Abstract
The sperm whale, made famous by Moby Dick, is one of the most fascinating of all ocean-dwelling species given their unique life history, novel physiological adaptations to hunting squid at extreme ocean depths, and their position as one of the earliest branching toothed whales (Odontoceti). We assembled the sperm whale (Physeter macrocephalus) genome and resequenced individuals from multiple ocean basins to identify new candidate genes for adaptation to an aquatic environment and infer demographic history. Genes crucial for skin integrity appeared to be particularly important in both the sperm whale and other cetaceans. We also find sperm whales experienced a steep population decline during the early Pleistocene epoch. These genomic data add new comparative insight into the evolution of whales.
Keywords: sperm whale, cetaceans, genome
Introduction
The sperm whale, made famous by Moby Dick, makes some of the deepest and longest dives of any marine mammal: >73 min long and up to 2,035 m deep (Watkins et al. 1993; Watwood et al. 2006) to feed on squid, including the infamous giant and colossal squids (Best 1979; Whitehead 2003). Previous comparative genomic analyses of cetaceans indicated genic adaptation to a marine existence (Foote et al. 2015; Yim et al. 2014), including convergent pathways of metabolism regulation for deep diving (Foote et al. 2015). However, to date, the sperm whale—one of the deepest diving and earliest branching toothed whales (Odontoceti; Whitehead 2003)—has been excluded from these comparisons. We sequenced and assembled multiple sperm whale genomes to explore genic adaptation. Given the important and broad physiological roles played by proteases, our explorations mostly focused on examining protease loss-of-function (LoF) events important in sperm whale, and cetacean, evolution. We also sought to discover which genes showed signs of positive selection shared with other cetaceans or unique to sperm whale. Finally, as previous analyses suggested that sperm whales experienced a global expansion <80,000 years ago (Alexander et al. 2016), we examine the estimated historical effective population size using samples from throughout the sperm whale’s range.
Materials and Methods
We sequenced a Gulf of Mexico female sperm whale (GMX) to high coverage (72×) using short-insert and mate-pair libraries of 100 bp length (detailed in the supplementary material S1, Supplementary Material online) on an Illumina HiSeq2000. We assembled the draft genome of all sequences with ALLPATHS (Gnerre et al. 2011) using default parameter settings, subjecting assembly input reads to quality control as detailed in the ALLPATHS documentation (Gnerre et al. 2011). We obtained RNAseq data from skin biopsies of a different GMX sperm whale to aid gene annotation as described in the supplementary material S1, Supplementary Material online. Gene annotation was performed according to the NCBI gene annotation pipeline as described here: http://www.ncbi.nlm.nih.gov/books/NBK169439/. After aligning genes from the sperm whale with other taxa (detailed in supplementary material S1, Supplementary Material online) to establish 1:1 gene orthology, positive selection was detected using PAML4.0 (Yang 2007) and impact on protein structure tested with Provean (Choi and Chan 2015). Canonical pathway enrichment of gene clusters under positive selection was established as detailed in the supplementary material S1, Supplementary Material online. Protease genes were manually annotated and validated for loss/duplication events using BATI (http://degradome.uniovi.es/downloads.html). Four additional sperm whale individuals (supplementary table S1, Supplementary Material online) were resequenced to moderate depth (21–28×) and reads were mapped to the draft genome as described in the supplementary material S1, Supplementary Material online. We calculated heterozygosity on a per-individual basis using VCFtools (Danecek et al. 2011). Effective population size was reconstructed with PSMC (Li and Durbin 2011) using the parameters specified in the supplementary material S1, Supplementary Material online.
Results and Discussion
Our sperm whale total assembled sequence was similar in size to other assembled cetacean genomes (supplementary table S2, Supplementary Material online). Using our GMX individual reference assembly (Genbank assembly accession GCA_000472045.1) we inferred 18,686 protein-coding genes—second only to the baiji (Lipotes vexillifer) among sequenced cetaceans at 18,906 genes. Using a core eukaryotic mapping method (Simao et al. 2015) we also demonstrated >94.7% of conserved genes were complete in our assembly (supplementary table S3, Supplementary Material online). Of the 18,686 protein-coding genes, 12,717 had single-copy orthologs in both human and other cetartiodactyls (supplementary table S4, Supplementary Material online; additional methods/results can be found in the supplementary material S1, Supplementary Material online). A total of 45 genes found across eight taxa were identified as being under positive selection in the sperm whale lineage; these genes also passed our stringent functional impact tests (default cutoff < −2.5) using Provean (Choi and Chan 2015; supplementary file S1, Supplementary Material online). Several significant pathways emerged from enrichment analyses, which included genes associated with blood-circulation and skin stress responses (table 1). Cetaceans, including the sperm whale, exhibit molting or skin sloughing (Amos et al. 1992), potentially as an adaptive response to fouling by barnacles and other organisms. However, sperm whales face the additional challenge of maintaining skin integrity and blood homeostasis at high water pressures during deep foraging dives.
Table 1.
Genes under Positive Selection Enriched by Pathway, Phenotype, or Protein Interactions
| Pathway | Source | Genes | Ratio of Enrichment | Adjusted P-value |
|---|---|---|---|---|
| Focal adhesion | Wiki | CHAD, COL1A2, THBS2, TNC, FLT1 | 6.9 | 0.015 |
| Focal adhesion | KEGG | CHAD, COL1A2, PARVG, THBS2, TNC, FLT1 | 7.5 | 0.0076 |
| Pemphigus | Disease | PPL, EVPL, DSP, DSG3 | 38.8 | 0.0006 |
| Calcium signaling | KEGG | PTK2B, ADCY3, RYR1, NOS2, P2RX3, PHKB | 7.6 | 0.0076 |
| Blood circulation | GO | ALOX5, CHD7, WNK1, EPAS1, CX3CL1, PPP1R13L, COL1A2, AZU1, DSP, GUCY1A3, MYBPC3, TBC1D8 | 3.4 | 0.036 |
| Cornified envelope | Reactome | DSP, TGM1, KRT4, PKP1, DSG3, PPL, EVPL | NA | aFDR 3.65 × 10−11 |
The false discovery rate (FDR) calculated within the Reactome software (Croft et al. 2014) is the probability corrected for multiple comparisons. Adjusted P values are not provided.
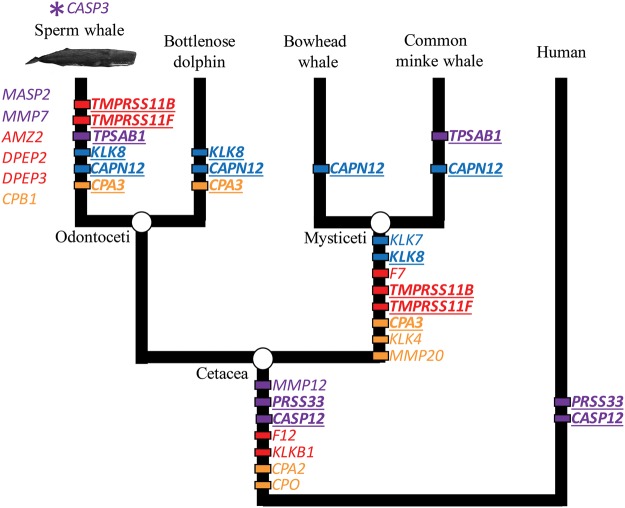
To complement the analysis of genes under positive selection, we manually annotated the complete set of proteases (i.e., degradome) of the sperm whale. This independent analysis identified several proteases involved in skin function and blood homeostasis that showed LoF events along the lineage leading to sperm whales (fig. 1; additional methods/results in supplementary material S1, Supplementary Material online). We also detected LoF in proteases involved in inflammation, immunity and metabolism within cetaceans, and specifically within the sperm whale. A loss of several proteases in cetaceans suggests a trend towards a milder inflammatory response relevant to Peto’s paradox: A theory postulated to explain the lower relative incidence of cancer in large mammals (Caulin and Maley 2011). In addition, MMP7—which promotes metastasis when expressed at high levels (Li et al. 2014; Koskensalo et al. 2011)—contains a premature stop codon in sperm whales, a putative sperm whale-specific mechanism to reduce cancer incidence. We also found that CASP12 and PRSS33 were independently lost in cetaceans and some hominoids, suggesting a case of convergent evolution of the immune system in very different environments. Several proteases involved in digestion (CPA2, CPA3, CPO) were also lost in cetaceans (fig. 1). In some cases these losses were independent, suggesting convergent evolution driven by trophic level. As expected, odontocetes retain functional orthologs of proteases involved in dentition (KLK4, MMP20), which were lost in mysticetes, who use baleen—not teeth—to filter food (Keane et al. 2015).
Fig. 1.
—Cetacean-specific losses of protease genes. Proteases that have undergone loss-of-function in sperm whales, specifically, are shown to the left of the phylogeny whereas those that are inferred to be convergent, or inferred to have occurred in ancestral lineages, are mapped on to the phylogeny. Each event is depicted along the branch where loss events have been inferred to occur. Genes expected to impact skin function are colored blue; immune system: purple; blood homeostasis: red; digestion: orange, and those showing convergent loss-of-function as underlined bold. The unique duplication of sperm whale CASP3 is shown above the phylogeny and marked by an asterisk.
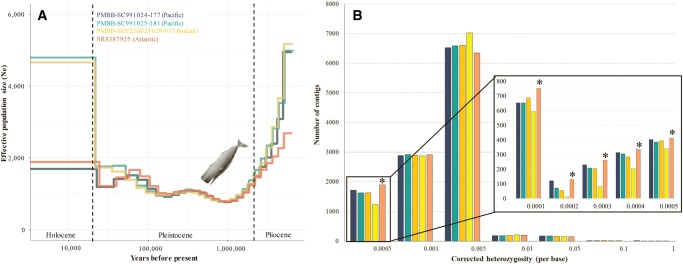
To better understand patterns of genetic diversity among sperm whales from different ocean basins, we carried out medium-coverage resequencing of individuals from the Pacific Ocean and Indian Ocean. Average genome-wide heterozygosity per base, corrected for callable sequence space, was 0.0011. This value is low in comparison with the fin whale (0.0015) and bottlenose dolphin (0.0014; Yim et al. 2014), suggesting the sperm whale has a smaller effective population size (Ne). A pairwise sequentially Markovian coalescent (PSMC) analysis (Li and Durbin 2011) indicated a rapid decline in Ne during the transition from the Pliocene to Pleistocene epoch, inferred consistently regardless of the ocean origin of samples (fig. 2A). The increase in upwelling associated with the Pliocene and/or cycles of glaciation within the Pleistocene have been implicated in the evolution of gigantism in mysticetes (Slater et al. 2017), as well as the diversification of marine dolphins (do Amaral et al. 2016). This suggests that changes in ocean dynamics during this time period have had a strong impact on cetaceans in general, and we suggest are also the likely cause of the inferred sperm whale population decline. The GMX sample had significantly lower heterozygosity than Pacific and Indian Ocean samples (fig. 2B, supplementary table S1, Supplementary Material online). Future sequencing will clarify whether lower diversity is restricted to GMX, or characteristic of the entire Atlantic. However, the isolation of GMX due to high levels of female philopatry (inferred from differentiation of the maternally-inherited mitochondrial DNA, Engelhaupt et al. 2009; Alexander et al. 2016), and the limited census size (763 sperm whales in 2009, Waring et al. 2013), suggest that GMX could be subjected to greater levels of genetic drift associated with a small and maternally-isolated population. The ability of the sperm whale to respond to future selective pressures, including climate change, in the face of such reduced genetic diversity should be a focus of ongoing study.
Fig. 2.
—Estimated effective population size history and heterozygosity of sperm whales from different ocean basins. Samples are color coded by the key, with blue/green = Pacific, orange/yellow = Indian Ocean, and dark orange = Gulf of Mexico, Atlantic. (A) PSMC reconstruction of effective population size through time by sample (excluding SEY420021031-063, see supplementary material S1, Supplementary Material online), dashed lines represent the estimated start dates for each epoch; (B) Genome wide distribution of heterozygosity for each sample, by contig/scaffold. The Gulf of Mexico sample—characterized by low heterozygosity—is marked by an asterisk where it has the largest number of contigs in a category. The insert emphasizes that this sample has the largest number of contigs with low heterozygosity (<0.0005). Bright yellow in panel (b) is additional Indian ocean sample
Overall, our results suggest positive selection has differentially affected localized portions of the sperm whale genome. In particular, the complex pattern of convergent gene evolution involving skin-related genes suggests they have played an important role in aquatic adaptation, possibly influenced by the somewhat contradictory requirements of heat insulation, buoyancy and deep diving. In comparison to the localized effects of selection on the genome, we infer that the sperm whale experienced a rapid population decline, potentially in response to glaciation, which had a broad effect on genome-wide diversity. Given the apparent influence of past climate change, monitoring the on-going response of sperm whales to anthropogenically mediated climate change will be paramount.
Authors Contributions
S.S.W., J.W.J., J.W.S. isolated genomic DNA and sexed all samples. L.K., T.M.B. performed all population history analyses. J.G.P., C.L.O., V.Q. performed all protease analyses. C.T., P.M. completed genome assembly and curation. J.C., T.J.K., T.G. analyzed population sequences. M.J.M., F.H.F. analyzed gene selection. R.M.W. completed all gene orthology analyses. W.C.W., A.A. wrote the paper and reviewed all analyses. All authors have read and approved the manuscript.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health grant 2R24OD011198-04A1 (W.C.W., PI); National Institute of Environmental Health Sciences [ES016893 (J.W. Sr, PI)]; Army Research Office [W911NF-09-1-0296 (J.W. Sr, PI)]; Swiss National Science Foundation grant PP00P3_170664 (R.M.W.); Ministerio of Economia and Competitividad (Spain); and European Union (ERC-Advanced Grant DeAge) (C.L.-O., PI); the Campbell Foundation; the Ocean Foundation; Ocean Alliance; and the many individual and anonymous Wise Laboratory donors. The authors declare no competing financial interests. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Sciences, the National Institutes of Health, the Army Research Office or the Department of Defense. Work was conducted under National Marine Fisheries Service permit #1008-1637-03 (J.W. Sr, PI) and permit #751-1614 (I.K., PI). We thank Kyung Kim for computational support. We would like to thank Catherine Wise, James Wise, Chris Gianios, and all the Wise Laboratory/Odyssey science team volunteers for their help with technical support, whale spotting and sample collection. We thank Iain Kerr, Roger Payne, Bob Wallace, Derek Walker, and all of the Odyssey boat crew for their help with sample collection and logistics. We thank C. Scott Baker for comments on this manuscript. Finally, we thank all the volunteers who supported us in this project.
Literature Cited
- Alexander A, et al. 2016. What influences the worldwide genetic structure of sperm whales (Physeter macrocephalus)? Mol Ecol. 2512:2754–2772. [DOI] [PubMed] [Google Scholar]
- Amos W, et al. 1992. Restrictable DNA from sloughed cetacean skin; its potential for use in population analysis. Mar Mamm Sci. 83:275–283. [Google Scholar]
- Best PB. 1979. Social organization in sperm whales, Physeter macrocephalus. In Behavior of marine animals. pp. 227–289. Springer US.
- Caulin AF, Maley CC.. 2011. Peto's Paradox: evolution's prescription for cancer prevention. Trends Ecol Evol. 264:175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Chan AP.. 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 3116:2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, et al. 2014. The Reactome pathway knowledgebase. Nucleic Acids Res. 42(Database issue):D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 2715:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Amaral KB, Amaral AR, Fordyce RE, Moreno IB.. 2016. Historical biogeography of delphininae dolphins and related taxa (Artiodactyla: Delphinidae). J Mamm Evol. 1–19. https://link.springer.com/article/10.1007/s10914-016-9376-3 [Google Scholar]
- Engelhaupt D, et al. 2009. Female philopatry in coastal basins and male dispersion across the North Atlantic in a highly mobile marine species, the sperm whale (Physeter macrocephalus). Mol Ecol. 1820:4193–4205. [DOI] [PubMed] [Google Scholar]
- Foote AD, et al. 2015. Convergent evolution of the genomes of marine mammals. Nat Genet. 473:272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 1084:1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 101:112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskensalo S, Louhimo J, Nordling S, Hagström J, Haglund C.. 2011. MMP-7 as a prognostic marker in colorectal cancer. Tumour Biol. 322:259–264. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 4757357:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. 2014. Prediction of peritoneal recurrence by the mRNA level of CEA and MMP-7 in peritoneal lavage of gastric cancer patients. Tumour Biol. 354:3463–3470. [DOI] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 3119:3210–3212. [DOI] [PubMed] [Google Scholar]
- Slater GJ, Goldbogen JA, Pyenson ND.. 2017. Independent evolution of baleen whale gigantism linked to Plio-Pleistocene ocean dynamics. Proc Biol Sci. 2841855:20170546.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring GTJE, Maze-Foley K, Rosel PE.. 2013. NOAA Tech Memo NMFS-NE-223. (ed. NOAA).
- Watkins WA, Daher MA, Fristrup KM, Howald TJ, di Sciara GN.. 1993. Sperm whales tagged with transponders and tracked underwater by sonar. Mar Mamm Sci. 91:55–67. [Google Scholar]
- Watwood SL, Miller PJ, Johnson M, Madsen PT, Tyack PL.. 2006. Deep-diving foraging behaviour of sperm whales (Physeter macrocephalus). J Anim Ecol. 753:814–825. [DOI] [PubMed] [Google Scholar]
- Whitehead H. 2003. Sperm whales: social evolution in the ocean. Chicago: The University of Chicago Press. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yim HS, et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 461:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.