Abstract
Candida albicans is among the most prevalent fungal species of the human microbiota and asymptomatically colonizes healthy individuals. However, it is also an opportunistic pathogen that can cause severe, and often fatal, bloodstream infections. The medical impact of C. albicans typically depends on its ability to form biofilms, which are closely packed communities of cells that attach to surfaces, such as tissues and implanted medical devices. In this Review, we provide an overview of the processes involved in the formation of C. albicans biofilms and discuss the core transcriptional network that regulates biofilm development. We also consider some of the advantages that biofilms provide to C. albicans in comparison with planktonic growth and explore polymicrobial biofilms that are formed by C. albicans and certain bacterial species.
Microbial biofilms are communities of cells that adhere to solid surfaces or are present at liquid–air interfaces, and they are considered the most common state of growth for many microbial species1–3. Cells within biofilms have properties that are distinct from their planktonic (free-floating) counterparts; for example, they are often more resistant to drugs and to physical perturbations4. Microbial biofilms have been observed in aquatic environments, on artificial industrial structures, on implanted medical devices, and on plant and mammalian tissues1. Although many microorganisms are capable of forming single-species biofilms, it is much more common to find two or more bacterial and/or fungal species in a biofilm; these polymicrobial biofilms often provide specific advantages to each species when compared with single-species biofilms5. In several programme announcements for funding (PA-03-047 and PA-07-288), the National Institutes of Health (NIH) estimates that biofilms are responsible for a ~80% of microbial infections in humans1,6.
Candida species are the predominant fungi isolated from infected medical devices and account for ~15% of hospital-acquired cases of sepsis1. Candida albicans is the most commonly identified Candida species in clinical contexts and is one of the leading causes of hospital-acquired infections. However, in healthy humans, C. albicans is usually a harmless member of the native microbiota and asymptomatically colonizes many niches, including the gastrointestinal tract, reproductive tract, mouth and skin7–11. Disturbances caused by shifts in pH, nutritional alterations, shifts in oxygen levels, antibiotic use, diseases, or immunosuppressant therapy can promote the over-proliferation of C. albicans and often lead to severe symptoms. C. albicans infections range from superficial mucosal and dermal infections to disseminated bloodstream infections with mortality rates above 40%12–14. C. albicans infections are particularly serious in immunocompromised individuals, such as patients with AIDS, patients undergoing chemotherapy and individuals receiving immunosuppressant therapies. In addition, individuals who have implanted medical devices are also particularly susceptible15,16.
C. albicans biofilms are highly structured; they contain yeast-form cells, pseudohyphal cells and hyphal cells surrounded by an extracellular matrix 6,17,18 (FIG. 1). In addition to forming biofilms on implanted medical devices (for example, catheters, pacemakers, heart valves, joint pros-theses and dentures), C. albicans biofilms also form on host surfaces, including mucosal surfaces, epithelial cell linings and parenchymal organs19,20. Existing antifungal drugs, at concentrations effective against planktonic C. albicans, are largely ineffective against C. albicans cells in biofilms. Although much higher concentrations can be effective against biofilms, these doses often cause serious side effects to the host (that is, kidney or liver damage). Resistance to antifungal drugs associated with C. albicans biofilms and the ability to colonize implanted medical devices have been linked to increased medical costs and negative patient outcomes19–24. C. albicans biofilms function as reservoirs of drug-resistant cells that can detach, multiply and seed bloodstream infections. If a recalcitrant biofilm infection is suspected, removal of the infected device is typically the standard procedure25,26; however, depending on the device, the removal can require invasive and potentially dangerous surgical procedures (for example, the treatment of heart valves is especially problematic). Critically ill patients are often unable to tolerate these procedures, leaving few, if any, available treatment options in these cases19,26.
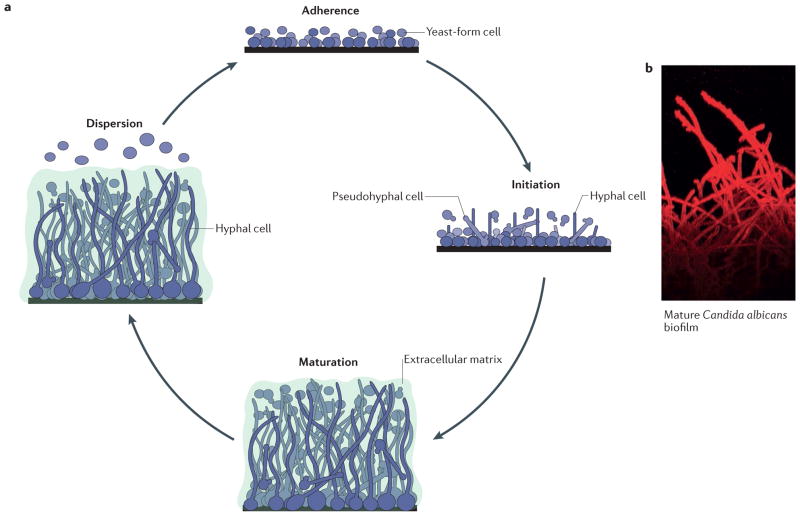
Figure 1. Formation of Candida albicans biofilms.
a | The formation of Candida albicans biofilms has been divided into four major stages: adherence of round yeast-form cells to a surface; initiation of biofilm formation, during which the cells adhered to the surface form a basal layer that contains yeast-form, pseudohyphal and hyphal cells (also known as the proliferation stage); maturation into a complex, structured biofilm, in which cells are encased in the extracellular matrix; and dispersion of yeast-form cells from the biofilm to seed new sites. b | A confocal laser-scanning microscopy image of the side view of a mature C. albicans biofilm is shown, with the long hyphal cells clearly visible. The dye (concanavalin A–Alexa Fluor 594 conjugate) used for imaging does not penetrate to the bottom of the biofilm; hence, the yeast-form cells attached to the solid surface are not readily visible. The extracellular matrix is also not visible, as it does not bind the dye. Part a modified with permission from the Annual Review of Microbiology, Volume 69 © 2015 by Annual Reviews, http://www.annualreviews.org.
In this Review, we provide an overview of the processes involved in the formation of a C. albicans biofilm and discuss the transcriptional circuitry that regulates biofilm development. We also consider some of the advantages that biofilms provide to C. albicans compared with planktonic growth and explore polymicrobial bio-films that are formed by C. albicans and certain bacterial species. Finally, we discuss several recent advances in the field, including the identification of new regulators of biofilm formation, the identification and characterization of biofilm-specific proteolysis, the proteomic characterization of persister cells, the analyses of naturally occurring C. albicans strains with differing abilities to form biofilms and the protection that biofilms confer against the host immune system.
C. albicans biofilm formation
The current understanding of C. albicans biofilm development is based on in vitro experiments, in vivo animal models of infection, and examination of biofilms that formed on medical devices in the clinic (BOX 1). On the basis of those observations, C. albicans biofilm development has been divided into four stages: adherence, initiation (also referred to as proliferation), maturation and dispersion17,27–31 (FIG. 1). In vitro results suggest that biofilm formation begins with the adherence of yeast-form cells to a solid surface and the formation of a basal layer that functions to anchor the biofilm. C. albicans cells adhere to both biotic and abiotic surfaces with consistencies ranging from hard to soft. This feature, coupled with the ability to adhere to other C. albicans cells, contributes to the structural integrity of biofilms and is the crucial first step in biofilm formation. Following adherence, these cells proliferate as yeast-form cells and remain attached to the anchor layer. Subsequently, pseudohyphal and hyphal cells begin to form from these dividing yeast-form cells, and they continue to elongate and proliferate throughout the completion of biofilm formation. Under most in vitro experimental conditions, the biofilm is considered mature by ~24 h and is characterized by a structured mixture of yeast-form, pseudohyphal and hyphal cells that are surrounded by an extracellular matrix that is composed of proteins, carbohydrates, lipids and nucleic acids. The biofilm matrix functions as a protective physical barrier from the environment and provides structural integrity to the biofilm, and it is critical in the resistance of mature biofilms to mechanical disruption. The presence of the matrix is most pronounced during and after the maturation stage of biofilm development. At those stages, the bio-film has a thick (several hundred micrometres), structured appearance with distinct layers (yeast-form cells towards the solid surface, hyphal and pseudohyphal cells extending away from the surface, and the extracellular matrix encompassing all cells). The hyphae that form in the later stages of C. albicans biofilm development form a scaffold that supports the different components of the biofilm, thereby contributing to the overall architectural stability of the biofilm structure. Once fully matured, the biofilm slowly disperses predominantly yeast-form cells that bud off from hyphae, thus contributing to the dissemination of the infection1,32. Although the general progression of biofilm development and the properties of the mature structure have been studied primarily in vitro, many of the same features (for example, a several-hundred- micrometre-thick biofilm consisting of yeast-form cells at the base, hyphal and pseudohyphal cells throughout the structure, and a thick extracellular matrix) have also been observed in vivo in rabbit, mouse and rat catheter models, in vivo in rat denture models, and in vivo and ex vivo in mouse vaginal models. C. albicans biofilms have also been extensively observed in clinical settings, for example on catheters and dentures of patients33–42.
Box 1. Biofilm assays.
In vitro biofilm assays typically involve adherence of cells to a solid surface (generally for 1–1.5 h), a wash to remove non-adherent or weakly adherent cells, and a maturation step (typically 24–48 h) with either shaking or continuous flow across the surface. Candida albicans biofilms can form under a range of environmental conditions in vitro, such as different surface materials (for example, polystyrene plates or silicone squares), different growth media, different temperatures and different shaking or flow rates151. Whereas biofilms can be directly observed using some in vitro assays (for example, confocal laser-scanning microscopy and biofilm formation on silicone squares)46,48, other methods rely on correlated readouts for biofilm formation (for example, dry weight and optical density)46,61,68,152. Measurements of cell viability (for example, metabolic reduction of the tetrazolium salt reagent XTT)153,154 and dye uptake (for example, crystal violet incorporation into the biofilm)155 are also commonly used as readouts to evaluate biofilm formation in vitro.
Regarding in vivo assays, rabbit, mouse and rat catheter models, avascular subcutaneous catheter models, rat denture models and mouse vaginal models have all been used to study biofilm formation in a host33–35,38,39,42,156,157. In the future, we can anticipate the results from additional model systems currently in development, such as those involving live imaging of animals and optical scanners149,150,158.
Regulation of biofilm development
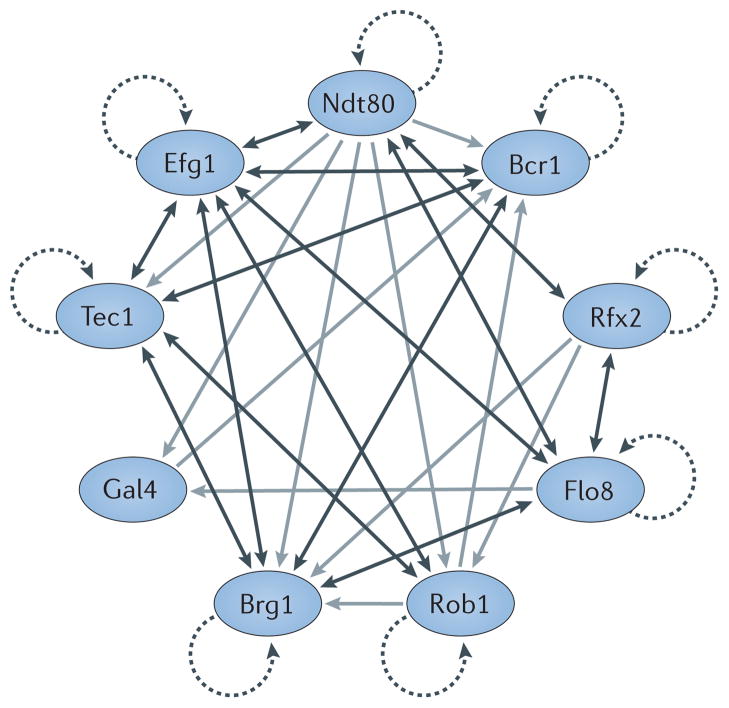
In light of the large number of differences between biofilm cells and planktonic cells, it is not surprising that more than 50 transcriptional regulators have been linked to the formation of C. albicans biofilms1,43–45 (FIG. 2; TABLE 1). Cells in which those genes have been deleted exhibit a range of phenotypes with regard to biofilms, including moderately reduced thickness of the mature biofilm (for example, an ndt80 deletion mutant), extremely thin biofilms (for example, an efg1 deletion mutant), abnormally thick biofilms (for example, an rfx2 deletion mutant), lack of hyphal cells within the biofilm structure (for example, a bcr1 deletion mutant) and reduced adherence to the solid substrate (for example, an rfg1 deletion mutant)46,47. Genetic screens that were carried out with collections of C. albicans deletion mutants identified a ‘core’ set of nine regulators (Bcr1, Brg1, Efg1, Flo8, Gal4, Ndt80, Rob1, Rfx2 and Tec1) that is required for biofilm development both in vitro on polystyrene plates and on silicone squares, and in vivo in the rat catheter and denture models46–48. Chromatin immunoprecipitation experiments revealed that these nine regulators form a highly interconnected transcriptional network (FIG. 2) in which the individual regulators control each other and, in addition, ~1,000 target genes46,47. The complex, highly interconnected structure of this network is similar to those of regulatory networks controlling complex processes such as white–opaque switching in C. albicans49, pseudohyphal growth in Saccharomyces cerevisiae50 and even development of mouse pluripotent embryonic stem cells51. Some of the target genes in the biofilm network themselves encode transcriptional regulators, several of which were subsequently shown to also be required for biofilm formation (for example, Grf10 and Sfp1) (TABLE 1). Other regulatory targets are required for specific processes in biofilm formation, such as hyphal growth (for example, Hwp1), the production of the extracellular matrix (for example, Gsc1 and Mnn1) and drug resistance (for example, Cdr1 and Mdr1). However, the functions of most target genes remain to be determined, as most of them lack clear homology with characterized genes from other organisms46. Consistent with this observation, orthology mapping suggests that the targets of the C. albicans biofilm network are enriched for genes that evolved more recently, after the Candida clade diverged from a non-pathogenic ancestor46. Thus, many of the target genes in the biofilm network are ‘novel’ genes (that is, genes that have rapidly evolved or were acquired by horizontal gene transfer) and their functions remain to be determined. In the following sections, we discuss recent updates to this regulatory circuit that are crucial for the individual steps of C. albicans biofilm formation.
Figure 2. The core transcriptional network controlling biofilm formation in Candida albicans.
More than 50 transcriptional regulators have been linked to the formation of Candida albicans biofilms. The proteins depicted are a ‘core’ set of nine regulators (Ndt80, Bcr1, Rfx2, Flo8, Rob1, Brg1, Gal4, Tec1 and Efg1) that is required for biofilm development. Autoregulation is indicated by dotted arrows, direct binding interactions between two regulators that each regulate the activity of the other are indicated by double-headed dark grey arrows, and direct binding interactions where one regulator controls another regulator are indicated by single-headed light grey arrows. The indicated interactions are based on previously reported chromatin immunoprecipitation data46,47.
Table 1.
Transcriptional regulators involved in Candida albicans biofilm formation
| ORF | Gene | Transcript levels in biofilm versus planktonic cells* | Core regulators binding to the control region of the gene‡ | Biofilm-related processes affected | Refs |
|---|---|---|---|---|---|
| orf19.6124 | ACE2 | 1.15 | None detected | Adherence, initiation and hyphal formation | 64,159 |
| orf19.2331 | ADA2 | 0.14 | None detected | Adherence and initiation | 64 |
| orf19.7381 | AHR1 | 1.15 | Bcr1, Brg1, Efg1, Ndt80, Tec1 | Adherence and initiation | 160 |
| orf19.4766 | ARG81 | −0.16 | None detected | Adherence and initiation | 64 |
| orf19.723 | BCR1 | 1.26 | Bcr1, Brg1, Efg1, Gal4, Ndt80, Rob1, Tec1§ | Adherence and initiation | 46,48 |
| orf19.6874 | BPR1 | −0.26 | Bcr1, Brg1, Efg1, Ndt80, Rob1, Tec1 | Adherence and initiation | 47 |
| orf19.4056 | BRG1 | 2.02 | Bcr1, Brg1, Efg1, Flo8, Ndt80, Rob1, Rfx2, Tec1§ | Adherence and initiation | 46 |
| orf19.4670 | CAS5 | −0.82 | Efg1, Ndt80 | Adherence and initiation | 64 |
| orf19.2356 | CRZ2 | 1.16 | Bcr1, Brg1, Efg1, Ndt80, Rob1, Tec1 | Adherence and initiation | 64 |
| orf19.3127 | CZF1 | 1.33 | Bcr1, Brg1, Efg1, Ndt80, Tec1 | Adherence, initiation and hyphal formation | 64 |
| orf19.3252 | DAL81 | 0.62 | None detected | Adherence and initiation | 64 |
| orf19.610 | EFG1 | −0.87 | Bcr1, Brg1, Efg1, Flo8, Ndt80, Rob1, Tec1§ | Adherence, initiation and hyphal formation | 46,76 |
| orf19.3193 | FCR3 | 0.03 | Bcr1, Brg1, Ndt80 | Adherence and initiation | 64 |
| orf19.6680 | FGR27 | 1.08 | None detected | Adherence and initiation | 64 |
| orf19.1093 | FLO8 | 1.04 | Brg1, Efg1, Flo8, Ndt80, Rfx2§ | Adherence, initiation and hyphal formation | 47 |
| orf19.5338 | GAL4 | 0.07 | Flo8, Ndt80§ | Adherence and initiation | 47 |
| orf19.1358 | GCN4 | 1.89 | Ndt80 | Adherence and initiation | 161 |
| orf19.4000 | GRF10 | 1.07 | Bcr1, Brg1, Efg1, Ndt80, Tec1 | Adherence, initiation and hyphal formation | 44 |
| orf19.2842 | GZF3 | 0.07 | Ndt80 | Adherence and initiation | 47 |
| orf19.4225 | LEU3 | 0.18 | None detected | Adherence and initiation | 64 |
| orf19.5312 | MET4 | 0.18 | None detected | Adherence and initiation | 64 |
| orf19.6309 | MSS11 | −0.10 | Rob1 | Adherence, initiation and hyphal formation | 162 |
| orf19.2119 | NDT80 | 0.49 | Efg1, Flo8, Ndt80, Rfx2§ | Adherence, initiation and hyphal formation | 46,163 |
| orf19.2012 | NOT3 | −0.68 | None detected | Adherence and initiation | 64 |
| orf19.7150 | NRG1 | −0.82 | Bcr1, Brg1, Efg1, Ndt80, Rob1, Tec1 | Dispersion and hyphal formation | 82 |
| orf19.4093 | PES1 | 0.12 | None detected | Dispersion | 28 |
| orf19.2823 | RFG1 | 0.14 | Bcr1, Brg1, Efg1, Ndt80 | Adherence and initiation | 47 |
| orf19.4590 | RFX2 | 1.07 | Flo8, Ndt80, Rfx2§ | Adherence and initiation | 47 |
| orf19.7247 | RIM101 | 1.18 | Bcr1, Brg1, Efg1, Tec1 | Adherence, initiation and hyphal formation | 47 |
| orf19.4662 | RLM1 | 0.60 | None detected | Production of the extracellular matrix | 73 |
| orf19.4998 | ROB1 | 0.01 | Efg1, Ndt80, Rob1, Rfx2, Tec1§ | Adherence, initiation and hyphal formation | 46 |
| orf19.5953 | SFP1 | 0.21 | Brg1, Ndt80 | Adherence and initiation | 45 |
| orf19.5871 | SNF5 | 1.24 | None detected | Adherence and initiation | 64 |
| orf19.7319 | SUC1 | 0.34 | None detected | Adherence and initiation | 64 |
| orf19.798 | TAF14 | −0.52 | None detected | Adherence and initiation | 64 |
| orf19.5908 | TEC1 | 1.28 | Bcr1, Brg1, Efg1, Ndt80, Rob1, Tec1§ | Adherence, initiation and hyphal formation | 46,48 |
| orf19.4062 | TRY2 | −0.14 | None detected | Adherence and initiation | 64 |
| orf19.1971 | TRY3 | −0.15 | None detected | Adherence and initiation | 64 |
| orf19.5975 | TRY4 | 1.38 | Ndt80 | Adherence and initiation | 64 |
| orf19.3434 | TRY5 | 0.00 | Bcr1, Brg1, Ndt80 | Adherence and initiation | 64 |
| orf19.6824 | TRY6 | 1.17 | Ndt80 | Adherence and initiation | 64 |
| orf19.4941 | TYE7 | 0.14 | Bcr1, Brg1, Efg1, Ndt80 | Hyphal formation | 164 |
| orf19.7317 | UGA33 | 1.86 | None detected | Adherence and initiation | 64 |
| orf19.1822 | UME6 | 1.20 | Bcr1, Brg1, Efg1, Ndt80, Tec1 | Dispersion and hyphal formation | 28,82 |
| orf19.1035 | WAR1 | 0.72 | None detected | Adherence and initiation | 64 |
| orf19.3794 | ZAP1 | 1.43 | Bcr1, Brg1, Efg1 | Production of the extracellular matrix | 64,72,165 |
| orf19.4767 | ZCF28 | 0.75 | None detected | Adherence and initiation | 64 |
| orf19.5924 | ZCF31 | −0.11 | Bcr1, Brg1, Efg1, Ndt80, Tec1 | Adherence and initiation | 64 |
| orf19.5940 | ZCF32 | −0.53 | None detected | Adherence, initiation and hyphal formation | 43 |
| orf19.6182 | ZCF34 | −0.25 | None detected | Adherence and initiation | 64 |
| orf19.7583 | ZCF39 | −0.48 | None detected | Adherence and initiation | 64 |
| orf19.1718 | ZCF8 | 0.04 | Brg1, Efg1, Ndt80 | Adherence and initiation | 64 |
| orf19.6781 | ZFU2 | 1.36 | None detected | Adherence and initiation | 64 |
| orf19.3187 | ZNC1 | 0.34 | None detected | Adherence and initiation | 64 |
Transcriptional expression levels comparing biofilm and planktonic cells (as determined by microarrays) are indicated; values are on a log2 scale and are taken from previous reports46. Positive values indicate genes whose expression is upregulated in biofilms and negative values indicate genes whose expression is downregulated in biofilms.
Based on previously published studies of genome-wide chromatin immunoprecipitation followed by DNA microarray (ChIP–chip) data for Bcr1, Brg1, Efg1, Ndt80, Rob1, Tec1 (REF. 46) and chromatin immunoprecipitation coupled with quantitative PCR (ChIP–qPCR) data for Flo8, Gal4, Rfx2 (REF. 47).
Binding by Flo8, Gal4 and Rfx2 has been evaluated only for the control regions of the nine core regulators; all other genes have been evaluated for binding by Bcr1, Brg1, Efg1, Ndt80, Rob1 and Tec1.
Adherence
Although we lack detailed understanding of how C. albicans cells adhere to surfaces, current evidence indicates that the nature of the surface, molecules involved in quorum sensing, host hormones and the presence of other interacting microorganisms can influence this initial step of biofilm formation52–59. The specific genes involved in adherence, both in the early stages of biofilm formation and in the later maturation phases, thus have important roles in biofilm formation and maintenance. For example, a group of ten genes (including ALS1, ALS2, ALS4 and PGA6) is upregulated early in biofilm formation, whereas a different set of ten genes (including IFF4, IFF6, PGA32 and PGA55) is upregulated at later time points47. Most of these genes encode proteins that exhibit homology to documented fungal adhesins52,60 and contain glycosylphosphatidylinositol (GPI) anchors; they were designated ‘adhesion genes’ based on experiments that established their requirement for adhesion of C. albicans in at least one assay measuring cellular adherence (of note, not necessarily in assays of biofilm formation). Of the ten ‘early’ adhesion genes, five (ALS1, ALS2, ALS3, EAP1 and MSB2) have been shown to be directly required for biofilm formation based on genetic studies in which they have been deleted46,61–65. Considerably less is known about the ‘late’ adhesion genes, as most have not been directly tested for either adhesion or biofilm formation46,66. Several models have been proposed to explain the chemical basis for adhesion-mediated adherence to biotic and abiotic surfaces and were recently reviewed67.
However, the process of adherence involves many more genes than those encoding adhesins. Using a silicone surface in an in vitro flow cell, more than 30 transcriptional regulators were shown to be required for adherence64. At least four of these regulators (Bcr1, Ace2, Snf5 and Arg81) are also required for adherence to polystyrene, and one of them (Bcr1) is required for biofilm formation under all conditions reported46,48,64. The role of Bcr1 is not surprising given its regulatory role in the expression of several cell wall proteins necessary for adherence (for example, Als1, Als3 and Hwp1)48,61,68,69. It may seem counterintuitive that so many other transcriptional regulators are also required for the initial adherence step in biofilm formation; however, this observation indicates how little we currently understand about this step in biofilm formation.
Production of the extracellular matrix
A mature C. albicans biofilm is encased in an extracellular matrix that is a mixture of glycoproteins (55%), carbohydrates (25%; largely α-mannan and β-1,6-glucan polysaccharides, and, to a lesser extent, β-1,3-glucan), lipids (15%) and nucleic acids (5%)70. More than 500 proteins have been identified by mass spectrometry in the matrix, and many of them are enzymes70. It has been noted that many of the enzymes found in the matrix lack canonical secretion sequences and are thus unlikely to be secreted enzymes, which suggests that the matrix contains a small fraction of lysed cells in addition to other components70. Although not yet experimentally validated, these extra-cellular enzymes could have several functions, including the digestion of cellular components to provide nutrients for C. albicans, promotion of cell dispersion and the protection of fungal cells from destruction by host immune cells70. Very little is known about the presence of extra-cellular DNA (eDNA) in the C. albicans biofilm matrix, but similar to bacterial biofilms, there is evidence that eDNA contributes to the stability of C. albicans biofilms. In agreement with this notion, one study has shown that eDNA is present at all stages of biofilm development and that treatment with DNase decreases biofilm biomass at later time points in biofilm formation71.
Two transcriptional regulators, Rlm1 and Zap1 (also known as Csr1), have been implicated in matrix production. Deletion of ZAP1 was shown to increase matrix formation, potentially through the upregulation of Gca1 and Gca2 (REF. 72), which are glucoamylase enzymes that convert long-chain polysaccharides into smaller-chain polysaccharides. Deletion of RLM1 reduces matrix formation, potentially through downregulation of the Gsc1 (also known as Fks1) β-1,3-glucan synthase subunit73, which catalyses the formation of β-1,3-glucan. Mutations in genes encoding enzymes that produce polysaccharide components of the matrix (ALG11, MNN4-4, MNN9, MNN1, PMR1, VAN1 and VRG4 involved in mannan synthesis, BIG1 and KRE5 involved in β-1,6-glucan synthesis, and GSC1 involved in β-1,3-glucan synthesis) result in broad defects in matrix synthesis74, which highlights their crucial role in the production of the matrix and biofilm development.
Formation of hyphal cells
C. albicans can form hyphal cells both in planktonic cultures and during the maturation step of biofilm formation1,32,75. Many of the same gene products, including transcriptional regulators and structural proteins, are important for hyphal formation in both of these contexts. For example, several of the previously mentioned core regulators of biofilm formation (for example, Efg1, Ndt80, Rob1 and Tec1) are also involved in the regulation of hyphal growth in planktonic cells; deletion of any one of these genes reduces or even eliminates the formation of hyphae under planktonic conditions46,76,77. Further support for the link between hyphal growth in planktonic cells and biofilms comes from a screen of kinase deletion mutants; those with defects in hyphal formation in suspension cultures were also defective in biofilm formation78. Furthermore, transcriptional profiling studies of C. albicans clinical isolates with varying abilities to form biofilms in vitro revealed that strains that formed thicker biofilms had increased expression of hyphal-specific genes (for example, HWP1) compared with the strains that formed thinner biofilms79. Finally, although the core biofilm regulator Bcr1 is not required for hyphal formation per se, it is required for hyphae to adhere to one another48, a critical property for maintaining the strength of the biofilm. Consistent with this idea, deletion of either BCR1 or HWP1 resulted in reduced retention of cells within a biofilm61,68. Thus, hyphae in the C. albicans biofilm seem to provide a scaffold for other cells and the matrix, resulting in a resilient structure that is several hundred micrometres thick. The extensive hyphal networks observed in mature C. albicans biofilms are a hallmark that distinguish C. albicans biofilms from those of closely related species such as Candida parapsilosis, which tend to have fewer and shorter hyphae80; this difference may result in the increased strength of C. albicans biofilms relative to those of other species1.
Dispersion
The dispersal of C. albicans into the surrounding environment (primarily as round, yeast-form cells) occurs throughout biofilm formation, with greater numbers of cells being dispersed once the biofilm reaches maturity28,81. Dispersed cells can initiate the formation of new biofilms, lead to systemic infections in the bloodstream of the host, or disseminate into host tissues, thus spreading the infection. The transcriptional regulators Ume6, Nrg1 and Pes1 (also known as Nop7) have all been linked to dispersal; overexpression of UME6 reduces dispersal, whereas overexpression of either NRG1 or PES1 increases the number of cells released from the biofilm28,82. Set3, a component of a chromatin-modifying complex, is also required for dispersal, probably by being recruited to specific genes by the transcriptional regulator Nrg1 (REFS 81,83). In addition, deletion of the molecular chaperone Hsp90 and the cell wall protein Ywp1 reduced dispersal84–86. Moreover, although both nutrient limitation and nutrient excess have been hypothesized to influence dispersion87, recent evidence indicates that more cells are dispersed in nutrient-rich medium than in nutrient-poor medium28,88. However, future studies are required to determine how these environmental changes lead to changes in dispersion.
Specific properties of biofilm cells
Cells in C. albicans biofilms have several properties that are distinct from those of planktonic cells. A combination of genetic screens, genome-wide transcriptional profiling and proteomics has been used to study C. albicans bio-film development46–48,64,70,89–91, and these approaches have uncovered major differences between cells of biofilms and those of exponentially growing suspension cultures. For example, RNA-sequencing studies identified ~1,600 genes that were upregulated and ~600 genes that were downregulated in cells in biofilms compared with cells grown in suspension cultures46, accounting for approximately 35% of the protein-coding genes in the haploid genome. When gene expression patterns were compared between biofilms and suspension-cultured cells in the stationary phase, additional differences were uncovered. For example, some metabolic pathways that are down-regulated in cells during stationary growth (compared with exponentially growing cells) have been reported to be further downregulated in cells of biofilms. However, other pathways that are downregulated in stationary growth (compared with exponential growth) are highly active in biofilm cells47,92. The differences between bio-film and planktonic cells extend beyond their metabolic activities; for example, several secreted proteases are upregulated in biofilm cells but not in planktonic cells, which results in distinct proteolytic cleavage profiles for each growth state91. Moreover, differential gene expression profiles have also been noted for specific stages of biofilm formation; for example, ~250 genes were upregulated and ~150 genes were downregulated in cells that had just adhered to a surface compared with the cells that are not adhered in the same medium47. Finally, when compared with ‘standard’ planktonic cells, the yeast-form cells that disperse from mature biofilms seem to be more virulent and to have increased abilities to adhere to surfaces to form new biofilms28. In the following section, we discuss two of these differences, increased drug resistance and altered detection by the host immune system.
Antifungal drug resistance
C. albicans biofilms are resistant to currently available antifungal drugs, although this resistance is less pronounced for the echinocandin class of antifungals93,94. This resistance is due to a combination of factors, including increased expression of drug efflux pumps (even in the absence of drugs), protective features of the extracellular matrix, and the existence of ‘persister’ cells in the biofilm (see below) (FIG. 3). Drug resistance is a serious issue for treating biofilm-based infections; in fact, the azole class of drugs, a common initial therapeutic for C. albicans infections, is largely ineffective against biofilms (BOX 2). Given the limited options, when a device-localized biofilm infection is suspected, removal of the medical device is typically recommended instead of antifungal treatments95.
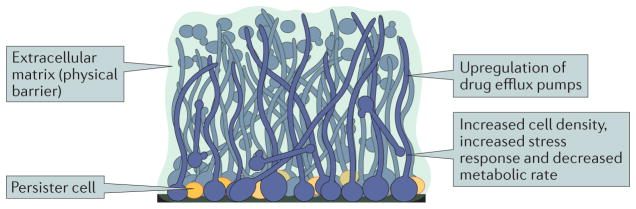
Figure 3. Overview of Candida albicans biofilm antifungal drug resistance.
Characteristics of Candida albicans biofilms that contribute to resistance to antifungal drugs. C. albicans biofilms are complex structures that contain round, yeast-form cells, pseudohyphal cells and hyphal cells (shown in blue) that are encased in an extracellular matrix (shown in green). The extracellular matrix functions as a physical barrier to antifungal drugs. Cells within the biofilm also exhibit increased cell density, increased stress response and decreased metabolic activity, which all contribute to antifungal drug resistance. A minority cell population, called ‘persister’ cells (shown in orange), can exist in the basal layer of the biofilm. Persister cells are non-dividing cells with decreased metabolic activities, making them highly resistant to antimicrobial drugs and likely to seed new biofilm infections after drug treatment. Efflux pumps export drug molecules from the inside the cell to the environment. Expression of these efflux pumps is highly upregulated in C. albicans biofilms, even in the absence of antifungal drugs, which contributes to the overall drug-resistant nature of biofilms.
Box 2. Antifungal drugs.
Four major classes of antifungal drugs are in common use to treat invasive fungal infections25,100. The oldest class of antifungal drugs is the polyenes (for example, amphotericin B), which are fungicidal against Candida albicans. Polyenes interact with ergosterol in the fungal cell membrane, forming pores that destabilize the membrane and induce lethal leakage of the cytoplasm. Azoles (for example, fluconazole and voriconazole) inhibit a demethylase enzyme (Erg11 in C. albicans) in the ergosterol biosynthesis pathway, which results in the depletion of ergosterol. Azoles also lead to the accumulation of toxic methylated sterol intermediates. Azoles, the most commonly used class of antifungals, have a fungistatic effect on C. albicans. Antifungals of the newest class, the echinocandins (for example, caspofungin), have a fungicidal effect on C. albicans. They inhibit the synthesis of β-1,3-glucan, which is important for cell wall crosslinking; this inhibition affects cell wall integrity and ultimately causes osmotic lysis of the cell. The final, less commonly used class of antifungal drugs is made up of the pyrimidine or nucleoside analogues, of which flucytosine (5-FC) is a common example. 5-FC is converted by the fungal cell into compounds that mimic nucleotides that become incorporated into DNA or RNA, interfering with fungal DNA replication and protein synthesis.
Common mechanisms of azole antifungal drug resistance include upregulation of (or point mutations within) genes whose products are directly inhibited by antifungal drugs (for example, Erg11), compensatory mutations or regulatory changes elsewhere in targeted pathways (for example, Erg3 in the ergosterol biosynthesis pathway), and upregulation of efflux pumps that export drugs from the cell (for example, Cdr1 and Mdr1)100.
C. albicans has two main classes of efflux pumps, the major facilitator (MF) and ATP binding cassette (ABC) transporter superfamilies96–99. Several efflux pumps are upregulated in the presence of antifungal drugs in planktonic cells, and mutations that lead to the constitutive upregulation of efflux pumps such as mutations in the promoters of the genes encoding Cdr1, Cdr2 or Mdr1 (or in transcriptional regulators that control their expression) are frequently associated with increased resistance to antifungal drugs100. The upregulation of genes encoding efflux pumps (even in the absence of drugs) seems to be a core part of the biofilm transcriptional programme46,47; several efflux pumps are upregulated early during the biofilm developmental process, within 6 h of adherence. Some of these efflux pumps are only transiently upregulated (for example, Nag3 and Nag4)47, whereas others (for example, Cdr1, Cdr2 and Mdr1) remain upregulated in mature biofilms46,98,99,101–103.
In addition to contributing to the structural features of the biofilm itself, the extracellular matrix is thought to sequester antifungal drugs and provide a physical barrier. Both eDNA and polysaccharides in the matrix have been found to contribute to antifungal resistance; compromising either of these components (for example by treatment with DNase I or β-1,3-glucanase, or by deletion of genes that encode enzymes involved in the synthesis of mannan, β-1,6-glucan or β-1,3-glucan) were shown to increase sensitivity to antifungal drugs74,104–106. Likewise, the addition of exogenous β-1,3-glucan to medium was found to increase the resistance of planktonic cells to fluconazole104. Moreover, the matrix component β-1,3-glucan has also been shown to bind to and sequester the antifungal drug amphotericin B107.
During C. albicans biofilm development, but not during planktonic growth, persister cells develop stochastically by an unknown mechanism108. These persister cells can revert back to metabolically active cells and contribute to the reformation of the biofilm once the antifungal drug has been removed109. Indeed, following treatment of a C. albicans biofilm with high doses of amphotericin B, a fraction of cells remained viable at drug concentrations well above those that killed most cells in the biofilm108,110,111. Based on extrapolations from persister cells in other microbial species, two hypotheses have been advanced: C. albicans persister cells exhibit low metabolic activity, and persister cells are stochastically upregulated for the stress response at the time the drug is added. Consistent with both hypotheses, a recent proteomics study found that the protein levels of many metabolic enzymes (including several enzymes involved in ergosterol biosynthesis, which is the target of azole class of drugs) were downregulated in persister cells compared with control cells in a biofilm, while the levels of proteins involved in the stress response and proteins maintaining the integrity of the cell wall were highly upregulated92. The fact that a number of genes and metabolic pathways involved in glycolysis, the electron transport chain and the tricarboxylic acid cycle are downregulated in biofilm cells, even below the levels observed in planktonic cells growing in the stationary phase, is also consistent with a link between low rates of metabolism and drug resistance in biofilms47. Finally, a recent study found that levels of the alkyl hydroperoxide reductase Ahp1 (a cell wall peroxidase involved in the response to oxidative stress) directly correlated with the fraction of persister cells that survived exposure to amphotericin B112, a result consistent with the persister cells expressing higher levels of the gene.
Evasion of host immunity and detection
A range of host immune cells, including epithelial cells, neutrophils, macrophages and dendritic cells, have key roles in the recognition of and elimination of C. albicans (Supplementary information S1 (box)). C. albicans uses several strategies to evade the host immune response, many of which are linked to biofilms. For example, the hyphal cells in the top layer of mature biofilms ‘mask’ their β-glucans (a cell wall feature recognized by the immune system), have the ability to penetrate epithelial cell layers during invasive growth, and mediate escape from phagocytic cells by physically piercing the phagocytic cell113. The differential expression of genes in cells in biofilms compared with planktonic cells has been linked to immune evasion mechanisms of the former. The activation of the host complement system is blocked by C. albicans Pra1 (an antigenic, zinc-binding cell surface protein) and Gpd2 (a glycerol-3-phosphate dehydrogenase that is found at the cell surface), as well as members of the secreted aspartyl protease (Sap) family114; these proteins are all highly expressed during biofilm formation. Moreover, Msb2, a mucin that functions as a cell wall damage sensor and is also highly expressed in biofilms, protects C. albicans from host-secreted antimicrobial peptides115,116.
Although neutrophils may surround biofilms in vivo, they often fail to engulf biofilm cells, possibly because of the protective matrix117. In the presence of biofilm cells, neutrophils also fail to trigger a reactive oxygen species response, although the mechanisms for this are unknown118. Building on this observation, it has also been shown that the extracellular matrix of C. albicans biofilms helps to block the release of neutrophil extracellular traps (NETs), thus reducing neutrophil- mediated killing119. In support of this, decreased production of the extracellular matrix leads to increased NET levels and decreased fungal proliferation119.
Mature C. albicans biofilms consist of a mixture of distinct cell types (including yeast-form, hyphal and pseudo-hyphal cells; FIG. 1) and distinct microenvironments (for example, hypoxic niches within the depths of the biofilm and aerobic niches near the biofilm surface); this heterogeneity also extends to the transcriptomes and proteomes of individual cells. This broad cellular diversity may be a key aspect that contributes to the resistance of biofilms to antifungal agents and the host immune system because any threat to the biofilm must address multiple targets rather than just a single type of cell. Coupled with the extracellular matrix and the physical architecture of the biofilm itself, given that its large, interconnected nature functions as a barrier to innate immune cells, it is not surprising that the biofilm state is the preferred state of C. albicans in stressful environments, such as in the presence of antifungal drugs or the host immune response.
Polymicrobial biofilms
The human microbiota includes bacteria, archaea and fungi, providing numerous opportunities for physical and chemical interactions between different species and even different kingdoms. Dental caries (oral cavities), periodontitis (gum disease), otitis media (ear infections), diabetic wound infections, chronic pulmonary infections (for example, cystic fibrosis), urinary tract infections and medical device infections are among the many ailments that are caused by polymicrobial biofilm formation120 (FIG. 4a). Polymicrobial infections are observed at an even higher frequency in immunocompromised patients32,120,121.
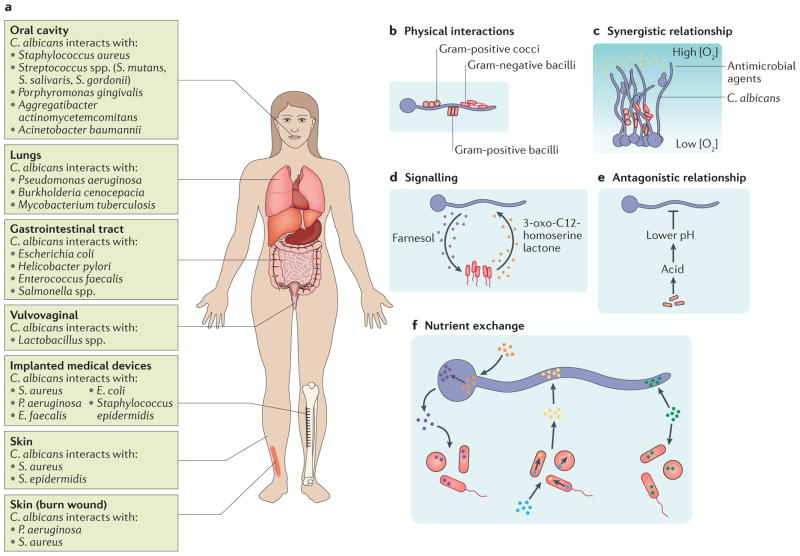
Figure 4. Multi-species biofilm formation.
a | Bacterial species most frequently isolated with Candida albicans from specific niches of the human body, including the oral cavity, lungs, gastrointestinal tract, vulvovaginal region and skin are listed. Infections resulting from the presence of a burn wound or an implanted medical device are often sites for bacterial–fungal infections; bacterial species most commonly isolated from these infections are listed. b–f | Common ways in which C. albicans interacts with various bacterial species in the context of a biofilm.b | Bacterial cells (Gram-positive cocci and bacilli are shown in red with a black outline; Gram-negative bacilli are shown in red with a red outline) can directly bind to C. albicans hyphal cells (shown in blue). c | C. albicans–bacterial interactions can be synergistic; for example, aC. albicans biofilm can physically protect bacteria from antimicrobial agents (shown in yellow) or can protect anaerobic bacteria from high oxygen concentrations by providing a low oxygen niche within the depths of the biofilm.d | Signalling molecules produced by bacterial species and C. albicans enable inter-kingdom communication within mixed-species biofilms. For example, in C. albicans–Pseudomonas aeruginosa mixed-species biofilms, the P. aeruginosa-secreted signalling molecule 3-oxo-C12-homoserine lactone (shown in orange) can influence the behaviour ofC. albicans, and the C. albicans secreted signalling molecule farnesol (shown in purple) can influence the behaviour of P. aeruginosa. e | C. albicans–bacterial interactions can be antagonistic; for example, acid produced byLactobacillus spp. results in a lower local pH, which inhibits C. albicans hyphal formation. f | Cells within mixed-species biofilms can also exchange nutrients. For example, cells can share nutrients (shown in green), or one species can utilize available nutrients (blue and orange) and in turn produce nutrients (yellow and purple) needed by the other species. S. gordonii, Streptococcus gordonii; S. mutans, Streptococcus mutans; S. salivaris, Streptococcus salivaris.
C. albicans is the fungal pathogen most frequently isolated from mixed bacterial–fungal infections (FIG. 4a). For example, Staphylococcus aureus and C. albicans are often co-isolated from neonatal polymicrobial bloodstream infections122. Biofilms containing C. albicans and Streptococcus mutans or Streptococcus gordonii have been isolated from denture stomatitis and dental carries123–126, whereas C. albicans and Pseudomonas aeruginosa are commonly co-isolated from skin and lung infections127. Scanning electron microscopy studies of dual-species biofilms has revealed that cells from species such as Staphylococcus epidermidis, Acinetobacter baumannii, Pseudomonas aeruginosa and various Streptococcus species bind to C. albicans hyphal cells present in a biofilm121,128 (FIG. 4b). Other studies have demonstrated that C. albicans adhesion proteins (for example, Als1, Als2 and Als3), hyphal wall proteins (for example, Hwp1) and transcription regulators (for example, Bcr1 and Tec1) play crucial roles in the interactions between C. albicans and bacterial species such as S. epidermidis, S. gordonii and S. aureus129–131; deletion of these proteins results in fewer bacteria bound to C. albicans as well as weaker adhesion of interacting bacteria. Several of these in vitro findings have also been observed in a murine model of oral candidiasis132.
Some relationships between C. albicans and bacterial species are synergistic, providing protection to one or both species in the context of dual-species biofilms133 (FIG. 4c). For example, when C. albicans and methicillin-resistant S. aureus (MRSA) strains were grown together in a biofilm, the presence of C. albicans seemed to protect the MRSA strain from elimination by vancomycin, which is an antibiotic that would normally be effective against MRSA134,135. A recent study linked this protection to the C. albicans extracellular matrix; both increased β-1,3-glucan production and exogenous β-1,3-glucan supplementation increased the survival of MRSA in the presence of vancomycin, and this protective effect was reduced if β-1,3-glucan was disrupted or if its synthesis was blocked136. In another study, dual-species biofilms were formed between C. albicans and either Bacteroides fragilis or Clostridium perfringens, which are two anaerobic bacterial species that are present in the human gut. C. albicans biofilms provided a protective hypoxic microenvironment that enabled the growth of these strictly anaerobic bacteria within the biofilm, even though the external environment was aerobic137 (FIG. 4c). Moreover, when cultured together under planktonic conditions in the presence of ambient oxygen, these anaerobic bacteria induced C. albicans to form ‘mini-biofilms’, which are free-floating cellular aggregates resembling miniature biofilms, that incorporated the bacteria within these structures, enabling B. fragilis and C. perfringens to proliferate under otherwise toxic conditions137. Thus, the presence of a C. albicans biofilm enables the proliferation of anaerobic pathogens in otherwise-hostile oxygen-rich environments; moreover, the bacteria seem to induce the formation of these protective structures. However, C. albicans–bacteria interactions are not confined to cooperation; for example, P. aeruginosa has been shown to kill C. albicans cells after attachment to C. albicans hyphae138, although the underlying mechanism is not understood in detail.
Interspecies interactions are not limited to direct physical contact; some rely on the secretion and diffusion of signalling molecules and others on local environmental changes (for example, alterations in pH, oxygen or carbon dioxide levels) to influence the behaviour of one species towards the other32,133 (FIG. 4d,e). For example, studies on the vaginal microbiota have revealed that Lactobacillus species lower the local pH (by releasing lactic acid), which results in the inhibition of C. albicans initial adherence to the vaginal mucosal surface139 (FIG. 4e). Extensive research on the crosstalk between P. aeruginosa and C. albicans has revealed that each species secretes a similarly structured quorum sensing molecule that can be detected by the other species. For example, the 12-carbon acyl homoserine lactone, which is secreted by P. aeruginosa, inhibits C. albicans hyphal growth140,141 (FIG. 4d). Similarly, Enterococcus faecalis secretes the bacteriocin EntV, which inhibits C. albicans hyphal and biofilm formation and disrupts mature C. albicans bio-films142. Finally, the LuxS quorum sensing system in S. gordonii is needed for the increase in biomass observed when S. gordonii and C. albicans form a dual-species bio-film125. The S. gordonii luxS mutant strain is less capable of inducing C. albicans hyphal growth than the wild-type S. gordonii strain and causes an overall reduction in biomass of the dual-species biofilm formed. This reduction in biomass is not due to a general biofilm defect in the luxS strain; the single-species biofilms formed by the luxS and wild-type strains are similar in biomass125. Other fascinating, but less explored, interactions include the sharing of nutrients and the exchange of electrons between microbial species during biofilm formation143,144 (FIG. 4f). The latter interaction is particularly intriguing in the context of polymicrobial biofilms, as it enables microorganisms of different species to gain energy from a series of reactions that a single species might lack. This strategy has been well documented for methane-producing and anaerobic polymicrobial biofilms found in the environment145. To our knowledge, the positive feedback loop between phenazine production in P. aeruginosa and ethanol production in C. albicans is the only instance in which such a strategy has been documented for polymicrobial biofilms that are relevant to human health146,147. Exposure to sub-toxic concentrations of phenazines results in the increased production of fermentation products, such as ethanol, by C. albicans. In response to the resulting increased ethanol levels, P. aeruginosa increases production of phenazine and promotes its own biofilm formation. This interaction has been shown to increase P. aeruginosa biofilm formation on bronchial epithelial cells and could, in principle, lead to increased virulence for both species in the host146,147.
Other interactions between C. albicans and bacterial partners can also affect virulence during an infection133. For example, when C. albicans and specific oral Streptococcus species, such as Streptococcus oralis, S. sanguinis, or S. gordonii, were placed in a flow-cell-based oral-cavity infection model, the two species demonstrated a cooperative interaction that increased tissue invasion and colonization compared with monotypic infections148. Similar synergies in virulence have been observed for other C. albicans–bacterial dual-species interactions; for example, higher host mortality was observed when S. aureus and C. albicans were introduced together at sub-lethal doses in a mouse peritonitis infection model compared with infections with either species alone123.
The broad range of synergies that have been observed between two or more microorganisms has implications for the treatment of infections. For example, treatment of a polymicrobial biofilm in the catheter of a patient is particularly difficult, often leading to the removal of the device instead. Further studies are needed to understand the complexity of polymicrobial biofilm infections and interspecies interactions to guide the development of new therapeutic strategies.
Conclusion
The medical impact of C. albicans on its human host depends on its ability to form biofilms, which are densely packed communities of cells that adhere to a range of biotic and abiotic surfaces. The specific characteristics of biofilms, especially their resistance to existing antifungal drugs, their ability to evade components of the host immune response, their stability to mechanical forces, and their ability to seed new infections, make them an important clinical problem. Work to date has identified numerous potential starting points for more focused examinations of specific stages of the biofilm life cycle. More targeted studies of the initial adhesion of C. albicans cells to solid surfaces, the subsequent development of mature biofilms, the formation of the extracellular matrix, the formation of persister cells, and the mechanism of dispersion have identified numerous gene products required for each of these steps28,47,70,92, although the function of many of them is unknown. Recent work on dual-species biofilms and the protection that they can provide for each species has revealed fascinating ways in which distantly related microorganisms work closely together.
Despite these recent advances, we still lack a clear, comprehensive understanding of the processes involved in the development of a biofilm, even in the simplest case of a pure C. albicans biofilm forming under controlled, in vitro conditions. For example, we do not know how environmental signals and proteins other than transcriptional regulators (such as kinases and cell wall proteins) feed into the biofilm regulatory circuitry. Likewise, more information is needed about the temporal regulation of biofilms, particularly how cells ‘decide’ to advance to later stages of the life cycle. We also do not understand how the different cell types in a biofilm are produced and organized in a biofilm. Another intriguing question concerns whether biofilm cells have a ‘cell memory’ and whether it persists in cells that are dispersed from bio-films. Finally, and perhaps most importantly, the roles of the great majority of genes upregulated during biofilm formation are simply not known.
Although not strictly a developmental process, bio-film formation could be profitably studied in the future using many of the tools used in developmental biology, such as lineage tracers and distinct fluorescent makers for each cell type. Widespread use of recently developed assays to track biofilm development in live rodent hosts in real time will undoubtedly challenge and change much of our current understanding of biofilms149,150. Finally, the study of mixed C. albicans–bacterial biofilms, although only beginning, has already revealed unanticipated synergies that further complicate the treatment of biofilms in the clinic. In the end, it is hoped that these studies will lead to new therapies to prevent, disrupt and otherwise render harmless the peculiar ability of C. albicans to form biofilms on almost any surface in the human body.
Supplementary Material
Acknowledgments
The authors thank Sheena Singh-Babak for comments on the manuscript. Research in the authors’ laboratories related to this work is supported by NIH grants R41AI112038 (to C.J.N.) and R01AI083311 (to A.D.J.), and by a Pew Biomedical Scholar Award (to C.J.N.) from the Pew Charitable Trusts. The funders had no role in planning and writing the Review or in the decision to submit the work for publication.
Glossary
- Yeast-form cells
Spherical fungal cells that form daughter cells, which bud off from the parent cell.
- Pseudohyphal cells
Ovoid chains of fungal cells that contain constrictions (rather than septa) at the cell junctions.
- Hyphal cells
Elongated, cylindrical fungal cells that contain complete septa at the cell junctions.
- Extracellular matrix
A protective physical barrier that surrounds cells in a biofilm and is composed of proteins, carbohydrates, lipids and nucleic acids.
- Persister cells
Non-dividing fungal cells with decreased metabolic activity that are resistant to antimicrobial agents.
- White–opaque switching
The ability for Candida albicans cells to switch between the ‘white’ and ‘opaque’ phenotypic cell types. The switch occurs epigenetically; that is, without a change in the primary DNA sequence of the genome.
- Horizontal gene transfer
The process through which genetic material is transferred between microorganisms through mechanisms such as transformation, conjugation and transduction. This process is distinct from vertical gene transfer, in which genetic material is transferred from mother cells to daughter cells.
- Quorum sensing
A method of communication that allows microorganisms to sense cell density and microbial community composition and respond as a group. The process involves the production and detection of soluble quorum sensing molecules.
- Glycosylphosphatidylinositol (GPI) anchors
Post-translational modifications of proteins in which a glycolipid is covalently attached and anchors the protein in the plasma membrane.
- Flow cell
A light microscopy method for observing biofilm formation in vitro under laminar flow conditions.
- Glucoamylase
An enzyme that catalyses the hydrolysis of glucosidic linkages in starch, which releases glucose.
- Glucan synthase
A glucosyltransferase enzyme that catalyses the synthesis of glucans, which are critical polysaccharide components of the fungal cell wall and the extracellular matrix.
- Chromatin-modifying complex
A protein complex that alters the chromatin structure.
- Complement system
A group of proteins that, when activated, mediate the innate immune response and inflammatory response to a pathogen.
- Mucin
A glycosylated protein that is the major component of mucus.
- Ergosterol
A sterol component of the fungal cell membrane necessary for membrane fluidity.
- Bacteriocin
A pore-forming peptide produced by some bacterial and archaeal species that is toxic to other microorganisms.
Footnotes
Author contributions:
M.B.L., M. G. and C.J.N. researched data for the article; M.B.L., M.G., A.D.J. and C.J.N. made substantial contributions to discussions of the content, wrote the article, reviewed and edited the manuscript before submission.
Competing interests statement
The authors declare competing interests: see Web version for details.
References
- 1.Nobile CJ, Johnson AD. Candida albicans biofilms and human disease. Annu Rev Microbiol. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolter R, Greenberg EP. Microbial sciences: The superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 5.Wolcott R, Costerton JW, Raoult D, Culter SJ. The polymicrobial nature of biofilm infection. Clin Microbiol Infect. 2013;19:107–112. doi: 10.1111/j.1469-0691.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 6.Fox EP, Nobile CJ. A sticky situation: untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription. 2012;3:315–322. doi: 10.4161/trns.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Achkar JM, Fries BC. Candida infections of the genitourinary tract. Clin Microbiol Rev. 2010;23:253–273. doi: 10.1128/CMR.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganguly S, Mitchell AP. Mucosal biofilms of Candida albicans. Curr Opin Microbiol. 2011;14:380–385. doi: 10.1016/j.mib.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy MJ, Volz PA. Ecology of Candida albicans gut colonization: inhibition of Candida adhesion, colonization, and dissemination from the gastrointestinal tract by bacterial antagonism. Infect Immun. 1985;49:654–663. doi: 10.1128/iai.49.3.654-663.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumamoto CA. Candida biofilms. Curr Opin Microbiol. 2002;5:608–611. doi: 10.1016/s1369-5274(02)00371-5. [DOI] [PubMed] [Google Scholar]
- 11.Kumamoto CA. Inflammation and gastrointestinal Candida colonization. Curr Opin Microbiol. 2011;14:386–391. doi: 10.1016/j.mib.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;9:327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- 13.Pappas PG, et al. Guidelines for treatment of candidiasis. Clin Infect Dis. 2004;38:161–189. doi: 10.1086/380796. [DOI] [PubMed] [Google Scholar]
- 14.Wenzel RP. Nosocomial candidemia: risk factors and attributable mortality. Clin Infect Dis. 1995;20:1531–1534. doi: 10.1093/clinids/20.6.1531. [DOI] [PubMed] [Google Scholar]
- 15.Kullberg BJ, Oude Lashof AM. Epidemiology of opportunistic invasive mycoses. Eur J Med Res. 2002;7:183–191. [PubMed] [Google Scholar]
- 16.Weig M, Gross U, Mühlschlegel F. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 1998;6:468–470. doi: 10.1016/s0966-842x(98)01407-3. [DOI] [PubMed] [Google Scholar]
- 17.Chandra J, et al. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramage G, Mowat E, Jones B, Williams C, Lopez-Ribot J. Our current understanding of fungal biofilms. Crit Rev Microbiol. 2009;35:340–355. doi: 10.3109/10408410903241436. [DOI] [PubMed] [Google Scholar]
- 19.Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramage G, Martínez JP, López-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 2006;6:979–986. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- 21.Donlan RM. Biofilm formation: a clinically relevant microbiological process. Clin Infect Dis. 2001;33:1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- 22.Tumbarello M, et al. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J Clin Microbiol. 2007;45:1843–1850. doi: 10.1128/JCM.00131-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tumbarello M, et al. Risk factors and outcomes of candidemia caused by biofilm-forming isolates in a tertiary care hospital. PLoS ONE. 2012;7:e33705. doi: 10.1371/journal.pone.0033705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebeaux D, Ghigo JM, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox EP, Singh-Babak SD, Hartooni N, Nobile CJ. In: Antifungals: From genomics to resistance and the development of Novel Agents. Coste AT, Vandeputte P, editors. Caister Academic Press; 2015. pp. 71–90. [Google Scholar]
- 26.Andes DR, et al. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis. 2012;54:1110–1122. doi: 10.1093/cid/cis021. [DOI] [PubMed] [Google Scholar]
- 27.Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 28.Uppuluri P, et al. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. This work provides an examination of cell dispersion from biofilms, including regulation, nutrient dependence and the characteristics of dispersed cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawser SP, Douglas LJ. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 1994;62:915–921. doi: 10.1128/iai.62.3.915-921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 2006;8:1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- 31.Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–679. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- 32.Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 2016;18:310–321. doi: 10.1016/j.micinf.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andes D, et al. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. This study describes a commonly used and well-established in vivo model of biofilm formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson CC, Yu A, Lee H, Fidel PL, Noverr MC. Development of a contemporary animal model of Candida albicans-associated denture stomatitis using a novel intraoral denture system. Infect Immun. 2012;80:1736–1743. doi: 10.1128/IAI.00019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nett JE, et al. Rat indwelling urinary catheter model of Candida albicans biofilm infection. Infect Immun. 2014;82:4931–4940. doi: 10.1128/IAI.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harriott MM, et al. Candida albicans forms biofilms on the vaginal mucosa. Microbiology. 2010;156:3635–3644. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. Characterization of mucosal Candida albicans biofilms. PLoS ONE. 2009;4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Řičicová M, et al. Candida albicans biofilm formation in a new in vivo rat model. Microbiology. 2010;156:909–919. doi: 10.1099/mic.0.033530-0. [DOI] [PubMed] [Google Scholar]
- 39.Schinabeck MK, et al. Rabbit model of Candida albicans biofilm infection: liposomal amphotericin B antifungal lock therapy. Antimicrob Agents Chemother. 2004;48:1727–1732. doi: 10.1128/AAC.48.5.1727-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuford JA, Rouse MS, Piper KE, Steckelberg JM, Patel R. Evaluation of caspofungin and amphotericin B deoxycholate against Candida albicans biofilms in an experimental intravascular catheter infection model. J Infect Dis. 2006;194:710–713. doi: 10.1086/506452. [DOI] [PubMed] [Google Scholar]
- 41.Ramage G, Tomsett K, Wickes BL, López-Ribot JL, Redding SW. Denture stomatitis: a role for Candida biofilms. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004;98:53–59. doi: 10.1016/j.tripleo.2003.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Lazzell AL, et al. Treatment and prevention of Candida albicans biofilms with caspofungin in a novel central venous catheter murine model of candidiasis. J Antimicrob Chemother. 2009;64:567–570. doi: 10.1093/jac/dkp242. [DOI] [PubMed] [Google Scholar]
- 43.Kakade P, Sadhale P, Sanyal K, Nagaraja V. ZCF32, a fungus specific Zn(II)2 Cys6 transcription factor, is a repressor of the biofilm development in the human pathogen. Candida albicans Sci Rep. 2016;6:31124. doi: 10.1038/srep31124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh AK, Wangsanut T, Fonzi WA, Rolfes RJ. The GRF10 homeobox gene regulates filamentous growth in the human fungal pathogen Candida albicans. FEMS Yeast Res. 2015;15:fov093. doi: 10.1093/femsyr/fov093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen HF, Lan CY. Role of SFP1 in the regulation of Candida albicans biofilm formation. PLoS ONE. 2015;10:e0129903. doi: 10.1371/journal.pone.0129903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nobile CJ, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. This study defines the core transcriptional network regulating C. albicans biofilms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fox EP, et al. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol. 2015;96:1226–1239. doi: 10.1111/mmi.13002. This work expands the core biofilm regulatory circuit while also providing insight into the transcriptional changes that occur temporally between different stages of biofilm development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 49.Hernday AD, et al. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol. 2013;90:22–35. doi: 10.1111/mmi.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borneman AR, et al. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–438. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorrells TR, Johnson AD. Making sense of transcription networks. Cell. 2015;161:714–723. doi: 10.1016/j.cell.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alves CT, et al. Effect of progesterone on Candida albicans vaginal pathogenicity. Int J Med Microbiol. 2014;304:1011–1017. doi: 10.1016/j.ijmm.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Sandini S, et al. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol. 2011;11:106. doi: 10.1186/1471-2180-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frade JP, Arthington-Skaggs BA. Effect of serum and surface characteristics on Candida albicans biofilm formation. Mycoses. 2011;54:e154–e162. doi: 10.1111/j.1439-0507.2010.01862.x. [DOI] [PubMed] [Google Scholar]
- 56.Salgado PS, et al. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 2011;108:15775–15779. doi: 10.1073/pnas.1103496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li F, Palecek SP. Distinct domains of the Candida albicans adhesin Eap1p mediate cell-cell and cell-substrate interactions. Microbiology. 2008;154:1193–1203. doi: 10.1099/mic.0.2007/013789-0. [DOI] [PubMed] [Google Scholar]
- 58.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Desai JV, Mitchell AP. Candida albicans biofilm development and its genetic control. Microbiol Spectr. 2015 doi: 10.1128/microbiolspec.MB-0005-2014. http://dx.doi.org/10.1128/microbiolspec.MB-0005-2014. [DOI] [PMC free article] [PubMed]
- 60.Sundstrom P. Adhesion in Candida spp. Cell Microbiol. 2002;4:461–469. doi: 10.1046/j.1462-5822.2002.00206.x. [DOI] [PubMed] [Google Scholar]
- 61.Nobile CJ, et al. Critical role of Bcr1-dependent adhesins in C albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao X, Oh SH, Yeater KM, Hoyer LL. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology. 2005;151:1619–1630. doi: 10.1099/mic.0.27763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li F, et al. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot Cell. 2007;6:931–939. doi: 10.1128/EC.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finkel JS, et al. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012;8:e1002525. doi: 10.1371/journal.ppat.1002525. This study provides a comprehensive examination of the regulation of adherence, the first step in biofilm formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swidergall M, Filler SG. Oropharyngeal candidiasis: fungal invasion and epithelial cell responses. PLoS Pathog. 2017;13:e1006056. doi: 10.1371/journal.ppat.1006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kempf M, et al. Disruption of Candida albicans IFF4 gene involves modifications of the cell electrical surface properties. Colloids Surf B Biointerfaces. 2007;58:250–255. doi: 10.1016/j.colsurfb.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 67.Hoyer LL, Cota E. Candida albicans agglutinin-like sequence (Als) family vignettes: a review of Als protein structure and function. Front Microbiol. 2016;7:280. doi: 10.3389/fmicb.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell. 2006;5:1604–1610. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nobile CJ, et al. Complementary adhesin function in C albicans biofilm formation. Curr Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zarnowski R, et al. Novel entries in a fungal biofilm matrix encyclopedia. mBio. 2014;5:e01333–4. doi: 10.1128/mBio.01333-14. This study provides a detailed breakdown of the composition of the biofilm extracellular matrix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martins M, et al. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia. 2010;169:323–331. doi: 10.1007/s11046-009-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nobile CJ, et al. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell. 2011;10:1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mitchell KF, et al. Community participation in biofilm matrix assembly and function. Proc Natl Acad Sci USA. 2015;112:4092–4097. doi: 10.1073/pnas.1421437112. This study examines the role of many polysaccharide components of the biofilm extracellular matrix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 76.Ramage G, Vande Walle K, López-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 77.Schweizer A, Rupp S, Taylor BN, Röllinghoff M, Schröppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol. 2000;38:435–445. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- 78.Konstantinidou N, Morrissey JP. Co-occurence of filamentation defects and impaired biofilms in Candida albicans protein kinase mutants. FEMS Yeast Res. 2015;15:fov092. doi: 10.1093/femsyr/fov092. [DOI] [PubMed] [Google Scholar]
- 79.Rajendran R, et al. Integrating Candida albicans metabolism with biofilm heterogeneity by transcriptome mapping. Sci Rep. 2016;6:35436. doi: 10.1038/srep35436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holland LM, et al. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 2014;10:e1004365. doi: 10.1371/journal.ppat.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nobile CJ, et al. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. mBio. 2014;5:e01201–14. doi: 10.1128/mBio.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uppuluri P, et al. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot Cell. 2010;9:1531–1537. doi: 10.1128/EC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hnisz D, et al. A Histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 2012;8:e1003118. doi: 10.1371/journal.pgen.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robbins N, et al. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 2011;7:e1002257. doi: 10.1371/journal.ppat.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shapiro RS, et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 2009;19:621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Granger BL. Insight into the antiadhesive effect of yeast wall protein 1 of Candida albicans. Eukaryot Cell. 2012;11:795–805. doi: 10.1128/EC.00026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blankenship JR, Mitchell AP. How to build a biofilm: a fungal perspective. Curr Opin Microbiol. 2006;9:588–594. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 88.Sellam A, et al. A Candida albicans early stage biofilm detachment event in rich medium. BMC Microbiol. 2009;9:25. doi: 10.1186/1471-2180-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinez-Gomariz M, et al. Proteomic analysis of cytoplasmic and surface proteins from yeast cells, hyphae, and biofilms of Candida albicans. Proteomics. 2009;9:2230–2252. doi: 10.1002/pmic.200700594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Richard ML, Nobile CJ, Bruno VM, Mitchell AP. Candida albicans biofilm-defective mutants. Eukaryot Cell. 2005;4:1493–1502. doi: 10.1128/EC.4.8.1493-1502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Winter MB, et al. Global identification of biofilm-specific proteolysis in Candida albicans. mBio. 2016;7:e01514–16. doi: 10.1128/mBio.01514-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li P, Seneviratne CJ, Alpi E, Vizcaino JA, Jin L. Delicate metabolic control and coordinated stress response critically determine antifungal tolerance of Candida albicans biofilm persisters. Antimicrob Agents Chemother. 2015;59:6101–6112. doi: 10.1128/AAC.00543-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fiori B, et al. In vitro activities of anidulafungin and other antifungal agents against biofilms formed by clinical isolates of different Candida and Aspergillus species. Antimicrob Agents Chemother. 2011;55:3031–3035. doi: 10.1128/AAC.01569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jacobson MJ, Steckelberg KE, Piper KE, Steckelberg JM, Patel R. In vitro activity of micafungin against planktonic and sessile Candida albicans isolates. Antimicrob Agents Chemother. 2009;53:2638–2639. doi: 10.1128/AAC.01724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pappas PG, et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6:187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 97.Anderson JB. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat Rev Microbiol. 2005;3:547–556. doi: 10.1038/nrmicro1179. [DOI] [PubMed] [Google Scholar]
- 98.Ramage G, Bachmann S, Patterson TF, Wickes BL, Lopez-Ribot JL. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother. 2002;49:973–980. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- 99.Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun. 2003;71:4333–4340. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Prasad R, Shah AH, Rawal MK. Antifungals: mechanism of action and drug resistance. Adv Exp Med Biol. 2016;892:327–349. doi: 10.1007/978-3-319-25304-6_14. [DOI] [PubMed] [Google Scholar]
- 101.Mateus C, Crow SA, Ahearn DG. Adherence of Candida albicans to silicone induces immediate enhanced tolerance to fluconazole. Antimicrob Agents Chemother. 2004;48:3358–3366. doi: 10.1128/AAC.48.9.3358-3366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nett JE, Lepak AJ, Marchillo K, Andes DR. Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis. 2009;200:307–313. doi: 10.1086/599838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yeater KM, et al. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology. 2007;153:2373–2385. doi: 10.1099/mic.0.2007/006163-0. [DOI] [PubMed] [Google Scholar]
- 104.Nett J, et al. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother. 2007;51:510–520. doi: 10.1128/AAC.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nett JE, Sanchez H, Cain MT, Andes DR. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J Infect Dis. 2010;202:171–175. doi: 10.1086/651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Taff HT, et al. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 2012;8:e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vediyappan G, Rossignol T, D’Enfert C. Interaction of Candida albicans biofilms with antifungals: transcriptional response and binding of antifungals to beta-glucans. Antimicrob Agents Chemother. 2010;54:2096–2111. doi: 10.1128/AAC.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 110.Mathe L, Van Dijck P. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet. 2013;59:251–264. doi: 10.1007/s00294-013-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khot PD, Suci PA, Miller RL, Nelson RD, Tyler BJ. A small subpopulation of blastospores in Candida albicans biofilms exhibit resistance to amphotericin B associated with differential regulation of ergosterol and β-1,6-glucan pathway genes. Antimicrob Agents Chemother. 2006;50:3708–3716. doi: 10.1128/AAC.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Truong T, et al. Comparative ploidy proteomics of Candida albicans biofilms unraveled the role of the AHP1 gene in the biofilm persistence against Amphotericin B. Mol Cell Proteom. 2016;15:3488–3500. doi: 10.1074/mcp.M116.061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ghosh S, et al. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect Immun. 2009;77:1596–1605. doi: 10.1128/IAI.01452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gropp K, et al. The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol Immunol. 2009;47:465–475. doi: 10.1016/j.molimm.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 115.Szafranski-Schneider E, et al. Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog. 2012;8:e1002501. doi: 10.1371/journal.ppat.1002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Swidergall M, Ernst AM, Ernst JF. Candida albicans mucin Msb2 is a broad-range protectant against antimicrobial peptides. Antimicrob Agents Chemother. 2013;57:3917–3922. doi: 10.1128/AAC.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hirschfeld J. Dynamic interactions of neutrophils and biofilms. J Oral Microbiol. 2014;6:26102. doi: 10.3402/jom.v6.26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xie Z, et al. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J Infect Dis. 2012;206:1936–1945. doi: 10.1093/infdis/jis607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Johnson CJ, et al. The extracellular matrix of Candida albicans biofilms impairs formation of neutrophil extracellular traps. PLoS Pathog. 2016;12:e1005884. doi: 10.1371/journal.ppat.1005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 122.Pammi M, Zhong D, Johnson Y, Revell P, Versalovic J. Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BMC Infect Dis. 2014;14:390. doi: 10.1186/1471-2334-14-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peters BM, Noverr MC. Candida albicans-Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect Immun. 2013;81:2178–2189. doi: 10.1128/IAI.00265-13. This study demonstrates that the combination of C. albicans and S. aureus results in dramatic increases in microbial burden and mortality in a mouse model of peritonitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jarosz LM, Deng DM, van der Mei HC, Crielaard W, Krom BP. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 2009;8:1658–1664. doi: 10.1128/EC.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bamford CV, et al. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jack AA, et al. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161:411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lindsay AK, Hogan DA. Candida albicans: molecular interactions with Pseudomonas aeruginosa and Staphylococcus aureus. Fungal Biol Rev. 2014;28:85–96. [Google Scholar]
- 128.Bamford CV, Nobbs AH, Barbour ME, Lamont RJ, Jenkinson HF. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology. 2015;161:18–29. doi: 10.1099/mic.0.083378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Beaussart A, et al. Single-cell force spectroscopy of the medically important Staphylococcus epidermidis-Candida albicans interaction. Nanoscale. 2013;5:10894–10900. doi: 10.1039/c3nr03272h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hoyer LL, Oh SH, Jones R, Cota E. A proposed mechanism for the interaction between the Candida albicans Als3 adhesin and streptococcal cell wall proteins. Front Microbiol. 2014;5:564. doi: 10.3389/fmicb.2014.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Peters BM, et al. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology. 2012;158:2975–2986. doi: 10.1099/mic.0.062109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schlecht LM, et al. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology. 2015;161:168–181. doi: 10.1099/mic.0.083485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Förster TM, et al. Enemies and brothers in arms: Candida albicans and gram-positive bacteria. Cell Microbiol. 2016;28:1709–1715. doi: 10.1111/cmi.12657. [DOI] [PubMed] [Google Scholar]
- 134.Harriott MM, Noverr MC. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother. 2009;53:3914–3922. doi: 10.1128/AAC.00657-09. This work shows that co-incubation of C. albicans and S. aureus results in increased biofilm formation and vancomycin resistance in S. aureus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Harriott MM, Noverr MC. Ability of Candida albicans mutants to induce Staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob Agents Chemother. 2010;54:3746–3755. doi: 10.1128/AAC.00573-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kong EF, et al. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. mBio. 2016;7:e01365–16. doi: 10.1128/mBio.01365-16. This study identifies the component of the C. albicans extracellular matrix that protects S. aureus from vancomycin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fox EP, et al. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr Biol. 2014;24:2411–2416. doi: 10.1016/j.cub.2014.08.057. This study demonstrates that bacteria can induce C. albicans biofilm formation and use the resulting biofilms for protection from the environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. This is a foundational study of interactions between C. albicans and bacterial species. [DOI] [PubMed] [Google Scholar]
- 139.Strus M, et al. The in vitro activity of vaginal Lactobacillus with probiotic properties against Candida. Infect Dis Obstet Gynecol. 2005;13:69–75. doi: 10.1080/10647440400028136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hogan DA, Vik A, Kolter RA. Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 141.McAlester G, O’Gara F, Morrissey JP. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol. 2008;57:563–569. doi: 10.1099/jmm.0.47705-0. [DOI] [PubMed] [Google Scholar]
- 142.Graham CE, Cruz MR, Garsin DA, Lorenz MC. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc Natl Acad Sci USA. 2017;114:4507–4512. doi: 10.1073/pnas.1620432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Seth EC, Taga ME. Nutrient cross-feeding in the microbial world. Front Microbiol. 2014;5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shrestha PM, Rotaru AE. Plugging in or going wireless: strategies for interspecies electron transfer. Front Microbiol. 2014;5:237. doi: 10.3389/fmicb.2014.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stams AJ, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol. 2009;7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 146.Morales DK, et al. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. mBio. 2013;4:e00526–12. doi: 10.1128/mBio.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen AI, et al. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog. 2014;10:e1004480. doi: 10.1371/journal.ppat.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Diaz PI, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80:620–632. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vande Velde G, Kucharíková S, Schrevens S, Himmelreich U, Van Dijck P. Towards non-invasive monitoring of pathogen-host interactions during Candida albicans biofilm formation using in vivo bioluminescence. Cell Microbiol. 2014;16:115–130. doi: 10.1111/cmi.12184. This study describes an in vivo live-animal imaging model of infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vande Velde G, Kucharíková S, Van Dijck P, Himmelreich U. Bioluminescence imaging of fungal biofilm development in live animals. Methods Mol Biol. 2014;1098:153–167. doi: 10.1007/978-1-62703-718-1_13. [DOI] [PubMed] [Google Scholar]
- 151.Tournu H, Van Dijck P. Candida biofilms and the host: models and new concepts for eradication. Int J Microbiol. 2012;2012:845352. doi: 10.1155/2012/845352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lohse MB, et al. Assessment and optimizations of Candida albicans in vitro biofilm assays. Antimicrob Agents Chemother. 2017;61:e02749–16. doi: 10.1128/AAC.02749-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ramage G, Vande Walle K, Wickes BL, López-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 2001;45:2475–2479. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nett JE, Cain MT, Crawford K, Andes DR. Optimizing a Candida biofilm microtiter plate model for measurement of antifungal susceptibility by tetrazolium salt assay. J Clin Microbiol. 2011;49:1426–1433. doi: 10.1128/JCM.02273-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jin Y, Yip HK, Samaranayake YH, Yau JY, Samaranayake LP. Biofilm-forming ability of Candida albicans is unlikely to contribute to high levels of oral yeast carriage in cases of human immunodeficiency virus infection. J Clin Microbiol. 2003;41:2961–2967. doi: 10.1128/JCM.41.7.2961-2967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nett JE, Marchillo K, Spiegel CA, Andes DR. Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun. 2010;78:3650–3659. doi: 10.1128/IAI.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang X, Fries BC. A murine model for catheter-associated candiduria. J Med Microbiol. 2011;60:1523–1529. doi: 10.1099/jmm.0.026294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Doyle TC, Nawotka KA, Kawahara CB, Francis KP, Contag PR. Visualizing fungal infections in living mice using bioluminescent pathogenic Candida albicans strains transformed with the firefly luciferase gene. Microb Pathog. 2006;40:82–90. doi: 10.1016/j.micpath.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 159.Kelly MT, et al. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 2004;53:969–983. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- 160.Askew C, et al. The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol Microbiol. 2011;79:940–953. doi: 10.1111/j.1365-2958.2010.07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.García-Sánchez S, et al. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell. 2004;3:536–545. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tsai PW, et al. The role of Mss11 in Candida albicans biofilm formation. Mol Genet Genom. 2014;289:807–819. doi: 10.1007/s00438-014-0846-0. [DOI] [PubMed] [Google Scholar]
- 163.Sellam A, et al. Role of transcription factor CaNdt80p in cell separation, hyphal growth, and virulence in Candida albicans. Eukaryot Cell. 2010;9:634–644. doi: 10.1128/EC.00325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bonhomme J, et al. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol Microbiol. 2011;80:995–1013. doi: 10.1111/j.1365-2958.2011.07626.x. [DOI] [PubMed] [Google Scholar]
- 165.Ganguly S, et al. Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot Cell. 2011;10:1448–1454. doi: 10.1128/EC.05196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.