Abstract
Background
Human pegiviruses (HPgV)—formerly known as hepatitis G virus or GB virus C (GBV-C)—are common single-stranded RNA viruses that may have a beneficial impact on slowing HIV disease progression. The data on HPgV in resource-limited regions such as Sub-Saharan Africa are scarce. Thus, we conducted the first study of HPgV in Botswana as part of a natural history study of HIV subtype C disease progression.
Methods
Plasma samples from 133 HIV-positive adults were evaluated for HPgV RNA, and the 5’UTR was sequenced to determine the HPgV genotype.
Results
HPgV RNA was detected in 41 (30.8%) individuals. While the presence of HPgV RNA had no impact on baseline HIV viral load, a significant difference in baseline CD4 cell count was observed. HPgV genotypes were determined for 27 individuals and included 5 individuals (18.5%) with genotype 1 and 22 (81.5%) with genotype 5. Baseline CD4 cell counts were significantly higher for persons infected with HPgV genotype 5 compared with genotype 1.
Conclusions
These data suggest that HPgV infection is common among HIV-positive individuals in Botswana and has a significant impact on CD4 cell count. This difference in CD4 cell count based on HPgV genotype suggests that HPgV genotype should be evaluated as a possible predictor of HIV disease progression and highlights the need for additional studies of this virus in resource-limited settings.
Keywords: Africa, Botswana, hepatitis G virus, HIV, human pegivirus (HPgV), GB virus C (GBV-C), genotype
The human pegivirus (HPgV)—originally described as hepatitis G virus or GB virus C (GBV-C)—is a positive-strand RNA virus that is distantly related to hepatitis C virus. HPgV has gained notoriety due to its possible beneficial impact on HIV disease progression. The prevalence of HPgV RNA ranges from 14% to 45% in HIV-positive persons (reviewed in [1]). Several groups have reported beneficial effects of HPgV viremia on HIV disease progression, as indicated by higher CD4 cell counts, lower HIV viral loads, and longer AIDS-free survival times [2–6]. In contrast, loss of HPgV RNA is associated with accelerated HIV disease [3, 7, 8]. In vitro studies demonstrate that HIV replication is inhibited by HPgV co-infection in peripheral blood mononuclear cell cultures [2]. Nevertheless, this beneficial effect of HPgV has not been observed in all studies [7–11]. Other studies conducted during the highly active antiretroviral therapy (HAART) era observed that a complete virologic response was more frequent in patients co-infected with HPgV, independent of CD4 cell count and HIV RNA level, although this was not uniformly found [12–14].
Country-specific data on HPgV are available from 13 Sub-Saharan African countries; the majority reported prevalence data and/or analysis of HPgV diversity [15]. Data regarding the impact of HPgV on HIV disease in this region are limited. Among HIV-positive women in Gambia, HPgV had no significant impact on HIV load, CD4 cell count, or mortality [16]. However, in HIV-positive South Africans, HPgV co-infection was associated with higher CD4 cell counts and lower HIV viral loads before HAART initiation, as well as faster viral load declines during HAART [17]. In Uganda, HIV/HPgV co-infected participants experienced slower CD4 cell decline and increased survival compared with HIV-positive adults who were HPgV-negative [18]. These data suggest that the beneficial effect of HPgV is not limited to the HIV subtypes predominant in the United States and Western Europe. Therefore, in resource-limited settings in which many HIV-infected individuals may not have access to antiretroviral therapy, a better understanding of the anti-HIV effects of HPgV infection may ultimately result in novel therapeutic strategies. Botswana has one of the highest HIV prevalence rates in the world [19, 20]. Thus, we evaluated the impact of HPgV infection in a natural history cohort of HIV disease progression in Botswana.
METHODS
Study Participants
In 2005, the Botsogo Study was established among HIV-infected antiretroviral therapy (ART)–naïve individuals in Gaborone, Botswana, to observe disease progression among individuals infected with HIV subtype C who did not qualify for ART according to the Botswana national guidelines (CD4+ T cell count ≥200 per mm3 and a World Health Organization clinical stage I or II) at the time of enrollment [21]. Exclusion criteria included any AIDS-defining illness requiring the initiation of HAART or previous ART use or exposure, except for use as part of the prevention of mother-to-child transmission program, the presence of an AIDS-related malignancy, patients requiring chronic corticosteroid use, less than 3 months postpartum, and/or participation in any study that provides immune-modulating agents. During follow-up, participants visited clinics quarterly, including 1 month after enrollment. The study was approved by the Human Research Development Committee at the Botswana Ministry of Health and Wellness (protocol number HRDC #00667) and the Harvard School of Public Health’s Office of Human Research Administration (protocol number 10366-127).
Detection of HPgV and Classification of HPgV Genotype
A secondary analysis of HPgV was performed in a convenience sampling of individuals enrolled in the Botsogo Study. As reported previously [22, 23], viral RNA was extracted from serum with the QIAmp Ultrasens Virus Kit (QIAGEN, Valencia, CA). HPgV RNA was detected by amplification of the 5’ untranslated region (UTR) with the antisense primer 5’ – ATG CCA CCC GCC CTC ACC CGA A – 3’ (nucleotides [nt] 494–473 according to GenBank accession number AY196904) and the sense primer 5’ – AAA GGT GGT GGA TGG GTG ATG – 3’ (nt 67–87) via OneStep RT-PCR (QIAGEN). Amplification conditions were 50°C for 59 minutes, 10 minutes at 94°C, then 35 cycles of 30 seconds at 94°C, 1 minute at 55°C, and 1 minute at 72°C, followed by 20 minutes at 72°C. First-round polymerase chain reaction (PCR) products were used in nested PCR with the antisense primer 5’ – CCC CAC TGG TCY TTG YCA ACT C – 3’ (nt 362–341) and sense primer 5’ – AAT CCC GGT CAY AYT GGT AGC CAC T – 3’ (nt 107–131). After 35 cycles of 30 seconds at 94°C, 30 seconds at 55°C, and 1 minute at 72°C, PCR products were analyzed by agarose gel electrophoresis for the presence of a 256-nt band. Population-based sequencing of amplicons was conducted, and 5’UTR sequences were aligned with GenBank accession numbers U59540, U59543, U59549, and U59555 (genotype 1); HGU59518, D90600, HGU59534, and HGU59535 (genotype 2); U59538 and U59539 (genotype 3); AB018667 and AB021287 (genotype 4); AY949771, AF092894, LT009490, KC618398, KC618400, KC618401, AY032965, AF172508, and KP710606 (genotype 5); AB003292 and AF177619 (genotype 6); and HQ331234 and HQ331235 (genotype 7). Phylogenetic inference was performed using a Bayesian Markov chain Monte Carlo (MCMC) approach executed in the Bayesian Evolutionary Analysis by Sampling Trees v1.8.4 [24] with an uncorrelated log-normal relaxed molecular clock, generalized time reversible model, and nucleotide site heterogeneity estimated with a gamma distribution. MCMC analysis was run for a chain length of 1 000 000 000. All effective sample sizes were >200, indicating sufficient sampling. The maximum clade credibility tree was selected from the posterior tree distribution after a 10% burn-in using TreeAnnotator v1.8.4. HPgV sequences were deposited in GenBank using accession numbers MF398545–MF398571.
Assessment of Liver Injury
The aspartate aminotransferase (AST) to platelet ratio index (APRI) and fibrosis 4 (FIB-4) score represent 2 noninvasive indices of liver damage (reviewed in [25]). APRI is equal to 100 * (AST/40) / platelet, while FIB-4 is calculated as age [years] × AST [IU/L] / √ (PLT [109/L] × (ALT [IU/L]). The APRI and FIB-4 indices were validated initially for hepatitis C virus and are now utilized during HIV mono-infection and chronic HBV as well [26–29].
Statistical Analysis
Sociodemographic and clinical data available at baseline were evaluated for the Botsogo Study. Fisher’s exact test was used to evaluate the difference in proportions for dichotomous variables, and the Wilcoxon rank sum test was used to compare select categories. All statistical analyses were performed using STATA 14.1 (College Station, TX).
RESULTS
The Botsogo Study followed 436 participants for 5 years, of whom 356 (82%) were female [21]. The median age was 33 years (interquartile range [IQR], 27–39 years); 133 participants had baseline plasma available for the current analysis of HPgV. HPgV RNA was detectable in 41 (30.8%). HPgV-positive and HPgV-negative individuals did not differ with respect to age or gender (Table 1). ALT levels were lower for HPgV-positive compared with HPgV-negative individuals (15.4 vs 16.5 cells/uL; P < .001), although AST levels were not significantly different.
Table 1.
Baseline Demographic and Clinical Data for HPgV-Positive and HPgV-Negative Individuals Enrolled in the Botsogo Study
| HPgV-Positive (n = 41) |
HPgV-Negative (n = 92) |
P Value | |
|---|---|---|---|
| Age, median (Q1, Q3), y | 34 (29, 41) | 32 (28, 41) | .444 |
| Male gender, n (%) | 10/23 (43.5) | 13/23 (56.5) | |
| Female gender, n (%) | 31/110 (28.2) | 79/110 (71.8) | .213* |
| Platelets, 109/L | 267 (227, 310) | 252 (224, 303) | .778 |
| Hemoglobin, median (Q1, Q3), g/dL | 12.8 (12.1, 13.7) | 12.5 (11.3, 13.5) | .306 |
| ALT , median (Q1, Q3), U/L | 15.4 (12.3, 21.5) | 16.5 (11.1, 24.6) | <.001 |
| AST, median (Q1, Q3), U/L | 22.0 (16.5, 27.3) | 23.0 (18.3, 28.5) | .444 |
| FIB-4 score, median (Q1, Q3) | 0.71 (0.525, 0.931) | 0.74 (0.58, 0.98) | .589 |
| APRI score, median (Q1, Q3) | 0.19 (0.15, 0.29) | 0.22 (0.18, 0.31) | .342 |
The data represent medians (interquartile ranges in parentheses) except as noted. *Comparisons are made between the HPgV-positive and HPgV-negative groups using the Wilcoxon rank sum test, with the exception of male gender and HPgV status, for which the chi-square test was used.
Abbreviations: ALT, alanine aminotransferase; APRI, AST to platelet ratio index; AST, aspartate aminotransferase; FIB-4, fibrosis 4.
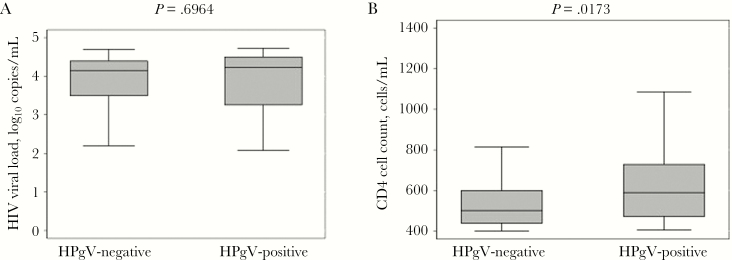
While HIV is known to impact liver disease progression in the presence and absence of viral hepatitis, the effect of HPgV infection on liver disease is unknown. Using 2 noninvasive indices of liver damage—APRI and FIB-4—there were no observed differences in liver disease between the 2 groups based on HPgV status. As shown in Figure 1A, the presence of HPgV RNA had no statistically significant impact on HIV viral loads. Baseline viral loads were 4.23 log10 copies/mL in HPgV-positive individuals and 4.15 log10 copies/mL in HPgV-negative individuals. However, the median CD4 cell count was higher for HPgV-positive compared with HPgV-negative individuals (589 cells/uL vs 501 cells/uL; P = .0173) (Figure 1B).
Figure 1.
(A) Baseline HIV viral load (log10 copies/mL) and (B) CD4 cell count (cells/uL) were evaluated for human pegivirus (HPgV)–positive and HPgV-negative individuals. P values are shown for the Wilcoxon rank sum test.
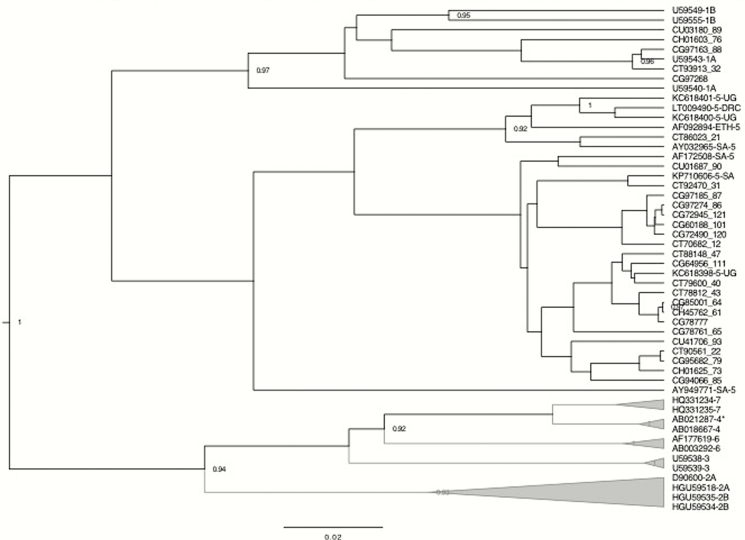
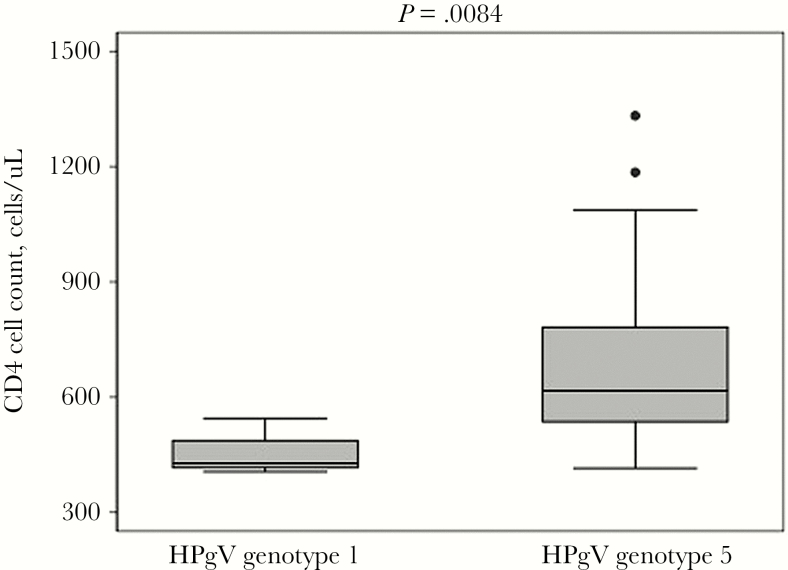
HPgV genotypes were available for 27 individuals (65.9% of those with detectable HPgV RNA) and included 5 (18.5%) with genotype 1 and 22 (81.5%) with genotype 5 (Figure 2). Individuals with HPgV genotype 5 had significantly higher baseline CD4 cell counts than those with HPgV genotype 1 (617 cells/uL vs 428 cells/uL; P = .0084) (Figure 3).
Figure 2.
Bayesian phylogenetic analysis of the 27 HPgV 5’UTR sequences from this study (indicated by a 6-digit study ID + a 2- to 3-digit sequence number) compared with GenBank references (indicated by their accession numbers and genotype). Full-length HPgV sequences with evidence of recombination are denoted by an asterisk. Posterior probability values >0.90 are indicated at tree nodes.
Figure 3.
Baseline CD4 cell count (cells/uL) was evaluated for HPgV-positive individuals with genotypes 1 and 5. Closed circles represent outliers. The P value shown is for the Wilcoxon rank sum test.
DISCUSSION
These data represent the first study of HPgV prevalence conducted in Botswana. Data on HPgV are scarce in Sub-Saharan Africa and largely limited to small prevalence studies (reviewed in [15]). Cross-sectional studies conducted in South Africa demonstrate a prevalence range of 10.2% to 41.2% in blood donors, hemodialysis patients, transplant patients, hemophiliacs, or patients with chronic liver disease [30–34]. Thus, the current finding of an HPgV prevalence of 30.8% is in agreement with other studies conducted in southern Africa.
Multiple HPgV genotypes have been described at the population level [35, 36]. Genotypes 1 and 2 are common throughout the Americas and northern and central Africa. Genotypes 3 and 4 are present in Asia. Genotype 5 circulates within central and southern Africa. Genotype 6 has been identified in Southeast Asia, while a putative genotype 7 has only been reported in China [36]. This study is the first to evaluate HPgV genotypes in Botswana and suggests that genotype 5 is the predominant circulating genotype. HPgV genotype 5 has also been reported in Uganda, the Democratic Republic of the Congo, Tanzania, and Ethiopia [37–43]. In South Africa, HPgV genotype 5 is most common, although genotypes 1 and 2 have also been reported [34, 40–42, 44, 45]. However, several limitations require cautious interpretation of these findings, including a modest sample size and lack of genotype data for all HPgV-positive individuals. The lack of a statistically significant difference in HIV viral load by HPgV genotype could have been due to low sample size. As with other studies, there is no information about the mode of HPgV transmission or the timing of infection. While the correlation between HPgV RNA levels and CD4 cell count or HIV viral load was not evaluated in this analysis, previous studies have reported an inverse correlation between HPgV and HIV levels [4, 46].
The possible impact of distinct HPgV genotypes on HIV disease progression has been evaluated in other studies outside of Africa. For instance, Muerhoff et al. reported that CD4 cell counts tended to be lower in HIV-positive patients co-infected with HPgV genotype 2a compared with those with HPgV genotype 2b [47]. In US patients with HIV/HCV/HPgV triple infection, higher CD4 cell counts were associated with HPgV genotype 2 compared with genotype 1 [23]. Similar findings were observed in Brazil, although no difference in CD4 cell count based on HPgV genotype was reported in Australia [48, 49]. In Brazil, HPgV RNA levels also differed by genotype [50]. Unfortunately, studies designed to evaluate the potential influence of HPgV genotype on HIV disease progression have not been conducted in Africa to date. To date, only a single functional study has included HPgV genotype 5 isolates. Xiang et al. evaluated South African samples and found that genotype 1 and 5 isolates replicated in lymphocyte cultures, inhibited X4 and R5 HIV isolates, and induced the chemokines RANTES/CCL5 and stromal-derived factor–1 (SDF-1) in vitro [34]. However, too few HPgV isolates were included to compare their ability to suppress HIV replication based on HPgV genotype.
The high prevalence of HPgV in Botswana, its beneficial impact on HIV disease progression, and the impact of HPgV genotype on CD4 cell count all suggest an immediate need to expand significantly the research on HPgV in resource-limited settings such as Sub-Saharan Africa.
Acknowledgements
The authors would like to thank the study participants and staff of the Botsogo study, the Models of Care Project, sponsored by the Ministry of Health and Wellness and the African Comprehensive HIV/AIDS Partnerships (ACHAP), and the Pathobiology and Molecular Medicine PhD program at the University of Cincinnati College of Medicine for their generous support.
Financial support. The Botsogo study was funded by ACHAP, a country-led, public–private development partnership between the Government of Botswana, the Bill and Melinda Gates Foundation, and the MSD/Merck Company Foundation. The authors were supported by the Sub-Saharan Africa Network for TB/HIV Research Excellence (grant 107752/Z/15/Z) from the Wellcome Trust, the Fogarty HIV Research Training Program for Low- and Middle-Income Country Institutions supported by the NIH Fogarty International Center (D43 TW009610), and the Oak Foundation (OUSA-12–025). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. All authors: no reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Berzsenyi MD, Bowden DS, Roberts SK. GB virus C: insights into co-infection. J Clin Virol 2005; 33:257–66. [DOI] [PubMed] [Google Scholar]
- 2. Xiang J, Wünschmann S, Diekema DJ et al. . Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med 2001; 345:707–14. [DOI] [PubMed] [Google Scholar]
- 3. Williams CF, Klinzman D, Yamashita TE et al. . Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med 2004; 350:981–90. [DOI] [PubMed] [Google Scholar]
- 4. Tillmann HL, Heiken H, Knapik-Botor A et al. . Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med 2001; 345:715–24. [DOI] [PubMed] [Google Scholar]
- 5. Lefrère JJ, Roudot-Thoraval F, Morand-Joubert L et al. . Carriage of GB virus C/hepatitis G virus RNA is associated with a slower immunologic, virologic, and clinical progression of human immunodeficiency virus disease in coinfected persons. J Infect Dis 1999; 179:783–9. [DOI] [PubMed] [Google Scholar]
- 6. Heringlake S, Ockenga J, Tillmann HL et al. . GB virus C/hepatitis G virus infection: a favorable prognostic factor in human immunodeficiency virus-infected patients? J Infect Dis 1998; 177:1723–6. [DOI] [PubMed] [Google Scholar]
- 7. Björkman P, Flamholc L, Nauclér A et al. . GB virus C during the natural course of HIV-1 infection: viremia at diagnosis does not predict mortality. AIDS 2004; 18:877–86. [DOI] [PubMed] [Google Scholar]
- 8. Birk M, Lindbäck S, Lidman C. No influence of GB virus C replication on the prognosis in a cohort of HIV-1-infected patients. AIDS 2002; 16:2482–5. [DOI] [PubMed] [Google Scholar]
- 9. Quiros-Roldan E, Maroto MC, Torti C et al. . No evidence of benefical effect of GB virus type C infection on the course of HIV infection. AIDS 2002; 16:1430–1. [DOI] [PubMed] [Google Scholar]
- 10. Van der Bij AK, Kloosterboer N, Prins M et al. . GB virus C coinfection and HIV-1 disease progression: The Amsterdam Cohort Study. J Infect Dis 2005; 191:678–85. [DOI] [PubMed] [Google Scholar]
- 11. Toyoda H, Fukuda Y, Hayakawa T et al. . Effect of GB virus C/hepatitis G virus coinfection on the course of HIV infection in hemophilia patients in Japan. J Acquir Immune Defic Syndr Hum Retrovirol 1998; 17:209–13. [DOI] [PubMed] [Google Scholar]
- 12. Rodriguez B, Woolley I, Lederman MM et al. . Effect of GB virus C coinfection on response to antiretroviral treatment in human immunodeficiency virus-infected patients. J Infect Dis 2003; 187:504–7. [DOI] [PubMed] [Google Scholar]
- 13. Brumme ZL, Chan KJ, Dong WW et al. . No association between GB virus-C viremia and virological or immunological failure after starting initial antiretroviral therapy. AIDS 2002; 16:1929–33. [DOI] [PubMed] [Google Scholar]
- 14. Sheng WH, Hung CC, Wu RJ et al. . Clinical impact of GB virus C viremia on patients with HIV type 1 infection in the era of highly active antiretroviral therapy. Clin Infect Dis 2007; 44:584–90. [DOI] [PubMed] [Google Scholar]
- 15. Singh S, Blackard J. Human pegivirus (HPgV) infection in Sub-Saharan Africa—a call for a renewed research agenda. Rev Med Virol 2017. [DOI] [PubMed] [Google Scholar]
- 16. Kaye S, Howard M, Alabi A et al. . No observed effect of GB virus C coinfection on disease progression in a cohort of African woman infected with HIV-1 or HIV-2. Clin Infect Dis 2005; 40:876–8. [DOI] [PubMed] [Google Scholar]
- 17. Mosam A, Sathar MA, Dawood H et al. . Effect of GB virus C co-infection on response to generic HAART in African patients with HIV-1 clade C infection. AIDS 2007; 21:1377–9. [DOI] [PubMed] [Google Scholar]
- 18. Yirrell DL, Wright E, Shafer LA et al. . Association between active GB virus-C (hepatitis G) infection and HIV-1 disease in Uganda. Int J STD AIDS 2007; 18:244–9. [DOI] [PubMed] [Google Scholar]
- 19. Department of HIV and AIDS Prevention, Ministry of Health. Botswana Second Generation HIV Antenatal Sentinel Surveillance Technical Report . Gaborone, Botswana: Government of Botswana; 2009. [Google Scholar]
- 20. National AIDS Coordinating Agency. Preliminary Results: Botswana AIDS Impact Survey IV (BAIS IV), 2013 . Gaborone, Botswana: National AIDS Coordinating Agency; 2013. [Google Scholar]
- 21. Farahani M, Novitsky V, Wang R et al. . Prognostic value of HIV-1 RNA on CD4 trajectories and disease progression among antiretroviral-naive HIV-infected adults in Botswana: a joint modeling analysis. AIDS Res Hum Retroviruses 2016; 32:573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blackard JT, Ma G, Welge JA et al. . GB Virus C (GBV-C) infection in hepatitis C virus (HCV) seropositive women with or at risk for HIV infection. PLoS One 2014; 9:e114467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwarze-Zander C, Blackard JT, Zheng H et al. ; AIDS Clinical Trial Group A5071 Study Team GB virus C (GBV-C) infection in hepatitis C virus (HCV)/HIV-coinfected patients receiving HCV treatment: importance of the GBV-C genotype. J Infect Dis 2006; 194:410–9. [DOI] [PubMed] [Google Scholar]
- 24. Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012; 29:1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baranova A, Lal P, Birerdinc A, Younossi ZM. Non-invasive markers for hepatic fibrosis. BMC Gastroenterol 2011; 11:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Orasan OH, Iancu M, Sava M et al. . Non-invasive assessment of liver fibrosis in chronic viral hepatitis. Eur J Clin Invest 2015; 45:1243–51. [DOI] [PubMed] [Google Scholar]
- 27. Zhang Z, Wang G, Kang K et al. . The diagnostic accuracy and clinical utility of three noninvasive models for predicting liver fibrosis in patients with HBV infection. PLoS One 2016; 11:e0152757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tahiri M, Sodqi M, Lahdami FE et al. . Risk factors for liver fibrosis among human immunodeficiency virus monoinfected patients using the FIB4 index in Morocco. World J Hepatol 2013; 5:584–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blackard JT, Welge JA, Taylor LE et al. . HIV mono-infection is associated with FIB-4—a noninvasive index of liver fibrosis—in women. Clin Infect Dis 2011; 52:674–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Casteling A, Sim J, Vardas E et al. . Hepatitis GBV-C in South Africa. S Afr Med J 1997; 87:182–3. [PubMed] [Google Scholar]
- 31. Casteling A, Song E, Sim J et al. . GB virus C prevalence in blood donors and high risk groups for parenterally transmitted agents from Gauteng, South Africa. J Med Virol 1998; 55:103–8. [DOI] [PubMed] [Google Scholar]
- 32. Lightfoot K, Skelton M, Kew MC et al. . Does hepatitis GB virus-C infection cause hepatocellular carcinoma in black Africans? Hepatology 1997; 26:740–2. [DOI] [PubMed] [Google Scholar]
- 33. Sathar MA, Soni PN, Naicker S et al. . GB virus C/hepatitis G virus infection in KwaZulu Natal, South Africa. J Med Virol 1999; 59:38–44. [PubMed] [Google Scholar]
- 34. Xiang J, Sathar MA, McLinden JH et al. . South African GB virus C isolates: interactions between genotypes 1 and 5 isolates and HIV. J Infect Dis 2005; 192:2147–51. [DOI] [PubMed] [Google Scholar]
- 35. Muerhoff AS, Dawson GJ, Desai SM. A previously unrecognized sixth genotype of GB virus C revealed by analysis of 5’-untranslated region sequences. J Med Virol 2006; 78:105–11. [DOI] [PubMed] [Google Scholar]
- 36. Feng Y, Zhao W, Feng Y et al. . A novel genotype of GB virus C: its identification and predominance among injecting drug users in Yunnan, China. PLoS One 2011; 6:e21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ghai RR, Sibley SD, Lauck M et al. . Deep sequencing identifies two genotypes and high viral genetic diversity of human pegivirus (GB virus C) in rural Ugandan patients. J Gen Virol 2013; 94:2670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iles JC, Abby Harrison GL, Lyons S et al. . Hepatitis C virus infections in the Democratic Republic of Congo exhibit a cohort effect. Infect Genet Evol 2013; 19:386–94. [DOI] [PubMed] [Google Scholar]
- 39. Stark K, Poggensee G, Höhne M et al. . Seroepidemiology of TT virus, GBC-C/HGV, and hepatitis viruses B, C, and E among women in a rural area of Tanzania. J Med Virol 2000; 62:524–30. [DOI] [PubMed] [Google Scholar]
- 40. Muerhoff AS, Leary TP, Sathar MA et al. . African origin of GB virus C determined by phylogenetic analysis of a complete genotype 5 genome from South Africa. J Gen Virol 2005; 86:1729–35. [DOI] [PubMed] [Google Scholar]
- 41. Sathar MA, York DF. Group 5: GBV-C/HGV isolates from South Africa. J Med Virol 2001; 65:121–2. [PubMed] [Google Scholar]
- 42. Sathar MA, Soni PN, Pegoraro R et al. . A new variant of GB virus C/hepatitis G virus (GBV-C/HGV) from South Africa. Virus Res 1999; 64:151–60. [DOI] [PubMed] [Google Scholar]
- 43. Luk KC, Berg MG, Naccache SN et al. . Utility of metagenomic next-generation sequencing for characterization of hiv and human pegivirus diversity. PLoS One 2015; 10:e0141723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tucker TJ, Smuts H, Eickhaus P et al. . Molecular characterization of the 5’ non-coding region of South African GBV-C/HGV isolates: major deletion and evidence for a fourth genotype. J Med Virol 1999; 59:52–9. [PubMed] [Google Scholar]
- 45. Hardie D, Smuts H. Human pegivirus-1 in the CSF of patients with HIV-associated neurocognitive disorder (HAND) may be derived from blood in highly viraemic patients. J Clin Virol 2017; 91:58–61. [DOI] [PubMed] [Google Scholar]
- 46. Björkman P, Flamholc L, Molnegren V et al. . Enhanced and resumed GB virus C replication in HIV-1-infected individuals receiving HAART. AIDS 2007; 21:1641–3. [DOI] [PubMed] [Google Scholar]
- 47. Muerhoff AS, Tillmann HL, Manns MP et al. . GB virus C genotype determination in GB virus-C/HIV co-infected individuals. J Med Virol 2003; 70:141–9. [DOI] [PubMed] [Google Scholar]
- 48. Berzsenyi MD, Bowden DS, Roberts SK, Revill PA. GB virus C genotype 2 predominance in a hepatitis C virus/HIV infected population associated with reduced liver disease. J Gastroenterol Hepatol 2009; 24:1407–10. [DOI] [PubMed] [Google Scholar]
- 49. Alcalde R, Nishiya A, Casseb J et al. . Prevalence and distribution of the GBV-C/HGV among HIV-1-infected patients under anti-retroviral therapy. Virus Res 2010; 151:148–52. [DOI] [PubMed] [Google Scholar]
- 50. Giret MT, Miraglia JL, Sucupira MC et al. . Prevalence, incidence density, and genotype distribution of GB virus C infection in a cohort of recently HIV-1-infected subjects in Sao Paulo, Brazil. PLoS One 2011; 6:e18407. [DOI] [PMC free article] [PubMed] [Google Scholar]