Abstract
Spectrally resolved fluorescence lifetime imaging1–3 and spatial multiplexing1,4,5 have offered information content and collection-efficiency boosts in microscopy, but efficient implementations for macroscopic applications are still lacking. An imaging platform based on time-resolved structured light and hyperspectral single-pixel detection has been developed to perform quantitative macroscopic fluorescence lifetime imaging (MFLI) over a large field of view (FOV) and multiple spectral bands simultaneously. The system makes use of three digital micromirror device (DMD)-based spatial light modulators (SLMs) to generate spatial optical bases and reconstruct N by N images over 16 spectral channels with a time-resolved capability (~40 ps temporal resolution) using fewer than N2 optical measurements. We demonstrate the potential of this new imaging platform by quantitatively imaging near-infrared (NIR) Förster resonance energy transfer (FRET) both in vitro and in vivo. The technique is well suited for quantitative hyperspectral lifetime imaging with a high sensitivity and paves the way for many important biomedical applications.
Optical imaging techniques are becoming central to the molecular investigation of samples at the macroscopic scale, with spectrally resolved imaging used to facilitate medical diagnosis6,7 and operations, such as in guided surgery8,9, because of the spectrum-dependent nature of optical contrasts. Especially, MFLI offers the advantage of high sensitivity as well as the unmixing of spectra and of fluorescence lifetime-based biomarkers. As an intrinsic characteristic of a fluorophore and its state, fluorescence lifetime can be used to investigate the molecular environment (for example, pH, ion concentration, pO2, temperature)10,11 with robustness because lifetime measurements are independent of intensity, which can be significantly altered by tissue heterogeneities and depth location. Spectrally resolved microscopic lifetime-imaging systems have been reported based on a frequency-domain Hadamard transform multiplexing1, raster scanning2 and Fourier multiplexing3. However, hyperspectral MFLI in preclinical and clinical settings is still challenging because of the complexity, relatively high instrumentation cost and long acquisition times for joint temporal and spectral measurements.
In parallel, in the past decade interests in transform-based single-pixel imaging systems have emerged in numerous applications, which include biological-information investigation4,5, profilometry12–14, terahertz imaging15,16 and remote sensing17. These systems allow for data-dimension reduction and data compression, without compromising the information content for sparse signals, based on orthogonal-basis sampling under the assumption of a band-limited signal or the well-established compressive-sensing theory18. Single-pixel imaging is well suited for situations in which sensitive two-dimensional (2D) array detectors are either technically or financially unavailable at the target wavelength bands19. It can also be readily coupled with light-dispersion devices for multispectral or hyperspectral data acquisition4,20,21. Therefore, wide-field single-pixel imaging is a promising technique for optical molecular imaging applications in which sensitive and multiplexed wide-field fluorescence detection is required.
This letter demonstrates experimentally a time-resolved single-pixel MFLI system that achieves simultaneously spatially and spectrally resolved wide-field intensity- and lifetime-based imaging. This approach, unlike previous time-resolved MFLI systems22,23, does not require scanning or the sequential acquisition of spectral data and is highly scalable in terms of sample size, spectral band and temporal resolution. Furthermore, the implementation benefits from highly sensitive detectors as well as efficient photon-counting techniques. This is critical for photon-starved conditions such as in vivo applications. Herein, the potential of the proposed approach is demonstrated by performing wide-field FRET–MFLI imaging both in vitro and in vivo, with application in drug delivery assessment.
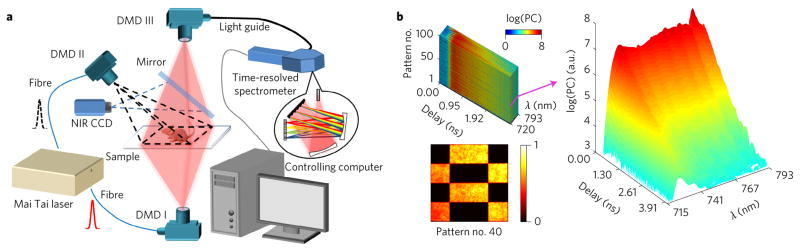
The implementation diagram of the proposed approach is shown in Fig. 1a (Supplementary Information gives a detailed description). An ultrafast Ti:sapphire pulsed laser is fibre-coupled to a DMD-based SLM to generate wide-field structured light on the sample (both reflectance and transmission illumination modes are available; the appropriate mode is selected for each sample). A second SLM collects the transmitted or reflected light over a matched FOV. Modulating patterns are generated on either the illumination or the detection side. The spatially integrated light after the second SLM is coupled to a time-resolved Czerny–Turner spectrophotometer based on a 16-channel photomultiplier tube via an aperture-converting fibre light guide.
Figure 1. System diagram and typical hyperspectral TPSF example.
a, Diagram of the time-resolved hyperspectral single-pixel imager with transmission mode (formed by DMD I and III) and reflectance mode (formed by DMD II and III). The NIR CCD and mirror are only used during calibration. The laser-power control and fibre-coupling modules are not shown. b, Visualizations of an acquired data cube (top left, measurements under the first 100 Hadamard patterns acquired by the system) and hyperspectral TPSFs (right) under the illumination of pattern no. 40 (bottom left). PC, photon count; a.u., arbitrary units.
To perform single-pixel imaging, we employ the classical Hadamard spatial base by reshaping and scaling rows of the Hadamard matrix Hn×n (n = 1,024) into square matrices (i = 1, …, n) and optically projecting complementary binary pattern pairs and (i = 1, …, n) to maintain orthogonality and suppress noise24. A NIR CCD (charge-coupled device) camera is employed to record the spatial distribution of the patterns for calibration. Spectrally resolved temporal point spread functions (TPSFs) (Fig. 1b) for each Hadamard pattern are acquired, and the data acquisition using m Hadamard patterns at emission wavelength λ and time point t can be expressed as:
| (1) |
where Hm×n, and are the calibrated Hadamard matrix, target fluorophore distribution and measurement, respectively. To retrieve , the inverse problem is solved using the least-squares method or regularized minimization methods. Finally, quantitative lifetime values at each pixel and wavelength are retrieved via biexponential fitting of the TPSFs that are pre-processed with the Anscombe transform25 and Gaussian filter.
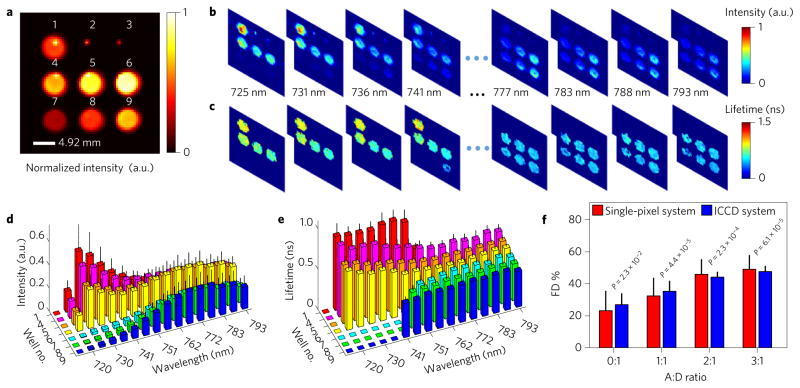
To demonstrate the potential of our system for lifetime imaging, we first performed an in vitro FRET–MFLI study. The FOV (3.5 × 3.5 cm2) was set to cover nine wells of a 96-well microplate, with Hadamard patterns imposed on DMD I in transmission geometry. Alexa Fluor 700 (mouse antibody-conjugated donor) and Alexa Fluor 750 (goat anti-mouse antibody-conjugated acceptor) were diluted with phosphate buffer solution (PBS) to create three different conditions: no fluorescence, mono-lifetime fluorescence and multi-lifetime fluorescence emission over the nine wells (Fig. 2a). The microplate was excited at 695 nm using 1,024 Hadamard patterns with an optical power density of ~5.49 mW cm−2 and the emission signals were detected within the 715–793 nm range using a long-pass filter. To establish the quantitative accuracy of the proposed approach, the same sample was also imaged using a well-established gated intensified CCD (ICCD)-based MFLI system23 at the same excitation wavelength, but detected at the donor emission peak band (~720 nm).
Figure 2. Results of in vitro study.
a, Well-plate sample intensity captured using the NIR CCD (rescaled to a 64 × 64 resolution). The layout of the plate in terms of the relative acceptor:donor (A:D) ratio from well no. 1 to no. 9 is as follows: 0:1, 0:0, 0:0, 1:1, 2:1, 3:1, 1:0, 2:0 and 3:0. Wells 2 and 3 are used as the control with PBS only. b,c, Reconstructed spectrally resolved intensity and mean-lifetime spatial maps via the single-pixel method. d,e, Spectrally resolved intensity and mean-lifetime distributions for each well. f, FD% estimated via a bi-exponential decay fitting of wells 1 and 4–6 at the 725 nm channel and comparison with results obtained using an ICCD-based imaging system. Equivalence tests show that their mean FD% values are within a ±6% absolute margin at the 0.05 significance level (P = 0.023). Error bars represent s.d.
Time- and spectrum-resolved data were processed to yield intensity (Fig. 2b) and lifetime (Fig. 2c) distributions over the entire wavelength range. The means and standard deviations of the intensity and lifetime over each well are reported in Fig. 2d,e. As shown, the spectrally resolved intensities demonstrate the expected trend over each well. First, in the cases of donor or acceptor only, the spectral shapes depict the fluorescence spectra of donor (well 1) and acceptor (wells 7–9), respectively, and the average relative intensities between wells 7–9 are within 10% of the expected values from CCD measurements. Additionally, in the cases of wells 1 and 4–6, the spectral intensity trend depicts the increase in energy transfer caused by the increase of acceptor concentration. Similarly, the mean-lifetime estimates follow the expected trends (spectrally and by well). In the cases of wells with donor or acceptor only, the estimated lifetimes are τAF700 = 0.90 ± 0.07 ns and τAF750 = 0.46 ± 0.07 ns at their emission peaks. For FRET samples (wells 4–6), the donor mean lifetime is reduced with increased quenching, whereas the acceptor mean lifetime is affected minimally. Moreover, a bi-exponential decay model to retrieve the Förster resonance energy transferring (FRETing) donor fraction (FD%) (associated FRETing donors with a short lifetime component) was applied to the donor emission peak channel in both ICCD and single-pixel measurements. Equivalence tests showed that the means of the FD% at each condition are within a ±6% absolute margin at a 0.05 significance level (P = 0.023) (Fig. 2f ). Overall, this in vitro study demonstrates the potential of our approach to retrieve dense 4D data cubes (x, y, t and λ) for quantitative intensity and lifetime imaging.
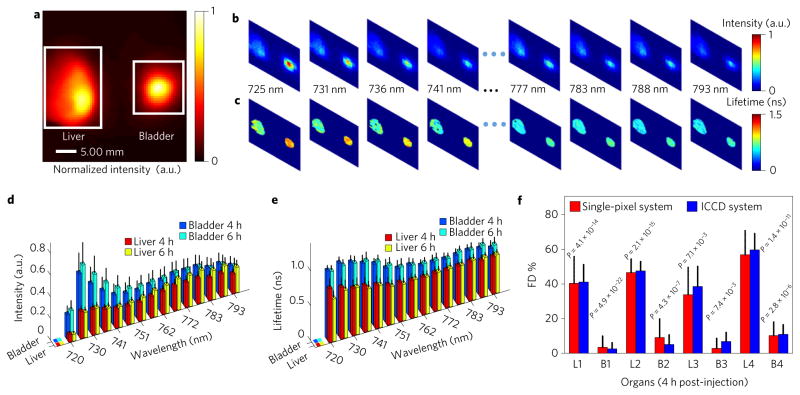
Furthermore, an in vivo FRET–MFLI mouse study was carried out to demonstrate the potential of our system for quantitative preclinical imaging. Four distinct mice were injected with transferrin-conjugated Alexa Fluor 700 (40 μg) and Alexa Fluor 750 (80 μg) via the tail vein (120 μl total volume) following previously published protocols26. Measurements were acquired four hours and six hours post-injection under the same excitation and detection spectral conditions as in the in vitro study. The subjects were illuminated through reflectance geometry (DMD II) with adaptive greyscale patterns to compensate for the high dynamic range in preclinical studies23. In this demonstration, we focused on two major organs of fluorescence accumulation, the liver and the urinary bladder (Fig. 3a). The whole-body signals were detected with Hadamard patterns applied to the detection channel (DMD III). The subjects were also imaged using the ICCD-based MFLI system under the same conditions (detection at the donor emission peak) for validation.
Figure 3. Results of the in vivo study.
a, Fluorescence signals captured using the NIR CCD (FOV, 40 mm× 40 mm) at 4 h post-injection of NIR FRET-labelled transferrin. b,c, Examples of the spectrally resolved single-pixel spatial reconstruction of intensity (b) and mean lifetime (c) at 4 h post-injection for subject no. 1. d,e, Means and standard deviations of intensity (d) and lifetime (e) over the liver and the urinary bladder as retrieved from single-pixel processing (4 and 6 h post-injection of subject no. 1). f, FD% of the two organs at the donor emission peak channel for four subjects and comparison with the results obtained from the ICCD-based system at 4 h post-injection. Here ‘Lm’ and ‘Bm’ (m= 1, 2, 3 and 4) represent the liver and the urinary bladder, respectively, for subject no. m. Equivalence tests show that their mean FD% values are within a ±6% absolute margin at the 0.05 significance level (P = 0.007). Error bars represent s.d.
As in the in vitro experiments, the single-pixel data sets were processed to yield spectrally resolved spatial maps of both fluorescence intensity and mean lifetime (Fig. 3b,c gives the spectrally resolved intensity and mean-lifetime maps at four hours for subject no. 1). These data are summarized further in Fig. 3d,e for each organ. Donor and acceptor mean-lifetime reductions were observed in the liver at both time points, in contrast to the lack of lifetime reduction in the urinary bladder. Additionally, the FD% values of the two organs were computed and validated for the four subjects (Fig. 3f), and no significant difference (±6% absolute margin) was observed between the single-pixel and ICCD quantification (P = 0.007) at the 0.05 significance level. A strong contrast between the results in the liver and the urinary bladder was observed. The liver is known to have a high density of transferrin receptor27 and thus demonstrates a strong FRET signal. The urinary bladder, as a pathway for excretion, is not expected to be associated with such receptors, and the FRET signal was minimal despite a strong fluorescence intensity. These in vivo results demonstrate that the proposed method enables a quantitative investigation of in vivo fluorescence signals over the whole body of the animal, even in challenging conditions such as FRET imaging (quenched donor).
In conclusion, we report the instrumentation design of a wide-field time-resolved hyperspectral single-pixel system to perform spectrally resolved MFLI. Its robustness was demonstrated by establishing the accuracy of FRET quantification in vitro and in vivo. The quantification of NIR FRET faces well-known challenges because of its bi-exponential behaviour with very short lifetimes. Hence, we expect that this implementation is well suited for a wide array of optical molecular applications that leverage lifetime sensing, such as multiplexed in vivo receptor engagement and/or high dimensional optical tomography based on structured light strategies28. The in vivo imaging protocols reported were conducted in about 14 minutes, and acquisition times from over ten minutes to hours are needed for the state-of-the-art commercial MFLI system, eXplore Optix, to scan a similar area of a mouse for only one wavelength29–31. However, the data acquisition time of our method can be considerably reduced if the spatial bases are optimized, less-dense temporal data sets are acquired, the fluorescence decay curve is temporally gated for an enhanced SNR or the spectral band integrated. Additionally, this approach can be easily scaled in space by adapting the optics, in time by changing the time-correlated single-photon-counting (TCSPC) configuration and in spectrum by adjusting the dispersive elements.
Methods
Pattern-implementation method
The Hadamard patterns were arranged in ascending order of their absolute difference summations of all the pixels along two orthogonal directions (sorted according to their spatial frequency content). Given that Hadamard patterns are formed by ‘+1’ and ‘−1’ only, the projection of matrices (i = 1, …, n) requires data acquisition under complementary binary pattern pairs, and (i = 1,…, n). To account for the deviations of experimental patterns from the computer-fed input because of optical distortion, the pattern calibration was completed using a NIR camera (UI-5240CP-NIR-GL (IDS)) and a silver plane mirror M1 (ME2S-P01 (Thorlabs)) before data acquisition. Pattern generation and data acquisition were achieved via LabVIEW-based (National Instruments) control software and data reconstruction was performed using MATLAB R2015a (MathWorks).
In vitro and in vivo studies
In the in vitro well-plate study, Alexa Fluor 700 (mouse IgG1 conjugated, MG129 (Thermo Fisher Scientific)) and Alexa Fluor 750 (goat anti-mouse IgG secondary antibody conjugated, A-21006 (Thermo Fisher Scientific)) were used and diluted with PBS (Sigma-Aldrich). The base concentration was set to 20 μg ml−1 (that is, A:D = 2:1 means 40 μg ml−1 of acceptor molecules mixed with 20 μg ml−1 of donor molecules). Each well was filled with 250 μl total volume. In the in vivo studies, four six-week-old athymic nude mice (Charles River Laboratories) were used. The subjects were anaesthetized with isoflurane and taped to the imaging bed (to prevent motions that could modify their posture and compromise the image quality). The illumination power density was ~2.8 mW cm−2 and the total imaging time for each subject at one time point was ~14 min for 800 measurements. The body temperature of the mouse was maintained by a warm-air blower during the image acquisition. The respiratory rate and pain reflex were monitored for the depth of anaesthesia. As is customary in macroscopic preclinical optical imaging, small physiological motions, such as breathing, were not taken into account. The animals were monitored during the whole imaging sessions to ensure they were not under stressful conditions. The use of animals in this study was approved by the Institutional Animal Care and Use Committee at Rensselaer Polytechnic Institute. To perform a quantitative comparison between the proposed method and a previously published ICCD-based method for validation, samples were imaged at similar time points and temporal settings (ICCD-based system, 40 ps time-gate step, 300 ps time-gate width, 150 gates in total, 20 ms exposure time for in vitro studies and 400 ms exposure time for in vivo studies; single-pixel system, 8.34 ns detection temporal range, 32.6 ps channel width under an eight-bit analogue-to-digital converter resolution and 1 s integration time per measurement for both studies). The ICCD-based system was built on two DMDs (transmission geometry, D4110 plus S2+ VIS optics module, f/2.6~2.9 (Digital Light Innovations); reflectance geometry, DLPDLCR3010EVM (Texas Instruments)) and a PicoStar HR 12 ICCD camera (LaVision) with a f = 50 mm objective (AF Nikkor 50 mm f/1.8D (Nikon)). Two filters (FF01-720/13-25 and FF01-715/LP-25 (Semrock)) were applied in ICCD-based studies. The data-acquisition times for the in vitro study were 1,024 patterns × 1 s/pattern = 1,024 s in the single-pixel MFLI system for 16 wavelength channels, and 150 time gates × 20 ms/time gate = 3 s for one wavelength channel using the ICCD-based system. As for the in vivo study, the data acquisition of one subject at one time point took 800 patterns × 1 s/pattern = 800 s for the single-pixel MFLI system for 16 wavelength channels, and 150 time gates × 400 ms/time gate = 60 s for one wavelength channel using the ICCD-based system.
Equivalence test
To compare the results obtained using the ICCD-based system and the single-pixel system for the same subject and establish the quantitative accuracy of the proposed method, equivalence tests were performed on the acquired pixel groups of the same well or organ area to quantify the differences in mean FD% values from a statistical point of view. The null (H0) and alternative hypothesis (H1) of the equivalence test are:
where and are the mean FD% obtained for the same subject (well/organ) using the single-pixel system and ICCD-based system, respectively, and εL and εU are the lower and upper margins, respectively. The significance level α was set as 0.05 and margins (εL, εU) were set as (−6%, 6%) for both in vitro and in vivo studies. Reconstructed images from the single-pixel imaging system were matched with the corresponding ICCD images through coordinate shift, rotation and stretch to determine the regions of interest for the equivalence tests. The equivalence test was achieved with two one-sided hypothesis tests. Given that the normal distribution assumption does not hold for the FD% of certain pixel groups, the Wilcoxon rank sum test was applied in the equivalence tests.
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
This work is supported by the National Science Foundation through Career Award CBET 1149407, and National Institute of Health Grants R01 EB19443 and R01 CA207725. The authors acknowledge the support from M. Barroso and A. Rudkouskaya for the sample preparation in animal studies, and the insightful discussions with W. Cong and G. Wang.
Footnotes
Author contributions
X.I. conceived the original idea. Q.P. contributed to the platform instrumentation, control software design and experimental validation. R.Y. contributed to the pattern implementation, initial simulations and image reconstruction. N.S. contributed to the animal study protocol and experiments. All the authors contributed to the analysis of the experimental results and to writing the manuscript.
Supplementary information is available in the online version of the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Hanley QS, Arndt-Jovin DJ, Jovin TM. Spectrally resolved fluorescence lifetime imaging microscopy. Appl Spectrosc. 2002;56:155–166. [Google Scholar]
- 2.Bird DK, Eliceiri KW, Fan CH, White JG. Simultaneous two-photon spectral and lifetime fluorescence microscopy. Appl Opt. 2004;43:5173–5182. doi: 10.1364/ao.43.005173. [DOI] [PubMed] [Google Scholar]
- 3.Zhao M, Li Y, Peng L. Parallel excitation-emission multiplexed fluorescence lifetime confocal microscopy for live cell imaging. Opt Express. 2014;22:10221–10232. doi: 10.1364/OE.22.010221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Studer V, et al. Compressive fluorescence microscopy for biological and hyperspectral imaging. Proc Natl Acad Sci USA. 2012;109:E1679–E1687. doi: 10.1073/pnas.1119511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizuno T, Iwata T. Hadamard-transform fluorescence-lifetime imaging. Opt Express. 2016;24:8202–8213. doi: 10.1364/OE.24.008202. [DOI] [PubMed] [Google Scholar]
- 6.Roblyer D, et al. Multispectral optical imaging device for in vivo detection of oral neoplasia. J Biomed Opt. 2008;13:024019. doi: 10.1117/1.2904658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu G, Fei B. Medical hyperspectral imaging: a review. J Biomed Opt. 2014;19:010901. doi: 10.1117/1.JBO.19.1.010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panasyuk SV, et al. Medical hyperspectral imaging to facilitate residual tumor identification during surgery. Cancer Biol Ther. 2007;6:439–446. doi: 10.4161/cbt.6.3.4018. [DOI] [PubMed] [Google Scholar]
- 9.Akbari H, Kosugi Y, Kojima K, Tanaka N. Detection and analysis of the intestinal ischemia using visible and invisible hyperspectral imaging. IEEE Trans Biomed Eng. 2010;57:2011–2017. doi: 10.1109/TBME.2010.2049110. [DOI] [PubMed] [Google Scholar]
- 10.Marcu L. Fluorescence lifetime techniques in medical applications. Ann Biomed Eng. 2012;40:304–331. doi: 10.1007/s10439-011-0495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suhling K, et al. Fluorescence lifetime imaging (FLIM): basic concepts and some recent developments. Med Photon. 2015;27:3–40. [Google Scholar]
- 12.Sun B, et al. 3D computational imaging with single-pixel detectors. Science. 2013;340:844–847. doi: 10.1126/science.1234454. [DOI] [PubMed] [Google Scholar]
- 13.Pham QD, Hayasaki Y. Optical frequency comb profilometry using a single-pixel camera composed of digital micromirror devices. Appl Opt. 2015;54:A39–A44. doi: 10.1364/AO.54.000A39. [DOI] [PubMed] [Google Scholar]
- 14.Yu WK, Yao XR, Liu XF, Li LZ, Zhai GJ. Three-dimensional single-pixel compressive reflectivity imaging based on complementary modulation. Appl Opt. 2015;54:363–367. [Google Scholar]
- 15.Chan WL, et al. A single-pixel terahertz imaging system based on compressed sensing. Appl Phys Lett. 2008;93:121105. [Google Scholar]
- 16.Withayachumnankul W, Abbott D. Terahertz imaging: compressing onto a single pixel. Nat Photon. 2014;8:593–594. [Google Scholar]
- 17.Ma J. Single-pixel remote sensing. IEEE Geosci Remote Sens Lett. 2009;6:199–203. [Google Scholar]
- 18.Donoho DL. Compressed sensing. IEEE Trans Inf Theory. 2006;52:1289–1306. [Google Scholar]
- 19.Duarte MF, et al. Single-pixel imaging via compressive sampling. IEEE Signal Process Mag. 2008;25:83–91. [Google Scholar]
- 20.Welsh SS, et al. Fast full-color computational imaging with single-pixel detectors. Opt Express. 2013;21:23068–23074. doi: 10.1364/OE.21.023068. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg J, Krishnamurthy K, Brady D. Compressive single-pixel snapshot X-ray diffraction imaging. Opt Lett. 2014;39:111–114. doi: 10.1364/OL.39.000111. [DOI] [PubMed] [Google Scholar]
- 22.Ma G, Gallant P, McIntosh L. Sensitivity characterization of a time-domain fluorescence imager: eXplore Optix. Appl Opt. 2007;46:1650–1657. doi: 10.1364/ao.46.001650. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L, et al. Spatial light modulator based active wide-field illumination for ex vivo and in vivo quantitative NIR FRET imaging. Biomed Opt Express. 2014;5:944–960. doi: 10.1364/BOE.5.000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Streeter L, Burling-Claridge GR, Cree MJ, Künnemeyer R. Optical full Hadamard matrix multiplexing and noise effects. Appl Opt. 2009;48:2078–2085. doi: 10.1364/ao.48.002078. [DOI] [PubMed] [Google Scholar]
- 25.Mäkitalo M, Foi A. Optimal inversion of the Anscombe transformation in low-count Poisson image denoising. IEEE Trans Image Process. 2011;20:99–109. doi: 10.1109/TIP.2010.2056693. [DOI] [PubMed] [Google Scholar]
- 26.Abe K, Zhao L, Periasamy A, Intes X, Barroso M. Non-invasive in vivo imaging of near infrared-labeled transferrin in breast cancer cells and tumors using fluorescence lifetime FRET. PLoS ONE. 2013;8:e80269. doi: 10.1371/journal.pone.0080269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian ZM, Li H, Sun H, Ho K. Targeted drug delivery via the transferrin receptor-mediated endocytosis pathway. Pharmacol Rev. 2002;54:561–587. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 28.Pian Q, Yao R, Zhao L, Intes X. Hyperspectral time-resolved wide-field fluorescence molecular tomography based on structured light and single-pixel detection. Opt Lett. 2015;40:431–434. doi: 10.1364/OL.40.000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wessels JT, et al. In vivo imaging in experimental preclinical tumor research—a review. Cytom A. 2007;71A:542–549. doi: 10.1002/cyto.a.20419. [DOI] [PubMed] [Google Scholar]
- 30.Christian NA, et al. In vivo dendritic cell tracking using fluorescence lifetime imaging and near-infrared-emissive polymersomes. Mol Imaging Biol. 2009;11:167–177. doi: 10.1007/s11307-008-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall DJ, et al. Dynamic optical imaging of metabolic and NADPH oxidasederived superoxide in live mouse brain using fluorescence lifetime unmixing. J Cereb Blood Flow Metab. 2012;32:23–32. doi: 10.1038/jcbfm.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author on reasonable request.