Abstract
Epigenetics processes may play a vital role in the biological embedding of early environmental adversity and the development of psychopathology. Accumulating evidence suggests that maltreatment is linked to methylation of the glucocorticoid receptor gene, nuclear receptor subfamily 3, group C, member 1 (NR3C1), which is a key regulator of the hypothalamic–pituitary–adrenal axis. However, prior work has been exclusively cross-sectional, greatly constraining our understanding of stress-related epigenetic processes over time. In the current study, we examined the effect of maltreatment and other adversity on change in NR3C1 methylation among at-risk preschoolers to begin to characterize within-child epigenetic changes during this sensitive developmental period. Participants were 260 preschoolers (3–5 years-old, 53.8% female), including 51.5% with moderate-severe maltreatment in the past six months. Child protection records, semi-structured interviews, and parent reports were used to assess child stress exposure. Methylation of exons 1D and 1F of NR3C1 via saliva DNA were measured at two time points approximately 6 months apart. Results indicate that maltreated children evidence higher baseline levels of NR3C1 methylation, significant decreases in methylation over time, and then at follow-up, lower levels of methylation, relative to non-maltreated preschoolers. Findings from the current study highlight the complex nature of stress related epigenetic processes during early development.
Keywords: epigenetics, NR3C1, maltreatment, early adversity, longitudinal
Each year in the United States four million reports of abuse involving over seven million children are made to child protective services (US Department of Health and Human Services, Administration of Children, Youth and Families, Children’s Bureau, 2017). Exposure to early environmental adversity (e.g., abuse, trauma, or contextual stressors) confers substantial risk for the development of psychopathology and lifelong risks of chronic disorders of health and well-being (see Cicchetti & Toth, 2016; Cohen, Janicki-Deverts, & Miller, 2007; Norman et al., 2012; Shonkoff, Boyce & McEwen, 2009, for reviews). Despite the prevalence of early environmental adversity and clear scientific consensus linking this adversity to detrimental health outcomes, the mechanisms underlying this connection are not well understood (McCrory & Viding, 2015).
One process by which maltreatment and other early adversities may influence the development of later psychopathology is through modification of the physiologic stress response system, and in particular, the hypothalamic–pituitary–adrenal (HPA) axis (Heim & Binder, 2012; Tyrka, Burgers, Philip, Price, & Carpenter, 2013). In response to stressful stimuli, the HPA axis is activated, and glucocorticoids are released, exerting cellular responses by binding at the intracellular glucocorticoid receptor (GR). GRs are distributed throughout the body and brain, where they regulate basal physiologic function and promote adaptive responses to acute stressors (de Kloet, Joels, & Holsboer, 2005; Kadmiel & Cidlowski, 2013). Further, activation of the GR through cortisol binding at the hypothalamus and pituitary engages a negative feedback loop that inhibits further release of cortisol and prevents damaging effects of extreme or chronic activation (Herman, McKlveen, Solomon, Carvalho-Netto, & Myers, 2012 ; Laryea, Muglia, Arnett, & Muglia, 2015). However, studies of rodents, children, and adults demonstrate that severe (e.g., child maltreatment) or chronic (e.g., contextual stressors associated with poverty) early life stress produces long-term alterations in glucocorticoid signaling that contribute to “wear and tear” on this system across the life span (McEwen et al., 2015; Tyrka, Ridout, & Parade, 2016). This stress-induced dysregulation of the HPA-axis has been implicated in the pathogenesis of stress-related psychiatric disorders (Barden, 2004; Braquehais, Picouto, Casas, & Sher, 2012; Doom & Gunnar, 2013).
Substantial research has been aimed at determining whether early-life stress is associated with epigenetic changes to the promoter region of the GR gene, nuclear receptor subfamily 3, group C, member 1 (NR3C1), that may alter GR expression and HPA axis homeostasis and responses to stress (see Turecki & Meaney, 2016; Tyrka, Ridout et al., 2016, for reviews). Epigenetic modifications to the genome allow for altered gene expression but do not change the DNA sequence and thus permit elaboration of the genome beyond what is determined by DNA base coding. The most highly studied and best characterized epigenetic modification, DNA methylation (DNAm), usually involves a direct covalent, chemical modification of a cytosine base lying sequentially adjacent to a guanine base (thus a CpG dinucleotide; G/GC). DNAm is associated with the silencing of gene transcription that appears to be mediated in one of two ways (Bird, 2002). First, wide swaths of DNA can be methylated, and the shear density of methylation precludes transcription factor binding to DNA sites, thus silencing gene expression. Second, when methylation levels are low, as in the case of NR3C1, small changes result in redistributing the transcriptional landscape, affect translational isoform production, and orchestrating the final proteomic landscape that results in gene silencing (see Leenen, Muller, & Turner, 2016; Meaney, 2010, for reviews).
Maltreatment and other early adversities have been linked in a majority of studies to increased DNAm within the NR3C1 gene, resulting in reduced gene expression and decreased hippocampal GR (e.g., Weaver et al., 2004; McGowan et al., 2009), and increased cortisol reactivity (e.g., Oberlander et al., 2008; Stroud et al., 2014). Further, methylation of NR3C1 in rodents has been shown to be responsive to many forms of early life stress including prenatal stress (Lillycrop et al., 2007; Szyf, 2013) and postnatal stress (Kundakovic, Lim, Gudsnuk, & Champagne, 2013; Witzmann, Turner, Meriaux, Meijer, & Muller, 2012). For example, low levels of maternal care in rodents causes greater methylation of the promoter region of the hippocampal GR gene, which interferes with binding of the transcription factor nerve growth factor inducible protein A (NGFI-A), resulting in reduced NR3C1 gene expression (Kosten & Nielsen, 2014; Weaver et al., 2004). These biological effects of early environmental stress have also been found in humans with studies finding prenatal (e.g., Oberlander et al., 2008; Ostlund, Conradt, Crowell, Tyrka, Marsit, & Lester, 2016) and early childhood (e.g., Romens, McDonald, Svaren, & Pollak, 2015; Tyrka et al., 2015) adversity linked with increased NR3C1 methylation evidenced in early development (e.g., Conradt et al., 2016; Parade et al., 2016) and long-lasting into adulthood (e.g., Tyrka, Price, Marsit, Walters, & Carpenter, 2012). In addition, these associations have been found across a number of human cell and tissue types including DNA from postmortem hippocampal tissue (e.g., Labonte et al., 2012; McGowan et al., 2009), placenta (e.g., Conradt et al., 2013; Kertes et al., 2016), umbilical cord blood (e.g., Hompes et al., 2013; Mulligan, D’Errico, Stees, & Hughes, 2012), peripheral blood (e.g., Romens et al., 2015; van der Knaap et al., 2014), and saliva (e.g., Melas et al., 2013; Parade et al., 2016). In sum, the association between early adversity and methylation of NR3C1 has been consistently observed across species and tissues (Vinkers et al., 2015) with replication of findings across multiple laboratories (Palma-Gudiel, Cordova-Palmera, Leza, & Fananas, 2015; Turecki & Meaney, 2016).
Emerging evidence also suggests that methylation of NR3C1 is associated with behavior and psychopathology (e.g., Conradt et al., 2013; Kosten, Huang, & Nielsen, 2014; Na et al., 2014; Weaver et al., 2004) as well as potentially functioning as a mediator for the link between early adversity and increased internalizing problems (Parade et al., 2016). However, findings as to the direction of NR3C1 methylation effects on behavior and psychopathology have been mixed with some studies finding positive associations with internalizing problems (e.g., Dadds, Moul, Hawes, Diaz, & Brennan, 2015; van der Knaap, van Oort, Verhulst, Oldehinkel, & Riese, 2015) while others have found negative associations with internalizing (Tyrka, Parade et al., 2016) and post-traumatic stress (Labonte, Azoulay, Yerko, Turecki, & Brunet, 2014; Vukojevic et al., 2014; Yehuda et al., 2015). One explanation for differential effects may be a developmental progression from dysregulation in early childhood to psychopathology in adulthood. Specifically, early acute stress-related hypermethylation results in emotional and behavioral dysregulation in early childhood (Parade et al., 2016) but then chronic environmental adversity throughout development may result in hypomethylation (Tyrka, Parade et al., 2016) as an adaptation to repeated stress exposures that, for some, results in sustained psychopathology in adulthood (Labonte et al., 2014; Vukojevic et al., 2014).
There has been substantial progress in behavioral epigenetics over the last decade (Lester, Conradt, & Marsit, 2016). A strong emphasis of behavioral epigenetics research has been on understanding how epigenetic processes play a vital, potentially explanatory, role in the biological embedding of early environmental adversity and the genesis of adaptive and maladaptive development (Boyce & Kobor, 2015). A major limitation, however, is that a basic understanding of how methylation of glucocorticoid signaling genes change over time has yet to be achieved, with little understanding of the role of risk factors that impact these developmental trajectories (Tyrka, Ridout et al., 2016; Vinkers et al., 2015). In fact, prior work examining the effect of early stress on methylation NR3C1 has been exclusively cross-sectional, greatly constraining our understanding of epigenetic processes over time. In addition, the extant cross-sectional data does not allow for examination of how maladaptive patterns of stress-related methylation changes are altered or maintained over time in association with adversity and the development of psychopathology. Such questions are crucial to understanding pathophysiology and informing the next generation of prevention and intervention efforts.
The primary aim of the current study was to begin to address the lack of longitudinal designs in environmental epigenetic research. We utilized a prospective longitudinal design with repeated assessments (two waves approximately 6 months apart) of methylation of saliva NR3C1 to understand how child maltreatment and other environmental adversity (e.g., stressor associated with living in poverty other traumatic events) contributes to within-child change in NR3C1 methylation over time. Specifically, using a sample of preschoolers, approximately half of which have documented maltreatment within 6 months of the baseline assessment, we examined the effect of maltreatment and other environmental adversity on short-term change in methylation to begin to characterize within-child epigenetic changes during this sensitive developmental period. We assessed DNA methylation of the well characterized exon 1F, and extended this research to exon 1D which also has support from cross-sectional studies for a role in these processes (Hompes et al., 2013; Parade et al., 2016; van der Knaap et al., 2014; Weder et al., 2014).
Method
Participants
Two hundred and sixty families residing in the Northeast enrolled in this study. One child from each family was included in the study. Children ranged in age from 3 to 5 years (M= 4.2, SD= 0.74), and 52.3% were female. The sample was racially and ethnically diverse (45.6% Hispanic, 27.7% White non-Hispanic, 16.3% Black, 21.9% biracial, 2.7% other races). Nearly all children were living in poverty (91%). Most caregivers (95%) were biological mothers, 60% of caregivers had less than or equal to a high school diploma, 53.3% were single parents, 55% of caregivers were unemployed, and 86.5% of the families qualified for public assistance. Approximately half of the children (53.3%) had substantiated cases of moderate to severe child maltreatment within the past 6 months as described below. Baseline results from a subsample (n = 184, 70.7%) the present sample were reported previously elsewhere (Tyrka et al., 2015).
Procedure
Families with a maltreated child were identified from the local child welfare agency and an emergency maltreatment assessment service via record review. Families of children with no indicated case of maltreatment within the past 6 months were recruited at a pediatric medical clinic during a well-child visit as well as at childcare centers. Based on review of available medical records and parent report, children with a chronic illness, medication use, obesity, and failure-to-thrive were excluded. Those with acute illness or medication use were included no less than 2 weeks following resolution of illness and medication use. Families completed a baseline set of assessments at the time of initial study enrollment and a follow-up set of assessments 6 months following enrollment (M = 6.42 months, SD = 0.67 months). At each wave of assessment, families completed two home visits and questionnaires between the visits. The current report focuses on data from the first home visit during the baseline assessment during which caregivers completed interviews on child stress exposure and a baseline saliva sample for DNA isolation was collected from the children, as well as the first home visit during the follow-up assessment during which caregivers completed an interview on service utilization and a follow-up saliva sample for DNA isolation was collected from children.
Measures
Child maltreatment status
All families consented to examination of child welfare records to determine maltreatment status. Trained research staff coded the records using the System for Coding Subtype and Severity of Maltreatment in Child Protective Records (Barnett, Manly, & Cicchetti, 1993). Five maltreatment subtypes and severity scores ranging from 1 (least severe) to 5 (most severe) were derived. Children with a case of moderate to severe levels of maltreatment (score of 3–5) within the last 6 months were considered as part of the maltreated group (n=134). Within the maltreated group, 41% of the sample had substantiated cases of multiple subtypes of maltreatment, 5 children had substantiated cases of physical abuse, 32 sexual abuse, 16 physical neglect/failure to provide, 36 physical neglect/lack of supervision, and 85 emotional maltreatment. The comparison group included children who had never had a substantiated case of maltreatment.
Additional adversity markers
Contextual stress interview
Caregivers completed a semi-structured interview developed in our laboratory to assess contextual stressors experienced in the child’s lifetime. Categories were: death of a caregiver, separation from a caregiver, housing instability, inadequate food or clothing, and other stressful events which included witnessing neighborhood violence or parental arrest. Interviews were conducted and scored by trained clinical social workers and PhD-level psychologists. The project coordinator reviewed each interview to ensure compliance with the scoring protocol. Each domain was scored positive if at least one episode occurred, and domains were summed to determine the number of contextual stressor categories the child experienced in their lifetime. Possible scores ranged from 0 (no stressors) to 5 (stressors in all five domains). In the current sample the number of stressor categories ranged from 0 to 5 (M = 1.34, SD = 1.20).
Traumatic life events
The Diagnostic Infant and Preschool Assessment (Scheeringa & Haslett, 2010) interview was conducted with caregivers to assess child experiences of traumatic life events. Interviews were conducted by trained clinical social workers and PhD-level psychologists, reviewed in a group supervision format, and scored based upon group consensus. Traumatic events in each domain were dichotomized (no trauma vs. ≥ 1 trauma), then summed to create a scale for number of types of traumas experienced in the child’s lifetime. Physical and sexual abuse were not included because they were assessed as maltreatment (above). Possible scores range from are 0 to 9 and in the present sample a mean of 1.01 and SD of 1.05 were observed.
Other Adversity Composite
Confirmatory factor analysis using Mplus was conducted to estimate a single factor latent adversity variable based on model fit and theoretical interpretability with the following indicators: the number of lifetime contextual stressors, the number of traumatic life events, single parent status (coded as 0 or 1), and the child’s lifetime experiences of homelessness (8%). Single parent status and experiences of homelessness were collected as part of the demographic questionnaire. A robust weighted least squares estimator using a diagonal weight matrix was used as it provides the best option for modelling categorical or ordered data. Model fit was excellent, χ2 (2, N = 260) = 1.44, p > .15, root mean square error of approximation = 0.00, 95% confidence interval (CI) [0.00 −0.11], comparative fit index = 1.0, weighted root mean square residual = 0.245, and standardized factor loadings were as follows: homelessness (.71), single parent status (.44), lifetime stressors (.89), and the number of traumatic life events (.62). The factor score of the latent adversity variable was saved and used as observed in primary analyses.
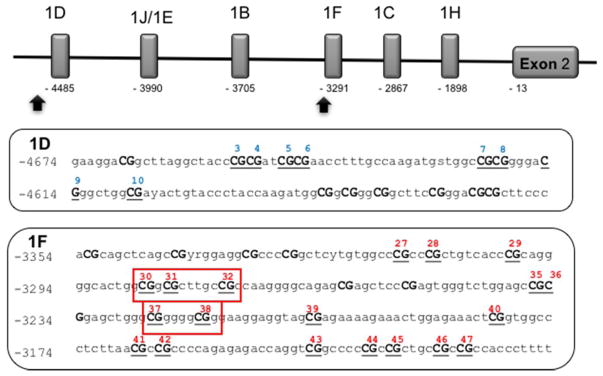
NR3C1 Methylation
Saliva samples were obtained using the Oragene DISCOVER kits (OGR-575) for Assisted Collections (DNA Genotek, Kanata, Ontario, Canada) at baseline and follow-up visits, and DNA was isolated following the manufacturer’s instructions. Sodium bisulfite modification was performed with 500 ng of DNA using the EZ DNA methylation Kit (Zymo Research, Irvine, CA, USA). For DNA methylation detection, bisulfite pyrosequencing was employed in two locations within the NR3C1 region: promoter of exon 1D and promoter of exon 1F (three assays; Figure 1) at the two time points. CpG numbering was taken from Palma-Gudiel et al. (2015) to improve comparability of results to past studies. PyroMark Assay Design software version 2.1.15 (Qiagen) was used to design the pyrosequencing assays. Amplification polymerase chain reactions and sequencing primers (Integrated DNA Technologies, Inc, Coralville, IA) and the genomic locations of the assays are provided in Table 1. The PyroMark polymerase chain reaction kit and forward and reverse primers were used to amplify specific regions of the NR3C1 promoter. Four forward pyrosequencing assays covering a total of 27 CpG loci were performed in triplicate using the PyroMark MD (Qiagen). Percent DNA methylation at each CpG locus was quantified with the PyroMark CpG software, version 1.11 (Qiagen). All procedures were performed following manufacturer’s protocols. The percent of alleles that were methylated was used in statistical analyses.
Figure 1. NR3C1 region: promoter of exon 1D and promoter of exon 1F.
Note. CpG numbering taken from Palma-Gudiel et al. (2015). Boxes around CpG site numbers represent NGFI-A transcription factor binding sites according to McGowan et al. (2009).
Table 1.
Pyrosequencing Primers and Assay Sequences.
| NR3C1 assay name (CpG positions) | Primersa | Sequence to analyze (converted) chromosome location |
|---|---|---|
| 1D (8) |
PCR forward GGATAAGAGGTTTGTTGAAAGTTTATT PCR reverse Biotin-ACTCCCCCTACTCTAACAT Forward sequencing 1D AGGAAGGAAGGTTTAGGT |
5:142785061-142785008 TATTYGG/AYGATYGYGAATTTTTGTTAAGATGKTGGTYGYGGGGAYGGGTTGGYGA TATTGTATTTTATTAAGATGG |
|
1F 3 pyrosequencing assays (19) |
PCR forward TTTTTTTTTTGAAGTTTTTTT PCR reverse Biotin-CCCCCAACTCCCCAAAAA Forward sequencing F0 GAGGAGTTTAGGTTTTTGTG Forward sequencing F1 GAGTGGTTTGGAGT Forward sequencing F2 AGAAAAGAATTGGAGAAATT |
Sequencing F0 (CpG 27–32) 5:142783720-142783676 GTTYGTTYGTTGTTATTYGTAGGGGTATTGGYGG/AYGTTTGTYGTTAAGGGGTAGAG Sequencing F1 (CpG 35–39) 5:142783640-142783588 YGYGGAGTTGGGYGGGGGYGGGAAGGAGGTAGYGAGAAAAGAAATTGGAGAAA Sequencing F2 (CpG 40–47) 5:142783585-142783501 YGGTGGTTTTTTTAAYGTYGTTTTAGAGAGATTAGGTYGGTTTTYGTYGTTGTYGTYGTTATTTTTTTTTTTGGGGAGTTGGGGG |
Note.
Primer Sequences are given 5′ to 3′ direction.
Covariates
Child age in months, child sex, and the number of days between baseline and the follow-up assessment were all examined as predictors of NR3C1 methylation at baseline and change in methylation between baseline and the 6-month follow-up. If significant associations emerged those variables were included as covariates in primary analyses.
Modeling Ancestry Differences Using Principal Component Analysis
Allele frequency differences due to systematic ancestry differences could cause spurious associations. Thus, we used PCA to model ancestry differences in the current study using genome-wide single nucleotide polymorphism (SNP) markers. Genotyping was conducted using the Illumina Infinium PsychArray-24 beadchip (over 588,000 autosomal SNPs). Genotypes were called using GenomeStudio software V2011.1 and genotyping module version 1.8.4 (Illumina), and cleaned using standard quality control procedures. We first conducted linkage disequilibrium-based pruning to lessen overweight of principle components (PCs) of genetic variation by the contribution of correlated SNPs, and followed by PCA using PLINK (Purcell et al., 2007). The first two PCs obtained using the PLINK were used for controlling the potential population stratification (Price et al., 2006).
Data analytic plan
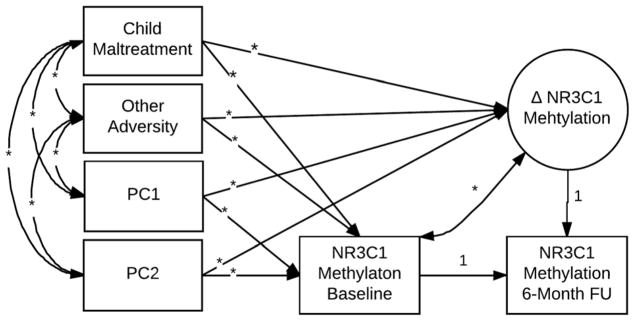
Prior to analyses, outliers, defined as values more than 3 SD from the mean, were Winsorized by setting them to the next highest value within three standard deviations. Mplus 7.31 software (Muthén & Muthén, 1998–2012) was used to conduct all analyses. To assess whether maltreatment and the other adversity composite predict baseline level and within-child changes over time in DNA methylation, we utilized latent change score (LCS) models. Of relevance to our main research questions, the flexibility of LCS allows for a simultaneous analysis of the predictors of individual differences in the initial level of and subsequent changes in key variables similar to a latent growth curve model but with two time points (McArdle, 2009). Primary LCS models examined maltreatment and the adversity composite simultaneously as predictors of baseline level and change in NR3C1 methylation. See Figure 2 for a depiction of the primary LCS model.
Figure 2. Primary latent change score models.
Note. * represents freely estimated parameter. PC1–2 = DNA-based Principal Components used to account for potential population stratification.
Missing data for methylation of single CpG sites was a maximum of 4.2% at baseline and 30.4% at 6 months with methylation data available for 86.5% of the sample at follow-up. Data were missing completely at random, Little’s missing completely at random test p > .15, and full information maximum likelihood estimation techniques were used for inclusion of all available data. To account for non-normality, maximum likelihood estimation with robust standard errors was used. The following fit statistics were employed to evaluate model fit: Chi-square, χ2: p > .05 excellent, Comparative Fit Index ( >0 .90 acceptable, > 0.95 excellent), root mean square error of approximation (< 0.08 acceptable, < 0.05 excellent) and the standardized root mean square residual (< 0.08 acceptable, < 0.05 excellent; Hu & Bentler, 1999).
Results
Preliminary analyses
Covariates
In individual predictor LCS models child sex, child age, and the number of days between baseline and follow-up assessments were examined as predictors of baseline level and change in mean methylation of NR3C1 exon 1D and 1F. None of the three potential covariates were significantly related to baseline or change in methylation of 1D or 1F. Thus, these variables were not considered further in analyses, and only the top two genetic PCs were included as covariates to account for potential population stratification in primary LCS analyses.
CpG correlations
Correlations between CpG sites within 1D were mostly small to medium in magnitude, similar at baseline and follow-up, with no clear clustering within the 1D CpG sites. For correlations between CpG sites within 1F, some CpG sites were more closely associated, including modest correlations between immediately adjacent CpG sites from 27 to 32 (e.g., CpG 28 and 29 r = .35 whereas CpG 28 and 30 r = .16), and large correlations between putative NGFI-A binding region CpG sites 30–32 (average r = .66), and between CpG sites 40–47 (average r = .62). In other words, methylation at some CpG sites across the 1F promoter region fluctuate in a coordinated fashion where as others are more loosely associated.
Primary analyses
We conducted primary analyses in two stages. First, LCS models were conducted with mean methylation across exons 1D and 1F. Next, based on the correlation results above and CFA models that did not support a single-factor structure for NR3C1 exon 1F (poor model fit for each model, complete results available upon request), LCS analyses were conducted with CpG bundles for 1F (i.e., average methylation across several sites) based on the literature and the above results: CpG sites 27–29 given higher than average interrelations (mean r = .42), CpGs 30–32 and 37–38 as they are known NGFI-A binding sites (McGowan et al., 2009), and 40–47 given higher than average interrelations (mean r = .59).
Mean methylation of NR3C1 exons 1D and 1F
First, an unconditional LCS model was estimated for each exon to characterized unconditional change over time. Across LCS models for exon 1D and 1F, the covariance of intercept (i.e., baseline) and change factors were significant and negative suggesting that preschoolers who had higher methylation values at baseline tended to decrease more rapidly across 6 months for exon 1D, β = −0.52, 95% CI [−0.63, −0.42], and 1F, β = −0.55, 95% CI [−0.65, −0.45]. In addition, mean rate of change and variances of intercept and change factors for exons 1D and 1F significantly differed from zero, all ps < .01, indicating potentially important individual variability in both starting-point and change over time.
A summary of primary conditional LCS model results are presented in Table 2 including unstandardized path coefficients and 95% CI. Model fit for the LCS model for exon 1D and 1F was excellent. In regard to the mean 1D methylation model (Model 1, Table 2), maltreatment status, but not other adversity, positively predicted baseline level and negatively predicted change in methylation, such that maltreated preschoolers evidenced higher baseline levels of exon 1D methylation but then significant decreases in 1D methylation between baseline and follow-up assessments. When the LCS model was reversed to examine follow-up methylation levels as the intercept, maltreated preschoolers evidenced marginally (p < .10) lower methylation levels at follow-up. Results for predicting change in 1D were consistent whether maltreatment and other adversity were examined simultaneously or separately as predictors.
Table 2.
Summary of latent change score analyses for CpG bundles.
| Model 1: Mean 1D |
Model 2: Mean 1F |
Model 3: 1F 27–29 |
Model 4: 1F NGFI-A |
Model 5: 1F 40–47 |
|
|---|---|---|---|---|---|
|
| |||||
| b [95% CI] | b [95% CI] | b [95% CI] | b [95% CI] | b [95% CI] | |
| Predicting Baseline | |||||
| Maltreatment | .07* [.02, .12] | −.02 [−.10, .06] | .07 [−.09, .22] | −.10 [−.25, .06] | −.02 [−.17, .14] |
| Adversity | .00 [−.05, .05] | .07* [.00, .13] | .11* [.001, .22] | −.01 [−.14, .12] | .12† [−.01, .25] |
| PC1a | .12 [−.32, .55] | −.63* [−.1.2, −.07] | .72 [−.63, 2.1] | .57 [−.49, 1.6] | −1.5* [−2.5, −.52 |
| PC2 a | .00 [−.45, .46] | −.49 [−1.2, .21] | −.41 [−2.7, 1.9] | −.45 [−1.3, .41] | −.54 [−1.7, .56] |
| Predicting Change | |||||
| Maltreatment | −.14* [−.24, −.05] | −.01 [−.15, .14] | −.27* [−.48, −.05] | .03 [−.20, .27] | .04 [−.25, .32] |
| Adversity | .03 [−.05, .11] | −.04 [−.16, .08] | −.10 [−.25, .05] | −.07 [−.27, .13] | −.01 [−.21, .20] |
| PC1a | −.48 [−1.1, .12] | .90 [−.20, 2.0] | −1.4 [−3.2, .33] | −.31 [−1.9, 1.3] | 2.2* [.32, 4.1] |
| PC2 a | −.19 [−.85, .47] | .83 [−.44, 2.1] | −.64 [−3.5, 2.2] | .85 [−.66, 2.4] | 1.4 [−.26, 3.0] |
| Predicting Follow-up | |||||
| Maltreatment | −.07† [−.15, .01] | −.03 [−.16, .10] | −.20* [−.35, −.06] | −.07 [−.26, .13] | .02 [−.23, .28] |
| Adversity | .03 [−.03, .10] | .02 [−.07, .12] | .01 [−.09, .11] | −.08 [−.24, .08] | .12 [−.08, .32] |
| PC1a | −.36 [−.89, .17] | .27 [−.66, 1.2] | −.70 [−1.6, .24] | .25 [−1.1, 1.6] | .74 [−1.1, 2.5] |
| PC2 a | −.19 [−.74, .36] | .34 [−.58, 1.3] | −1.1* [−1.9, −.19] | .40 [−.99, 1.8] | .82 [−.75, 2.4] |
|
| |||||
| Model Fit | |||||
| χ2 (df), p value | .01 (1), .96 | .02 (1), .90 | .02 (1), .88 | .001 (1), .98 | .00 (1), .99 |
| RMSEA [95% CI] | .00 [.00, .00] | .00 [.00, .07] | .00 [.00, .08] | .00 [.00, .00] | .00 [.00, .00] |
| CFI | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| SRMR | .001 | .001 | .002 | .000 | .000 |
Note. 1F NGFI-A = transcription factor binding sites as identified by McGowan et al. (2009): 30–32 & 37–38.
p < .05,
p < .10.
Bolded cells represent p value < .10.
DNA-based Principal Components used to account for potential population stratification. Predicting Follow-up paths are from reverse LCS models where follow-up functions as the intercept, results from these models had near identical model fit.
Results for the mean exon 1F methylation model (Model 2, Table 2) differed from those of 1D such that other adversity, but not maltreatment status, positively predicted baseline methylation levels and neither predictor was significantly related to change in 1F methylation. Results for predicting change in 1F were consistent whether maltreatment and other adversity were examined simultaneously or separately as predictors.
NR3C1 exon 1F CpG bundles
LCS model results for exon 1F bundles are also presented in Table 2. Model fit for all models was excellent and comparable to model fit statistics reported above. For the CpG 27–29 bundle (Model 3, Table 2), other adversity was positively related to baseline levels of methylation such that children who faced more adversity, including contextual stress and trauma exposure, evidenced higher levels of methylation in this region. Further, maltreatment status was negatively related to change in methylation for the 27–29 region such that maltreated preschoolers evidenced decreases in methylation. Once the LCS model was reversed, maltreated preschoolers evidenced significantly lower methylation levels at follow-up, b = −0.20, 95% CI [−0.35, −0.06]. In other words, for this region, maltreated and comparison children did not significantly differ in methylation level at baseline but maltreated children had significant decreases in methylation over time and ended with lower levels of methylation compared to comparison children. However, though maltreatment status was not a statistically significant predictor of baseline methylation for exon 1F 27–29, the effect size was nearly identical to that of 1D, where the difference was statistically significant. Finally, for the NGFI-A binding site bundle (Model 4, Table 2) and the CpG 40–47 bundle (Model 5, Table 2), neither maltreatment nor adversity significantly predicted baseline or change in methylation (though the effect of adversity on baseline methylation levels of 40–47 approached significance).
Post-hoc analyses
Following primary LCS analyses, post-hoc analyses were conducted using individual CpG sites to examine whether specific sites drove significant effects and because this study is the first to examine prospective short-term within-child change in NR3C1 methylation.
LCS analysis of individual CpG sites
Given the number of models (27) for LCS analyses with individual CpG sites, p values are not interpreted in favor of examining the pattern of results across CpG sites via effect sizes and 95% bootstrapped CI. A summary of LCS model results for individual CpG sites is presented in Table 3. Each row of Table 3 represents a different model based on an individual CpG site. The maltreatment and adversity columns present results of maltreatment or adversity predicting baseline levels and change in the methylation outcome for each model. Model fit for all models was excellent and comparable to model fit statistics reported above.
Table 3.
Summary of latent change score analyses for individual CpG sites.
| IV → | Maltreatment | Adversity | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| DV → | Baseline | Δ | Baseline | Δ | ||||
| ↓ | b | 95% CI | b | 95% CI | b | 95% CI | b | 95% CI |
| 1D_3 | .10 | −.002, .20 | −.11 | −.29, .07 | .03 | −.05, .11 | .01 | −.14, .15 |
| 1D_4 | .10 | .001, .20 | −.10 | −.21, .02 | .02 | −.06, .09 | −.01 | −.10, .08 |
| 1D_5 | .11 | .01, .20 | −.23 | −.38, −.07 | .01 | −.08, .08 | .16 | .03, .30 |
| 1D_6 | .02 | −.06, .10 | −.02 | −.14, .09 | .06 | −.002, .12 | −.07 | −.15, .02 |
| 1D_7 | .08 | −.13, .30 | −.19 | −.59, .21 | .03 | −.13, .30 | −.14 | −.50, .23 |
| 1D_8 | .10 | .01, .19 | −.18 | −.32, −.05 | −.05 | −.12, .01 | .10 | −.01, .21 |
| 1D_9 | .05 | −.04, .13 | −.12 | −.26, .01 | −.01 | −.08, .06 | .12 | .007, .23 |
| 1D_10 | .03 | −.08, .14 | −.18 | −.36, .006 | −.07 | −.16, .02 | .08 | −.08, .23 |
| 1F_27 | .21 | −.06, .49 | −.45 | −.80, −.10 | .09 | −.10, .28 | −.03 | −.27, .20 |
| 1F_28 | −.06 | −.22, .10 | −.12 | −.40, .16 | .12 | .001, .22 | −.15 | −.34, .05 |
| 1F_29 | .07 | −.06, .21 | −.26 | −.49, −.02 | .14 | .04, .25 | −.14 | −.32, .04 |
| 1F_30 | −.15 | −.40, .10 | .09 | −.30, .48 | .07 | −.15, .30 | −.18 | −.50, .15 |
| 1F_31 | −.19 | −.41, .03 | .15 | −.19, .50 | −.01 | −.18, .17 | −.06 | −.33, .20 |
| 1F_32 | −.20 | −.51, .12 | .05 | −.40, .51 | −.10 | −.34, .15 | .00 | −.38, .37 |
| 1F_35 | .06 | .03, .31 | −.11 | −.30, .09 | .07 | −.06, .19 | .10 | −.05, .25 |
| 1F_36 | .06 | −.06, .17 | −.05 | −.23, .14 | .06 | −.04, .15 | .01 | −.13, .14 |
| 1F_37 | −.03 | −.19, .14 | .03 | −.22, .24 | −.04 | −.17, .06 | −.06 | −.24, .11 |
| 1F_38 | .07 | −.05, .18 | −.14 | −.30, .01 | −.02 | −.10, .07 | .01 | −.10, .11 |
| 1F_39 | −.13 | −.21, −.04 | .14 | −.04, .32 | .03 | −.04, .10 | .06 | −.07, .18 |
| 1F_40 | −.05 | −.18, .08 | .03 | −.16, .22 | .08 | −.03, .19 | −.07 | −.22, .09 |
| 1F_41 | −.06 | −.21, .09 | −.05 | −.36, .25 | .06 | −.06, .18 | .08 | −.14, .31 |
| 1F_42 | −.09 | −.25, .08 | .21 | −.08, .51 | .12 | −.004, .25 | −.03 | −.26, .20 |
| 1F_43 | −.11 | −.35, .12 | .21 | −.25, .67 | .21 | .008, .40 | .05 | −.29, .40 |
| 1F_44 | .01 | −.19, .22 | .07 | −.27, .40 | .11 | −.06, .27 | −.09 | −.33, .16 |
| 1F_45 | .04 | −.18, .25 | .11 | −.30, .52 | .18 | .003, .35 | −.05 | −.34, .25 |
| 1F_46 | .02 | −.17, .21 | −.08 | −.40, .24 | .08 | −.07, .22 | −.01 | −.23, .22 |
| 1F_47 | .12 | −.15, .38 | −.19 | −.62, .25 | .13 | −.10, .36 | .10 | −.24, .43 |
Note. Light gray shading and bold represent 95% CI that either did not include zero or had one end approximately at zero but an effect size similar to other nearby CpG sites. Δ = latent change. Each row represents a separate model with both maltreatment and other adversity as simultaneous predictors.
Overall, maltreated children evidenced higher baseline levels of methylation across most exon 1D CpG sites but few 1F CpG sites. Further, higher levels of adversity were related to higher baseline levels of methylation across much of the 1F, but not 1D, promoter region, though most of this effect was outside of known NGFI-A binding sites (comparable effect sizes for CpG 27–29 and 40–47). With regard to prediction of change over time, maltreated children evidenced decreases in methylation relative to comparison children across most 1D CpG sites (CpG 6 as the exception), but for 1F this effect was only observed consistently at CpG sites 27–29 with the largest difference observed for 1F CpG 27, which was nearly twice as large as the next largest difference. Finally, adversity rarely predicted change in methylation with wide confidence intervals observed for nearly all CpG sites. Intriguingly, for CpG sites 5, 8, and 9 of exon 1D, maltreatment status predicted decreases in methylation over time whereas other adversity predicted increases in methylation over time.
Post-hoc CpG site bundle
To better characterize the effect of maltreatment on baseline level and change in NRC31 methylation we combined CpG sites that evidenced consistency across analyses (exon 1D CpGs 3–5, 7–10 and exon 1F CpGs 27–29). The results of this LCS model served as the basis of Figure 3, which displays mean methylation at baseline and follow-up across these regions. As is shown in Figure 3, across these sites, maltreated children evidence higher baseline levels of methylation, β = 0.15, 95% CI [0.01, 0.29], significant decreases in methylation over time, β = −0.23, 95% CI [−0.38, −0.08], and then at follow-up, lower levels of methylation, β = −0.19, 95% CI [−0.35, −0.02], relative to non-maltreated preschoolers (whose methylation levels remained stable) after accounting for the effects of other adversity and potential population stratification.
Figure 3. Depiction of NR3C1 change over time separately by maltreatment and comparison children.
Note. Average methylation across multiple CpG sites (exon 1D CpGs 3–5, 7–10 & exon 1F CpGs 27–29) is presented separately for maltreated and comparison children. * = p < .05, ns = p > .05. The graph represents multiple analyses to better understand study results. Maltreated children n = 134, Comparison group n = 126.
Discussion
Results from the current study suggest that child maltreatment is associated with higher initial levels of NR3C1 promoter methylation within 6 months of documented maltreatment as well as the rate of within-child change in NR3C1 methylation approximately 1 year after documented maltreatment. The primary aim of the current study was to begin to address the lack of longitudinal methylation designs in environmental epigenetic research by utilizing a short-term longitudinal model to examine predictors of change in methylation of saliva DNA NR3C1 in a sample of children. While others have examined change in NR3C1 DNAm after non-randomized psychological intervention with small samples (Roberts et al., 2015; Yehuda et al., 2013), the current study is the first to examine early environmental adversity as a predictor of NR3C1 DNAm change. Further, though several studies have examined the influence of child maltreatment and other early environmental adversity on later NR3C1 DNAm in adulthood (e.g., Tyrka, Parade et al., 2016; Tyrka et al., 2012), the current study is the first to explicitly model NR3C1 promoter methylation change in early childhood proximal to a documented incidence of child maltreatment or current adversity inherent within environments of poverty. Establishing trajectories of short-term change following child maltreatment and temporal precedence before the onset of significant psychopathology is crucial to drawing inferences about the development of psychopathology (Jones, Moore, & Kobor, in press).
In the current study, baseline associations between child maltreatment status and other adversities with NR3C1 promoter methylation were largely consistent with those previously reported using a sub-sample of participants in the current study (Tyrka et al., 2015). Specifically, preschoolers with documented maltreatment evidenced higher mean baseline methylation at NR3C1 region 1D, but not 1F, relative to non-maltreated but demographically similar comparison children. In addition, other early life adversity (e.g., contextual stress associated with poverty, trauma exposure), but not maltreatment, was associated with higher baseline mean methylation at region 1F. Results from the current study extend these previous results by examining child maltreatment and other adversity as predictors of change in methylation at these same regions while accounting for PCs used to adjust for potential population stratification. Findings indicated that though maltreated children evidenced higher baseline levels of methylation, they also had significant decreases in methylation over time, and at the 6-month follow-up their methylation levels were lower than those of non-maltreated preschoolers (whose methylation levels remained stable over time). These results were observed across most of the 1D CpG sites examined and one region of 1F (CpGs 27 and 29), but limited support was found at other 1F sites including the NGFI-A transcription factor binding sites identified by McGowan et al. (2009).
Baseline associations found in the current study were consistent with a large body of research examining the influence of early environmental adversity on NR3C1 methylation that has been replicated across species and cell populations as diverse as peripheral blood cells, salivary DNA (which is primarily of leukocyte origin), and central nervous system-derived cells (Turecki & Meaney, 2016; Tyrka, Ridout et al., 2016; Vinkers et al., 2015). The baseline effect sizes in the current study were small to medium in magnitude, which is consistent with effect sizes found in other studies. One of the most pressing problems in environmental epigenetics is that we currently do not know whether these small effect sizes constitute biologically meaningful differences (Vinkers et al., 2015). Nevertheless, there is evidence that small effect sizes have significant downstream effects on gene expression (Breton et al., 2017), and, specifically for NR3C1, significant effects of methylation on post-mortem human GR expression are found in the hippocampus (Labonte et al., 2012; McGowan et al., 2009).
The current study is the first to test this association longitudinally and, thus, results are in need of replication and extension to longer time intervals. The observation of dynamic stress-related changes in DNAm during a sensitive developmental period highlights the importance of longitudinal designs for environmental epigenetic research. Despite the lack of extant literature for interpretation of this longitudinal association, results from the current study are consistent with previous findings from two longitudinal twin studies that examined intraindividual longitudinal change in DNAm. First, though not examining NR3C1, Wong et al. (2010) observed changes in DNAm between ages 5 and 10 among monozygotic twin pairs at the promoter/regulatory regions of the dopamine receptor D4(DRD4), serotonin transport (SERT), and monoamine oxidase A (MAOA) genes that were primarily attributable to environmental, and not heritable, influences. Second, Levesque et al. (2014) examined the stability of genome-wide DNA methylation patterns over 3–6 months in a sub-sample of 8 adolescent monozygotic twins and found that NR3C1 methylation was part of a highly variable statelike gene network that may be highly responsive to changes in the environment. The results of the current study support and extend those of these two previous studies by finding that child maltreatment predicts changes in NR3C1 promoter methylation in early childhood.
However, the directionality of stress-related DNAm change was unexpected, and DNAm differences between maltreated and comparison children reversed between assessments for some CpG sites in the current study (see Figure 3). Previously reported associations between early adversity and NR3C1 promoter methylation have not been universal in direction. Though the majority of published studies find higher levels of methylation of NR3C1 with early environmental stress (Turecki & Meaney, 2016), some studies have found the opposite (Daskalakis & Yehuda, 2014; Turecki & Meaney, 2016; Tyrka, Parade et al., 2016). One hypothesis for the dynamic stress-related DNAm changes depicted in Figure 3 is that early acute stress-related hypermethylation results in emotional and behavioral dysregulation in early childhood (Parade et al., 2016) but then chronic or severe environmental adversity throughout development results in hypomethylation (Tyrka, Parade et al., 2016) as an adaption to repeated stress exposures that, for some, results in sustained psychopathology (Labonte et al., 2014; Vukojevic et al., 2014). In line with this developmental progression of biological embedding hypothesis, the DNAm difference at baseline in the current study may represent an acute stress response associated with concurrent emotional and behavioral dysregulation as reported by Parade et al. (2016) using a sub-sample (n = 174) of the current studies participants. After the initial abuse is stopped or reduced after state agency involvement, this initial hypermethylation and dysregulation may transition, for some, to hypomethylation.
The developmental progression hypothesis offered above is supported by results from the current study finding reductions in methylation over time as well as lower levels at follow-up, approximately 1 year after documented abuse, for maltreated children relative to non-maltreated comparison children. In addition, this hypothesis is supported by research with adult populations consistently finding that hypomethylation is associated with posttraumatic stress disorder (e.g., Labonte et al., 2014; Vukojevic et al., 2014; Yehuda et al., 2015). Furthermore, this hypothesis is also supported by recent research finding that within-subject decreasing DNAm levels over time at several genes, though not NR3C1, were related to the increasing levels of posttraumatic stress disorder symptoms over time among military veterans (Rutten et al., 2017). If the developmental progression hypothesis is correct, we would expect the current trajectory of change in NR3C1 promoter methylation to continue into middle childhood and adolescence with accompanying onset of posttraumatic stress disorder or severe mood dysregulation. Thus, further research utilizing longitudinal designs with follow-ups across developmental stages are needed to support or refute this hypothesis.
An alternative hypothesis for explaining the observed reduction in NR3C1 promoter methylation in the current study is that this change is an intervention-induced return to baseline after state agency involvement. The higher levels of NR3C1 methylation and emotional dysregulation seen in maltreated preschoolers might be reversed with early intervention. According to this hypothesis, elevated levels of DNAm would have persisted without intervention, and the observed reduction in DNAm is because of intervention in early childhood before stabilization of the epigenome later in development. However, this hypothesis was not supported by posthoc analyses in the current study that found no association between service utilization (assess via an interview at the follow-up assessment) and change in NR3C1 methylation among maltreated preschoolers nor that service utilization moderated the association between maltreatment and change in methylation. Yet, the services interview in the current study was limited, without indictors of quality, frequency, or intensity of services nor effectiveness of intervention services on treatment targets. Therefore, future longitudinal research would benefit from designs that more effectively parse the contextual factors that might promote or inhibit methylation following severe adversity.
There are several limitations of the current study that should be noted. It is possible that the longitudinal results observed in the present study could be accounted for by unmeasured confounders. Baseline associations in the current study are consistent with numerous other studies across species and tissues, which increase our confidence in these results. However, the longitudinal stress-related associations in the present study are the first such results reported, and as such, replication studies are necessary prior to integration into established conceptualizations of the effects of adversity on NR3C1 methylation. Specifically, one potentially important unmeasured confounder in the current study is cell composition. Because our young children were not able to provide saliva via passive drool, we used a standardized method that involves using a sponge to collect pooled saliva beneath the tongue and at the intersection of the cheek and gum, so that in addition to various types of leukocytes, it is likely to yield some epithelial cells. Cell type within a tissue has been found to be the second biggest contributor to DNAm variation after tissue type (Farré et al. 2015). However, despite this cell type issue, Smith et al. (2015) found DNAm from saliva may be more similar to that of brain tissues on average than DNAm from blood. Further, the best method to correct for interindividual variability in cell type is still highly debated (McGregor Labbe, & Greenwood, 2017) which is further complicated by the lack of well-characterized reference profiles for young child saliva. Regardless, future environmental epigenetics studies will benefit from controlling for this important type of variation (Jones et al., in press).
An additional limitation of the current study is the lack of accompanying gene expression data. Although there is prior evidence that NR3C1 promoter DNAm is associated with gene expression (Turecki & Meaney, 2016), future longitudinal epigenetics research will benefit from assessing whether the DNAm changes observed alter gene expression (Jones et al., in press).
The current study also had several significant strengths that should be noted. First, the current study utilized a diverse sample of preschoolers exposed to a range of adversities including documented maltreatment while also controlling for potential population stratification in all primary analyses. Second, the current study is the first to explicitly model change in NR3C1 methylation signifying an important step in this line of research. Third, we report CpG site-specific effects given their utility in guiding basic molecular research on the biological and functional effects of DNAm differences observed (Daskalakis & Yehuda, 2014). Fourth, the inconsistent numbering of NR3C1 CpG sites have contributed to difficulty comparing results across multiple laboratories (Turecki & Meaney, 2016), so we employed a universal CpG number system developed by Palma-Gudiel et al. (2015)
In summary, epigenetic marks, such as DNAm, appear to functionally mold genetically guided developmental plasticity in response to early environmental experiences, thus providing a molecular basis for the enduring effects of early adversity exposures via biological embedding (Jones et al., in press). The current study is the first to examine maltreatment and other adversities inherent within environments of poverty as predictors of intra-individual change in NR3C1 methylation. Results from the current study highlight the complex nature of stress-related epigenetic variation during early development. Future studies that model early environmental adversity-induced DNAm changes into later developmental stages will continue to add to our understanding of the molecular mechanisms underlying the development of psychopathology.
Acknowledgments
This research was supported by grant R01 MH083704 (to A.R.T) and R25 MH101076 (K.K.R) from the National Institute of Mental Health. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIMH. We are grateful to the children and families who participated in this study, and we thank Hasbro Children’s Hospital, Rhode Island Head Start, and the Rhode Island Department of Children, Youth, and Families for assisting in recruitment of study participants. We also thank Brittney Josefson and the numerous other research assistants who contributed to this project, and Asi Polly Gobin for data management. Isolation of DNA and genotyping were done in the laboratory of Joel Gelernter, M.D., and we are grateful to Dr. Gelernter and his staff for their contribution.
References
- Barden N. Implication of the hypothalamic-pituitary-adrenal axis in the physiopathology of depression. Journal of Psychiatry and Neuroscience. 2004;29:185–193. [PMC free article] [PubMed] [Google Scholar]
- Barnett D, Manly JT, Cicchetti D. Defining child maltreatment: the interface between policy and research. In: Cicchetti D, Toth SL, editors. Child abuse, child development, and social policy. Norwood, NJ: Ablex; 1993. pp. 7–73. [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Boyce WT, Kobor MS. Development and the epigenome: the ‘synapse’ of gene environment interplay. Developmental Science. 2015;18:1–23. doi: 10.1111/desc.12282. [DOI] [PubMed] [Google Scholar]
- Braquehais MD, Picouto MD, Casas M, Sher L. Hypothalamic–pituitary–adrenal axis dysfunction as a neurobiological correlate of emotion dysregulation in adolescent suicide. World Journal of Pediatrics. 2012;8:197–206. doi: 10.1007/s12519-012-0358-0. [DOI] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PloS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, … Yousefi P. Small-magnitude effect sizes in epigenetic end points are important in children’s environmental health studies: The Children’s Environmental Health and Disease Prevention Research Center’s Epigenetics Working Group. Environmental health perspectives. 2017;125:511–526. doi: 10.1289/EHP595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti D, Toth SL. Child maltreatment and developmental psychopathology: A multilevel perspective. In: Cicchetti D, editor. Developmental Psychopathology. 3. Vol. 3. New York: Wiley; 2016. pp. 457–512. [Google Scholar]
- Children’s Bureau, U.S. Department of Health & Human Services, Administration for Children and Families, Administration on Children, Youth and Families. Child Maltreatment 2015. 2017 Available from http://www.acf.hhs.gov/programs/cb/research-data-technology/statistics-research/child-maltreatment.
- Cohen S, Janicki-Deverts D, Miller GE. Psychological Stress and Disease. JAMA. 2007;298:1685–1687. doi: 10.1001/jama.298.14.1685. [DOI] [PubMed] [Google Scholar]
- Conradt E, Hawes K, Guerin D, Armstrong DA, Marsit CJ, Tronick E, et al. The contributions of maternal sensitivity and maternal depressive symptoms to epigenetic processes and neuroendocrine functioning. Child Development. 2016;87:73–85. doi: 10.1111/cdev.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt E, Lester BM, Appleton AA, Armstrong DA, Marsit CJ. The roles of DNA methylation of NR3C1 and 11beta-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics. 2013;8:1321–1329. doi: 10.4161/epi.26634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadds MR, Moul C, Hawes DJ, Mendoza Diaz A, Brennan J. Individual differences in childhood behavior disorders associated with epigenetic modulation of the cortisol receptor gene. Child Development. 2015;86:1311–1320. doi: 10.1111/cdev.12391. [DOI] [PubMed] [Google Scholar]
- Doom JR, Gunnar MR. Stress physiology and developmental psychopathology: Past, present, and future. Development and Psychopathology. 2013;25:1359–1373. doi: 10.1017/S0954579413000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré P, Jones MJ, Meaney MJ, Emberly E, Turecki G, Kobor MS. Concordant and discordant DNA methylation signatures of aging in human blood and brain. Epigenetics & chromatin. 2015;8:19. doi: 10.1186/s13072-015-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, Binder EB. Current research trends in early life stress and depression: Review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Experimental Neurology. 2012;233:102–111. doi: 10.1016/j.expneurol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- Herman JP, McKlveen JM, Solomon MB, Carvalho-Netto E, Myers B. Neural regulation of the stress response: glucocorticoid feedback mechanisms. Brazilian Journal of Medical and Biological Research. 2012;45:292–298. doi: 10.1590/S0100-879X2012007500041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hompes T, Izzi B, Gellens E, Morreels M, Fieuws S, Pexsters A, et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. Journal of Psychiatric Research. 2013;47:880–891. doi: 10.1016/j.jpsychires.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Hu L, Bentler P. Cutoff criteria for fit indices in covariance structure analysis: conventional criteria versus new alternatives. Structural Equation Modeling. 1999;6:1–55. doi: 10.1080/10705519909540118. [DOI] [Google Scholar]
- Jones MJ, Moore SR, Kobor MS. Principles and challenges of applying epigenetic epidemiology to psychology. Annual Review of Psychology. :69. doi: 10.1146/annurev-psych-122414-033653. (in press) [DOI] [PubMed] [Google Scholar]
- Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends in Pharmacological Sciences. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kloet ER, Joëls M, Holsboer F. Stress and the brain: From adaptation to disease. Nature Reviews Neuroscience. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- Kertes DA, Kamin HS, Hughes DA, Rodney NC, Bhatt S, Mulligan CJ. Prenatal maternal stress predicts methylation of genes regulating the hypothalamic-pituitary-adrenocortical system in mothers and newborns in the Democratic Republic of Congo. Child Development. 2016;87:61–72. doi: 10.1111/cdev.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosten TA, Huang W, Nielsen DA. Sex and litter effects on anxiety and DNA methylation levels of stress and neurotrophin genes in adolescent rats. Developmental Psychobiology. 2014;56:392–406. doi: 10.1002/dev.21106. [DOI] [PubMed] [Google Scholar]
- Kosten TA, Nielsen DA. Litter and sex effects on maternal behavior and DNA methylation of the Nr3c1 exon 17 promoter gene in hippocampus and cerebellum. International Journal of Developmental Neuroscience. 2014;36:5–12. doi: 10.1016/j.ijdevneu.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Lim S, Gudsnuk K, Champagne FA. Sex-specific and strain dependent effects of early life adversity on behavioral and epigenetic outcomes. Frontiers in Psychiatry. 2013;4:78. doi: 10.3389/fpsyt.2013.00078. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Azoulay N, Yerko V, Turecki G, Brunet A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Translational Psychiatry. 2014;4:e368. doi: 10.3389/fpsyt.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, et al. Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biological Psychiatry. 2012;72:41–48. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- Laryea G, Muglia L, Arnett M, Muglia LJ. Dissection of glucocorticoid receptor mediated inhibition of the hypothalamic-pituitary-adrenal axis by gene targeting in mice. Frontiers in Neuroendocrinology. 2015;36:150–164. doi: 10.1016/j.yfrne.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenen FA, Muller CP, Turner JD. DNA methylation: conducting the orchestra from exposure to phenotype? Clinical Epigenetics. 2016;8:92. doi: 10.1186/s13148-016-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester BM, Conradt E, Marsit C. Introduction to the special section on epigenetics. Child Development. 2016;87:29–37. doi: 10.1111/cdev.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. British Journal of Nutrition. 2007;97:1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle JJ. Latent variable modeling of differences and changes with longitudinal data. Annual Review of Psychology. 2009;60:577–605. doi: 10.1146/annurev.psych.60.110707.163612. [DOI] [PubMed] [Google Scholar]
- McCrory EJ, Viding E. The theory of latent vulnerability: Reconceptualizing the link between childhood maltreatment and psychiatric disorder. Development and Psychopathology. 2015;27:493–505. doi: 10.1017/S0954579415000115. [DOI] [PubMed] [Google Scholar]
- McEwen BS, Bowles NP, Gray JD, Hill MN, Hunter RG, Karatsoreos IN, Nasca C. Mechanisms of stress in the brain. Nature neuroscience. 2015;18:1353–1363. doi: 10.1038/nn.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature Neuroscience. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor K, Labbe A, Greenwood CM. Response to: Correcting for cell-type effects in DNA methylation studies: reference-based method outperforms latent variable approaches in empirical studies. Genome biology. 2017;18:25. doi: 10.1186/s13059-017-1149-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ. Epigenetics and the biological definition of gene x environment interactions. Child Development. 2010;81:41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Melas PA, Wei Y, Wong CC, Sjoholm LK, Aberg E, Mill J, et al. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. International Journal of Neuropsychopharmacology. 2013;16:1513–1528. doi: 10.1017/S1461145713000102. [DOI] [PubMed] [Google Scholar]
- Mersha TB, Abebe T. Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Human Genomics. 2015;9:1. doi: 10.1186/s40246-014-0023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan CJ, D’Errico NC, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7:853–857. doi: 10.4161/epi.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén LK, Muthén BO. Mplus user’s guide. 6. Los Angeles: Muthén & Muthén; 1998–2012. [Google Scholar]
- Na KS, Chang HS, Won E, Han KM, Choi S, Tae WS, et al. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One. 2014;9:e85425. doi: 10.1371/journal.pone.0085425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RE, Byambaa M, De R, Butchart A, Scott J, Vos T. The long-term health consequences of child physical abuse, emotional abuse, and neglect: A systematic review and meta-analysis. PLoS Medicine. 2012;9:e1001349. doi: 10.1371/journal.pmed.1001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Ostlund BD, Conradt E, Crowell SE, Tyrka AR, Marsit CJ, Lester BM. Prenatal stress, fearfulness, and the epigenome: exploratory analysis of sex differences in dna methylation of the glucocorticoid receptor gene. Frontiers in behavioral neuroscience. 2016:10. doi: 10.3389/fnbeh.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Gudiel H, Cordova-Palomera A, Leza JC, Fananas L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: A critical review. Neuroscience & Biobehavioral Reviews. 2015;55:520–535. doi: 10.1016/j.neubiorev.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Parade SH, Ridout KK, Seifer R, Armstrong DA, Marsit CJ, McWilliams MA, et al. Methylation of the glucocorticoid receptor gene promoter in preschoolers: Links with internalizing behavior problems. Child Development. 2016;87:86–97. doi: 10.1111/cdev.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N, Paoloni-Giacobino A, Prada P, Olie E, Salzmann A, Nicastro R, et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Translational Psychiatry. 2011;1:e59. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, … Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Roberts S, Keers R, Lester KJ, Coleman JR, Breen G, Arendt K, … Havik OE. HPA axis related genes and response to psychological therapies: Genetics and epigenetics. Depression and anxiety. 2015;32:861–870. doi: 10.1002/da.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romens SE, McDonald J, Svaren J, Pollak SD. Associations between early life stress and gene methylation in children. Child Development. 2015;86:303–309. doi: 10.1111/cdev.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, … Kenis G. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Molecular Psychiatry. 2017 doi: 10.1038/mp.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeringa MS, Haslett N. The reliability and criterion validity of the Diagnostic Infant and Preschool Assessment: a new diagnostic instrument for young children. Child Psychiatry and Human Development. 2010;41:299–312. doi: 10.1007/s10578-009-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonkoff JP, Boyce WT, McEwen BS. Neuroscience, molecular biology, and the childhood roots of health disparities building a new framework for health promotion and disease prevention. JAMA. 2009;301:2252–2259. doi: 10.1001/jama.2009.754. [DOI] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Klengel T, Mercer KB, Bradley B, Conneely KN, … Binder EB. DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2015;168:36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud LR, Papandonatos GD, Rodriguez D, McCallum M, Salisbury AL, Phipps MG, et al. Maternal smoking during pregnancy and infant stress response: Test of a prenatal programming hypothesis. Psychoneuroendocrinology. 2014;48:29–40. doi: 10.1016/j.psyneuen.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. The genome- and system-wide response of DNA methylation to early life adversity and its implication on mental health. Canadian Journal of Psychiatry. 2013;58:697–704. doi: 10.1177/070674371305801208. [DOI] [PubMed] [Google Scholar]
- Turecki G, Meaney MJ. Effects of the social environment and stress on glucocorticoid receptor gene methylation: A systematic review. Biological Psychiatry. 2016;79:87–96. doi: 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Burgers DE, Philip NS, Price LH, Carpenter LL. The neurobiological correlates of childhood adversity and implications for treatment. Acta Psychiatrica Scandinavica. 2013;128:434–447. doi: 10.1111/acps.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Parade SH, Eslinger NM, Marsit CJ, Lesseur C, Armstrong DA, et al. Methylation of exons 1D, 1F, and 1H of the glucocorticoid receptor gene promoter and exposure to adversity in pre-school aged children. Development and Psychopathology. 2015;27:577–585. doi: 10.1017/S0954579415000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Parade SH, Welch ES, Ridout KK, Price LH, Marsit C, et al. Methylation of the leukocyte glucocorticoid receptor gene promotor in adults: associations with early adversity and depressive, anxiety and substance-use disorders. Translational Psychiatry. 2016;6:e848. doi: 10.1038/tp.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Price LH, Marsit C, Walters OC, Carpenter LL. Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PLoS One. 2012;7:e30148. doi: 10.1371/journal.pone.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Ridout KK, Parade SH. Childhood adversity and epigenetic regulation of glucocorticoid signaling genes: Associations in children and adults. Development and psychopathology. 2016;28:1319–1331. doi: 10.1017/S0954579416000870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap LJ, Riese H, Hudziak JJ, Verbiest MMPJ, Verhulst FC, Oldehinkel AJ, et al. Glucocorticoid receptor gene (NR3C1) methylation following stressful events between birth and adolescence. The TRAILS study. Translational Psychiatry. 2014;4:e381. doi: 10.1038/tp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap LJ, van Oort FV, Verhulst FC, Oldehinkel AJ, Riese H. Methylation of NR3C1 and SLC6A4 and internalizing problems. The TRAILS study. Journal of Affective Disorders. 2015;180:97–103. doi: 10.1016/j.jad.2015.03.056. [DOI] [PubMed] [Google Scholar]
- Vinkers CH, Kalafateli AL, Rutten BP, Kas MJ, Kaminsky Z, Turner JD, et al. Traumatic stress and human DNA methylation: a critical review. Epigenomics. 2015;7:593–608. doi: 10.2217/epi.15.11. [DOI] [PubMed] [Google Scholar]
- Vukojevic V, Kolassa IT, Fastenrath M, Gschwind L, Spalek K, Milnik A, et al. Epigenetic modification of the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. Journal of Neuroscience. 2014;34:10274–10284. doi: 10.1523/JNEUROSCI.1526-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A, … O’Loughlin K. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. Journal of the American Academy of Child & Adolescent Psychiatry. 2014;53:417–424. doi: 10.1016/j.jaac.2013.12.025. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzmann SR, Turner JD, Meriaux SB, Meijer OC, Muller CP. Epigenetic regulation of the glucocorticoid receptor promoter 17 in adult rats. Epigenetics. 2012;7:1290–1301. doi: 10.4161/epi.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CCY, Caspi A, Williams B, Craig IW, Houts R, Ambler A, … Mill J. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–526. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F, … Meaney MJ. Lower methylation of glucocorticoid receptor gene promoter 1 F in peripheral blood of veterans with posttraumatic stress disorder. Biological psychiatry. 2015;77:356–364. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Desarnaud F, Makotkine I, Lehrner AL, Koch E, … Bierer LM. Epigenetic biomarkers as predictors and correlates of symptom improvement following psychotherapy in combat veterans with PTSD. Frontiers in psychiatry. 2013;4:118. doi: 10.3389/fpsyt.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]