Abstract
Pseudomonas aeruginosa (P. aeruginosa) is one of the most common nosocomial pathogens worldwide. Although the emergence of multidrug-resistant (MDR) P. aeruginosa is a critical problem in medical practice, the key features involved in the emergence and spread of MDR P. aeruginosa remain unknown. This study utilized whole genome sequence (WGS) analyses to define the population structure of 185 P. aeruginosa clinical isolates from several countries. Of these 185 isolates, 136 were categorized into sequence type (ST) 235, one of the most common types worldwide. Phylogenetic analysis showed that these isolates fell within seven subclades. Each subclade harbors characteristic drug resistance genes and a characteristic genetic background confined to a geographic location, suggesting that clonal expansion following antibiotic exposure is the driving force in generating the population structure of MDR P. aeruginosa. WGS analyses also showed that the substitution rate was markedly higher in ST235 MDR P. aeruginosa than in other strains. Notably, almost all ST235 isolates harbor the specific type IV secretion system and very few or none harbor the CRISPR/CAS system. These findings may help explain the mechanism underlying the emergence and spread of ST235 P. aeruginosa as the predominant MDR lineage.
Keywords: multidrug-resistance, Pseudomonas aeruginosa, whole genome sequence, population structure
Introduction
Antimicrobial resistance (AMR) is a global concern, as it threatens the effective treatment of infectious diseases (http://www.who.int/antimicrobial-resistance/en/; last accessed November 27, 2017). Pseudomonas aeruginosa is a representative nosocomial pathogen showing AMR, and a major cause of death in patients with cystic fibrosis (Hoban et al. 2003; Rodriguez-Rojas et al. 2012). In addition to having intrinsic drug resistance mechanisms, this bacterium is able to acquire exogenous genes, resulting in the emergence of multidrug-resistant (MDR) strains of P. aeruginosa, which are resistant to carbapenems, aminoglycosides, and fluoroquinolones. As MDR P. aeruginosa has a major impact on medical practice (Viedma et al. 2009), knowledge of the mechanisms underlying multidrug-resistance to antibiotics and the epidemiology of MDR P. aeruginosa will be necessary to overcome infections with these bacteria.
Pseudomonas aeruginosa resistance to carbapenems and aminoglycosides was shown to be mediated by the acquisition of drug resistance genes (Hancock and Speert 2000; Livermore 2002), whereas resistance to fluoroquinolones is mediated by gene mutations (Chen and Lo 2003). Acquisition of genes encoding several families of β-lactamases has been found to contribute to resistance to carbapenems, such as IMP and VIM (Queenan and Bush 2007). Similarly, acquisition of exogenous genes encoding several types of aminoglycoside-modifying enzymes, such as AAC (Poole 2005) and 16S rRNA methylases (Wachino and Arakawa 2012), was shown to contribute to aminoglycoside resistance. In contrast, mutations in target genes, including those encoding DNA gyrase (gyrA or gyrB) and topoisomerase IV (parC and parE), were found to contribute to fluoroquinolone resistance (Chen and Lo 2003). Interestingly, drug resistance genes acquired by MDR P. aeruginosa differ markedly among communities in various countries (Viedma et al. 2009; Seok et al. 2011).
Epidemiological studies utilizing multilocus sequence typing (MLST; http://pubmlst.org/perl/bigsdb/bigsdb.pl? page=pubmlst_paeruginosa_isolates; last accessed November 27, 2017), a powerful molecular method for typing bacteria, can help understand and identify current MDR P. aeruginosa strains worldwide. These epidemiological studies showed that P. aeruginosa sequence type (ST) 235 is the predominant global clinical isolate (Viedma et al. 2009; Giske et al. 2006; Samuelsen et al. 2010; Lepsanovic et al. 2008; Empel et al. 2007; Cholley et al. 2011; van Belkum et al. 2015; Kos et al. 2015), with 88.3% of MDR P. aeruginosa isolates resistant to carbapenems, aminoglycosides, and fluoroquinolones being classified as ST235 (Kitao et al. 2012). Despite the importance of this subgroup, and despite the first ST235 P. aeruginosa being isolated in 1997, the mechanism underlying its persistence remains largely unknown.
To address the key traits responsible for its emergence and to identify the novel features of MDR P. aeruginosa, whole genome sequence (WGS) analysis was performed on P. aeruginosa strains isolated from hospital patients between 2001 and 2013 throughout Japan, as well as in other countries. In this study, MDR P. aeruginosa was defined according to the criteria of the Ministry of Health, Labour, and Welfare of Japan (Kirikae et al. 2008), because the definition of MDR P. aeruginosa varied among previous studies (Falagas et al. 2006).
Materials and Methods
Pseudomonas aeruginosa Isolates
MDR P. aeruginosa strains were collected based on the criteria specified by the Ministry of Health, Labour, and Welfare of Japan, including resistance to fluoroquinolones (minimum inhibitory concentration [MIC] ≥ 4 µg/ml), carbapenems (MIC ≥ 16 µg/ml), and amikacin (MIC ≥ 32 µg/ml; Kirikae et al. 2008). Several nonMDR P. aeruginosa strains were also included in this study (supplementary data 1, Supplementary Material online). Of the 185 clinical isolates evaluated by WGS analyses, 158 were from Japan, one from Thailand, three from Vietnam, seven from Nepal, and 15 from Poland. Also tested was the nonMDR P. aeruginosa strain PAO1 (USA). These isolates were grown on LB medium, and genomic DNA was purified using DNeasy Blood & Tissue kits (Qiagen). This study also included 26 P. aeruginosa strains registered in the database through 2012. Data including MLST and the genes or regions carried by each isolate are summarized in supplementary data 1 and 4, Supplementary Material online. In addition, 150 nonMDR P. aeruginosa isolates were screened to assess the frequency of ST235 P. aeruginosa among these nonMDR isolates.
WGS and Secondary Analyses
Nextera paired-end multiplex libraries of the isolates were generated and sequenced on a Genome Analyzer IIx (Illumina), according to the manufacturer’s instructions, to generate 93-bp paired-end reads. More than 110-fold coverage was archived for each isolate. In some experiments, MiSeq (Illumina) was used to generate 251-bp paired end reads. The sequence data have been registered in the DNA Data Bank of Japan (DDBJ) under accession numbers DRA002419, DRA001216, and DRA001252. To identify SNPs among the isolates, all reads were assembled de novo into contigs using CLC Genomics Workbench (CLC bio), and the resulting contigs were concatenated by 60 × “n” gap filling and used as a reference genome. To identify SNPs among the 136 ST235 isolates, reads from each were aligned against the genome sequence of a reference ST235 strain, NCGM2.S1 (Miyoshi-Akiyama et al. 2011), excluding regions of prophages and integrons (supplementary table 1, Supplementary Material online), to obtain high-quality concatenated SNP sequences. The average quality of the SNPs identified was Qv35, covered by > 95% of the total reads.
MLST Analysis, O-type Analysis, and Analyses of Gene Distributions
MLST typing of all isolates was performed using the MLST plug-in of CLC Genomics Workbench with the MLST scheme for P. aeruginosa (Curran et al. 2004). The O-type of each O-type sequence was also determined using CLC Genomics Workbench. To analyze the distribution of drug resistance genes contributing to resistance against β-lactams, carbapenems, and aminoglycosides, as well as the distribution of genes contributing to oprD disruption and to the acquisition of exogenous genetic materials, Illumina reads of each isolate were assembled de novo and the resulting contigs were searched with the BLAST algorithm (Altschul et al. 1990) for each gene or region using CLC Genomics Workbench. The numbers of CIRSPRs and spacers were analyzed using CRISPRfinder (http://crispr.i2bc.paris-saclay.fr; last accessed November 27, 2017; Grissa et al. 2007). Genes for CRISPR-associated proteins (CASs) harbored by P. aeruginosa retrieved from the NCBI database (supplementary table 2, Supplementary Material online) were used as queries to search for genes encoding CAS in P. aeruginosa strains used in this study. These results are presented in supplementary data 1, Supplementary Material online.
Phylogenetic Analyses
Concatenated SNP sequences were aligned with MAFFT (Katoh and Frith 2012). Evolutionary models (TVM + I + G for analysis of the 185 isolates and TVM + G for analysis of the 136 ST235 isolates) were chosen based on the results obtained with jModelTest 2.1.2 (Posada 2008) and convergence of the trees during preliminary phylogenetic analyses. Maximum-likelihood phylogenetic trees were constructed for the total 185 isolates and for the 136 ST235 isolates by concatenated SNPs with PhyML 3.0 (Guindon et al. 2010). The probability for node branching was evaluated with 100 bootstraps. In BEAST phylogeny of the 136 ST235 isolates, a clock model was chosen based on preliminary analyses showing better convergence of the tree. All other parameters were set to default, with chain lengths of 895,564,000 states and resampling every 10,000 states. Effective sample sizes (ESS) were >200 for all parameters. Time from the appearance of the most recent common ancestor was estimated using BEAST (Drummond and Rambaut 2007) and Path-O-Gen v1.4 (http://tree.bio.ed.ac.uk/software/pathogen/; last accessed November 27, 2017) programs.
Results
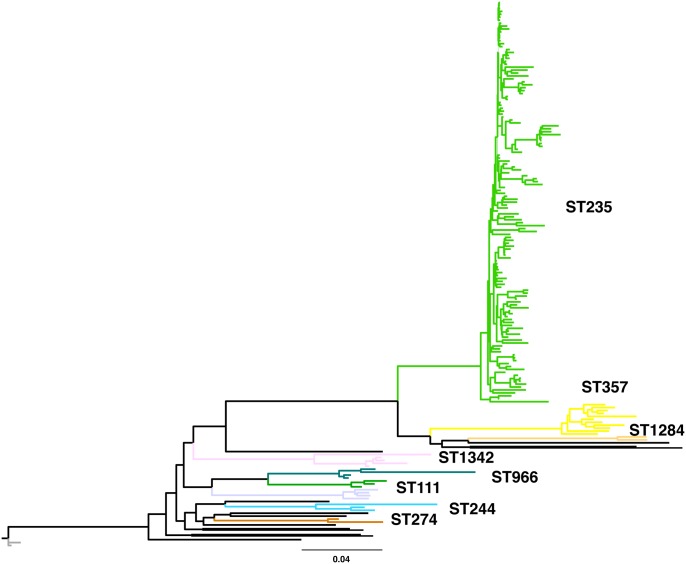
WGS analysis was performed on 156 MDR and 29 sensitive or nonMDR P. aeruginosa isolates (total 185 isolates) collected from patients throughout Japan and in other countries, including Thailand, Vietnam, Poland, and Nepal (supplementary data 1, Supplementary Material online). To determine the genetic relationships among these isolates, phylogenetic analysis was performed based on SNP concatenation. As the P. aeruginosa genome shows a high degree of plasticity, it is not sufficient to map Illumina reads to a particular P. aeruginosa genome. Thus, to determine sequence information on all of these isolates, the reads from all 185 libraries were assembled de novo, yielding 8,930 contigs (17,592,198 bp). These contigs were concatenated and used as the reference sequence to map reads from each isolate and to determine high-quality SNPs. A total of 249,067 SNPs were identified, and these concatenated SNP sequences were used to reconstruct a maximum-likelihood phylogenetic tree by MEGA5 (Tamura et al. 2011; fig. 1). One hundred thirty six of the 185 isolates (73.5%) were clustered into a ST235 clade, with 125 of these 136 isolates (80.1%) found to be MDR P. aeruginosa, consistent with findings of previous studies (Cholley et al. 2011; Kitao et al. 2012; Kirikae et al. 2008; Yoo et al. 2012), indicating that ST235 was the predominant clade in these P. aeruginosa isolates. Strikingly, screening of 150 nonMDR P. aeruginosa clinical isolates identified only two ST235 isolates, indicating that ST235 isolates are significantly enriched in the MDR P. aeruginosa population (chi-square test, P < 0.001).
Fig. 1.
—Phylogenic tree of the 185 Pseudomonas aeruginosa strains. The unrooted phylogeny of all P. aeruginosa strains was based on the maximum-likelihood method using PhyML 3.0 (Guindon et al. 2010). Each sequence type with more than one isolate [i.e. ST27 (n = 2), ST111 (n = 3), ST235 (n = 136), ST244 (n = 3), ST274 (n = 2), ST277 (n = 5), ST357 (n = 11), ST966 (n = 4), ST1284 (n = 2), and ST1342 (n = 4)] is indicated in color. Scale bar: 0.04 substitutions per variable site. Each main branch had >99% bootstrap support.
Divergence within Subclades of ST235
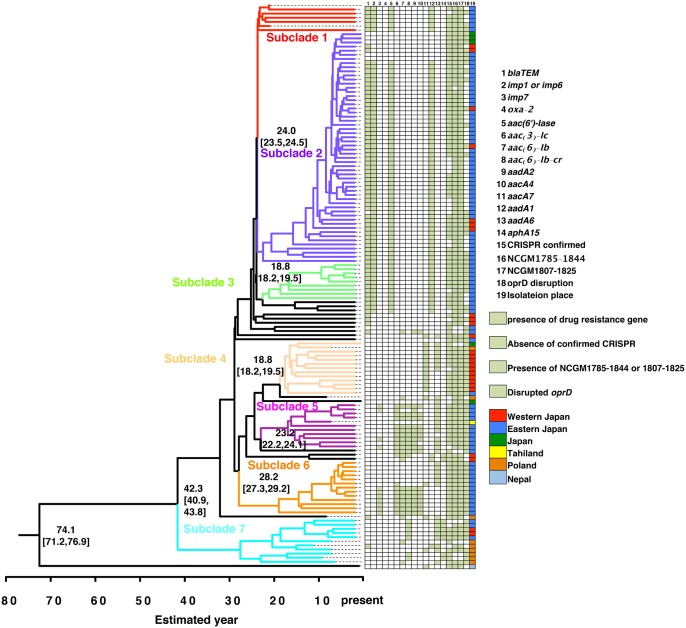
As ST235 was the dominant MDR P. aeruginosa isolates, we analyzed the detailed relationships of the 136 ST235 isolates based on a Bayesian phylogenetic tree (Drummond and Rambaut 2007) and a maximum-likelihood phylogenetic tree constructed using the concatenated SNP sequence of these 136 ST235 isolates (fig. 2 and supplementary fig. 1, Supplementary Material online). The SNPs of each ST235 strain were compared with a representative ST235 MDR P. aeruginosa strain, NCGM2.S1 (Miyoshi-Akiyama et al. 2011). To determine the genetic backbone of these isolates, SNPs located in mobile elements, such as prophages and integrons (supplementary table 1, Supplementary Material online), were omitted from the concatenations. Phylogenetic analysis therefore included 34,926 SNPs. Bayesian phylogeny was thought to be more robust than maximum likelihood phylogeny, based on posterior probability (values for all subclades were > 0.99) and bootstrap values, respectively. Therefore, each subclade was analyzed in detail based on Bayesian phylogeny.
Fig. 2.
—Phylogenic tree of the 136 ST235 Pseudomonas aeruginosa strains. The phylogeny of ST235 MDR P. aeruginosa strains was evaluated using the BEAST program (Drummond and Rambaut 2007). The seven subclades are shown in color. The estimated ages of the branches are shown as median values with 95% highest posterior density (HPD). The proportion of isolates from each location carrying conserved antibiotic resistance genes (>50% per subclade) and their oprD disruption status are also indicated. The posterior probability value for each main branch was >0.99. More detailed results are shown in supplementary figure 2 and supplementary data 1, Supplementary Material online. The posterior probability of each subclade designated was > 0.95.
The Bayesian phylogenetic tree resulted in the clustering of the 136 ST235 isolates into seven subclades (fig. 2). NP9, which was isolated from a patient in Nepal, was not included in any clade and was far separated from the other clades. Interestingly, NP9 was the only isolate expressing New Delhi metallo-beta-lactamase-1 (NDM1). The isolate from Thailand was clustered in subclade 5 and the isolates from Poland in subclades 4 and 7. None of the isolates from Vietnam was classified as ST235.
Assessments of the relationships between phylogenetic results and the geographic location of the isolates in Japan (fig. 2, and supplementary fig. 1, Supplementary Material online) showed that all isolates in subclade 3 were from one prefecture in Eastern Japan and most of the isolates in subclade 4 were from Western Japan. Six of the 11 isolates in subclade 7 were from Poland, with most of the remaining subclades being from Eastern Japan. The geographical distribution of isolates in each subclade suggests that the ancestors of each clade had spread on a global scale. Comparison of unique SNPs in each subclade with the genome of NCGM2.S1 (supplementary table 3 and supplementary data 2, Supplementary Material online) showed that subclades 3 and 4 had specific regions with unique SNPs; for example, 1,956,904_1,968,105 (NCGM2_1807 to NCGM2_1819) and 4,195,981_4,221,917 (NCGM2_3853 to NCGM2_3884) in subclades 3 and 5,270,138_5,299,359 (NCGM2_4888 to NCGM_4926) and 5,796,989_5,802,393 (NCGM2_5407 to NCGM2_5412) in subclade 4. The geographic location and the accumulated unique SNPs in regions in each subclade, especially in subclades 3 and 4, suggest that each subclade evolved independently following its emergence in different communities after separation from their ancestors. Because some isolates obtained from countries other than Japan were closely related with subclades that included isolates from Japan such as subclades 4 and 5, the ancestors of these isolates were likely present in different countries independently.
More than 50% of the drug-resistance genes were conserved in multiple subclades (fig. 2, supplementary fig. 1 and supplementary data 1, Supplementary Material online). The genes blaTEM, imp1 or 6, aac(6′)-Iae and aadA1 were conserved in subclades 1, 2, and 3; aac(3)-Ic, aac(6′)-Ib, aacA4 and aadA2 were conserved in subclades 5 and 6; and imp7 was conserved in subclade 6. In addition, oxa-2, aacA7, aacA8, and aadA6 were conserved in subclade 4, and aac(6′)-lb-cr, aacA4, aadA6, and aphA15 were conserved in subclade 7. These gene distribution patterns indicate that each subclade harbors specific drug resistance genes.
Subclades could also be distinguished by disruption of the oprD gene, which has been reported associated with carbapenem resistance (Lister 2002; Strateva and Yordanov 2009). In the prototype ST235 MDR strain, NCGM2.S1, which was clustered in subclade 1, oprD was found to be disrupted by insertion of an integron harboring imp-1 and aac(6′)-Iae(Miyoshi-Akiyama et al. 2011). Disruption of oprD was highly prevalent in isolates of subclades 1 and 2, with 52 of 62 (83.9%) harboring imp-1 and 55 of 62 (88.7%) harboring aac(6′)-Iae. The isolates in subclades 1 and 2 may have derived from a common ancestor containing an integron harboring imp-1 and aac(6′)-Iae and spread throughout Eastern Japan.
Bayesian phylogenetic analysis also suggested that the most recent common ancestor of ST235 appeared ∼74.1 (95% highest posterior density, 71.2–76.9) years ago and that branching within each subclade occurred ∼20–40 years ago (fig. 2). Additionally, Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/; last accessed November 27, 2017), a tool for investigating the temporal signal and “clocklikeness” of molecular phylogenies, suggested that the most recent common ancestor of the 136 ST235 isolates emerged ∼86.0 years ago. These results suggest that after emergence, the ancestors of these subclades were separated, with each acquiring particular drug resistance genes over the last 20–40 years, possibly coincident with the increased availability of antibiotics, such as aminoglycosides and carbapenems. The acquisition of drug resistance genes may have led to the expansion of each subclade over the past several decades.
Taken together, phylogenetic evidence suggests that several outbreaks accompanied the clonal expansion of ST235 MDR P. aeruginosa over the past several decades, and that these outbreaks may have been the driving force behind the generation of the population structure of ST235 MDR P. aeruginosa in Japan.
High Substitution Rate in ST235 MDR P. aeruginosa
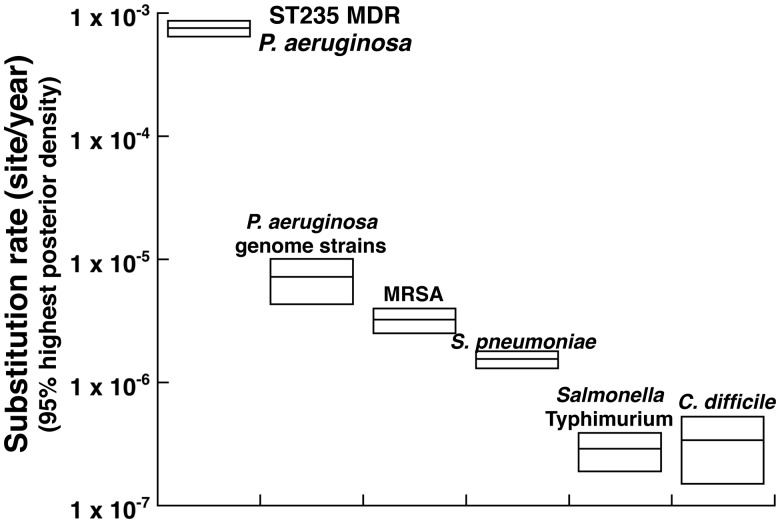
Bayesian analyses suggested that the estimated substitution rate of ST235 MDR P. aeruginosa was between 6.44 and 8.66 × 10−4 (95% highest posterior density) substitutions per site per year (fig. 3). This substitution rate is at least 100-fold higher than those of other bacterial species (Harris et al. 2010; Croucher et al. 2011; Okoro et al. 2012; He et al. 2013). We analyzed the genome sequences of P. aeruginosa isolates, determined at least at the scaffold level registered in the database (https://www.ncbi.nlm.nih.gov hereafter referred to as “genome strains,” supplementary table 4, Supplementary Material online) with the same parameters. The results showed a substitution rate of 4.3 ×10−6 to 1.0 × 10−5, which was lower than that of ST235 MDR P. aeruginosa (fig. 3). These results suggested that the substitution rate of genomes in the ST235 P. aeruginosa isolates was relatively high.
Fig. 3.
—Substitution rate of various bacteria, including ST235 Pseudomonas aeruginosa. Comparison of reported substitution rates of MRSA (Harris et al. 2010), Streptococcus pneumonia (Croucher et al. 2011), Salmonella typhimurium (Okoro et al. 2012), Clostridium difficile (He et al. 2013), ST235 MDR P. aeruginosa, and P. aeruginosa (supplementary 4, Supplementary Material online). The substitution rates of P. aeruginosa were estimated using the BEAST program (Drummond and Rambaut 2007).
Unique Features of ST235 MDR P. aeruginosa
To examine why ST235 isolates are dominant among MDR P. aeruginosa, we analyzed genes not having homologs in the representative nonMDR P. aeruginosa strains PAO1 and LESB58, but found uniquely in ST235 P. aeruginosa. Of the 827 genes found to be conserved in NCGM2.S1, but not in PAO1 or LESB58, 466 were present at frequencies >95% in the 136 ST235 P. aeruginosa strains (supplementary table 5 and supplementary data 3, Supplementary Material online). We identified five regions in ST235 isolates (NCGM2_1785 to NCGM2_1844, NCGM2_1949 to NCGM2_1963, NCGM2_3701 to NCGM2_3710, NCGM2_3761 to NCGM_3772, and NCGM2_5888 to NCGM2_5904) containing unique genes with high density. Although four of these regions contained genes that presumably contribute to metabolism or are annotated as hypothetical proteins, one region, NCGM2_1785 to NCGM2_1844, showed a high degree of similarity to a region previously identified as an ExoU island in P. aeruginosa 6077 (Kulasekara et al. 2006). This region encodes type IV secretion systems (T4SS), which have been extensively characterized and shown to mediate conjugation, uptake, and release of DNA, as well as effector translocation (Abajy et al. 2007; Alvarez-Martinez and Christie 2009; Wallden et al. 2010; Smillie et al. 2010; Stingl et al. 2010). The T4SS harbored by >95% of 136 ST235 strains analyzed in this study was the Tfc type (supplementary fig. 2 and table 6, Supplementary Material online, NCGM2.S1 was used as the representative), which contributes to conjugation (Mohd-Zain et al. 2004). PAO1 harbors nine genes or clusters, including competence-associated genes and T4SS, highly identical to genes in other P. aeruginosa strains (supplementary fig. 3 and supplementary data 4, Supplementary Material online). In contrast to PAO1, the additional T4SS found in ST235 isolates was not conserved among the P. aeruginosa strains, except for the ST235 strain 39016. Strains negative for the additional T4SS, such as PAO1 and LBSE58, did not harbor any drug resistance genes, suggesting that ST235 strains with additional T4SS are prone to acquisition of exogenous genetic material.
Absence of CRISPR/CAS System in ST235 P. aeruginosa
Additional genetic traits affecting the acquisition of exogenous genetic material have been reported to cluster in regularly-interspaced short palindromic repeats (CRISPRs) and in CASs (CRISPR/CAS; Bhaya et al. 2011; Wiedenheft et al. 2012). CRISPRs rely on small RNAs for sequence-specific detection and silencing of foreign nucleic acids, including bacteriophages and plasmids. Although the distributions of CRISPRs and CASs differed among the 26 P. aeruginosa genome strains, the ST235 isolates NCGM2.S1 and 39016 did not harbor more than one confirmed CRISPR and had relatively few CRISPR spacers (supplementary fig. 3 and supplementary data 4, Supplementary Material online). Furthermore, NCGM2.S1 and 39016 may not harbor recognizable CAS1 proteins. To confirm this tendency, the numbers of CRISPRs and spacers among the 136 ST235 P. aeruginosa strains were counted, using CRISPRfinder (http://crispr.i2bc.paris-saclay.frlast; accessed November 27, 2017; Grissa et al. 2007). Remarkably, most ST235 isolates did not harbor confirmed CRISPRs and spacers, with the numbers of both varying in other ST isolates. We also analyzed whether the 136 ST235 strains harbored the 149 genes encoding CASs of P. aeruginosa listed in supplementary table 2, Supplementary Material online. The results indicated that none of the ST235 isolates harbored genes encoding CAS except for only 2 strains (PA1604, PL3220) (for PA1604: csf1, csf2, csf3, and csf4; for PL3220: cas1 cas6/csy4, and csf2; supplementary data 1, Supplementary Material online).
These results suggest that ST235 strains without the CRISPR/CAS system are prone to acquisition of exogenous genetic material.
Discussion
In this study, the features of MDR P. aeruginosa were determined by WGS analyses of 185 P. aeruginosa isolates isolated from hospitalized patients. MLST analysis showed that ST235 was the predominant clonal ST lineage, being identified in 136 of the 185 strains. These 136 strains, consisting of isolates from Japan and other countries, could be divided into seven subclades, each of which had specific features, including conserved drug-resistance genes, geographic location and genomic background. Estimates suggested that the most recent common ancestor of these ST235 isolates arouse ∼80 years ago, with each subclade radiating over a few decades. These findings suggest that the ancestor of each subclade emerged over a few decades and spread in particular communities. WGS analysis also showed that ST235 MDR P. aeruginosa had a higher substitution rate, an additional T4SS, and no functional CRIPSR/Cas interference. These genetic traits may enable these bacteria to acquire exogenous genetic elements more efficiently, as well as increasing the intrinsic high competency of P. aeruginosa. Most currently used antibiotics were developed and used in patients from the 1960s to the 1980s. The acquisition of resistance to commonly used antibiotics, whether by substitution or the acquisition of drug resistance genes, may be the driving force behind the persistence and expansion of MDR ST235 P. aeruginosa in hospitals over the last several decades. Selection pressure from antibiotic treatment would allow the expansion of particular strains, such as ST235, that efficiently acquire substitution and drug resistance genes.
In conclusion, these results provide clues to the mechanism underlying the emergence and spread of MDR P. aeruginosa in particular communities and the key traits of the predominant ST235 MDR P. aeruginosa strain.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
T.M.A. designed the experiments and wrote the manuscript; T.T., N.O., and T.K. collected the P. aeruginosa isolates, contributed experimental suggestions, and strengthened the writing of the manuscript; N.V.H., P.T., B.M.P., and M.S. collected the P. aeruginosa isolates. All authors reviewed and provided comments to the text.
Supplementary Material
Acknowledgments
We thank M. Komiya, Y. Sakurai, and K. Shimada for their excellent technical assistance. We also thank Dr Oshiro at Tsukuba University for critical reading of the manuscript. This research project was supported by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases of the Ministry of Education, Culture, Sports, Science and Technology, Japan. T.T. was supported by JSPS KAKENHI Grant Number 16K19133. T.K. was supported by Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED). T.M.A., and N.O. were supported by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from the Ministry of Education, Culture, Sport, Science & Technology in Japan and AMED.
Literature Cited
- Abajy MY, et al. 2007. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J Bacteriol. 189(6):2487–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Alvarez-Martinez CE, Christie PJ.. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 73(4):775–808.http://dx.doi.org/10.1128/MMBR.00023-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R.. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 45:273–297.http://dx.doi.org/10.1146/annurev-genet-110410-132430 [DOI] [PubMed] [Google Scholar]
- Chen FJ, Lo HJ.. 2003. Molecular mechanisms of fluoroquinolone resistance. J Microbiol Immunol Infect. 36(1):1–9. [PubMed] [Google Scholar]
- Cholley P, et al. 2011. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J Clin Microbiol. 49(7):2578–2583.http://dx.doi.org/10.1128/JCM.00102-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331(6016):430–434.http://dx.doi.org/10.1126/science.1198545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG.. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 42(12):5644–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A.. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7(1):214..http://dx.doi.org/10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empel J, et al. 2007. Outbreak of Pseudomonas aeruginosa infections with PER-1 extended-spectrum beta-lactamase in Warsaw, Poland: further evidence for an international clonal complex. J Clin Microbiol. 45(9):2829–2834.http://dx.doi.org/10.1128/JCM.00997-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas ME, Koletsi PK, Bliziotis IA.. 2006. The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J Med Microbiol. 55(12):1619–1629.http://dx.doi.org/10.1099/jmm.0.46747-0 [DOI] [PubMed] [Google Scholar]
- Giske CG, et al. 2006. Establishing clonal relationships between VIM-1-like metallo-beta-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J Clin Microbiol. 44(12):4309–4315.http://dx.doi.org/10.1128/JCM.00817-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C.. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35(Web Server):W52–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.http://dx.doi.org/10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hancock RE, Speert DP.. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat. 3(4):247–255.http://dx.doi.org/10.1054/drup.2000.0152 [DOI] [PubMed] [Google Scholar]
- Harris SR, et al. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327(5964):469–474.http://dx.doi.org/10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, et al. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 45:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban DJ, Biedenbach DJ, Mutnick AH, Jones RN.. 2003. Pathogen of occurrence and susceptibility patterns associated with pneumonia in hospitalized patients in North America: results of the SENTRY Antimicrobial Surveillance Study (2000). Diagn Microbiol Infect Dis. 45(4):279–285.http://dx.doi.org/10.1016/S0732-8893(02)00540-0 [DOI] [PubMed] [Google Scholar]
- Katoh K, Frith MC.. 2012. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 28(23):3144–3146.http://dx.doi.org/10.1093/bioinformatics/bts578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirikae T, Mizuguchi Y, Arakawa Y.. 2008. Investigation of isolation rates of Pseudomonas aeruginosa with and without multidrug resistance in medical facilities and clinical laboratories in Japan. J Antimicrob Chemother. 61(3):612–615.http://dx.doi.org/10.1093/jac/dkm537 [DOI] [PubMed] [Google Scholar]
- Kitao T, et al. 2012. Emergence of a novel multidrug-resistant Pseudomonas aeruginosa strain producing IMP-type metallo-beta-lactamases and AAC(6′)-Iae in Japan. Int J Antimicrob Agents. 39(6):518–521. [DOI] [PubMed] [Google Scholar]
- Kos VN, et al. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother. 59(1):427–436.http://dx.doi.org/10.1128/AAC.03954-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara BR, et al. 2006. Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J Bacteriol. 188(11):4037–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepsanovic Z, et al. 2008. Characterisation of the first VIM metallo-beta-lactamase-producing Pseudomonas aeruginosa clinical isolate in Serbia. Acta Microbiol Immunol Hung. 55(4):447–454.http://dx.doi.org/10.1556/AMicr.55.2008.4.9 [DOI] [PubMed] [Google Scholar]
- Lister PD. 2002. Chromosomally-encoded resistance mechanisms of Pseudomonas aeruginosa: therapeutic implications. Am J Pharmacogenomics. 2(4):235–243.http://dx.doi.org/10.2165/00129785-200202040-00003 [DOI] [PubMed] [Google Scholar]
- Livermore DM. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 34(5):634–640. [DOI] [PubMed] [Google Scholar]
- Miyoshi-Akiyama T, Kuwahara T, Tada T, Kitao T, Kirikae T.. 2011. Complete genome sequence of highly multidrug-resistant Pseudomonas aeruginosa NCGM2.S1, a representative strain of a cluster endemic to Japan. J Bacteriol. 193(24):7010.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Zain Z, et al. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J Bacteriol. 186(23):8114–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoro CK, et al. 2012. Intracontinental spread of human invasive Salmonella typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 44(11):1215–1221.http://dx.doi.org/10.1038/ng.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 49(2):479–487.http://dx.doi.org/10.1128/AAC.49.2.479-487.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.http://dx.doi.org/10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Queenan AM, Bush K.. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 20(3):440–458.http://dx.doi.org/10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rojas A, Oliver A, Blazquez J.. 2012. Intrinsic and environmental mutagenesis drive diversification and persistence of Pseudomonas aeruginosa in chronic lung infections. J Infect Dis. 205(1):121–127. [DOI] [PubMed] [Google Scholar]
- Samuelsen O, et al. 2010. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob Agents Chemother. 54(1):346–352.http://dx.doi.org/10.1128/AAC.00824-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok Y, et al. 2011. Dissemination of IMP-6 metallo-beta-lactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea. J Antimicrob Chemother. 66(12):2791–2796.http://dx.doi.org/10.1093/jac/dkr381 [DOI] [PubMed] [Google Scholar]
- Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F.. 2010. Mobility of plasmids. Microbiol Mol Biol Rev. 74(3):434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B.. 2010. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci U S A. 107(3):1184–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strateva T, Yordanov D.. 2009. Pseudomonas aeruginosa – a phenomenon of bacterial resistance. J Med Microbiol. 58(Pt 9):1133–1148. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739.http://dx.doi.org/10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, et al. 2015. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. MBio 6(6):e01796–e01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viedma E, et al. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum beta-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother. 53(11):4930–4933.http://dx.doi.org/10.1128/AAC.00900-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J, Arakawa Y.. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat. 15(3):133–148.http://dx.doi.org/10.1016/j.drup.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Wallden K, Rivera-Calzada A, Waksman G.. 2010. Type IV secretion systems: versatility and diversity in function. Cell Microbiol. 12(9):1203–1212.http://dx.doi.org/10.1111/j.1462-5822.2010.01499.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA.. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482(7385):331–338.http://dx.doi.org/10.1038/nature10886 [DOI] [PubMed] [Google Scholar]
- Yoo JS, et al. 2012. Dissemination of genetically related IMP-6-producing multidrug-resistant Pseudomonas aeruginosa ST235 in South Korea. Int J Antimicrob Agents. 39:300–304.http://dx.doi.org/10.1016/j.ijantimicag.2011.11.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.