Abstract
Background
The antiapoptotic protein survivin has been demonstrated to play an important role in colorectal carcinogenesis. However it is unclear whether the upregulation of survivin is maintained through progressive stages of disease, or if other apoptosis-related genes are coexpressed and/or repressed. We sought to evaluate survivin expression in colonic neoplasia and identify relationships with additional regulators of apoptosis.
Patients and Methods
Tissue samples from 168 patients with primary colorectal cancer were profiled using the GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA) and evaluated for survivin expression. Immunohistochemical staining for survivin and a panel of apoptosis-associated proteins were used in 86 patients with tissue microarray (TMA) blocks; scoring was by stain intensity and percentage of positive cells (range, 0–9).
Results
Survivin mRNA was upregulated (1.8-fold increase) in primary colon cancers— irrespective of American Joint Committee on Cancer (AJCC) stage— and metastases compared with normal colonic tissue (P < .0001). Survivin staining was positive in 93% of adenocarcinomas (median immunohistochemistry [IHC] score: 2 [range, 1–6]), 100% of adenomas (1 [range,1–2]), and 43% of normal colonic mucosa (1, [range 1–2]) (P = .006). Survivin expression increased with worsening tumor grade (P < .05). In colon cancers, survivin expression positively correlated with the coexpression of PUMA (P = .001), TACE (P = .003), and MCL1 (P = .01), and trended toward an inverse correlation with BAX (P = .058).
Conclusions
Survivin expression increases during the normal mucosa-adenoma-carcinoma sequence and is maintained throughout progression of disease, which strengthens its appeal as a therapeutic target. Furthermore, we have demonstrated co-overexpression of several other apoptosis-related genes, which may in turn serve as additional and potentially synergistic therapeutic targets.
Keywords: Apoptosis, Colon cancer, Immunohistochemistry, PUMA, Survivin
Introduction
The ability to avoid apoptosis is one of the major oncogenic switches contributing to carcinogenesis.1 To date, 2 gene families of apoptosis regulators have been identified: the BCL2 family, which is composed of pro- and antiapoptotic members, and the inhibitor of apoptosis (IAP) proteins.2,3 Recently there has been increasing clinical interest in survivin, a member of the IAP protein family because it possesses inherent properties that make it an ideal tumor marker and potential therapeutic target.4–6
Survivin prevents apoptosis through the inhibition of caspase 3 and caspase 7 and also functions as a chromosome passenger protein, regulating the G2 and M phases of the cell cycle.7–10 Survivin is involved in spindle formation, cellular stress response, chemoresistance, and angiogenesis.11 Additionally, survivin appears to be globally deregulated in transformed cells but does not appear to be isolated to the proliferating fraction of cells within a given tumor.12 In comparison with corresponding normal tissues, survivin has been found to be 1 of the most highly differentially expressed genes in numerous human cancers including tumors of the lung, breast, stomach, esophagus, pancreas, liver, ovaries, and kidneys.13–20
In colorectal cancer, survivin overexpression has been demonstrated to occur in precursor lesions such as tubular adenomas, suggesting that survivin upregulation is an early event in tumorigenesis.21 However it remains unclear whether survivin upregulation is necessary or constitutive through progressive stages of disease, including distant metastases. Furthermore, although the biologic function of survivin is related to other apoptosis-associated genes, no consistent relationships with BCL2 or IAP family members have been demonstrated in colorectal cancers. We therefore sought to evaluate survivin expression in colonic neoplasia, from normal mucosa to invasive carcinoma, and furthermore to identify associations between the expression of survivin and other apoptosis-related genes.
Methods
This study was approved by the Institutional Review Board at the University of South Florida. Between 1990 and 2002, a pilot program of tumor gene expression profiling began at our institution and included colon cancer specimens resected from patients consenting to the institution’s tissue collection protocol. We retrospectively reviewed the gene expression profiles of patients with colonic adenocarcinoma for the expression of survivin. We subsequently constructed a tissue microarray (TMA) using all patients who underwent gene expression profiling and with available formalin-fixed paraffin-embedded (FFPE) tissues, which included adenomatous lesions that had not been specifically evaluated with expression profiling. Clinical information was obtained from our prospectively maintained colorectal cancer database. Tumors were staged according to the AJCC criteria, 6th edition. All patients who received neoadjuvant chemotherapy and/or radiation were excluded.
Tissue Preparation and Sample Processing for Microarray Analysis
Viable tumor was isolated at the time of surgery by macrodissection, and the specimen was flash frozen within 15 minutes. Frozen-section analysis was used to confirm that at least 80% of the tissue was composed of tumor. Total RNA from the excised tissue was isolated using TRIzol reagant (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The aqueous phase containing the RNA separated from the TRIzol reagent was further purified using the RNeasy cleanup procedure (Qiagen Inc, Valencia, CA). The quality of total RNA was then assessed by agarose gel electrophoresis and A260/280 ratio or by analysis on the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Santa Clara, CA).
Five micrograms of total RNA from each sample were processed for microarray analysis. The poly(A) RNA was specifically converted to cDNA and then amplified and labeled with biotin, following the procedure initially described by Van Gelder et al.22 Hybridization with the biotin-labeled RNA, staining, and scanning of the chips (U133A 2.0 Plus) followed the prescribed procedure outlined in the Affymetrix technical manual and has been previously described.23 Scanned output files were visually inspected for hybridization artifacts and then analyzed using the robust multiarray analysis method developed by Irizarry et al.24
Tissue Microarray Analysis
Colon cancer TMAs were prepared and constructed using a subset of patients with adequate FFPE tissue remaining in the histology section of the Tissue Core Facility at the Moffitt Cancer Center. All specimens were preserved in 10% buffered formalin before embedding in paraffin. Four-micrometer sections were cut with a Leica microtome (Leica Microsystems Inc, Bannockburn, IL) and transferred to adhesive-coated slides. The tissue array slides (4 slides, including 2 test duplicate slides and positive and negative controls) were stained for survivin, pAKT, BCl2, BAX, BCLXL, BAK, PUMA, XIAP, TACE, and MCL1. The slides were prepared using our standard protocol as previously published.25
The TMA stain results were examined by 2 pathologists (D.C. and A.H.). The positive reaction was scored into 4 grades according to the intensity of cytoplasmic staining: 0, 1+, 2+ and 3+. The percentages of positive cells were also scored into 4 categories: 0 (0%), 1+ (1–33), 2+ (36–66%), and 3+ (more than 66%). In cases with a discrepancy between duplicated cores, the higher score from the 2 tissue cores was taken as a final score. The product between intensity and percentage scores was used as a final TMA staining score. The final TMA staining score was defined as follows: 0, negative; 1–3 weak; 4–6 moderate; 7–9, strong.
Statistical Analysis
A student t test was used to assess the difference in gene expression between normal mucosa and cancerous tissue. Mean values with standard errors were used to visualize different categories of tissue (eg, normal, stage I). Analysis of variance was used to test whether categorical clinical parameters (eg, grade, gender, and histologic features) were associated with TMA scores. For clinical parameters with more than 2 groups (normal, adenoma, and carcinoma), the Tukey HSD (Honestly Significant Difference) test was used to adjust for a P value for pairwise comparison. Pearson correlation was used to measure linear relationship with TMA scores when appropriate. Survival curves were generated using the Kaplan-Meier method. The log-rank test was used to evaluate survival differences.
Results
Demographics
Survivin gene expression was evaluated in 254 samples from 168 patients (78 male/90 female) of median age 63 years (range, 24–92 years). Samples evaluated included normal mucosa (10), stage I (32), stage II (66), stage III (65), and stage IV (42) primary tumors, and liver (18), lung (4), and lymph node (7) metastases.
The expression of apoptosis-associated proteins was evaluated by TMA in 86 colonic adenocarcinomas, 7 colonic adenomas, and 7 slides of normal colonic mucosa. Patients with colonic adenocarcinomas included 54 men and 32 women, median age 67 years (range, 24–92). Samples evaluated included stage I (5), stage II (29), stage III (43), and stage IV (9) primary tumors of well (11), moderate (61), and poor (14) differentiation.
Survivin Gene Expression
At the mRNA level, survivin was upregulated (1.8-fold increase) in all stages of cancer, as well as in liver, lymph node, and lung metastases compared with normal colonic tissue (P ≤ .0001) (Figure 1). The level of survivin expression was not different among various stages of cancer or among various sites of metastatic disease. No correlation between the degree of survivin expression and overall survival was identified.
Figure 1.
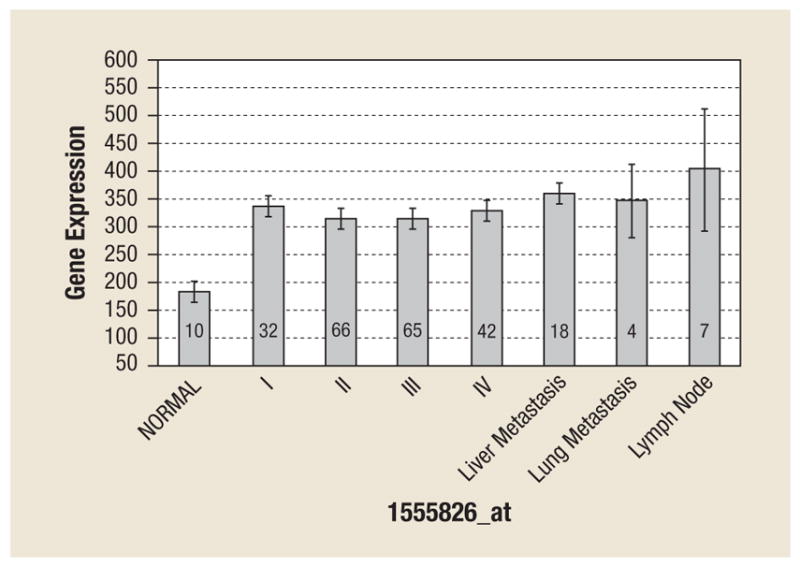
Microarray Gene Expression of Survivin in 254 Colorectal Samples. Mean Relative Expression Values for the Patient Cohort is Graphed with Standard Error Bars. The Number of Samples in Each Category is Provided. Survivin Expression is Significantly (P ≤ .0001) Upregulated in Cancer Vs. Normal Tissues
Tissue Microarray
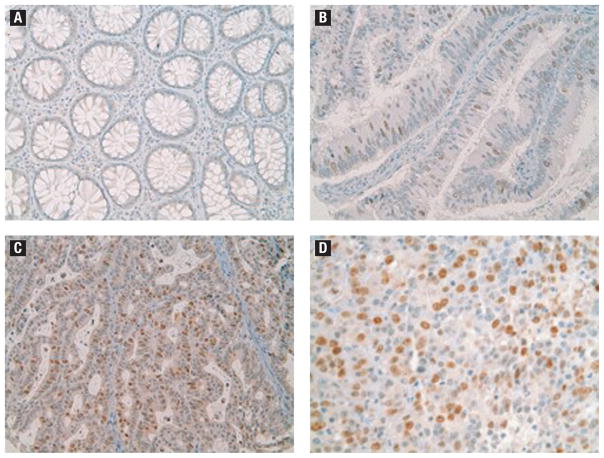
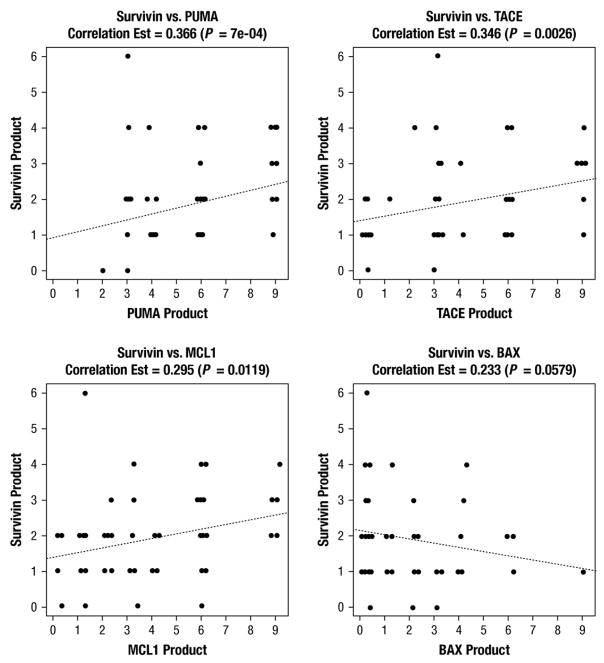
Survivin protein staining was positive in 80 of 86 (93%) adenocarcinoma tissue specimens, 7 of 7 (100%) adenoma tissue specimens, and 3 of 7 (43%) normal mucosa tissues. Adenocarcinoma tissues were more likely to stain positive for survivin than were normal colonic mucosal tissues (P =.025). Median survivin TMA staining scores for adenocarcinoma, adenoma, and normal colonic mucosa were 2 (range, 1–6), 1 (range, 1–2), and 1 (range, 1–2), respectively (P =.006). Figure 2 shows histopathologic images demonstrating TMA specimens with varying degrees of survivin staining. Increasing survivin scores correlated with increasing degrees of tumor dedifferentiation such as poor or undifferentiated cancers (P =.04), although no correlation was found with T stage, lymph node involvement, or presence of metastatic disease. Although previously reported to be of unclear significance,26 we also analyzed nuclear survivin staining separately and in doing so, found no correlation with tumor stage, lymph node involvement, presence of metastatic disease, or overall survival (Figure 3). Survivin protein expression was found to be significantly and positively correlated with the expression of PUMA (P <.001), TACE (P =.003), and MCL1 (P =.01), and demonstrated a trend toward an inverse correlation with the expression of BAX (P =.058) (Figure 4).
Figure 2.
Histopathologic Images of Survivin Staining. (A) Normal Colonic Mucosa Showing Undetectable Levels of Survivin Protein. (B) Tubulovillous Adenoma with Several Scattered Survivin-Positive Epithelial Cells. (C) A Well-Moderately Differentiated Adenocarcinoma Exhibiting Strong Survivin Protein Expression in Many of the Tumor Cells. (D) A Poorly Differentiated Adenocarcinoma shows Strong Survivin Staining in Most of the Tumor Cells
Figure 3.
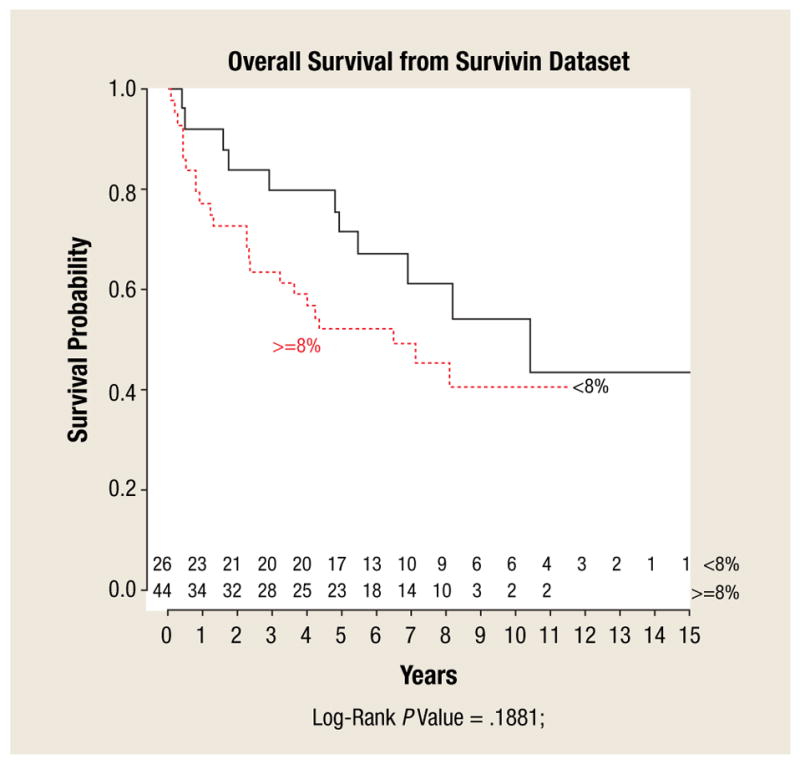
Plot of Kaplan-Meier Overall Survival Curves for Patients with Tumors Demonstrating High and Low Survivin Expression
Figure 4.
Box Plots of Survivin Expression Vs. PUMA, TACE, MCL1, and BAX, with Spearman Correlation Estimate and P values
Discussion
Survivin overexpression is an early event in the development of colonic adenocarcinoma and appears to increase during the normal mucosa-adenoma-carcinoma sequence and with the progressive loss of tumor differentiation. In an analysis of colonic adenocarcinomas, we identified a positive correlation between survivin overexpression and the concomitant upregulation of other antiapoptotic proteins (TACE and MCL1), as well as a trend toward an inverse correlation with the proapoptotic gene BAX. Our data support the notion that survivin-related antiapoptotic pathways are turned on early in tumorigenesis to promote proliferation and may therefore potentially be useful to stratify patients with precursor lesions such as adenomas. We have also demonstrated retained survivin upregulation across all stages of disease, including lymph node and distant metastases, suggesting targeting of survivin may be feasible in patients with advanced disease as well. Furthermore, we have identified additional potential therapeutic targets for inhibition that may act synergistically to increase the efficacy of survivin-directed therapy in colorectal cancer.
Current care guidelines from the American College of Gastroenterology (ACG), the American Society for Gastrointestinal Endoscopy (ASGE), and the American Cancer Society (ACS) do not take into account genetic alterations for sporadic adenomas in stratifying patients for surveillance or treatment. However, survivin may be a suitable candidate given the results of several recent studies. An analysis of 374 patients with sporadic colorectal adenomas found survivin expression to be an independent risk predictor for the development of metachronous colorectal cancer at locations distant from the originally identified adenoma.21 Survivin has been found to be significantly associated with the transition from low-grade dysplasia to high-grade dysplasia.27–30 These data support the suggestion that survivin overexpression is an early event in the pathogenesis of colo-rectal cancer and may therefore serve a prognostic role during the evaluation of colonic adenomas. However given that uniform over-expression of survivin with invasive cancer and the lack of correlation between stage and overall survival, survivin expression currently appears to have limited prognostic value in the evaluation of colonic adenocarcinomas.
Survivin is known to be highly expressed during fetal development but is nearly undetectable in normal adult tissues. An analysis of 3.5 million transcriptomes found survivin to be among the most highly upregulated proteins in cancer compared with normal tissue.31 Using TMA, a previous study of 230 patients found no survivin expression among normal samples.32 However, survivin is known to be expressed in a few normal adult tissue types, including basal colonic basal epithelial cells.11 We identified survivin expression in 43% of histologically normal colonic mucosal samples, albeit at levels significantly reduced compared with those observed in cancer tissues.
Survivin may serve as an effective therapeutic target given the following: (1) disabling survivin is expected to compromise multiple signaling networks, (2) survivin may be a uniquely flexible target, suitable for multiple targeting strategies, and (3) targeting survivin pathways is unlikely to affect normal cells or tissues.11 Several phase I and II trials have been completed with promising results using antisense, small-molecule antagonists and immunotherapy against survivin pathways.33–38 Targeting survivin may also act to sensitize cancer cells to subsequent therapies. For example, sequential treatment of prostate cancer cell lines with small interfering (si)RNA to survivin and 17-AAG (heat shock protein [HSP]90 inhibitor) demonstrated synergistic growth suppression with enhanced caspase 9-dependant apoptosis.39 More recently, survivin repression has been demonstrated to effectively induce apoptosis in colon cancer cell lines using multiple agents, including histone deacetylase (HDAC) inhibitors.40–44
We identified 4 members of the BCL-2 family (PUMA, TACE, MCL1, and BAX) that correlated with survivin expression. PUMA (a p53 upregulated modulator of apoptosis) is a proapoptotic family member known to be induced by nonsteroidal anti-inflammatory drugs (NSAIDs).45,46 Upregulation of PUMA in association with survivin overexpression could potentially be a reflex compensatory mechanism in response to cellular apoptotic imbalance or, as recently suggested, could be secondary to NSAID use in the study population. Interestingly, a recent randomized prospective trial demonstrated a 45% reduction in the incidence of any adenoma and of high-risk colorectal adenomas by 66% with twice-daily celecoxib, a cyclooxygenase (COX)-2 inhibitor (NSAID) known to suppress the expression of antiapoptotic proteins, including survivin.47 Upregulation of PUMA has also been demonstrated using a panHER/vascular endothelial growth factor receptor (VEGFR) inhibitor (BMS-690514) in non–small-cell lung cancer cell lines with somatic epidermal growth factor receptor (EGFR) mutations.48 BMS-690514 treatment resulted in the downregulation of both MCL1 (an antiapoptotic protein) and HSP90 (the association of survivin with HSP90 is required for its stability and function49), which led to antiproliferative and proapoptotic effects.48 Additionally, PUMA has been shown to mediate the apoptotic effects of EGFR inhibitors in small-cell carcinoma of the head and neck.50
TACE (TNF-α converting enzyme) functions in the cleavage of several EGFR ligands in a Zn-dependent manner, releasing them from the cell surface to activate the receptor and its downstream pathways.51 Silencing expression of TACE in HeLa cells decreases proliferation, adhesion, and migration and induces apoptosis.52 Similar effects have been observed in colorectal cancer using EGFR monoclonal antibodies in conjunction with TACE inhibitors.52,53 MCL1, another antiapoptotic BCL2 family member, has been shown to be activated by RAS/RAF/ERK-mediated pathways, which are downstream of EGFR signaling.54 MCL1 provides rapid short-term protection of cell viability and can do so in response to various chemo-therapeutic agents.54 In HCT116, a colon cancer cell line, MCL1 protects against cell death associated with BCL2/BCL-XL inhibition largely by antagonizing the effects of PUMA.55 Interestingly, the proapoptotic effects of PUMA are almost completely abolished in BAX null cells, suggesting PUMA function depends on its ability to activate BAX.55 We identified an inverse relationship between survivin and BAX expression. BAX, a proapoptotic member of the BCL2 family, is virtually required to initiate mitochondrial-dependant apoptosis (intrinsic pathway).56 Of note, survivin is able to block mitochondrial-induced, but not death receptor–induced apoptosis.12
Conclusion
Defects in apoptotic pathways augment cell survival and may facilitate cancer development by allowing time for the accrual of additional, potentially advantageous mutations. We have shown that survivin plays an integral role in colorectal cancer cell survival, being nearly uniformly overexpressed in this malignancy. In addition, survivin is upregulated early in the oncogenic process, before the development of invasive cancer. We have also demonstrated a relationship between survivin overexpression and expression of several other apoptosis-regulating proteins, including PUMA, TACE, MCL1 and BAX.
Targeted molecular therapy has been established for the treatment of colorectal cancer through several randomized prospective trials. Although agents such as those targeting VEGFR and EGFR have proved modestly efficacious, few colorectal cancers demonstrate dependence on a single pathway and/or receptor. Therefore simultaneous targeting of multiple genes in 1 or multiple pathways has been hypothesized as a means of increasing the efficacy of targeted therapy. Survivin-directed therapy in conjunction with simultaneous inhibition of other apoptosis-related regulatory proteins, deserves further investigation and may hold potential as a future therapeutic strategy.
Footnotes
Disclosures
All authors report that they have no relevant relationships to disclose.
References
- 1.Qiao L, Wong BC. Targeting apoptosis as an approach for gastrointestinal cancer therapy. Drug Resist Updat. 2009;12:55–64. doi: 10.1016/j.drup.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–56. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–10. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 4.Zaffaroni N, Pennati M, Daidone MG. Survivin as a target for new anticancer interventions. J Cell Mol Med. 2005;9:360–72. doi: 10.1111/j.1582-4934.2005.tb00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pennati M, Folini M, Zaffaroni N. Targeting survivin in cancer therapy: fulfilled promises and open questions. Carcinogenesis. 2007;28:1133–9. doi: 10.1093/carcin/bgm047. [DOI] [PubMed] [Google Scholar]
- 6.Pennati M, Folini M, Zaffaroni N. Targeting survivin in cancer therapy. Expert Opin Ther Targets. 2008;12:463–76. doi: 10.1517/14728222.12.4.463. [DOI] [PubMed] [Google Scholar]
- 7.Islam A, Kageyama H, Takada N, et al. High expression of survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617–23. doi: 10.1038/sj.onc.1203358. [DOI] [PubMed] [Google Scholar]
- 8.Kasof GM, Gomes BC. Livin, a novel inhibitor of apoptosis protein family member. J Biol Chem. 2001;276:3238–46. doi: 10.1074/jbc.M003670200. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki A, Ito T, Kawano H, et al. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346–53. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- 10.Tamm I, Wang Y, Sausville E, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315–20. [PubMed] [Google Scholar]
- 11.Altieri DC. Targeted therapy by disabling crossroad signaling networks: the survivin paradigm. Mol Cancer Ther. 2006;5:478–82. doi: 10.1158/1535-7163.MCT-05-0436. [DOI] [PubMed] [Google Scholar]
- 12.Grossman D, McNiff JM, Li F, et al. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J Invest Dermatol. 1999;113:1076–81. doi: 10.1046/j.1523-1747.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- 13.Ikeguchi M, Ueta T, Yamane Y, et al. Inducible nitric oxide synthase and survivin messenger RNA expression in hepatocellular carcinoma. Clin Cancer Res. 2002;8:3131–6. [PubMed] [Google Scholar]
- 14.Kato J, Kuwabara Y, Mitani M, et al. Expression of survivin in esophageal cancer: correlation with the prognosis and response to chemotherapy. Int J Cancer. 2001;95:92–5. doi: 10.1002/1097-0215(20010320)95:2<92::aid-ijc1016>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Lu CD, Altieri DC, Tanigawa N. Expression of a novel antiapoptosis gene, survivin, correlated with tumor cell apoptosis and p53 accumulation in gastric carcinomas. Cancer Res. 1998;58:1808–12. [PubMed] [Google Scholar]
- 16.Monzo M, Rosell R, Felip E, et al. A novel anti-apoptosis gene: re-expression of survivin messenger RNA as a prognosis marker in non-small-cell lung cancers. J Clin Oncol. 1999;17:2100–4. doi: 10.1200/JCO.1999.17.7.2100. [DOI] [PubMed] [Google Scholar]
- 17.Satoh K, Kaneko K, Hirota M, et al. Expression of survivin is correlated with cancer cell apoptosis and is involved in the development of human pancreatic duct cell tumors. Cancer. 2001;92:271–8. doi: 10.1002/1097-0142(20010715)92:2<271::aid-cncr1319>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K, Iwamoto S, Gon G, et al. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127–34. [PubMed] [Google Scholar]
- 19.Yoshida H, Ishiko O, Sumi T, et al. Survivin, bcl-2 and matrix metalloproteinase-2 enhance progression of clear cell- and serous-type ovarian carcinomas. Int J Oncol. 2001;19:537–42. doi: 10.3892/ijo.19.3.537. [DOI] [PubMed] [Google Scholar]
- 20.Parker AS, Kosari F, Lohse CM, et al. High expression levels of survivin protein independently predict a poor outcome for patients who undergo surgery for clear cell renal cell carcinoma. Cancer. 2006;107:37–45. doi: 10.1002/cncr.21952. [DOI] [PubMed] [Google Scholar]
- 21.Soreide K, Gudlaugsson E, Skaland I, et al. Metachronous cancer development in patients with sporadic colorectal adenomas—multivariate risk model with independent and combined value of hTERT and survivin. Int J Colorectal Dis. 2008;23:389–400. doi: 10.1007/s00384-007-0424-6. [DOI] [PubMed] [Google Scholar]
- 22.Van Gelder RN, von Zastrow ME, Yool A, et al. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87:1663–7. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobbin KK, Beer DG, Meyerson M, et al. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin Cancer Res. 2005;11:565–72. [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Kabra N, Li Z, Chen L, et al. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J Biol Chem. 2009;284:18210–7. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponnelle T, Chapusot C, Martin L, et al. Cellular localisation of survivin: impact on the prognosis in colorectal cancer. J Cancer Res Clin Oncol. 2005;131:504–10. doi: 10.1007/s00432-005-0682-z. [DOI] [PubMed] [Google Scholar]
- 27.Lin LJ, Zheng CQ, Jin Y, et al. Expression of survivin protein in human colorectal carcinogenesis. World J Gastroenterol. 2003;9:974–7. doi: 10.3748/wjg.v9.i5.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fogt F, Poremba C, Shibao K, et al. Expression of survivin, YB-1, and KI-67 in sporadic adenomas and dysplasia-associated lesions or masses in ulcerative colitis. Appl Immunohistochem Mol Morphol. 2001;9:143–9. doi: 10.1097/00129039-200106000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Alper M, Cukur S, Belenli O, et al. Evaluation of the immunohistochemical stain patterns of survivin, Bak and Bag-1 in colorectal cancers and comparison with polyps situated in the colon. Hepatogastroenterology. 2008;55:1269–73. [PubMed] [Google Scholar]
- 30.Mikami T, Yoshida T, Akino F, et al. Apoptosis regulation differs between ulcerative colitis–associated and sporadic colonic tumors. Association with survivin and bcl-2. Am J Clin Pathol. 2003;119:723–30. doi: 10.1309/YLX4-L4H3-6K54-X92H. [DOI] [PubMed] [Google Scholar]
- 31.Velculescu VE, Madden SL, Zhang L, et al. Analysis of human transcriptomes. Nat Genet. 1999;23:387–8. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 32.Abd El-Hameed A. Survivin expression in colorectal adenocarcinoma using tissue microarray. J Egypt Natl Canc Inst. 2005;17:42–50. [PubMed] [Google Scholar]
- 33.Li F, Ackermann EJ, Bennett CF, et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–6. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- 34.Zangemeister-Wittke U. Antisense to apoptosis inhibitors facilitates chemotherapy and TRAIL-induced death signaling. Ann N Y Acad Sci. 2003;1002:90–4. doi: 10.1196/annals.1281.019. [DOI] [PubMed] [Google Scholar]
- 35.Hirschowitz EA, Foody T, Kryscio R, et al. Autologous dendritic cell vaccines for non-small-cell lung cancer. J Clin Oncol. 2004;22:2808–15. doi: 10.1200/JCO.2004.01.074. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro GI. Preclinical and clinical development of the cyclin-dependent kinase inhibitor flavopiridol. Clin Cancer Res. 2004;10:4270s–5s. doi: 10.1158/1078-0432.CCR-040020. [DOI] [PubMed] [Google Scholar]
- 37.Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets. 2004;8:409–22. doi: 10.1517/14728222.8.5.409. [DOI] [PubMed] [Google Scholar]
- 38.Xia W, Gerard CM, Liu L, et al. Combining lapatinib (GW572016), a small molecule inhibitor of ErbB1 and ErbB2 tyrosine kinases, with therapeutic anti-ErbB2 antibodies enhances apoptosis of ErbB2-overexpressing breast cancer cells. Oncogene. 2005;24:6213–21. doi: 10.1038/sj.onc.1208774. [DOI] [PubMed] [Google Scholar]
- 39.Paduano F, Villa R, Pennati M, et al. Silencing of survivin gene by small interfering RNAs produces supra-additive growth suppression in combination with 17-allyl-amino-17-demethoxygeldanamycin in human prostate cancer cells. Mol Cancer Ther. 2006;5:179–86. doi: 10.1158/1535-7163.MCT-05-0132. [DOI] [PubMed] [Google Scholar]
- 40.Biran A, Brownstein M, Haklai R, et al. Downregulation of survivin and aurora a by histone deacetylase and RAS inhibitors: A new drug combination for cancer therapy. Int J Cancer. 2011;128:691–701. doi: 10.1002/ijc.25367. [DOI] [PubMed] [Google Scholar]
- 41.Kim EJ, Park SY, Lee JY, et al. Fucoidan present in brown algae induces apoptosis of human colon cancer cells. BMC Gastroenterol. 2010;10:96. doi: 10.1186/1471-230X-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu HF, Hu HC, Chao JI. Oxaliplatin down-regulates survivin by p38 MAP kinase and proteasome in human colon cancer cells. Chem Biol Interact. 2010;188:535–45. doi: 10.1016/j.cbi.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Sreevalsan S, Jutooru I, Chadalapaka G, et al. 1,1-Bis(3′-indolyl)-1-(p-bromophenyl)methane and related compounds repress survivin and decrease gamma-radiation-induced survivin in colon and pancreatic cancer cells. Int J Oncol. 2009;35:1191–9. doi: 10.3892/ijo_00000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun PC, Tzao C, Chen BH, Liu CW, Yu CP, Jin JS. Suberoylanilide hydroxamic acid induces apoptosis and sub-G1 arrest of 320 HSR colon cancer cells. J Biomed Sci. 2010;17:76. doi: 10.1186/1423-0127-17-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishihara T, Hoshino T, Namba T, et al. Involvement of up-regulation of PUMA in non-steroidal anti-inflammatory drug-induced apoptosis. Biochem Biophys Res Commun. 2007;356:711–7. doi: 10.1016/j.bbrc.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 46.Liu HF, Hsiao PW, Chao JI. Celecoxib induces p53-PUMA pathway for apoptosis in human colorectal cancer cells. Chem Biol Interact. 2008;176:48–57. doi: 10.1016/j.cbi.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Konturek PC, Rembiasz K, Burnat G, et al. Effects of cyclooxygenase-2 inhibition on serum and tumor gastrins and expression of apoptosis-related proteins in colorectal cancer. Dig Dis Sci. 2006;51:779–87. doi: 10.1007/s10620-006-3206-z. [DOI] [PubMed] [Google Scholar]
- 48.de La Motte Rouge T, Galluzzi L, et al. A novel epidermal growth factor receptor inhibitor promotes apoptosis in non-small cell lung cancer cells resistant to erlotinib. Cancer Res. 2007;67:6253–62. doi: 10.1158/0008-5472.CAN-07-0538. [DOI] [PubMed] [Google Scholar]
- 49.Fortugno P, Beltrami E, Plescia J, et al. Regulation of survivin function by Hsp90. Proc Natl Acad Sci U S A. 2003;100:13791–6. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Q, Ming L, Thomas SM, et al. PUMA mediates EGFR tyrosine kinase inhibitor-induced apoptosis in head and neck cancer cells. Oncogene. 2009;28:2348–57. doi: 10.1038/onc.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kenny PA. TACE: a new target in epidermal growth factor receptor dependent tumors. Differentiation. 2007;75:800–8. doi: 10.1111/j.1432-0436.2007.00198.x. [DOI] [PubMed] [Google Scholar]
- 52.Yan Y, Zhang J, Guo JL, et al. Multiple shRNA-mediated knockdown of TACE reduces the malignancy of HeLa cells. Cell Biol Int. 2009;33:158–64. doi: 10.1016/j.cellbi.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 53.Merchant NB, Voskresensky I, Rogers CM, et al. TACE/ADAM-17: a component of the epidermal growth factor receptor axis and a promising therapeutic target in colorectal cancer. Clin Cancer Res. 2008;14:1182–91. doi: 10.1158/1078-0432.CCR-07-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Townsend KJ, Trusty JL, Traupman MA, et al. Expression of the antiapoptotic MCL1 gene product is regulated by a mitogen activated protein kinase-mediated pathway triggered through microtubule disruption and protein kinase C. Oncogene. 1998;17:1223–34. doi: 10.1038/sj.onc.1202035. [DOI] [PubMed] [Google Scholar]
- 55.Gallenne T, Gautier F, Oliver L, et al. Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J Cell Biol. 2009;185:279–90. doi: 10.1083/jcb.200809153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zong WX, Lindsten T, Ross AJ, et al. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 2001;15:1481–6. doi: 10.1101/gad.897601. [DOI] [PMC free article] [PubMed] [Google Scholar]