Abstract
Here, we employed cDNA amplicon sequencing using a long-read portable sequencer, MinION, to characterize various types of mutations in cancer-related genes, namely, EGFR, KRAS, NRAS and NF1. For homozygous SNVs, the precision and recall rates were 87.5% and 91.3%, respectively. For previously reported hotspot mutations, the precision and recall rates reached 100%. The precise junctions of EML4-ALK, CCDC6-RET and five other gene fusions were also detected. Taking advantages of long-read sequencing, we conducted phasing of EGFR mutations and elucidated the mutational allelic backgrounds of anti-tumor drug-sensitive and resistant mutations, which could provide useful information for selecting therapeutic approaches. In the H1975 cells, 72% of the reads harbored both L858R and T790M mutations, and 22% of the reads harbored neither mutation. To ensure that the clinical requirements can be met in potentially low cancer cell populations, we further conducted a serial dilution analysis of the template for EGFR mutations. Several percent of the mutant alleles could be detected depending on the yield and quality of the sequencing data. Finally, we characterized the mutation genotypes in eight clinical samples. This method could be a convenient long-read sequencing-based analytical approach and thus may change the current approaches used for cancer genome sequencing.
Keywords: MinION, lung cancer cell lines, cancer mutations, phasing
1. Introduction
Sequencing technologies have substantially advanced the study of cancer genomics. Large-scale studies, such as The Cancer Genome Atlas (TCGA)1 and the International Cancer Genome Consortium (ICGC)2, have generated catalogs of various types of mutations in cancer cells, and this information is currently an invaluable resource for various purposes. In particular, these data collections provide critical information that can be used to identify the driver mutations in each cancer patient and inform initial therapeutic approaches, including molecular-targeted anti-cancer drugs. For example, several effective anti-cancer drugs are available for the treatment of lung adenocarcinoma. The EGFR tyrosine kinase inhibitors gefitinib and erlotinib have been used to treat EGFR-mutant tumors,3 while crizotinib and other ALK-kinase inhibitors can be a highly effective therapy for ALK-fusion positive patients.4,5 To determine whether these drugs should be administered, it is necessary to sequence the cancer nucleic acids and identify the driver mutation in each case. However, sequencing is limited by the following disadvantages: a large and very expensive instrument is needed; short-read sequencing occasionally overlooks the allelic background of the cancer mutations; and structural aberrations, such as fusion genes and splicing alterations, are typically difficult to detect.
MinION, which is a recently developed portable, disposable long-read sequencer,6 has the potential to address the drawbacks of the currently used cancer genome sequencing technologies. MinION is USB-sized and can be operated on a laptop PC. The template preparation takes only 2–3 h and does not require any specific experimental skills. In less than 48 h, hundreds of thousands of reads can be obtained, which are occasionally longer than 10 kb. Recent studies have applied MinION for several purposes focusing primarily on the detection and genotyping of pathogens. For example, one study reported the antibiotic genomic regions in Salmonella strains.7 In a more clinical setting, real-time sequencing using MinION and a phylogenic analysis of the obtained sequences revealed Salmonella outbreaks in a hospital.8 More recently, the first field application of MinION was used to diagnose the lethal virus Ebola in Africa.9 However, an analysis of genomic mutations in cancers using MinION has not been reported, except for a few pioneering studies.10,11 To the best of our knowledge, the application of MinION for the identification and characterization of various genomic aberrations in cancer cells remains elusive.
In this study, we assessed the ability of MinION sequencing technology to characterize various types of cancer mutations. We sequenced cDNA amplicons from cancer-related genes and detected single nucleotide variants (SNVs) in EGFR, KRAS and NRAS; a short deletion in EGFR; aberrantly spliced RNAs in NF1; and CCDC6-RET, EML4-ALK, and other gene fusion transcripts in lung adenocarcinoma cell lines.12 The phasing of the allelic information of the mutations was also examined using the long-read sequencing capability of MinION. We used cell lines as a model to apply a simple sequencing method to clinical cancer genomes.
2. Materials and Methods
2.1. Cell lines
The lung adenocarcinoma cell lines PC-9, LC2/ad, PC-7, RERF-LC-Ad2, H1437, H1975, H2228, H2347, A549 and H322 were used in this study (Supplementary Table S1) and have been previously described.12 Total RNA was extracted from frozen cancer cell pellets using the RNeasy Maxi kit (Qiagen). The total RNA was assessed using an Agilent BioAnalyzer (Agilent), and the total RNA samples satisfied an RNA Integrity Number (RIN)>9. For the H1975 and A549 cell lines, the genomic DNA was extracted from 1 × 107 cell pellets using the DNeasy Blood & Tissue Kit (Qiagen). The extracted genomic DNA was eluted into 200 μl Buffer AE.
2.2. Clinical samples
The RNA samples were obtained from eight Japanese lung adenocarcinoma patients after obtaining their appreciated informed consent and the institutional review at the National Cancer Center Japan. The total RNA was extracted from frozen tissues using TRIzol (Invitrogen) as previously reported.13 The RNA was assessed using an Agilent BioAnalyzer (Agilent). All samples satisfied RIN > 7.
2.3. RT-PCR
Prior to the cDNA synthesis, the total RNA (approximately 10 μg from the cell lines and 500 ng from the clinical samples) was treated with DNase I (Takara) in a buffer containing 8 mM MgCl2, 40 mM Tris-HCl (pH 7.5), 5 mM DTT and RNasin Ribonuclease Inhibitor (Promega) for 10 min at 37 °C. After the phenol-chloroform extraction and ethanol precipitation for RNA purification, first-strand cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen) in First-Strand buffer with 0.8 mM dNTPs, 12 mM DTT, 2.5 μl dT primer (5’-GCGGCTGAAGACGGCCTATGTGGCCTTTTTTTTTTTTTTTTT-3’, 10 pmol/μl) and RNasin Ribonuclease Inhibitor (Promega) at 42 °C overnight. After the phenol-chloroform extraction of 100 μl cDNA sample, 2 μl 0.5 M EDTA (pH 8.0) and 15 μl 0.1 M NaOH were added. The samples were incubated at 65 °C for 40 min, and 20 μl 1 M Tris-HCl (pH 7.0–7.5) were added to the sample. After the ethanol precipitation, the samples were dissolved in ≥50 μl (for cell lines) or 20 μl (for clinical samples) H2O. The cDNA samples were PCR amplified using 1 cycle of 30 s at 98 °C; 40 cycles of 10 s at 98 °C, 30 s at 60 °C and 5 min at 72 °C; and a final 10 min cycle at 72 °C. For the PCR reaction, 25 μl 2× Phusion Master Mix (Finnzymes), 16-19 μl H2O and 5 μl 5 μM forward and reverse primers were added to 1–4 μl cDNA template. For the EGFR analysis in the clinical samples (EGFR-iii), a second (nested) PCR was conducted with 1/10-diluted PCR products using 1 cycle of 30 s at 98 °C; 30 cycles of 10 s at 98 °C, 30 s at 60 °C and 5 min at 72 °C; and a final 10 min cycle at 72 °C. The PCR primers and expected product lengths are shown in Supplementary Fig. S1 and Supplementary Table S2A. The primers were designed using Primer3plus.14 After the PCR reaction, the amplicons were purified using a QIAquick PCR purification kit (Qiagen).
2.4. PCR of EGFR using genomic DNAs
For the PCR reaction, 1μl genomic DNA templates were mixed with 25 μl 2× Phusion Master Mix (Finnzymes), 19 μl nuclease-free water, 2.5 μl forward primer (10 μM) and 2.5 μl reverse primer (10 μM). The mixed samples were amplified using 1 cycle of 40 s at 98 °C; 40 cycles of 10 s at 98 °C, 30 s at 60 °C and 1 min at 72 °C; and a final 10 min cycle at 72 °C. The PCR primers are shown in Supplementary Table S2B.
2.5. MinION library preparation for the cell lines
Using a mixture of cDNA amplicons, MinION sequencing libraries were prepared using the Nanopore Sequencing Kit (SQK-MAP005, Oxford Nanopore Technologies) according to the manufacturer’s instructions. In total, 1 μg cDNA amplicons were prepared in 80 μl nuclease-free water and 5 μl DNA CS were added to the template. For the end-repair, the template was incubated at 20 °C for 20 min with 10 μl End-repair buffer and 5 μl End-repair enzyme mix (NEBNext End Repair Module, New England Biolabs). The template was purified using 100 μl Agencourt AMPure XP beads (Beckman Coulter, Inc.) and eluted into 25 μl H2O. To perform the dA tailing, 3 μl dA-tailing buffer and 2 μl dA-tailing enzyme (NEBNext dA-Taling Module) were added to the end-repaired DNA. The template was incubated at 37 °C for 10 min, purified by AMPure XP beads and eluted to 30 μl nuclease-free water. For the adapter ligation, the template was incubated for 10 min at room temperature with 10 μl Adapter Mix, 10 μl HP Adapter and 50 μl Blunt/TA adapter Ligase Master Mix (New England Biolabs). In total, 10 μl His-Tag beads (Dynabeads His-Tag Isolation and Pulldown, Life Technologies) were washed twice and resuspended with the Bead Binding Buffer. After the ligation, the adapter-ligated DNA was purified using the washed His-Tag beads and eluted in 25 μl Elution Buffer (‘pre-sequencing mix’). Before the sample loading, 325 μl priming buffer were mixed with 6.5 μl Fuel Mix, 162.5 μl 2× Running Buffer and 156 μl H2O. To prime the MinION R7.3 flow cell (FLO-MAP003, Oxford Nanopore technologies), 150 μl of the priming buffer were loaded twice with a 10-min waiting period before each loading. After preparing the flow cell, 75 μl 2× Running Buffer, 66 μl H2O, 3 μl Fuel Mix and 6 μl pre-sequencing mix were mixed, and then 150 μl of the resulting MinION sequencing library were loaded into the flow cell.
2.6. MinION library preparation for the clinical samples and dilution series
The Nanopore Sequencing Kit SQK-MAP006 (Oxford Nanopore Technologies) was used for the library preparation for the clinical samples and dilution series. Using a cDNA template (approximately 1 μg in 45 μl), 5 μl DNA CS were added. To perform the end-repair and dA tailing, 7 μl End-Prep buffer and 3 μl End-Prep enzyme mix (NEBNext Ultra II End-Repair/dA-tailing Module, New England Biolabs) were added. The template was incubated at 20 °C for 5 min and 65 °C for 5 min and was then purified using Agencourt AMPure XP beads (Beckman Coulter, Inc.). For the adapter ligation, 8 μl H2O, 10 μl Adapter Mix, 2 μl HP Adapter and 50 μl Blunt/TA adapter Ligase Master Mix (New England Biolabs) were added to the 30 μl DNA template. After incubating for 10 min at room temperature, 1 μl HP tether was added. The template was incubated for 10 min at room temperature. During the ligation, Dynabeads MyOne Streptavidin C1 beads (Life Technologies) were washed twice and suspended with 100 μl Bead Binding Buffer. After the ligation, the adapter-ligated DNA samples were purified using the washed beads and eluted using 25 μl Elution Buffer, followed by an incubation for 10 min at 37 °C. The eluted DNA sample was called the ‘pre-sequencing mix.’ Before loading the library, a mixture was prepared with 26.6 μl Fuel Mix, 500 μl 2× Running Buffer and 473.4 μl H2O. To prime the MinION R7.3 flow cell (FLO-MAP103, Oxford Nanopore Technologies), 500 μl of the mixture were loaded twice with a waiting period of 10 min before each loading. After preparing the flow cell, the MinION sequencing library was prepared with 75 μl 2× Running Buffer, 65 μl H2O, 4 μl Fuel Mix and 6 μl pre-sequencing mix and loaded to the flow cell.
2.7. MinION sequencing
Sequencing was performed for 48 h using MinKNOW. After the sequencing, base calling (2D Basecalling v1.24, SQK-MAP005 v1.34, SQK-MAP006 v1.62 and SQK-MAP006 v1.69) was performed via the ONT Metrichor (https://metrichor.com/s/index.shtml).
2.8. Alignment of MinION sequencing data of cDNA amplicons
The fastq files for the template, complement or 2D reads were converted from fast5 files using poretools version 0.5.1.15 Both ‘pass’ and ‘fail’ reads were used in this study. To compare the yield and qualities of the reads under different alignment conditions, LAST (version 658).16 and BWA (version 0.7.12-r1039).17 were used with the following sets of parameters; 1) BWA in ont2d mode; 2) LAST with the default parameter; 3) LAST with a match score of 1 (-r1), mismatch cost of 1 (-q1), gap existence cost of 1 (-a1) and gap extension cost of 1 (-b1); and 4) LAST with a gap existence cost of 12 (-a12), insertion existence cost of 15 (-A15), gap extension cost of 4 (-b4) and insertion extension cost of 4 (-B4) as determined according to the last-train.18 LAST uses the fastq quality data (with -Q1) to obtain more accurate alignments.19 The gap costs correspond to a statistical model of the specific probabilities of opening and extending insertions and deletions. The last-train determines the probabilities (and therefore costs) that fit the given sequence data. In these data, insertions are rarer than deletions; thus, their cost is higher.
Using LAST with the parameters -a12, -A15, -b4 and -B4, all 2D reads were mapped onto the 32,104 reference mRNA sequences based on the annotation file (refGene.txt) distributed by the UCSC Genome Browser (https://genome.ucsc.edu/).20 The alignments with the best score in each query were extracted and used for further analysis. When the scores were identical for two hits, the first hit was selected. When the read was aligned to different isoforms of the same gene with identical scores, the representative isoform was prioritized (representative isoforms selected were NM_005528 in EGFR, NM_004985 in KRAS, NM_002524 in NRAS, NM_000267 in NF1, NM_019063 in EML4, NM_020630 in RET, NM_000855 in GUCY1A2, NM_025202 in EFHD1, NM_004198 in CHRNA6 and NM_022464 in SIL1 according to Illumina RNA-Seq data previously published in the DNA Data Bank of Japan (DDBJ) under the accession number DRA00184612).
2.9. Alignment of MinION sequencing data of genomic DNA amplicons
Using LAST (version 658) with the parameters -a13, -A14, -b4 and -B3 which were determined by the last-train, all 2D reads were mapped onto chromosome 7 of the human genome UCSC hg38. The best alignments in each query were determined using last-split.
2.10. Detection of SNVs
Using the MinION reads, SNVs were detected as follows: 1) the reads aligned to the EGFR, KRAS, NRAS and NF1 target regions were extracted; 2) at each position, the depths of each base context were calculated considering only reads without errors ±3 bp of that position, and consensus sequences were constructed; 3) the consensus sequences were compared with the reference sequences and the SNVs were detected; and 4) the SNV candidates were verified using the Illumina whole-genome sequencing and RNA-Seq data obtained from the DDBJ under the accession numbers DRA001859 and DRA001846.12
Consensus sequences for the MinION reads were constructed as shown in Supplementary Fig. S2 (step2). Positions with a read depth of less than 100 were defined as ‘Low depth.’ Positions in which the types of bases could not be determined were defined as ‘Unknown.’ A base (A, T, G or C) in a given position was called if the number of supporting reads for that base was more than twice the sum of the other bases. For heterozygous sites, a second base was similarly defined. Reads without mismatches ±3 bp of the same position were used.
To verify the SNVs detected by the MinION consensus, the SNVs were also identified using the Illumina RNA-Seq data.12 The RNA-Seq data were mapped onto the human reference genome (UCSC hg19) using GSNAP.21 By scanning all the EGFR, KRAS, NRAS and NF1 target regions, 41 SNVs (variant allele frequency >10%) were detected as a validation dataset. Among the 41 true SNVs, 39 SNVs harboring sufficient depths in the MinION reads were considered true positives.
2.11. Detection of deletions and aberrant splicing
To detect short deletions in EGFR, the deletion depths were considered as the consensus sequences were constructed. The deletions were detected similarly to the SNV detection. After comparing to a known driver EGFR deletion, the false positive detections were removed. For aberrant NF1 exon skipping, the reads aligned to NF1 were re-aligned by split alignment using LAST. After calculating the depth of the split alignment, NF1 skipping was observed as site with no or few aligned reads.
2.12. Analysis of fusion transcripts
To detect the fusion transcripts and junction points in the MinION reads, the MinION reads were aligned to 32,104 reference mRNA sequences with split alignment22 using LAST (with the -m1 option to allow for multiple hits). Reads split between two different genes were extracted and counted. Gene pairs supported by more than 100 split reads were defined as fusion transcripts. A position at which the depth most significantly changed was considered a potential junction point. The junction points of known driver fusion transcripts (ALK and RET fusion transcripts) have been confirmed in previous studies.23–25
To assess the alignment accuracy and confirm the junction points, the MinION reads were also aligned to fused RNA sequences using LAST with the trained parameters described above. In addition, the MinION reads were aligned to 32,104 reference mRNA sequences to avoid a misalignment.
2.13.Phasing
To phase the two EGFR mutations (T790M and L858R) in the H1975 cells, the reads completely covering the EGFR kinase domain were extracted. Reads without any mismatches ±3 bp at both mutation sites were used for the phasing. The reads with deletions at the mutation sites were removed from this analysis.
2.14. EGFR dilution analysis for mutation detection
PCR amplicons of EGFR (EGFR-i, 3.4 kb) were prepared using cDNAs from H1975 (mutant) and RERF-LC-Ad2 (wild-type) cells. The H1975 amplicons were diluted with those from RERF-LC-Ad2 at the following ratios: 1:1, 1:4, 1:9, 1:19 and 1:99. The diluted samples were sequenced using MinION according to the manufacturer’s instructions. The MinION reads were processed for mutation phasing as described above.
2.15. Validation using TA cloning and Sanger sequencing
All driver mutations and junction points in the fusion transcripts were validated by direct Sanger sequencing. The results of the mutation phasing of EGFR in H1975 and junction sequences of EML4-ALK in H2228 were validated with TA cloning using the pMD20-T vector (Mighty TA-Cloning Kit, #6028, Takara). The PCR and sequencing primers are listed in Supplementary Table S3.
2.16. Data access
The MinION sequencing data from the cell lines were deposited in the DDBJ under the accession numbers DRA004627 and DRA005767. The MinION sequencing data from the clinical samples were published in the National Bioscience Database Center (NBDC) and DDBJ in Japan with the accession number JGAS00000000065 under controlled access.
3. Results
3.1. Nanopore sequencing of cancer-related genes
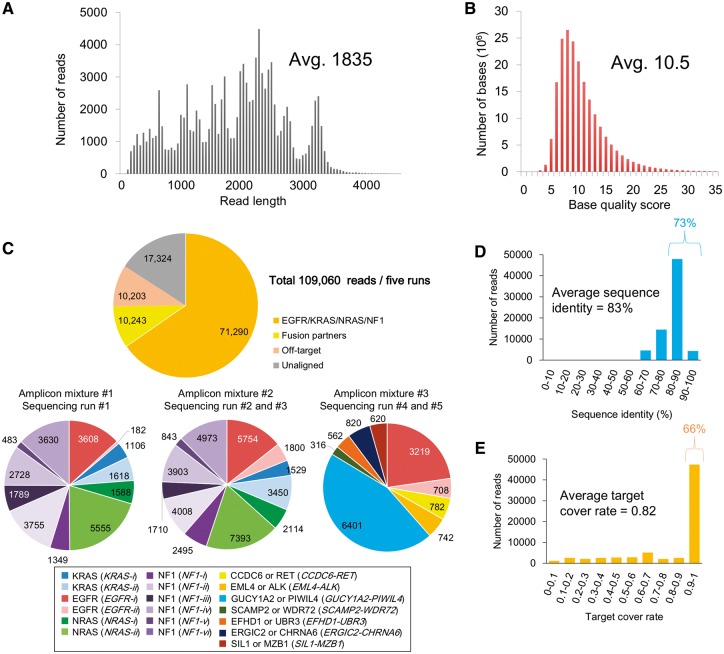
Using MinION, which was provided by Oxford Nanopore Technologies (ONT), we sequenced the reverse-transcription (RT)-PCR products from four genes, i.e., EGFR, KRAS, NRAS and NF1, as representative oncogenes and tumor-suppressor genes in lung cancers (Supplementary Table S1 and Supplementary Fig. S1; the primer sequences are shown in Supplementary Table S2A).26–30 These genes represent various mutation patterns. Specifically, single base substitutions, short deletions and exon skipping have been reported in the EGFR, KRAS and NRAS genes; the EGFR gene; and the NF1 gene, respectively. We also analyzed the ALK and RET fusion transcripts and several novel fusion transcripts identified in previous Illumina RNA-Seq studies.12 (Supplementary Table S1). In total, 33 PCR products were sequenced. We divided these products into three pools and performed five MinION sequencing runs. In each run, an average of 47,306, 25,948 and 21,812 reads were obtained as ‘template,’ ‘complement’ and ‘2D’ reads, respectively (Supplementary Table S4). In the following analysis, we used a total of 109,060 2D reads from the five runs.
As shown in Fig. 1A, the average read length was 1,835 bases. We expected that such generally long reads might sufficiently cover entire target regions, which ranged from 640 to 3,402 bases in length (see below). The base quality values (QV) were approximately 10.5 on average (Fig. 1B, statistics of the template and complement reads are also shown in Supplementary Fig. S3). We aligned the obtained reads to the reference human transcript sequences (32,104 transcripts; UCSC Genome Browser20). In the alignment, we compared two commonly used alignment programs BWA (ont2d mode)17 and LAST (with three sets of parameters).16 These programs were generally consistent with the aligned reads, although the target cover rates and sequence identities were dependent on the respective parameters (Supplementary Fig. S4, the parameters used in this analysis are shown in the Methods section and Supplementary Figure legends). Using LAST with the ‘trained’ parameters, on average 75% of the reads were aligned to the target mRNAs (Fig. 1C). Under these conditions, at least 150 reads were obtained for each of the bases in the target regions. For the EGFR, KRAS, NRAS and NF1 genes, the aligned reads showed 83% sequence identity on average, and 73% of the reads showed more than 80% sequence identity (Fig. 1D). We also examined the patterns of the sequencing errors. The error rates for the mismatches, deletions and insertions accounted for 7.3%, 7.8% and 2.5%, respectively, of the total error rate of 17% (Supplementary Fig. S5A). Cytosine or guanine bases were more likely to be miscalled. Deletions accumulated more at the homopolymer sites (Supplementary Fig. S5B and C). We also examined the length of each read that aligned to the target sequence. The results showed that on average, each read covered 82% of the target region, and 66% of the aligned reads covered more than 90% of the target regions, indicating that nearly the full-length of these amplicons was sequenced (Fig. 1E). These results suggested that the MinION reads obtained in this study are worthy for further analyses, such as mutation detection and phasing.
Figure 1.
Summary of the amplicon sequencing and alignment statistics. (A, B) Distribution of the read lengths (A) and QVs (B) in all five sequencing runs. The average number is shown in the inset. (C) The number of MinION reads aligned to each of the PCR target regions. For the alignment, we used LAST with tuned parameters as described in the Methods section. (D, E) Distributions of the sequence identity (D) and target cover rate (E) in each read. The average number is shown in the inset.
3.2.Detection of SNVs and other types of cancerous mutations
In the EGFR, KRAS, NRAS and NF1 genes, the expected types of mutations were detected, namely SNVs, short deletions and aberrantly spliced transcripts reflecting a splice site mutation. To detect the SNVs, we constructed consensus sequences for the MinION reads that were aligned to the target transcripts. As shown in Supplementary Fig. S2, an SNV was considered a possible homozygous SNV if the read count of a particular base (A, T, G or C) at a given position was larger than twice the sum of the other bases. We also considered possible heterozygous SNVs similarly using varying thresholds (see below; also see Supplementary Fig. S2). To further refine the detected SNVs, we only considered the reads with no mismatches in the ±3 bp region. This filter was useful, likely reflecting the base call scheme of MinION, based on ‘squiggle,’ a five-base window.
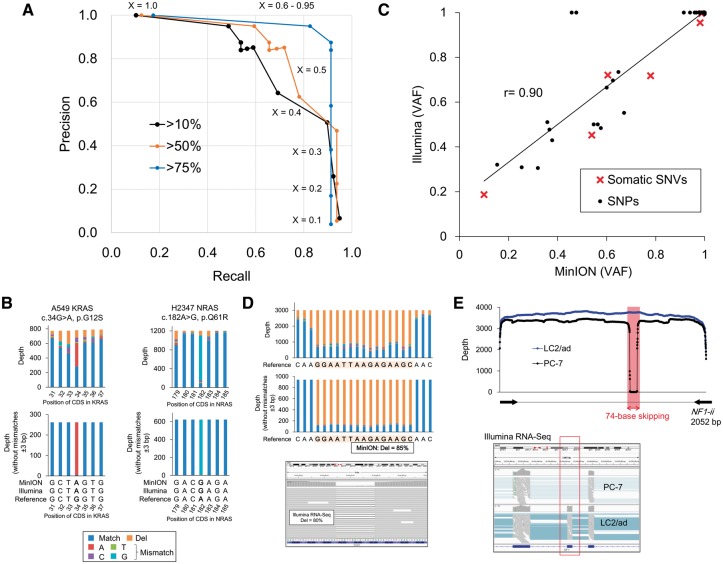
For the detected SNVs, we validated the correct identification rates by comparing these variants with the RNA-Seq data from Illumina HiSeq. The indicated precision and recall rates of the called SNVs were determined according to the category (putative homo, heterozygous SNVs or ‘minor’ SNVs reflecting minor cancer cell numbers or mutant alleles in the population). The detection depended on the threshold of the variant allele frequencies (VAFs) of the MinION reads, which is indicated as ‘X’ in Fig. 2A (also see Supplementary Tables S5 and S6). Indeed, the sensitivity and precision of detecting the SNVs primarily depended on the extent of the coverage of the minor SNVs. SNVs with high variant allele frequencies (>75% in the Illumina RNA-Seq data), including homogeneous SNPs and mutations, were detected with high precision and recall rates (87.5% and 91.3%, respectively, when X = 0.9; blue line; Fig. 2A). However, the performance declined when minor SNPs (>50% or >10% in the Illumina RNA-Seq data) were also considered (precision and recall rates were 85.2% and 71.9%, respectively, when X = 0.5; orange line; 50.7% and 89.7%, respectively, when X = 0.3; black line; Fig. 2A). Intriguingly, the variant allele frequencies of the known heterozygous SNPs in the MinION reads were highly correlated with those in the Illumina RNA-Seq reads (Fig. 2C). Notably, most of the representative driver mutations, such as KRAS G12S in the A549 cells and NRAS Q61R in the H2347 cells (Fig. 2B), were detected.
Figure 2.
Detection of SNPs and mutations in the MinION reads. (A) Precision and recall rates of SNV detection using MinION. Blue, orange and black lines represent three datasets of SNVs corresponding to different variant allele frequencies (VAF) of Illumina standards, which are more than 75% (targets are only homozygous variants), 50% (targets include heterozygous variants) and 10% (targets include minor population variant). ‘X’ represents one of the parameters for the SNV detection, which is the threshold of the VAFs of the MinION reads. Additional details regarding the procedure are described in Supplementary Fig. 2. (B) The depths and base patterns of KRAS G12S in the A549 cells (left) and NRAS Q61R in the H2347 cells (right). The pre-cleaned data are shown in the upper panel. The cleaned data in which the MinION reads without mismatches ±3 bp of the SNVs were used are shown in the lower panel. The color key for the base patterns is represented in the margin. (C) VAFs for the Illumina RNA-Seq and MinION sequencing at the 41 SNVs. SNPs and somatic SNVs are shown as black circles and red crosses, respectively. (D) The depths and base patterns of the 15-base EGFR deletion in the PC-9 cells in the MinION reads. The pre-cleaned and cleaned data are shown in the upper and middle panel. IGV visualization of the Illumina RNA-Seq data is represented in the lower panel. The color key is the same as that shown in B. (E) Exon skipping in exon 19 of NF1 in the PC-7 cells. In the upper panel, the sequence depths of the MinION reads aligned to the NF1-ii region with split alignment using LAST are shown in the PC-7 (black) and LC2/ad (blue, wild-type) cells. The exon skipping in the Illumina RNA-Seq data is also shown in the lower panel.
We further examined the patterns in the false SNV detection. For example, one SNP in the 3’ UTR of KRAS (c.*264C > T in A549 and H2228; rs4285970) was not detected using MinION. Upon further inspection, we observed that this SNP is sandwiched between a 4-base homopolymer GGGG and another 4-base homopolymer TTTT (Supplementary Fig. S6), and the deletions were called in most of the reads at this locus. In total, 521 false positive SNVs were called when the threshold ‘X’ (to 0.1) was lowered to consider the minor SNVs (Supplementary Fig. S7A). Among these false positive SNVs, nearly half were located within or adjacent to ≥3-base homopolymer sequences. In addition, more than 70% of the false positives were miscalls to a surrounding base (±1 bp) and/or C > G/G > C errors (Supplementary Fig. S7B and C). These results suggested that, for a more precise SNV detection, we should consider the presence of homopolymer sites in the surrounding regions. We also found that these errors could be decreased by examining longer matches of the surrounding bases (right, Supplementary Fig. S7C). A lower accuracy regarding homopolymers is one of the known problems with MinION sequencing. Base-callers occasionally call a wrong number of bases at homopolymer sites. Oxford Nanopore Technologies and several academic bioinformatics teams have been attempting to improve the accuracy by developing several options. Nanopolish (https://github.com/jts/nanopolish) is a software package for MinION data, which attempts to improve the read accuracy. One of the other options is a newly developed base-caller program, Scrappie. Using this program, a more precise estimation of the homopolymer lengths is enabled. Some other programs, which should be complementarily used for the original base-callers, are also under development. Indeed, the increasing noise-prone detection of SNVs with lower allelic frequencies is a concern that is not unique to MinION sequencing and is also an issue for Illumina sequencing, where one of the biggest advantages lies in its overwhelming sequencing depth. Further increasing the sequencing depth in MinION could, at least partially, address this issue.
In addition to the SNVs, other types of mutations were also detected. The PC-9 cells harbored a 15-base deletion in EGFR that was covered by 943 reads without mismatches of ±3 bp (Fig. 2D). No mutant reads were detected in the RERF-LC-Ad2 cells, in which the EGFR gene is not mutated. The precise identification and mutant allele frequency were validated using Illumina RNA-Seq (lower panel, Fig. 2D). We could also detect aberrantly spliced transcripts. We observed irregular alignments in the sequence reads of the NF1 gene (NF1-ii) in the PC-7 cells that covered a portion of the target length, leaving a long gap in the transcript. We further re-aligned the 3,755 reads to NF1-ii to obtain a split alignment.22 As shown in Fig. 2E, the split alignment revealed that aberrant exon skipping occurred in exon 19, which eliminated 74 bases from this gene.
These results showed that various categories of cancer mutations can be detected by MinION sequencing. All the driver aberrations were validated with Sanger sequencing (Supplementary Fig. S8). Additionally, we conducted MinION sequencing of genomic DNA amplicons of the EGFR gene (Supplementary Fig. S9 and Table S2). We could also detect mutations in genomic DNAs. However, the phasing analysis was challenging using the genomic DNA as a starting material because the mutations were occasionally separated by a long distance (e.g., 10.4 kb distance between EGFR T790M and L858R in the genome, Supplementary Fig. S9D). Particularly, it is practically very difficult or almost impossible to detect the fusion transcripts in the genomic DNA rather compared to the cDNA amplicons. The junction points of the fusion genes are frequently located in a large segment of an intron. PCR should cover very long DNA fragments that include such junction points when genomic DNA was used as the starting material. The detailed analysis of the mutation phasing and fusion genes are described in a later section. In summary, we could detect the driver mutations in the cell lines as follows: four somatic SNVs (KRAS G12S in A549; NRAS Q61R in H2347 and EGFR T790M/L858R in H1975) (Table 1), a deletion (EGFR E746_A750del in PC-9) and splice aberration (NF1 exon 19 skipping in PC-7), which have also been reported in a previous short-read sequencing study.12
Table 1.
Summary of the driver mutations in the MinION sequencing data
| Mutation | Cell line | Status in Illumina RNA-Seq | Thresholds of MinION; X (0.1 - 0.9) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | |||
| EGFR T790M | H1975 | Positive | TP | TP | TP | TP | TP | TP | TP | Unk | Unk |
| PC-9 | Negative | TN | TN | TN | TN | TN | TN | TN | TN | TN | |
| RERF-LC-Ad2 | Negative | TN | TN | TN | TN | TN | TN | TN | TN | TN | |
| EGFR L858R | H1975 | Positive | TP | TP | TP | Unk | Unk | Unk | Unk | Unk | Unk |
| PC-9 | Negative | TN | TN | TN | TN | TN | TN | TN | TN | TN | |
| RERF-LC-Ad2 | Negative | TN | TN | TN | TN | TN | TN | TN | TN | TN | |
| KRAS G12S | A549 | Positive | TP | TP | TP | TP | TP | TP | TP | TP | TP |
| H2228 | Negative | TN | TN | TN | TN | TN | TN | TN | TN | TN | |
| NRAS Q61R | H2347 | Positive | TP | TP | TP | TP | TP | TP | TP | TP | TP |
| H2228 | Negative | TN | TN | TN | TN | TN | TN | TN | TN | TN | |
| Concordance | Precision | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| Recall | 1.0 | 1.0 | 1.0 | 0.8 | 0.8 | 0.8 | 0.8 | 0.5 | 0.5 | ||
By only considering the positions of the 4 driver mutations described above, there were no false positive detections.
TP, true positive; TN, true negative; Unk, unknown (false negative).
3.3. Sequencing junction points in fusion transcripts
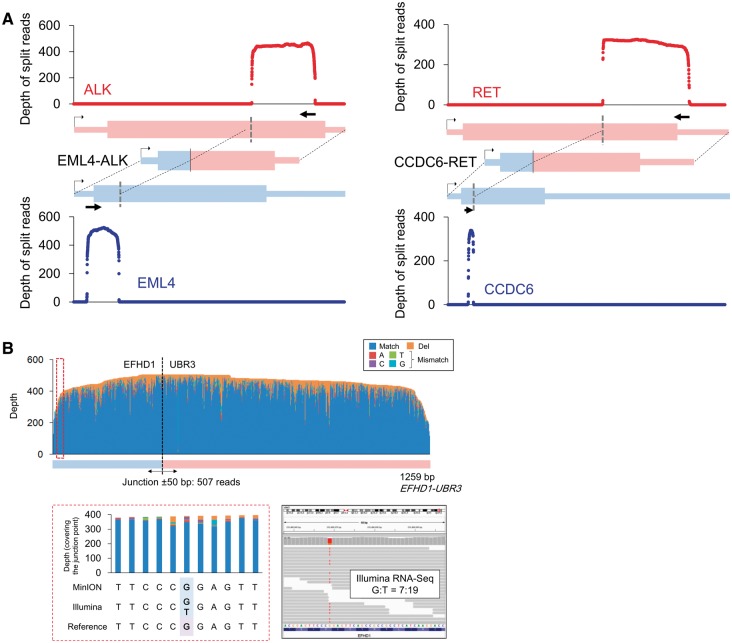
We also used MinION sequencing to identify fusion transcripts. First, we selected two representative oncogenic fusion transcripts, i.e., EML4-ALK in H2228 cells.25 and CCDC6-RET in LC2/ad cells.23,24 We designed PCR primers allowing for the resulting amplicon to sufficiently cover the potential junction points in the fusion transcripts (Supplementary Fig. S1). A split alignment to the sequence reads was conducted using LAST, which generated 540 and 341 reads bridging EML4-ALK and CCDC6-RET, respectively (Fig. 3A and Supplementary Table S7). We also sequenced five other fusion transcripts that were discovered in our previous RNA-Seq study of lung cancers12 (Supplementary Fig. S1). Using only the short reads, we could not obtain any information other than their junction points. Using the long reads in MinION, we could analyze their entire structures and obtain the phasing information of the junction points and the neighboring mutations/variations. More than 150 reads bridged the fusion partners in all cases (Supplementary Table S7). These reads accounted for 63%, on average, of the reads aligned to either of the fusion partners. We further re-aligned the MinION reads to the detected fusion transcript sequences (Supplementary Table S7). The aligned reads showed 82% sequence identity, and an average of 76% of the target regions were covered by the individual reads (Supplementary Fig. S10). Among these, more than 100 reads covered ±50 bases of the junction point of each of the fusion genes.
Figure 3.
Sequencing the fusion transcripts using the MinION reads.(A) Sequence depths of EML4-ALK and CCDC6-RET. The reads were split to both fusion partners with split alignment. (B) Allelic relevance between the SNP and the junction point of the EFHD1-UBR3 fusion transcript in the PC-9 cells. In the upper panel, the depths and base patterns of the MinION reads are shown in the EFHD1-UBR3 target region. The junction point is shown as a broken black line. One of the heterozygous SNPs in EFHD1, which is encircled with a red broken line and is referred to as G, co-occurred with the fusion junction in the MinION reads (left in the lower panel). This SNP was verified as heterozygous using Illumina RNA-Seq (right in the lower panel).
RNA-Seq and whole-genome Illumina sequencing data published by a previous study12 revealed that LC2/ad cells harbor a heterozygous SNP in CCDC6 near the junction point. This SNP was also detected as a homogeneous SNP using MinION, indicating that the gene fusion occurred in the same allele encoding this SNP. This SNP and the gene fusion appeared on the same Illumina reads because the distance between them was only seven bases; therefore, a particularly long MinION read was not necessary in this case. However, a heterozygous SNP (chr2:233498669, G/T) was identified in the fusion transcript EFHD1-UBR3 in PC-9 cells.12 In this case, the SNP was more than 300 bases from the fusion junction. The results of the MinION sequencing suggested that the gene fusion occurred at the G allele (reference allele of UCSC hg19) of this SNP. Such ‘phasing’ of multiple SNVs or SNVs with rearrangement points would be difficult using only Illumina short-read sequencing (Fig. 3B). All fusion junctions were validated by Sanger sequencing (Supplementary Fig. S11).
3.4. Phasing SNVs and evaluation of the detection limits
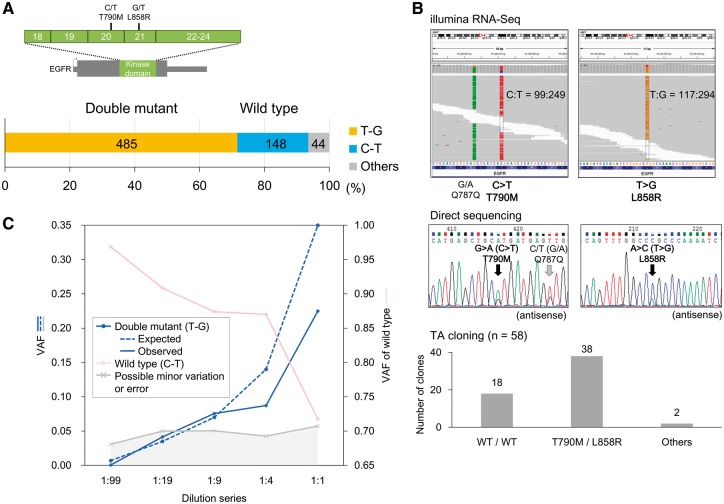
Using the long-read sequencing of MinION, we attempted to phase multiple SNVs. We selected the targets as allelic associations of multiple SNVs, which should have direct clinical relevance such as sensitivity to the therapeutic anti-cancer drugs. The Illumina reads revealed that the H1975 cells harbored two mutations, i.e., T790M and L858R, in the EGFR kinase domain. While anti-cancer drugs targeting EGFR mutants, such as gefitinib and erlotinib, are effective for the L858R mutation, the T790M secondary mutation on the same allele, if exists, would suppress the effect of the drugs.31 The Illumina sequencing also showed that both mutations were heterozygous SNVs, and we sought to determine whether these SNVs were in the same allele. The MinION sequencing revealed that 71% of the aligned reads (4089/5754 in EGFR-i) covered the kinase domain. We used 677 reads without mismatches ±3 bp at both sites. As shown in Fig. 4A, 72% of the reads harbored both mutations and 22% of the reads showed neither of the mutations. These results suggested that these mutations occurred in the same allele and that the remaining allele was not mutated. These results were also validated using TA cloning followed by Sanger sequencing (Fig. 4B). Small fractions of both the MinION and Sanger reads suggested additional very minor and complicated mutation patterns. Notably, we did not detect any other positions, particularly, in the protein kinase domain. A recently reported mutation, i.e., C797S, is known to render cancers resistant to the third-generation EGFR tyrosine kinase inhibitor AZD9291, which is effective against cancers with double mutations at L858R and T790M.32 For the prognosis of a possible novel drug resistance, it should be of remarkable clinical relevance to monitor the acquisition of novel mutations, which may occasionally scatter over a wide region beyond the reach of Illumina sequencing, in an allele-specific manner.
Figure 4.
Phasing cancer mutations. (A) Phasing of the EGFR mutations T790M and L858R in H1975 cells. The number of MinION reads called for each of the SNV patterns. The MinION reads without mismatches ±3 bp of the positions of both SNVs. (B) T790M and L858R mutations using Illumina RNA-Seq and Sanger sequencing. Both SNVs were called as heterozygous mutations by Illumina RNA-Seq and direct Sanger sequencing in the upper and middle panels, respectively. The pattern and variant tag frequencies of both SNVs in each DNA molecule were validated with TA cloning, followed by Sanger sequencing (lower panel). (C) Variant allele frequencies (VAFs) in the five-point EGFR-mutant dilution series. The expected and observed VAFs of the phased double mutant (T790M/L858R) are shown in blue solid and dashed lines, respectively. The VAFs in the wild-type and other patterns are also represented in pink and gray, respectively.
Before initiating clinical applications of MinION sequencing, we further investigated whether mutations present at low levels could be detected and phased. This analysis was performed to ensure that the clinical requirements can be met if the cancer cell population is occasionally low in a clinical sample. Using serially diluted samples containing mixtures of the EGFR mutant (H1975) and wild-type (RERF-LC-Ad2) cell lines (mutant:wild type = 1:99, 1:19, 1:9, 1:4 and 1:1), we assessed the number of reads required to obtain sufficient mutant reads and determined the precision at which mutations can be detected and phased in the dilution series (Supplementary Table S8). The mutations with the respective frequencies could be detected and phased at almost the expected rates (Fig. 4C). Indeed, we found that several percent of the mutants were dropped in our dilution analysis. It may be still difficult to detect mutations in very minor population (less than several percent) using MinION. However, the sequencing yields and quality of the MinION data have been continuously increasing; therefore, this ‘drop’ problem, which is represented by the gray-colored area in Fig. 4C, will become less significant. Indeed, we found the error rates were significantly decreased in the newer version of the flow cell (R9). Using the R9 flow cell, 23,640 2D pass reads were aligned and the reads showed an average of 88.4% sequence identity (Supplementary Fig. S12). More than 45% of the aligned reads harbored more than 90% sequence identity.
3.5. Sequencing clinical samples using MinION
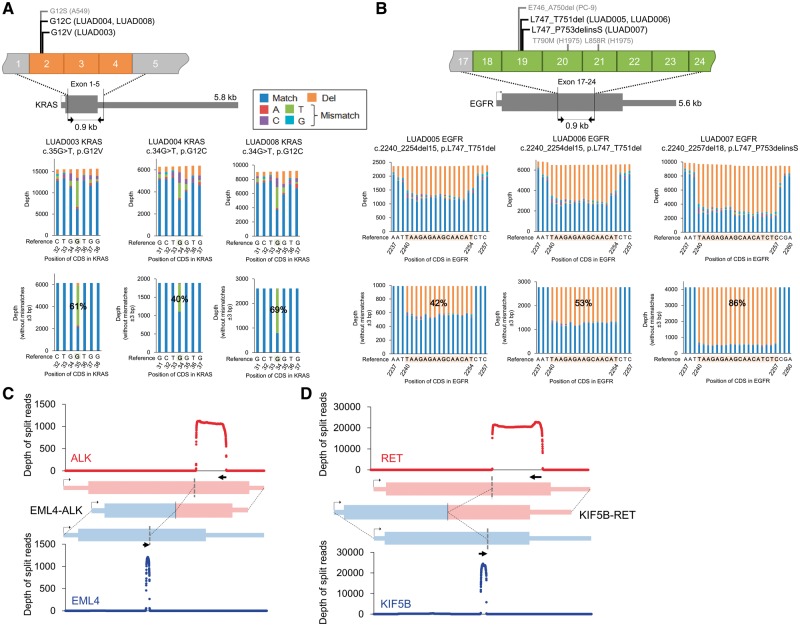
Finally, for clinical applications, we performed MinION sequencing using cDNA amplicons prepared from eight Japanese lung adenocarcinoma patients (Supplementary Fig. S13). Six patients harbored EGFR or KRAS mutations. To detect the driver mutations in the EGFR and KRAS genes, we amplified target regions of approximately 1 kb that covered the mutational hotspot. Three patients harbored KRAS G12 mutations (G12V in one patient and G12C in two patients, Fig. 5A) and three other patients harbored deletions in exon 19 of EGFR (L747_T751del in two patients and L747_P753delinsS in one patient, Fig. 5B). These mutations were further confirmed by Sanger sequencing (Supplementary Fig. S13). The variant allele frequencies were calculated by MinION using only reads without any mismatches in the 3-bp region surrounding the mutations, which is consistent with the cell line analysis. The results highly correlated with those obtained by the Sanger sequencing.
Figure 5.
Detection of driver alterations in clinical samples. (A, B) Detection of driver alterations in clinical samples using MinION sequencing. KRAS (A) and EGFR (B) mutations are shown for the mutation-positive patients. The PCR target regions are shown in the upper panel. The pre-cleaned and cleaned (without mismatches ±3 bp of the mutation) depths are shown in the middle and lower panel. (C, D) Sequence depths of split alignment for EML4-ALK (C) and KIF5B-RET (D).
The other two patients carried the EML4-ALK and KIF5B-RET gene fusions. By split alignment using LAST, 1,219 and 26,023 reads were aligned to EML4-ALK and KIF5B-RET (Fig. 5C and D), respectively, in these patients. Using previously reported gene fusion variants as references, we determined that EML4-ALK is variant E18;A20,4,33 and KIF5B-RET is variant K23;R12.13 The precise detection of the junction points was confirmed by Sanger sequencing (Supplementary Fig. S13D). We also aligned the MinION reads to the fused transcripts to further examine the sequencing accuracy and junction coverage. The reads showed 84% sequence identity on average, and 79% and 69% of the reads, respectively, aligned to the ALK and RET fusions covered the junction points of these gene fusions (±50 bp, Supplementary Table S9A). We also compared the consensus sequences of the MinION reads with those of the Illumina RNA-Seq reads. The overall concordances were consistent with those obtained from the cell line analysis (Supplementary Table S9B).
Furthermore, we conducted a phasing analysis of a short deletion and a heterozygous SNP in EGFR in one patient with EGFR mutations (Supplementary Fig. S14). In total, 88% of the reads showed two major patterns of allelic associations. Based on these results, we concluded that the precise detection of driver mutations by sequencing was also achieved using the clinical samples. The MinION sequencing data of the clinical samples are summarized in Table 2.
Table 2.
Summary of the MinION sequencing data of clinical samples
| Run | Sample ID | Target gene | Avg. QV | Avg. read length | Number of 2D reads |
Avg. identity | Avg. target cover rate | Driver mutation (%VAF) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Aligned | On-target | |||||||||
| #C1 | LUAD001 | EML4-ALK | 10.6 | 916 | 59,810 | 51,967 (87%) | 47,290 (79%) | 2,296 | 84%a | 0.65a | E18:A20 |
| LUAD002 | KIF5B-RET | 44,994 | K23;R12 | ||||||||
| #C2 | LUAD003 | EGFR | 9.8 | 754 | 144,229 | 126,038 (87%) | 115,999 (80%) | 88,751 | 85% | 0.74 | WT |
| KRAS | 27,248 | G12V (61%) | |||||||||
| #C3 | LUAD004 | EGFR | 9.2 | 600 | 63,112 | 50,881 (81%) | 45,062 (71%) | 33,177 | 84% | 0.59 | WT |
| KRAS | 11,885 | G12C (40%) | |||||||||
| #C4 | LUAD005 | EGFR | 9.6 | 672 | 36,535 | 28,359 (78%) | 19,586 (54%) | 5,895 | 84% | 0.76 | L747_T751del (42%) |
| KRAS | 13,691 | WT | |||||||||
| #C5 | LUAD006 | EGFR | 9,3 | 516 | 64,566 | 43,469 (67%) | 31,768 (49%) | 20,498 | 85% | 0.58 | L747_T751del (53%) |
| KRAS | 11,270 | WT | |||||||||
| #C6 | LUAD007 | EGFR | 9.3 | 631 | 41,537 | 32,207 (78%) | 27,910 (67%) | 21,141 | 84% | 0.63 | L747_P753delinsS (86%) |
| KRAS | 6,769 | WT | |||||||||
| #C7 | LUAD008 | EGFR | 9.4 | 827 | 98,312 | 77,847 (79%) | 65,238 (66%) | 51,805 | 82% | 0.82 | WT |
| KRAS | 13,433 | G12C (69%) | |||||||||
aReads aligned to the fusion RNA were used in the calculations.
4. Discussion
In this study, we used a convenient, long-read MinION sequencer for mutation detection and phasing in cancers. We successfully applied the developed approach to identify cancerous mutations in first cultured cell models and then in clinical samples. Despite the error-prone nature of the sequence data of MinION, in the case of homozygous mutant alleles, the cancerous mutations could be robustly detected. We observed that the minor mutant alleles were occasionally difficult to detect depending on the allele frequency. The detection of such a minor mutant allele is often important for many clinical cancers. Tumor cells are evolutionally diverse with genetic heterogeneity within the population and samples are occasionally mixed with normal cells. In the current study, we demonstrate that additional filtration of the data considering ±3 bp of the SNVs is useful for lowering the error rate and, thus, for increasing the detection limit. Further bioinformatics tools could address these concerns of accuracy and sensitivity. Moreover, we expect that the drawbacks of MinION should be lessened by rapid improvements in the sequencing technology.
One of the characteristics of MinION is its convenience for use, including its portability and easy settings for the library preparation and sequencing. Although second-generation sequencers have enabled us to easily conduct genotyping in clinical samples, we need to develop simple and cost-effective procedures to identify driver genes in each patient for personalized medicine for various kinds of cancers, particularly lung adenocarcinoma. In our study, we could detect the major driver genes, which have diverse patterns, including point mutations and fusions, using MinION. In lung adenocarcinoma, a number of molecular-targeting medicines are available, such as gefitinib, erlotinib and afatinib for EGFR;34 crizotinib, ceritinib and alectinib for ALK;35 and vandetanib and cabozantinib for RET.36 The detection of KRAS mutations is also required because a large portion of patients harbor these mutations, but there are no effective anti-cancer drugs targeting KRAS. The simple methods of MinION sequencing could possibly enable small/mid-scale research centers and hospitals to conduct research studies by genotyping driver genes and selecting suitable therapeutic approaches.
Following the initial success of this methodological development, the obvious next goal is to expand this approach to other cancers. Thus, additional primer sets should be designed for various genes that have clinical and diagnostic importance depending on the cancer types. Eventually, it would be particularly beneficial to design an array of primer sets covering the representative frequently mutated genes as depicted in some commercial cancer panels. Therefore, further technical developments are needed, such as designing accurate and robust PCR primers for each gene.
In addition to the detection of the cancerous mutations, using the long MinION reads, we could analyze the structural alterations and phasing of SNPs and mutations. It is important to elucidate the allelic background of mutations that are occasionally distantly located. For example, mutual associations between primary and secondary mutations, particularly mutations associated with drug resistance,31,32 have important clinical relevance. In addition to the cases of EGFR, various types of associations between primary and secondary/drug-resistant mutations have been reported. For example, for ALK-fusion-positive lung adenocarcinoma patients, drugs that molecularly target the tyrosine kinases, such as crizotinib, have been used. Similar to other effective anti-cancer drugs, a relapse frequently occurs as resistant clones are developed. Patients move to the second line tyrosine kinase inhibitor, which could also eventually become ineffective.37 However, intriguingly, very recent reports indicated that, in some cases, the cancer recovers the sensitivity for the previous round of drugs depending on the patterns of the initial and secondary resistant mutations and their allelic backgrounds.38,39 For these cases, long-read sequencing could provide indispensable information for selecting the next therapeutic approaches. This study is the first step toward a more widespread application of long-read sequence-based diagnosis in cancers in general clinical practice.
Supplementary Material
Acknowledgements
The authors would like to thank K. Imamura, T. Horiuchi, H. Wakaguri and K. Abe for their technical assistance in the library preparation, sequencing and data processing. The authors would also like to thank A. Kinase, A. Onai and Y. Ichinose for their assistance with the PCR experiments.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at DNARES online.
Funding
MEXT KAKENHI Grant Number 16H01582 and MEXT KAKENHI Grant Number 16H06279.
References
- 1. The Cancer Genome Atlas Research Network, Weinstein J.N., Collisson E.A., et al. 2013, The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet., 45, 1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. International Cancer Genome Consortium 2010, International network of cancer genome projects. Nature, 464, 993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharma S.V., Bell D.W., Settleman J., Haber D.A.. 2007, Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer, 7, 169–81. [DOI] [PubMed] [Google Scholar]
- 4. Soda M., Choi Y.L., Enomoto M., et al. 2007, Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature, 448, 561–6. [DOI] [PubMed] [Google Scholar]
- 5. Kwak E.L., Bang Y.J., Camidge D.R., et al. 2010, Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl J. Med., 363, 1693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jain M., Fiddes I.T., Miga K.H., Olsen H.E., Paten B., Akeson M.. 2015, Improved data analysis for the MinION nanopore sequencer. Nat. Methods, 12, 351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ashton P.M., Nair S., Dallman T., et al. 2015, MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol., 33, 296–300. [DOI] [PubMed] [Google Scholar]
- 8. Quick J., Ashton P., Calus S., et al. 2015, Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol., 16, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quick J., Loman N.J., Duraffour S., et al. 2016, Real-time, portable genome sequencing for Ebola surveillance. Nature, 530, 228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Norris A.L., Workman R.E., Fan Y., Eshleman J.R., Timp W.. 2016, Nanopore sequencing detects structural variants in cancer. Cancer Biol. Ther., 17, 246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Minervini C.F., Cumbo C., Orsini P., et al. 2016, TP53 gene mutation analysis in chronic lymphocytic leukemia by nanopore MinION sequencing. Diagn. Pathol., 11, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suzuki A., Makinoshima H., Wakaguri H., et al. 2014, Aberrant transcriptional regulations in cancers: genome, transcriptome and epigenome analysis of lung adenocarcinoma cell lines. Nucleic Acids Res., 42, 13557–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kohno T., Ichikawa H., Totoki Y., et al. 2012, KIF5B-RET fusions in lung adenocarcinoma. Nat. Med., 18, 375–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.. 2007, Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res., 35, W71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loman N.J., Quinlan A.R.. 2014, Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics, 30, 3399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kiełbasa S.M., Wan R., Sato K., Horton P., Frith M.C.. 2011, Adaptive seeds tame genomic sequence comparison. Genome Res., 21, 487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamada M., Ono Y., Asai K., Frith M.C.. 2016, Training alignment parameters for arbitrary sequencers with LAST-TRAIN. Bioinformatics, 33, 926–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frith M.C., Wan R., Horton P.. 2010, Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res, 38, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Speir M.L., Zweig A.S., Rosenbloom K.R., et al. 2016, The UCSC Genome Browser database: 2016 update. Nucleic Acids Res, 44, D717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu T.D., Nacu S.. 2010, Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics, 26, 873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frith M.C., Kawaguchi R.. 2015, Split-alignment of genomes finds orthologies more accurately. Genome Biol., 16, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsubara D., Kanai Y., Ishikawa S., et al. 2012, Identification of CCDC6-RET fusion in the human lung adenocarcinoma cell line, LC-2/ad. J. Thorac. Oncol., 7, 1872–6. [DOI] [PubMed] [Google Scholar]
- 24. Suzuki M., Makinoshima H., Matsumoto S., et al. 2013, Identification of a lung adenocarcinoma cell line with CCDC6-RET fusion gene and the effect of RET inhibitors in vitro and in vivo. Cancer Sci., 104, 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jung Y., Kim P., Keum J., et al. 2012, Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer, 51, 590–7. [DOI] [PubMed] [Google Scholar]
- 26. Ding L., Getz G., Wheeler D.A., et al. 2008, Somatic mutations affect key pathways in lung adenocarcinoma. Nature, 455, 1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Imielinski M., Berger A.H., Hammerman P.S., et al. 2012, Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell, 150, 1107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seo J.S., Ju Y.S., Lee W.C., et al. 2012, The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res., 22, 2109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suzuki A., Mimaki S., Yamane Y., et al. 2013, Identification and characterization of cancer mutations in Japanese lung adenocarcinoma without sequencing of normal tissue counterparts. PLoS One, 8, e73484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. The Cancer Genome Atlas Research Network 2014, Comprehensive molecular profiling of lung adenocarcinoma. Nature, 511, 543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pao W., Miller V.A., Politi K.A., et al. 2005, Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med., 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thress K.S., Paweletz C.P., Felip E., et al. 2015, Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat. Med., 21, 560–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Soda M., Isobe K., Inoue A., et al. 2012, A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin. Cancer Res., 18, 5682–9. [DOI] [PubMed] [Google Scholar]
- 34. Yang Z., Hackshaw A., Feng Q., et al. 2017, Comparison of gefitinib, erlotinib and afatinib in non-small cell lung cancer: A meta-analysis. Int. J. Cancer, 140, 2805–19. [DOI] [PubMed] [Google Scholar]
- 35. Katayama R., Friboulet L., Koike S., et al. 2014, Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res., 20, 5686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kohno T., Tsuta K., Tsuchihara K., Nakaoku T., Yoh K., Goto K.. 2013, RET fusion gene: translation to personalized lung cancer therapy. Cancer Sci., 104, 1396–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Camidge D.R., Pao W., Sequist L.V.. 2014, Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat. Rev. Clin. Oncol., 11, 473–81. [DOI] [PubMed] [Google Scholar]
- 38. Niederst M.J., Hu H., Mulvey H.E., et al. 2015, The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin. Cancer Res., 21, 3924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shaw A.T., Friboulet L., Leshchiner I., et al. 2016, Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N. Engl J. Med., 374, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.