: 10.1016/S1470-2045(16)30671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KJ, Husami A, Marsh R, Jordan MB. Identification of a phosphoinositide 3-kinase (PI-3K) p 110 δ (PIK3CD) deficient individual. J Clin Immunol. 2013;33:673–674. [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.