Abstract
Nucleophilic amino acids make important contributions to protein function, including performing key roles in catalysis and serving as sites for post-translational modification. Electrophilic groups that target amino-acid nucleophiles have been used to create covalent ligands and drugs, but have, so far, been mainly limited to cysteine and serine. Here we report a chemical proteomic platform for the global and quantitative analysis of lysine residues in native biological systems. We quantified, in total, more than 9000 lysines in human cell proteomes and identified several hundred residues with heightened reactivity that are enriched at protein functional sites and can frequently be targeted by electrophilic small molecules. We discovered lysine-reactive fragment electrophiles that inhibit enzymes by active site and allosteric mechanisms, as well as disrupt protein-protein interactions in transcriptional regulatory complexes, emphasizing the broad potential and diverse functional consequences of liganding lysine residues throughout the human proteome.
Small molecules can serve as versatile probes for perturbing the functions of proteins in biological systems and are a primary source of therapeutic agents to treat human disorders1. Nonetheless, most human proteins still lack selective chemical ligands and some classes of proteins are even considered undruggable2. Covalent ligands offer one strategy to expand the landscape of proteins amenable to targeting by small molecules. By combining features of recognition and reactivity, covalent ligands have the potential to target sites on proteins that are difficult to address by reversible binding interactions alone3. While original covalent probes often targeted essential catalytic residues within the active sites of enzymes, in particular, serine4 and cysteine5 residues of enhanced nucleophilicity, more recent successes in covalent ligand development include electrophilic small molecules that react with non-catalytic cysteines across diverse protein classes, including kinases6,7, GTPases8, and non-enzymatic proteins (e.g., nuclear export factors9). These efforts have culminated in the approval of several covalent kinase inhibitors as drugs for treating diverse cancers6,7.
In attempts to understand the scope of proteins that may be targeted by covalent ligands, we recently evaluated the proteome-wide reactivity of a diverse set of cysteine-directed electrophilic fragments, which were found, as a collection, to engage cysteine residues on hundreds of proteins in human cell systems10. These proteins originated from diverse classes, including those deemed historically challenging to target with small molecules (e.g., adaptor proteins, transcription factors). The total number of proteins harboring liganded cysteines, however, still accounted for only ~20% of all proteins quantified in the study, suggesting that the realization of a more complete ligandability map of the human proteome may require extending beyond cysteine as a source for covalent probe development.
Among proteinaceous amino acids, lysine represents a potentially attractive candidate for covalent ligand development, as the lysine ε-amine is intrinsically nucleophilic, and lysines are found at many functional sites, including enzyme active sites11,12 and at interfaces mediating protein-protein interactions13. Lysines also frequently serve as sites for post-translational regulation of protein structure and function through, for instance, acetylation14, methylation15,16, and ubiquitylation17. Individual lysine residues within functional protein pockets are also susceptible to modification by electrophilic small molecules, including natural products, such as Wortmannin18, which targets a lysine in the active sites of PI3K kinases, activated esters that react with a lysine in transthyretin (TTR)19, and boronic acid carbonyl antagonists of the apoptosis regulatory protein MCL-113. Additional electrophiles that have been shown to react with proteinaceous lysine residues include dichlorotriazines20,21, imidoesters22, 2-acetyl- or 2-formyl-benzeneboronic acids13,23, isothiocyanates24,25, pyrazolecarboxamidines26,27, sulfonyl fluorides28,29, and vinyl sulfonamides30.
Despite the aforementioned examples, the full spectrum of functional and ligandable lysines in the human proteome remains poorly understood. Building on previous work describing a chemical proteomic platform for assessing cysteine reactivity on a global scale31, initial attempts have been made to assess lysine reactivity in human proteomes, but these datasets, which were generated using aryl halide probes, were limited to quantifying a small number of lysines (< 100) in proteomes21. Given the frequency of lysine residues in human proteins (~6% of all residues32), we hypothesized that the development of more advanced chemical proteomic methods capable of quantifying a much larger number of lysines in human proteomes would provide a deeper and more complete portrait of lysine reactivity and ligandability, as well as the potential relationship between these two parameters. Here, we show that an amine-reactive pentynoic acid sulfotetrafluorophenyl ester probe provides access to a very rich content of lysines (> 9000 residues in total) in the human proteome. We use this probe to quantify lysine reactivity and ligandability on a global scale, leading to the discovery of functional lysines that can be targeted by covalent ligands to perturb the activities of a diverse range of proteins.
Results
A chemical proteomic method for assessing lysine reactivity
We have previously described a quantitative and site-specific chemical proteomic method termed isoTOP-ABPP (isotopic tandem orthogonal proteolysis-activity-based protein profiling) for measuring cysteine reactivity in native proteomes31. Here, we reasoned that exchanging the cysteine-directed iodoacetamide alkyne probe for a probe that shows preferential reactivity with amines would afford a platform for global lysine reactivity analysis (Fig. 1a). Among candidate amine-reactive groups, we considered activated esters as a good potential probe class, as they should show preferred reactivity with amines, display good solubility, and form stable, structurally simple adducts with proteinaceous lysines for characterization by MS methods. In an initial screen of alkyne-modified ester probes (1–15, Supplementary Fig. 1), we found that sulfotetrafluorophenyl (STP) and N-hydroxysuccinimide esters showed strong proteomic reactivity as evaluated by copper-catalyzed azide-alkyne cycloaddition (CuAAC, or click chemistry33) to a rhodamine-azide tag, SDS-PAGE, and in-gel fluorescence scanning (Supplementary Fig. 1). Considering that tetrafluorophenyl esters are more stable in aqueous solution compared to NHS esters34, we selected STP-alkyne 1 as a probe for proteomic profiling of lysine reactivity.
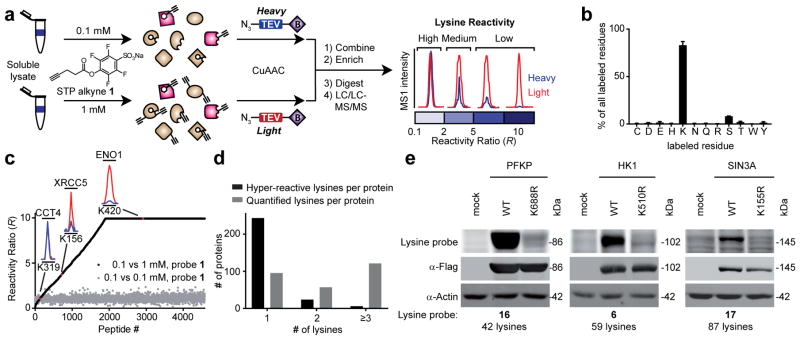
Figure 1.
Proteome-wide quantification of lysine reactivity. a, General protocol for lysine reactivity profiling by isoTOP-ABPP. Cellular lysates are labeled with an amine-reactive STP alkyne probe (1) at different concentrations. Labeled samples are conjugated to isotopically-differentiated TEV protease-cleavable biotin tags [heavy (blue) and light (red) for 0.1 and 1.0 mM probe 1 treatment groups, respectively] by CuAAC33) mixed, and 1-labeled proteins enriched by streptavidin-conjugated beads and digested stepwise with trypsin and TEV to yield 1-labeled peptides for LC-MS analysis. Isotopic ratios, or R values reflect the relative MS1 chromatographic peak intensities for 1-labeled peptides in light versus heavy samples. b, Probe 1 preferentially labels lysine residues in human cell proteomes. Residues labeled by 1 were assigned by differential modification analysis of all quantified peptides identified in three replicate experiments comparing 0.1 vs 0.1 mM probe 1 treatments of MDA-MB-231 cell lysates. Peptides were required to feature no missed cleavage sites on unmodified lysine residues. Data represent mean values ± standard deviation for three experiments. c, R values for probe 1-labeled peptides from human cancer cell proteomes (MDA-MB-231, Ramos and Jurkat) treated with 0.1 vs 1.0 mM (black) or 0.1 vs 0.1 mM (gray) of probe 1. Representative MS1 chromatographic peaks for lysines of different reactivity categories are shown as insets (high, or hyper-reactive, R < 2.0 (K319 of CCT4); medium, 2.0 < R < 5.0 (K156 of XRCC5); and low, > 5.0 (K420 of ENO1)). d, Number of hyper-reactive and quantified lysines per protein shown for proteins found to contain at least one hyper-reactive lysine. e, Hyper-reactive lysines are site-selectively labeled by activated ester probes. HEK 293T cells expressing representative proteins with hyper-reactive lysines (or the corresponding lysine-to-arginine mutant) as FLAG epitope-tagged proteins were treated with the indicated lysine-reactive probe and analyzed by gel-based ABPP.
We initially assessed the scope and selectivity with which 1 reacted with lysine residues in human cell proteomes. Two equal amounts of proteomic lysate from the human breast cancer cell line MDA-MB-231 were treated with 1 (100 μM, 1 h), CuAAC-conjugated to isotopically differentiated TEV-cleavable, azide-biotin tags (heavy and light, respectively), combined, and analyzed by isoTOP-ABPP. Measurement of the MS1 chromatographic peak ratios for isotopically-differentiated light/heavy peptide pairs provided an isoTOP-ABPP ratio or R value, which centered on ~1.0 for the more than 5000 quantified, probe 1-labeled peptides. As determined by tandem MS and differential modification analysis, > 52% of 1-labeled peptides were assigned as being uniquely modified on lysine residues, with 54% of the remaining 1-labeled peptides being assigned with lysine modifications as well as alternative residue modifications. Because lysine modification creates a missed trypsin cleavage site, we further assessed the fraction of alternative amino-acid modification assignments for their occurrence on peptides harboring a missed lysine cleavage site. We found that most of the predicted non-lysine modifications for 1 occurred on peptides with missed lysine cleavage sites (Supplementary Fig. 1), indicating that they likely represent mis-assignments of reactivity events that actually occurred on lysine. Once the isoTOP-ABPP data were filtered to remove peptide assignments with unmodified, missed lysine cleavage events, lysine accounted for the vast majority of all assignments for probe 1 modification (Fig. 1b). The remaining alternative probe 1 modifications were mostly assigned to serine (~8% of the total 1-labeled peptides), and these occurred on fully digested tryptic peptides (Fig. 1b), likely designating them as authentic modifications. These results, taken together, indicate that 1 shows broad reactivity and good selectivity for lysine residues in the human proteome.
Quantitative profiling of lysine reactivity in human cell proteomes
Previous isoTOP-ABPP studies have shown that the human proteome possesses a specialized set of hyper-reactive cysteine residues that are enriched in functional residues (e.g., catalytic residues, redox-active residues) compared to bulk cysteine content31. Here, we assessed the intrinsic reactivity of lysine residues in human cell proteomes by comparing their concentration-dependent labeling with probe 1, where highly reactive lysines would be expected to show nearly equivalent labeling intensities at low versus high concentrations of probe 1, with less reactive lysines displaying clear concentration-dependent increases in labeling intensity. In brief, we treated proteomes from three human cancer cell lines (MDA-MB-231, Ramos, and Jurkat cells) with low vs high concentrations of probe 1 (0.1 vs 1 mM, n = 4 per group) for 1 h and then analyzed the samples by isoTOP-ABPP, wherein high, medium, and low reactivity lysines were distinguished by their respective isotopic ratio values (R10:1 < 2, 2 < R10:1 < 5, R10:1 > 5, respectively). To minimize false quantification events, we also required that lysines were detected in control (0.1 vs 0.1 mM) experiments with R1:1 values of ~1.0 (see Supplementary Methods for additional details).
In total, ~4000 lysine residues were assessed for intrinsic reactivity across the three tested cell lines (Supplementary Fig. 2), and individual lysines showed consistent reactivity values for replicate experiments performed within (Supplementary Fig. 2) and across these cell lines (Supplementary Fig. 2). The majority of quantified lysines showed strong, concentration-dependent increases in reactivity with probe 1, indicative of residues with low intrinsic reactivity (Fig. 1c). In contrast, a rare subset of the quantified lysines (< 10%, or 310 total residues) exhibited heightened (hyper-) reactivity with probe 1 (R10:1 values < 2) (Fig. 1c). Most proteins contained only one hyper-reactive lysine among several quantified lysines (Fig. 1d), and the atypical hyper-reactivity of these lysines was further supported by comparing their R10:1 values to those of other lysines quantified on the same protein (Supplementary Fig. 2). We confirmed the lysine hyper-reactivity determinations made by isoTOP-ABPP by recombinantly expressing wild type and lysine-to-arginine mutant proteins and comparing their reactivity by gel-based ABPP using fluorescent or alkyne-tagged activated ester probes (Supplementary Fig 1). Each protein examined showed strong labeling with activated ester probes and the labeling of one or more of these probes was generally blocked, in many cases completely, by mutation of the hyper-reactive lysine to arginine (Fig. 1e, Supplementary Fig. 2, and Supplementary Table 1). Considering that there were, on average, 30 lysine residues per examined protein, the blockade of activated ester probe labeling by mutation of a single lysine in each protein underscores the unusual hyper-reactivity of these residues.
Features of hyper-reactive lysines
Hyper-reactive lysines were found on proteins from all major classes and showed a similar distribution to less reactive lysines (Fig. 2a). Hyper-reactive lysines were not, as a group, more conserved across organisms than lysines of lower reactivity, although this analysis was complicated by the high median conservation (~80%) of all 1-labeled lysines across the species examined (Supplementary Fig. 3). The primary sequence surrounding hyper-reactive lysines also did not show evidence of any obvious conserved motifs (Supplementary Fig. 3), indicating that higher-order structural features in proteins are likely imparting enhanced reactivity on these lysines. Consistent with this hypothesis, the frequency of lysines found in functional sites on proteins (e.g., enzyme active sites, ligand-binding sites), as assessed by analysis of three-dimensional protein structures, was positively correlated with reactivity (Fig. 2b). Protein pockets of uncharacterized function (as defined by AutoSite35 analysis of protein structures) also contained a greater percentage of hyper-reactive lysines compared to less reactive lysines (Supplementary Fig. 3). Interestingly, we observed a striking inverse correlation between lysine reactivity and evidence of ubiquitylation as reported in the PhosphoSitePlus® database36, (Fig. 2c), and a similar, albeit more tempered trend was found for lysine acetylation (Supplementary Fig. 3). These data, taken together, indicate that the localization of lysines to pockets on proteins may represent a prevalent mechanism for conferring heightened reactivity, and such distributions may further hinder post-translational modification of the lysines possibly due to limited surface exposure.
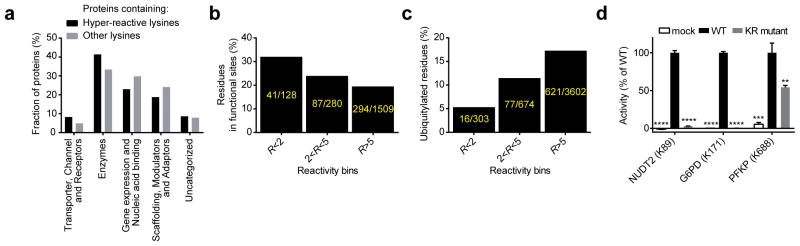
Figure 2.
Global and specific assessments of the functionality of lysine reactivity. a, Distribution of functional classes of proteins that contain hyper-reactive lysines compared to other quantified proteins lacking hyper-reactive lysines. b, Hyper-reactive lysines are enriched proximal to (within 10 Å of) annotated functional sites for proteins that have x-ray or NMR structures in the Protein Data Bank (see Supplementary Methods for further details). c, Hyper-reactive lysines are less likely to be ubiquitylated than lysines of lower reactivity (ubiquitylated lysines were defined as those with ≥ 10 reported ubiquitylation events in public databases36). d, Mutation of hyper-reactive lysines blocks the catalytic activity of NUDT2 and G6PD and reduces the activity of PFKP. Data represent mean values ± standard deviation for three experiments. Statistical significance was calculated with unpaired students t-tests in comparison to the WT activity; **, p < 0.01, ***, p < 0.001, ****, p < 0.0001.
We examined whether some of the hyper-reactive lysines located in functional pockets contributed to protein activity. NUDT2, which is a diadenosine tetraphosphate hydrolase implicated in cancer and immune cell metabolism37, possesses a hyper-reactive lysine (K89) that is highly conserved and predicted, based on an NMR structure of NUDT2, to coordinate alpha-phosphate substrate binding38. However, to our knowledge, the contributions of K89 to NUDT2 catalysis have not been investigated. We found that mutation of K89 to arginine dramatically reduced the hydrolytic activity of NUDT2 (Fig. 2d). A similar disruption of catalysis was observed by mutation of the conserved, hyper-reactive lysine (K171) in the pentose phosphate pathway enzyme glucose 6-phosphate 1-dehydrogenase (G6PD) (Fig. 2d), which is consistent with previous findings39. Both K89 of NUDT2 and K171 of G6PD are active-site residues (Supplementary Fig. 3), and we therefore wondered whether hyper-reactive lysines located in potential allosteric pockets might also affect enzyme function. As a case study, we examined the hyper-reactive lysine (K688) in platelet-type phosphofructokinase (PFKP), which is located in an allosteric pocket >22 angstroms away from the active site (Supplementary Fig. 3). Mutation of K688 to arginine in PFKP produced a partial, but significant reduction in PFKP activity (Fig. 2d), pointing to a role for this lysine in allosteric regulation of PFKP function.
Quantitative profiling of lysine ligandability in human cell proteomes
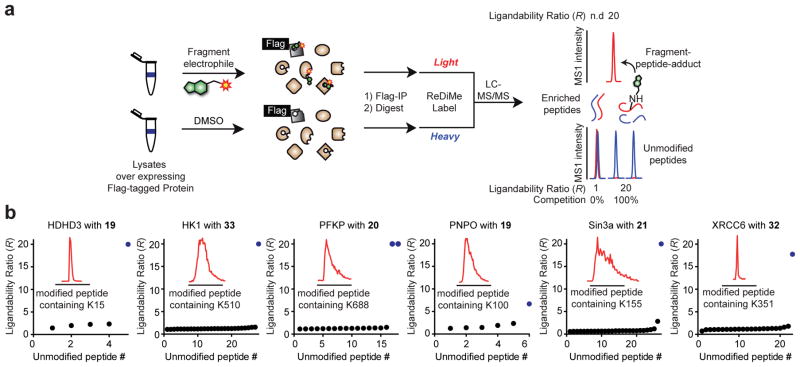
We next applied isoTOP-ABPP in a “competitive” format to assess the ligandability of lysines (Fig. 3a), where human cell proteomes were pre-treated with a small library (~30 member; 50–100 μM) of amine-reactive electrophilic fragments (activated esters, such as pentafluorophenyl- (19–28), dinitrophenyl- (29–45), and NHS esters (46), and N,N′-diacyl-pyrazolecarboxamidines (49,50)26,27) as well as one non-electrophilic control compound 51 (Fig. 3b and Supplementary Fig. 4) or DMSO control, followed by exposure to probe 1 (100 μM). Fragment-sensitive lysines were identified as those showing substantial reductions (≥ 75%) in enrichment by 1 in the presence of one or more fragments compared to the DMSO control (R values ≥ 4 for DMSO/fragment).
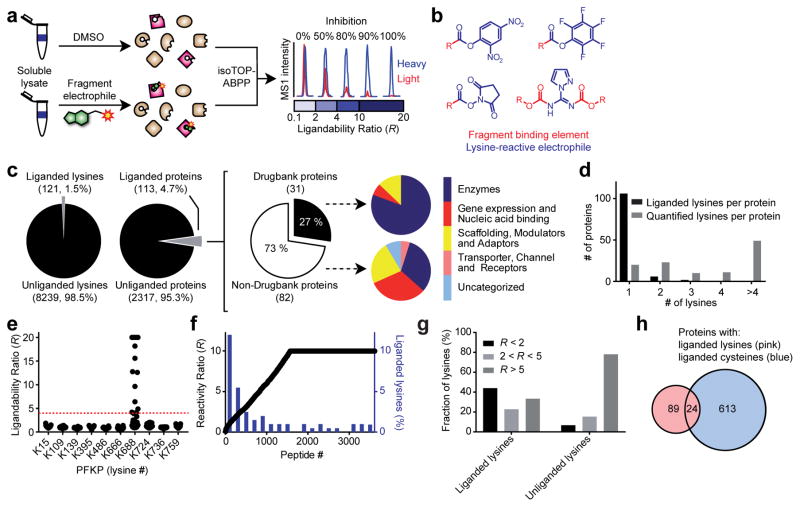
Figure 3.
Proteome-wide screening of lysine-reactive fragment electrophiles. a, General protocol for competitive isoTOP-ABPP. Competition ratios, or R values, are measured by quantifying the relative MS1 chromatographic peak intensities for 1-labeled peptides in DMSO- (heavy, or blue) versus fragment-treated (light, or red) samples. An R value of ≥ 4 was used to define a fragment liganding event for a quantified lysine. b, General structures of a lysine-reactive, electrophilic fragment library. See Supplementary Fig. 4 for chemical structures of library members. c, Fraction of total quantified lysines and proteins that were liganded by fragment electrophiles (left panel); of the liganded proteins, the fraction that is found in Drugbank (middle panel); functional classes of liganded Drugbank and non-Drugbank proteins (right panel). d, Number of liganded and quantified lysines per protein. Analysis was applied to proteins containing at least one liganded lysine. e, R values for ten lysines in PFKP, identifying K688 as the only liganded lysine in this protein. Each point represents a distinct fragment-lysine interaction quantified by isoTOP-ABPP. The red dashed line marks an R value of 4 used to define a fragment liganding event. f, Comparison of the ligandability of lysine residues as a function of reactivity with probe 1 (as measured in Fig. 1). Individual lysines are plotted on the x-axis sorted by reactivity, which is shown on the left y-axis, with lower R values correlating with elevated reactivity. A histogram with a bin-size of 200 is shown in blue for the percentage of liganded lysines within each reactivity bin (percent values shown on the right y-axis). g, Lysine reactivity distribution for both liganded and unliganded lysine residues labeled by probe 1. h, Overlap of proteins harboring liganded lysines and liganded cysteines. Cysteine ligandability was taken from reference 10.
We quantified, on average, > 2700 lysines per dataset and, in aggregate, > 8,000 lysines from 2,430 proteins across all datasets (Fig. 3c and Supplementary Table 2). Each lysine was quantified, on average, in 24 individual experiments (Supplementary Fig. 4 and Supplementary Table 2), providing a good initial assessment of ligandability potential. We identified, in total, 121 liganded lysines in 113 proteins (Fig. 3c). We quantified, on average, ~four lysines per protein that reacted with probe 1 (Fig. 3d), indicating that ligandability was a rare feature. A striking example is PFKP, where a single liganded lysine was identified – the aforementioned K688 that resides in an allosteric pocket – along with nine additional quantified lysines that showed no evidence of ligandability (Fig. 3e). Likewise, hexokinase-1 (HK1) possessed a single liganded lysine K510 among six quantified lysines (Supplementary Fig. 4). The majority of proteins harboring liganded lysines were not found in DrugBank (73%; Fig. 3c), and these proteins showed much broader class distribution than the smaller fraction of DrugBank proteins containing liganded lysines (27%), which were mostly enzymes (Fig. 3c). Prominent sub-groups of non-Drugbank proteins with liganded lysines included transcription factors and scaffolding proteins (Fig. 3c), which are considered challenging to target with small molecules.
Hyper-reactive lysines showed greater ligandability compared to less reactive lysines, although many liganded lysines were also found in the latter group (R10:1 > 2.0; Fig. 3f, g). Of note, only a small fraction (~20%) of proteins with liganded lysines were found to contain liganded cysteines in a previous study10 (Fig. 3h). These results, taken together, indicate that fragment electrophile interactions with lysines depend on both reactivity and recognition and canvas a distinct and complementary portion of the human proteome compared to covalent chemistries targeting other nucleophilic amino acids.
SAR analysis of lysine-fragment electrophile interactions
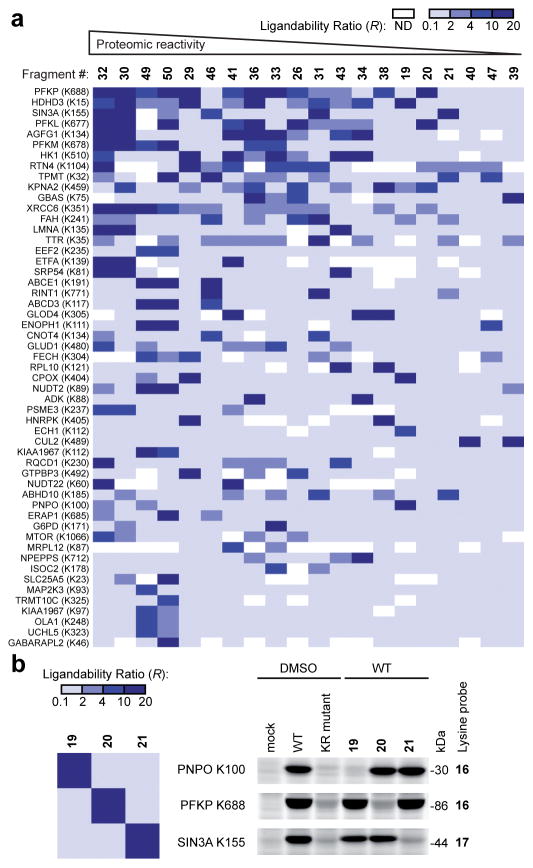
Most of the liganded lysines (69%) interacted with a limited fraction (< 10%) of the tested fragment electrophiles, although a small subset of lysines (8%) was targeted by a substantial portion of the compounds (≥ 25%) (Supplementary Fig. 5). Conversely, the fragment electrophiles showed large differences in proteomic reactivity towards lysines (Supplementary Fig. 5), ranging from 1% to 35% of the liganded residues (Supplementary Fig. 5). No lysine reactivity was observed for the non-electrophilic control fragment 51 (Supplementary Fig. 4 and 5). The dinitrophenyl esters showed somewhat greater overall reactivity compared to the corresponding pentafluorophenyl esters (Supplementary Fig. 5), which correlated with the faster solvolysis rates observed for the former class of compounds (Supplementary Fig. 5). Despite these general trends, individual lysines displayed markedly distinct structure-activity relationships (SARs) that, in some cases, directly opposed the overall reactivity profiles of the fragment electrophile library (Fig. 4a and Supplementary Table 2). The hyper-reactive lysine K35 in the hormone-binding protein transthyretin TTR, for instance, which has previously been shown to be modified selectively in human plasma by activated (thio)ester and sulfonyl fluoride ligands19,28, was preferentially targeted by the dinitrophenyl ester fragment 31 over fragments that showed much greater proteome-wide reactivity (e.g., 29 and 30) (Fig. 4a and Supplementary Fig. 5). Further evidence that recognition events make substantive contributions to fragment-lysine interactions was found in the distinct lysine reactivity profiles displayed by fragment electrophiles bearing a common leaving group (Fig. 4b, left panel). We confirmed these SAR assignments by gel-based ABPP with recombinantly expressed proteins (Fig. 4b, right panels, and Supplementary Fig. 5). The identity of the leaving group of activated ester fragments also influenced reactivity, as reflected by a subset of lysines that were preferentially liganded by pentafluorophenyl or dinitrophenyl esters bearing the same recognition group (Supplementary Fig. 5). The most distinctive lysine reactivity profiles were observed for the N,N′-diacyl-pyrazolecarboxamidine fragments 49 and 50, which, despite sharing several targets with activated esters, also reacted with 15 lysines in human cell proteomes that showed negligible cross-reactivity with activated esters (see representative proteins at the bottom of Fig. 4a and Supplementary Table 2). We confirmed the reactivity of one of these lysines (K89 of NUDT2) with N,N′-diacyl-pyrazolecarboxamidine fragments by recombinant expression of the parent protein and competitive gel-based ABPP (Supplementary Fig. 5).
Figure 4.
Analysis of fragment-lysine interactions. a, Heat-map showing R values for representative lysines and fragments organized by relative proteomic reactivity of the fragments (high to low, left to right) and number of fragment hits for individual lysines (high to low, top to bottom). b, Fragment SAR determined by competitive isoTOP-ABPP is recapitulated by gel-based ABPP of recombinant proteins. Left panel, heat-map depicts R values for the indicated fragment-lysine interactions determined by competitive isoTOP-ABPP. Right panel, HEK 293T cells recombinantly expressing representative liganded proteins (or the corresponding lysine-to-arginine (KR) mutants) as FLAG epitope-tagged proteins were treated with fragment electrophiles (50 μM, 1 h) followed by the indicated lysine-reactive probes and analyzed by gel-based ABPP. SIN3A corresponds to a.a. 1-400 of SIN3A.
We next set out to confirm fragment-lysine adducts by developing a quantitative, MS-based platform that simultaneously measured both fragment electrophile modification of lysines in individual proteins and the fractional occupancy and specificity of these reactions (Fig. 5a). Proteins containing liganded lysines discovered by isoTOP-ABPP were expressed with a Flag epitope tag in HEK 293T cells, treated with fragment electrophiles or DMSO, enriched by anti-Flag immunoprecipitation, proteolytically digested, and the tryptic peptides from fragment- and DMSO-treated samples then isotopically differentiated by reductive dimethylation (ReDiMe)40,41, combined pairwise, and analyzed by LC-MS/MS. This protocol yielded high average sequence coverage (> 40%) for the six tested proteins (PFKP, PNPO, HK1, HDHD3, XRCC6 and SIN3A), and, in each case, we obtained definitive evidence that the liganded lysine assigned by isoTOP-ABPP was directly adducted by the corresponding electrophilic fragment (Fig. 5b and Supplementary Table 2). We also observed depletion of the unmodified tryptic peptides containing the liganded lysines and/or adjacent peptides requiring the liganded lysine as a cleavage site (Fig. 5b, blue dots). Other tryptic peptides generated by a lysine cleavage event were unaffected by fragment electrophile treatment (Fig. 5b, black dots), indicating the specificity of fragment reactions with individual lysines on the tested proteins (as also predicted by isoTOP-ABPP; see Fig. 3d).
Figure 5.
Confirmation of site-specific fragment-lysine reactions by MS-based proteomics. a, Schematic workflow for direct measurement of lysine-fragment reactions on proteins by quantitative proteomics. Flag epitope-tagged proteins are recombinantly expressed in HEK 293T cells and the cellular lysates treated with DMSO or a fragment ligand and immunoprecipitated with anti-Flag agarose resin. The enriched proteins are eluted from the resin, digested with trypsin, and tryptic peptides from the DMSO and fragment-treated samples isotopically labeled by reductive dimethylation (ReDiMe) with heavy (blue) and light (red) formaldehyde40,41, respectively. The DMSO and fragment-treated samples are then combined and analyzed by LC-MS. Lysine-fragment reactions were confirmed by both: i) detection of the peptide-fragment adduct exclusively in the fragment-treated sample (top trace); and ii) depletion of the parent unlabeled tryptic peptides containing the indicated lysine or having the lysine at a proteolytic cleavage site (bottom trace). b, R values for all detected, unmodified lysine-containing tryptic peptides for representative liganded proteins after treatment with the indicated compounds at 50 μM for 1 h. Unmodified peptides that contain the liganded lysine or have it at a proteolytic cleavage site are shown as blue dots. MS1 chromatographic peaks for fragment-peptide adducts are shown in the inset traces.
Functional analysis of fragment-lysine interactions
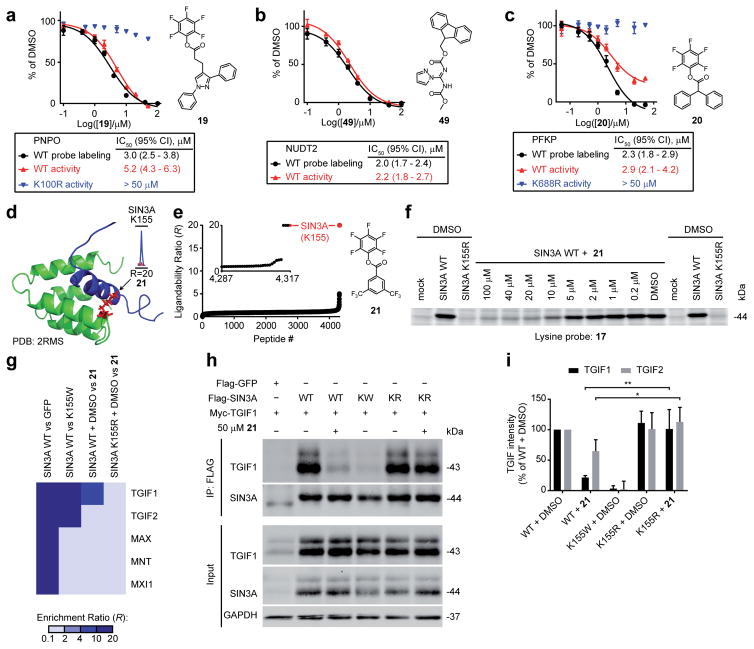
We next aimed to determine the functional impact of fragment-lysine interactions mapped by isoTOP-ABPP. As initial case studies, we selected two enzymes with liganded active-site lysines – pyridoxamine-5′-phosphate oxidase (PNPO) and NUDT2. PNPO catalyzes the FMN-dependent oxidation of pyridoxamine-5′-phosphate and pyridoxine-5′-phosphate to pyridoxal-5′-phosphate in vitamin B6 synthesis42. PNPO possesses a hyper-reactive lysine K100 (R10:1 = 0.7; Supplementary Table 1) located in the enzyme’s active site and shown in previous structural studies to interact with substrate42 (Supplementary Fig. 6). Competitive isoTOP-ABPP uncovered a highly restricted SAR for ligand engagement of K100, with only two fragments (19 and 22) fully blocking probe 1 labeling of this residue (Supplementary Fig. 6 and Supplementary Table 2). We confirmed by gel-based ABPP that fragment 19 blocked probe labeling of K100 in PNPO with an apparent IC50 value of 3 μM (Fig. 6a and Supplementary Fig. 6). A similar IC50 value (~5 μM) was measured for blockade of PNPO catalytic activity by 19 using a substrate assay43 (Fig. 6a). The inhibitory effect of 19 was not observed with a K100R mutant of PNPO (Fig. 6a), which also did not label with amine-reactive probes (Supplementary Fig. 6).
Figure 6.
Fragment-lysine reactions inhibit the function of diverse proteins. a–c, Fragments targeting active site (PNPO and NUDT2) and allosteric (PFKP) lysines in metabolic enzymes block enzymatic activity in a concentration-dependent manner with apparent IC50 values comparable to those measured by gel-based ABPP with lysine-reactive probes (probe labeling). Data represent mean values ± standard deviation for at least three experiments. CI, confidence interval. d, The liganded lysine K155 in SIN3A (red) is located at the protein-protein interaction site of the PAH1 domain (green) (shown in the NMR structure (PDB ID: 2RMS) to interact with the SID domain of SAP2564 (blue)). e, Fragment 21 (50 μM) fully competes probe 1 labeling of K155 of SIN3A as determined by isoTOP-ABPP of human cancer cell proteomes. See inset in part d, for a representative MS1 chromatographic peak of the tryptic peptide containing K155 (R = 20, >95% blockade of probe 1 labeling by 21). f, Gel-based ABPP confirms that 21 blocks probe 17 labeling of SIN3A at K155 in a concentration-dependent manner. g, Heat-map showing the enrichment of SIN3A-interacting proteins in co-immunoprecipitation-MS-based proteomic experiments and blockade of SIN3A-TGIF1 interaction by 21 (50 μM, 1 h). h, i, Western blot analysis of Flag-SIN3A or the indicated Flag-SIN3A mutants, or Flag-GFP, co-expressed in HEK 293T cells with Myc-TGIF1 or Myc-TGIF2. Cellular lysates were treated with DMSO or 21 (50 μM, 1 h), and proteins immunoprecipitated prior to western blot analysis (h). Quantification of western blotting data for four biological replicates (i). Data represent mean values ± standard deviation for four experiments. Statistical significance was calculated with unpaired students t-tests comparing SIN3A WT + 21 to K155R + 21 groups; *, p < 0.05, **, p < 0.01. For panels f–i, Flag-SIN3A or the indicated Flag-SIN3A mutants correspond to a.a. 1-400 of SIN3A.
NUDT2 is responsible for the catabolism of nucleotide cellular stress signals in human cells37 and was found to contain a hyper-reactive and liganded lysine K89 that is located proximal to the enzyme’s nucleotide-binding site (Supplementary Fig. 3). K89 also exhibited a restricted SAR by isoTOP-ABPP, preferentially reacting with the two N,N′-diacyl-pyrazolecarboxamidine fragments 49 and 50 (Supplementary Fig. 6 and Supplementary Table 2). We confirmed by gel-based ABPP that fragment 49 blocked probe labeling of NUDT2 with an apparent IC50 of 2 μM (Fig. 6b and Supplementary Fig. 6), and an equivalent IC50 value was measured for inhibition of NUDT2 activity using a substrate assay44, (Fig. 6b), which was also used to determine a kobs/[I] value for 49 of 46.3 ± 1.3 M−1s−1 (Supplementary Fig. 6). Since mutation of K89 to arginine (K89R) inactivated NUDT2 in the substrate assay (Fig. 2d), we could not test the inhibitory effect of 49 on the K89R mutant, but we did confirm by gel-based ABPP that the K89R mutant showed a substantial reduction in amine-reactive probe labeling equivalent to that observed following treatment of NUDT2 with 49 (Supplementary Fig. 6).
We next turned our attention to liganded lysines residing in more poorly characterized sites on proteins, specifically, a putative allosteric pocket in PFKP and a protein-protein interaction site in SIN3A. PFKP is responsible for the phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate, the committed step of glycolysis45. Probe 1 labeling of the hyper-reactive lysine K688 in PFKP was completely blocked by fragment 20, which otherwise exhibited limited reactivity across the proteome (Fig. 4a and Supplementary Fig. 5 and 6). Gel-based ABPP confirmed that 20 blocked probe labeling of recombinant PFKP with an apparent IC50 of 2 μM (Fig. 6c and Supplementary Fig. 6), and a similar loss in probe reactivity was observed for the K688R mutant of PFKP (Supplementary Fig. 6). Using an enzyme-coupled assay monitoring the conversion of NAD+ to NADH by UV absorbance46, we found that the activity of WT-PFKP, but not the K688R-PFKP mutant was inhibited by 20 with an apparent IC50 of 2.9 μM (Fig. 6c and Supplementary Fig. 6). Fragment 20 inhibition of the catalytic activity of WT-PFKP plateaued at ~80% reduction in substrate turnover (Fig. 6c and Supplementary Fig. 6), indicating that ligand reactivity at the K688 allosteric site substantially, but incompletely blocks enzyme function.
SIN3A is a multi-domain 145 kDa transcriptional repressor involved in histone deacetylase regulation47 and suppression of MYC-responsive genes48. We found that SIN3A contains a hyper-reactive lysine K155 (R10:1 = 1.2; Supplementary Table 1) located in the first paired amphipathic helix (PAH1) domain of the protein (Fig. 6d). Our isoTOP-ABPP experiments revealed that fragment 21 engages K155 in SIN3A (Fig. 6d, inset, and Fig. 6e), but otherwise shows low proteome-wide reactivity (Fig. 6e and Supplementary Fig. 5). We recombinantly expressed a Flag-tagged SIN3A variant containing the N-terminal PAH1 and PAH2 protein-protein interaction domains (a.a. 1-400) in HEK293T cells and found that treatment of cell lysates with 21 produced a site-specific and complete blockade of probe labeling of K155 with an apparent IC50 of 5 μM (Fig. 6f and Supplementary Fig. 7). We then used quantitative SILAC (Stable Isotopic Labeling with Amino acids in Cell culture49) proteomics to identify SIN3A-interacting proteins that were sensitive to mutation of K155 and/or treatment with 21. HEK293T cells metabolically labeled with isotopically differentiated amino acids were transfected with cDNA constructs for Flag-SIN3A (heavy-labeled cells) or Flag-GFP (light-labeled cells), harvested, lysed, and immunoprecipitated with anti-Flag antibodies. Heavy- and light-labeled immunoprecipitates were combined and subjected to tryptic digestion followed by LC-MS/MS analysis, which furnished a set of SIN3A-interacting proteins, defined as proteins that were substantially (> five-fold) enriched in the SIN3A-transfected compared to GFP-transfected samples (Fig. 6g and Supplementary Table 2). Similar quantitative proteomic experiments compared WT-SIN3A to a K155W-SIN3A mutant and DMSO-treated WT-SIN3A to 21-treated WT-SIN3A. The K155W mutant, which was generated to mimic incorporation of a bulky hydrophobic group into the 21-sensitive pocket of SIN3A, failed to substantially enrich two established SIN3-interacting proteins – TGIF1 and TGIF250,51 – that co-immunoprecipitated with WT-SIN3A (Fig. 6g and Supplementary Table 2). Treatment with 21 also strongly blocked the TGIF1-SIN3A interaction, but only produced a marginal effect on TGIF2-SIN3A interaction (Fig. 6g and Supplementary Table 2). Other known SIN3A-interacting proteins that co-immunoprecipitated with WT-SIN3A, such as MAX52, MNT52 and MXI153, were not affected by K155W mutation or 21 treatment (Fig. 6g).
We further evaluated the effect of 21 on SIN3A interactions with TGIF1/TGIF2 by co-expressing these proteins with complementary epitope tags (Flag and Myc, respectively). In this system, fragment 21 treatment, as well as K155W mutation, blocked the co-immunoprecipitation of TGIF1 as measured by anti-Myc blotting (Fig. 6h, i). The K155W mutant also strongly inhibited co-immunoprecipitation of TGIF2 with SIN3A, while 21 exerted a partial blockade of this association (Fig. 6i and Supplementary Fig. 7). Importantly, mutation of K155 to arginine (K155R) conferred resistance to the effects of 21 on the SIN3A-TGIF1 interaction (Fig. 6h, i and Supplementary Fig. 7). Taken together, these data demonstrate that covalent ligands targeting K155 in SIN3A can pharmacologically disrupt a select subset of protein-protein interactions implicated in gene regulation.
Discussion
Chemical proteomic technologies, such as ABPP, have proven valuable for covalent ligand/drug development by providing quantitative readouts of target engagement and selectivity in native biological systems10. Considering its nucleophilic side chain and prevalence in proteins, lysine is an attractive candidate amino acid for covalent ligand development. pKa-perturbed lysine residues play important functional roles in proteins54,55, and electrophilic compounds have been found to target lysines in diverse types of proteins (e.g., metabolic enzymes, such as PGAM156, hormone-binding proteins, such as TTR57, lipid kinases, such as PI3Ks18, and adaptor proteins, such as MCL-113). Nonetheless, our understanding of lysine reactivity and ligandability across the human proteome remains limited. We and others have used the chemical proteomic method isoTOP-ABPP to measure the reactivity31 and covalent ligand interactions of cysteine residues in native biological systems10,58–60. Here, we have extended this platform to globally profile the reactivity and ligandability of thousands of lysine residues in human cell proteomes. Key to success was selection of an electrophilic group – the STP ester – that displayed broad and selective reactivity with lysines over other proteinaceous amino acids, which likely accounted for the much deeper coverage of lysines compared to first-generation probes based on aryl halide reactive groups21.
When combined with previous chemical proteomic studies of cysteine reactivity31, our results provide further evidence that heightened reactivity of nucleophilic amino acids is a hallmark of functionality and ligandability. Cysteine, however, is a much less frequent amino acid in proteins compared to lysine, and, in this context, we find it noteworthy that hyper-reactive lysines could be site-selectively modified by activated ester probes in proteins that harbor 50+ other lysines (e.g., Fig. 1e and Supplementary Fig. 2). This feature enabled screening of these hyper-reactive lysines for ligandability using convenient gel-based assays (e.g., Supplementary Fig. 5). On the other hand, the greater frequency of lysine compared to cysteine in proteins presents a technical challenge for achieving a complete inventory of lysines in the proteome. This problem may not simply be overcome by raising the concentration of activated ester probe in chemical proteomic experiments, which we have found instead tends to increase the signals and coverage of lower reactivity lysines in abundant proteins (possibly at the expense of detecting high reactivity lysines on lower abundance proteins). More promising might be to perform additional upfront chromatography steps to better fractionate the proteome prior to enrichment and MS analysis of peptides containing probe-reactive lysines. Additionally, because probe reactivity with lysines blocks tryptic cleavage sites, the use of alternative proteases may uncover additional probe-lysine reactivity events that evade detection in conventional tryptic digest protocols. Finally, subsets of lysines can be selectively targeted with greater sensitivity using tailored electrophilic probes that leverage recognition elements to favor binding to functional protein pockets, such as the ATP-binding sites of kinases11,29.
Our chemical proteomic experiments have also provided valuable initial insights into the global ligandability potential of lysines in the human proteome. Most of the liganded lysines discovered herein were found in proteins lacking small-molecule probes, including proteins not present in DrugBank or targeted by cysteine-reactive fragments in a previous study10. We also demonstrated that lysine-reactive fragments can block the function of proteins, including inhibition of enzyme activity by both active site (PNPO, NUDT2) and allosteric (PFKP) mechanisms, as well as disruption of specific protein-protein interactions in transcriptional regulatory complexes (SIN3A-TGIFs). The SIN3A-TGIF1 interaction has been found to contribute to invasiveness of triple negative breast cancer50, suggesting that more optimized chemical probes targeting K155 in SIN3A may exert anti-tumorigenic effects.
Based on our competitive isoTOP-ABPP results, we believe that a broader effort to discover covalent ligands for lysines has the potential to substantially expand the druggable content of the human proteome. Success of such a program, however, may depend on identifying alternative amine-reactive chemotypes, as the activated esters tested herein are likely too prone to enzymatic and non-enzymatic hydrolysis for development into cellular or in vivo probes. Alternative amine-reactive electrophiles, such as sulfonyl fluorides28,29 or the N,N′-diacyl-pyrazolecarboxamidines explored herein may offer more suitable starting points for optimization of lysine-targeting covalent ligands that are more suitable for cell biological studies. Alternative electrophiles, when used as broad profiling probes, may also provide access to additional lysine residues in the proteome, although the chemoselectivity of such probes could present a challenge. While our manuscript was under review, for instance, Ward and colleagues characterized the proteomic reactivity of an NHS-ester probe and found that, while this activated ester labeled lysines, it also showed substantive reactivity with several other amino acid residues (serine, threonine, tyrosine, arginine, cysteine) across the mouse liver proteome61. These results are consistent with our initial gel-based profiling experiments studies of a similar NHS ester probe (8), which showed substantially higher overall proteomic reactivity compared to the STP probe 1 (Supplementary Fig. 1).
In summary, we have described a quantitative chemical proteomic platform to globally map the reactivity and ligandability of lysine residues in the human proteome. Projecting forward, it is interesting to speculate on the broader functional ramifications of lysines that display heightened reactivity. Minimally, this feature appears to correlate well with ligandability, which could reflect the enriched presence of hyper-reactive lysines in pockets, where the pKa of these residues can be presumably altered. On the other hand, the localization of hyper-reactive lysines to pockets could also restrict their access to post-translational machinery, such as ubiquitylation processes (see Fig. 2c), which may instead mostly target surface-exposed (i.e., less reactive) lysines. We also believe that our studies, despite having uncovered more than 100 lysines targeted by fragment electrophiles, almost certainly still underestimate the global ligandability of lysines in the human proteome. The development and evaluation of larger compound libraries displaying more diversified recognition and amine-reactive elements, including covalent-reversible electrophiles (e.g., aldehydes), in combination with surveying complementary cell types (e.g., primary immune cells62, metabolic organs63), should greatly enrich our understanding of functional and ligandable lysines in the human proteome and, through doing so, extend its druggable landscape for basic and translational research objectives
Methods
See Supporting Information for a detailed Methods section.
Data availability statement
The data supporting the findings discussed here are available within the paper, its Supplementary Information file and Supplementary Tables 1 and 2, as well as from the corresponding authors upon request.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (CA087660, CA132630 (B.F.C.), GM108208 (K.M.B.), GM069832 (S.F.)) and the Deutsche Forschungsgemeinschaft (S.M.H.). We thank Dr. Melissa Dix and Mihai Radu Suciu for providing assistance with proteomics data collection and analysis, respectively. We would like to acknowledge PhosphoSitePlus® (www.phosphosite.org) and the Scripps NMR and MS core facilities.
Footnotes
Contributions
S.M.H., K.M.B and B.F.C. conceived of the project, designed experiments and analyzed data. S.M.H and K.M.B performed mass spectrometry experiments and data analysis. S.M.H synthesized compounds, cloned, expressed and purified proteins and conducted inhibition studies. K.M.B, M.R.L. and B.E.C wrote software. S.M.H and K.M.B. compiled and analyzed mass spectrometry data. S.F. wrote software and conducted computational analyses. M.R.L conducted immunoprecipitation studies and data analysis. S.M.H, K.M.B. and B.F.C wrote the manuscript. S.M.H. and K.M.B. contributed equally to this work.
Competing financial interests
The authors declare competing financial interests. Dr. Cravatt is a founder and advisor to Vividion Therapeutics, a biotechnology company interested in using chemical proteomic methods to develop small-molecule drugs to treat human disease.
References
- 1.Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3:739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makley LN, Gestwicki JE. Expanding the Number of “Druggable” Targets: Non-Enzymes and Protein-Protein Interactions. Chem Biol Drug Des. 2013;81:22–32. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 4.Bachovchin DA, Cravatt BF. The Pharmacological Landscape and Therapeutic Potential of Serine Hydrolases. Nat Rev Drug Discov. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato D, et al. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 6.Pan Z, et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. ChemMedChem. 2007;2:58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 7.Li D, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neggers JE, et al. Identifying drug-target selectivity of small-molecule CRM1/XPO1 inhibitors by CRISPR/Cas9 genome editing. Chem Biol. 2015;22:107–116. doi: 10.1016/j.chembiol.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Backus KM, et al. Proteome-wide covalent ligand discovery in native biological systems. Nature. 2016;534:570–574. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patricelli MP, et al. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 12.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 13.Akcay G, et al. Inhibition of Mcl-1 through covalent modification of a noncatalytic lysine side chain. Nat Chem Biol. 2016;12:931–936. doi: 10.1038/nchembio.2174. [DOI] [PubMed] [Google Scholar]
- 14.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 15.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, Dent SY. Histone modifying enzymes and cancer: going beyond histones. J Cell Biochem. 2005;96:1137–1148. doi: 10.1002/jcb.20615. [DOI] [PubMed] [Google Scholar]
- 17.Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat Struct Mol Biol. 2014;21:308–316. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- 18.Wymann MP, et al. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi S, Connelly S, Reixach N, Wilson IA, Kelly JW. Chemoselective small molecules that covalently modify one lysine in a non-enzyme protein in plasma. Nat Chem Biol. 2010;6:133–139. doi: 10.1038/nchembio.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crawford LA, Weerapana E. A tyrosine-reactive irreversible inhibitor for glutathione S-transferase Pi (GSTP1) Mol Biosyst. 2016;12:1768–1771. doi: 10.1039/c6mb00250a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon DA, et al. Investigating the proteome reactivity and selectivity of aryl halides. J Am Chem Soc. 2014;136:3330–3333. doi: 10.1021/ja4116204. [DOI] [PubMed] [Google Scholar]
- 22.Hunter M, Ludwig M. The reaction of imidoesters with proteins and related small molecules. J Am Chem Soc. 1962;84:3491–3504. [Google Scholar]
- 23.Bandyopadhyay A, Gao J. Iminoboronate-Based Peptide Cyclization That Responds to pH, Oxidation, and Small Molecule Modulators. J Am Chem Soc. 2016;138:2098–2101. doi: 10.1021/jacs.5b12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, et al. Selective depletion of mutant p53 by cancer chemopreventive isothiocyanates and their structure-activity relationships. J Med Chem. 2011;54:809–816. doi: 10.1021/jm101199t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Kensler TW, Cho CG, Posner GH, Talalay P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc Natl Acad Sci U S A. 1994;91:3147–3150. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musiol HJ, Moroder L. N,N′-di-tert-butoxycarbonyl-1H-benzotriazole-1-carboxamidine derivatives are highly reactive guanidinylating reagents. Org Lett. 2001;3:3859–3861. doi: 10.1021/ol010191q. [DOI] [PubMed] [Google Scholar]
- 27.Kapp TG, Fottner M, Maltsev OV, Kessler H. Small Cause, Great Impact: Modification of the Guanidine Group in the RGD Motif Controls Integrin Subtype Selectivity. Angew Chem Int Ed Engl. 2016;55:1540–1543. doi: 10.1002/anie.201508713. [DOI] [PubMed] [Google Scholar]
- 28.Grimster NP, et al. Aromatic sulfonyl fluorides covalently kinetically stabilize transthyretin to prevent amyloidogenesis while affording a fluorescent conjugate. J Am Chem Soc. 2013;135:5656–5668. doi: 10.1021/ja311729d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Q, et al. Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes. J Am Chem Soc. 2017;139:680–685. doi: 10.1021/jacs.6b08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asano S, Patterson JT, Gaj T, Barbas CF., 3rd Site-selective labeling of a lysine residue in human serum albumin. Angew Chem Int Ed Engl. 2014;53:11783–11786. doi: 10.1002/anie.201405924. [DOI] [PubMed] [Google Scholar]
- 31.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tekaia F, Yeramian E, Dujon B. Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: a global picture with correspondence analysis. Gene. 2002;297:51–60. doi: 10.1016/s0378-1119(02)00871-5. [DOI] [PubMed] [Google Scholar]
- 33.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Lockett MR, Phillips MF, Jarecki JL, Peelen D, Smith LM. A Tetrafluorophenyl Activated Ester Self-Assembled Monolayer for the Immobilization of Amine-Modified Oligonucleotides. Langmuir. 2008;24:69–75. doi: 10.1021/la702493u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravindranath PA, Sanner MF. AutoSite: an automated approach for pseudo-ligands prediction-from ligand-binding sites identification to predicting key ligand atoms. Bioinformatics. 2016;32:3142–3149. doi: 10.1093/bioinformatics/btw367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marriott AS, et al. NUDT2 Disruption Elevates Diadenosine Tetraphosphate (Ap4A) and Down-Regulates Immune Response and Cancer Promotion Genes. PLoS One. 2016;11:e0154674. doi: 10.1371/journal.pone.0154674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge H, Chen X, Yang W, Niu L, Teng M. Crystal structure of wild-type and mutant human Ap4A hydrolase. Biochem Biophys Res Co. 2013;432:16–21. doi: 10.1016/j.bbrc.2013.01.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang YP, et al. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J. 2014;33:1304–1320. doi: 10.1002/embj.201387224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inloes JM, et al. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc Natl Acad Sci U S A. 2014;111:14924–14929. doi: 10.1073/pnas.1413706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson-Grady JT, Haas W, Gygi SP. Quantitative comparison of the fasted and re-fed mouse liver phosphoproteomes using lower pH reductive dimethylation. Methods. 2013;61:277–286. doi: 10.1016/j.ymeth.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 42.Musayev FN, Di Salvo ML, Ko TP, Schirch V, Safo MK. Structure and properties of recombinant human pyridoxine 5′-phosphate oxidase. Protein Sci. 2003;12:1455–1463. doi: 10.1110/ps.0356203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang JH, et al. Genomic organization, tissue distribution and deletion mutation of human pyridoxine 5′-phosphate oxidase. Eur J Biochem. 2004;271:2452–2461. doi: 10.1111/j.1432-1033.2004.04175.x. [DOI] [PubMed] [Google Scholar]
- 44.Hacker SM, Buntz A, Zumbusch A, Marx A. Direct Monitoring of Nucleotide Turnover in Human Cell Extracts and Cells by Fluorogenic ATP Analogs. ACS Chem Biol. 2015;10:2544–2552. doi: 10.1021/acschembio.5b00459. [DOI] [PubMed] [Google Scholar]
- 45.Schoneberg T, Kloos M, Bruser A, Kirchberger J, Strater N. Structure and allosteric regulation of eukaryotic 6-phosphofructokinases. Biol Chem. 2013;394:977–993. doi: 10.1515/hsz-2013-0130. [DOI] [PubMed] [Google Scholar]
- 46.Yi W, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochim Biophys Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nascimento EM, et al. The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis. Nat Cell Biol. 2011;13:1395–1405. doi: 10.1038/ncb2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 50.Kwon YJ, et al. Targeted interference of SIN3A-TGIF1 function by SID decoy treatment inhibits Wnt signaling and invasion in triple negative breast cancer cells. Oncotarget. 2016 doi: 10.18632/oncotarget.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melhuish TA, Wotton D. The Tgif2 gene contains a retained intron within the coding sequence. BMC Mol Biol. 2006;7:2. doi: 10.1186/1471-2199-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hurlin PJ, Queva C, Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 53.Rao G, et al. Mouse Sin3A interacts with and can functionally substitute for the amino-terminal repression of the Myc antagonist Mxi1. Oncogene. 1996;12:1165–1172. [PubMed] [Google Scholar]
- 54.Andre I, Linse S, Mulder FA. Residue-specific pKa determination of lysine and arginine side chains by indirect 15N and 13C NMR spectroscopy: application to apo calmodulin. J Am Chem Soc. 2007;129:15805–15813. doi: 10.1021/ja0721824. [DOI] [PubMed] [Google Scholar]
- 55.Karlstrom A, et al. Using antibody catalysis to study the outcome of multiple evolutionary trials of a chemical task. Proc Natl Acad Sci U S A. 2000;97:3878–3883. doi: 10.1073/pnas.97.8.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol. 2005;23:1303–1307. doi: 10.1038/nbt1149. [DOI] [PubMed] [Google Scholar]
- 57.Choi S, Ong DS, Kelly JW. A stilbene that binds selectively to transthyretin in cells and remains dark until it undergoes a chemoselective reaction to create a bright blue fluorescent conjugate. J Am Chem Soc. 2010;132:16043–16051. doi: 10.1021/ja104999v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roberts AM, et al. Chemoproteomic Screening of Covalent Ligands Reveals UBA5 As a Novel Pancreatic Cancer Target. ACS Chem Biol. 2017;12:899–904. doi: 10.1021/acschembio.7b00020. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Y, et al. Chemoproteomic Strategy to Quantitatively Monitor Transnitrosation Uncovers Functionally Relevant S-Nitrosation Sites on Cathepsin D and HADH2. Cell Chem Biol. 2016;23:727–737. doi: 10.1016/j.chembiol.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Meth. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward CC, Kleinman JI, Nomura DK. NHS-Esters as Versatile Reactivity-Based Probes for Mapping Proteome-Wide Ligandable Hotspots. ACS Chem Biol. 2017 doi: 10.1021/acschembio.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blewett MM, et al. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci Signal. 2016;9:rs10. doi: 10.1126/scisignal.aaf7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ford B, Bateman LA, Gutierrez-Palominos L, Park R, Nomura DK. Mapping Proteome-wide Targets of Glyphosate in Mice. Cell Chem Biol. 2017;24:133–140. doi: 10.1016/j.chembiol.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 64.Sahu SC, et al. Conserved themes in target recognition by the PAH1 and PAH2 domains of the Sin3 transcriptional corepressor. J Mol Biol. 2008;375:1444–1456. doi: 10.1016/j.jmb.2007.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings discussed here are available within the paper, its Supplementary Information file and Supplementary Tables 1 and 2, as well as from the corresponding authors upon request.