Abstract
Optogenetic control of individual neurons with high temporal precision, within intact mammalian brain circuitry, would enable powerful explorations of how neural circuits operate. Two-photon computer generated holography enables precise sculpting of light, and could in principle enable simultaneous illumination of many neurons in a network, with the requisite temporal precision to simulate accurate neural codes. We designed a high efficacy soma-targeted opsin, finding that fusing the N-terminal 150 residues of kainate receptor subunit 2 (KA2) to the recently discovered high-photocurrent channelrhodopsin CoChR restricted expression of this opsin primarily to the cell body of mammalian cortical neurons. In combination with two-photon holographic stimulation, we found that this somatic CoChR (soCoChR) enabled photostimulation of individual cells in intact cortical circuits with single cell resolution and <1 millisecond temporal precision, and use soCoChR to perform connectivity mapping on intact cortical circuits.
Introduction
Optogenetics is in widespread use for the activation and silencing of populations of neurons defined by their molecular profiles, activity patterns, or projection patterns1. Natural neural codes, however, vary from neuron to neuron even within a single class of cells, with neighboring neurons of the same type exhibiting potentially highly differing neural codes2, and the firing of even single neurons capable of altering mammalian behavior or brain state3,4. Thus, an intriguing question is whether it is possible to drive individual neurons, or sets of neurons, with neuron-specific, distributed activity patterns exhibiting the temporal precision requisite to mimic natural neural codes.
Recently, interest has arisen around the possibility of two-photon (2P) optogenetics, which due to the quadratic dependence of 2P excitation on light intensity, should enable light to be targeted to single neurons. 2P activation of opsins is robust at a biophysical level, with an excellent cross section for opsin photoactivation5, but a challenge is to generate enough photo-evoked current using the micrometer-sized excitation volume of conventional 2P microscopy. Two primary strategies have been developed in order to increase the 2P illumination volume while keeping micrometer axial resolution: scanning and parallel. Scanning a laser spot, axially elongated to cover cell membranes, around a circular area with approximately the diameter of a cell body enables sequential excitation of channels, each contributing to the total induced photocurrent5. Scanning approaches, however, have limited temporal resolution because of the time needed to sequentially scan the laser through multiple positions to cover a single cell body or multiple cells6
Alternatively, parallel stimulation approaches such as generalized phase contrast7 or computer generated holography (CGH)8 use phase modulation to precisely sculpt the illumination volume and simultaneously illuminate an entire cell or even multiple cells. In this case, simultaneous illumination of all channels enables efficient integration of light evoked currents allowing minimization of illumination time. In this condition millisecond temporal resolution7,9 and < 1 ms temporal jitter10,11 action potential (AP) generation can be achieved. Combined with temporal focusing, parallel stimulation also enables micrometer axial resolution and good preservation of axial resolution and spot shape after hundreds of µms of propagation in scattering tissues9,12,13. CGH also enables 3D light generation12,14–16, laying the groundwork for generating complex patterns with single cell spatial resolution and millisecond temporal precision.
Advances in 2P stimulation methods, however, do not directly address the problem of cell bodies being densely surrounded by neurites of neighboring cells, whose membranes are separated by very thin gaps of extracellular space, smaller than the diffraction limit of light17. Thus, if neurons within a region are bearing densely expressed opsins, even 2P stimulation of a single neuron’s cell body may excite opsins on dendrites or axons that are passing by, causing stray excitation of those nearby neurons. To address this crosstalk problem (Fig. 1a), we launched a screen to discover peptide sequences that could facilitate the localization of a high-photocurrent opsin to the cell body. We found that a short amino terminal segment of the kainate receptor KA2 subunit18, fused to the high-photocurrent channelrhodopsin CoChR19, could enable the selective trafficking of CoChR to the cell body of neurons in the mammalian cortex (Fig. 1b), and in combination with holographic 2P stimulation with a low repetition fiber laser, could support optogenetic stimulation of single cells in mammalian brain slices, with < 1 millisecond temporal resolution, and with zero-spike crosstalk activation of nearby cells, using low light powers. This fusion protein, which we call somatic CoChR (soCoChR), used in conjunction with optimized 2P stimulation approaches, may enable a diverse set of neural codes and computations to be probed in a causal fashion in systems and circuit neuroscience.
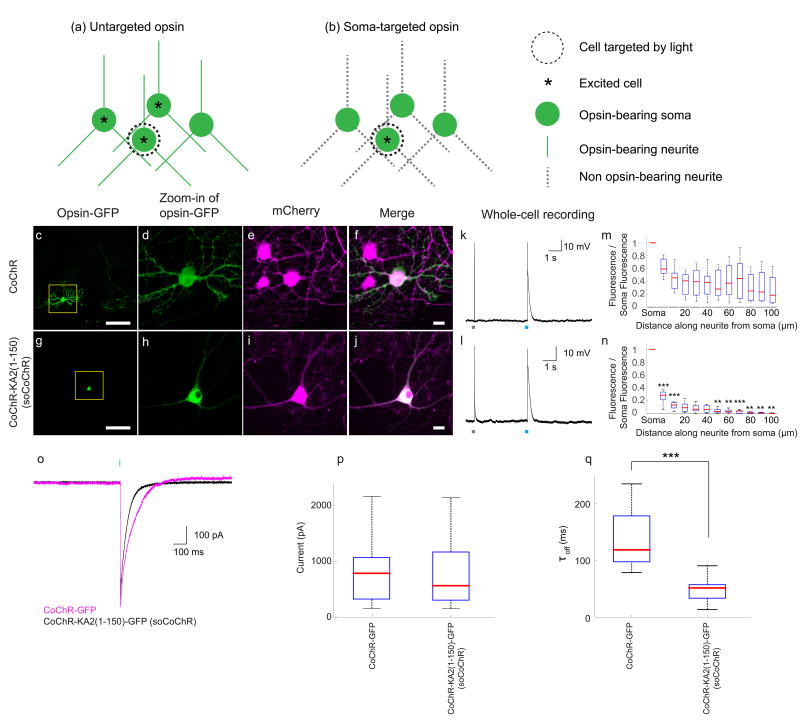
Figure 1. Soma-targeted optogenetics using the high-performance channelrhodopsin CoChR.
(a, b) Concept of soma-targeted optogenetics. Untargeted opsins (a) express over the entire neural membrane. One can aim light at a given neural soma, but each cell body is surrounded by opsin-bearing neurites from other cells, resulting in artifactual activation of those cells. Restricting opsin expression to the cell body would prevent such side effects, enabling single cell optogenetic stimulation (b). (c–j) Images are presented for cultured hippocampal neurons expressing wild-type vs. selectively trafficked CoChR fused to GFP, along with the cytosolic countermarker mCherry. (c) A hippocampal neuron in culture expressing CoChR-GFP and mCherry, seen in the GFP channel (scale bar: 100 µm). (d) Zoomed-in image from the yellow rectangle of c. (e) The neuron of c, seen in the mCherry channel (magenta), along with surrounding neurons. (f) Merge of d and e (scale bar for d–f: 20 µm). (g–j) As in c–f, but for a neuron expressing CoChR-KA2(1–150)-GFP (soma-targeted CoChR-GFP, abbreviated soCoChR-GFP). (k) Whole cell current clamp recording of a cultured hippocampal neuron expressing CoChR-GFP, under current injection (10 ms duration; gray rectangle) and optical stimulation (480nm, 34.84mW/mm2, 1 ms duration; blue rectangle). Rectangles not to scale. (l) As in k, but for a neuron expressing CoChR-KA2(1–150)-GFP (soCoChR-GFP). (m) Box-and-whiskers plot of GFP brightness versus position along a neurite, normalized to GFP brightness at the soma, extracted from neurites of cultured hippocampal neurons expressing CoChR-GFP (n = 7 neurites taken from 5 cells from 2 cultures). Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values respectively. (n) As in m, but for neurons expressing CoChR-KA2(1–150)-GFP (soCoChR-GFP; n = 9 neurites taken from 7 cells from 3 cultures). **P < 0.01 and ***P < 0.001, Bonferroni-corrected Kolmogorov-Smirnov test of brightness between wild-type and soma-targeted; see Supplementary Table 3 for full statistics for Figure 1). (o) Representative photocurrents for cultured hippocampal neurons expressing CoChR-GFP (magenta) and CoChR-KA2(1–150)-GFP (soCoChR-GFP; black), under voltage clamp conditions, using 5 ms light pulses (480nm, 34.84 mW/mm2; light blue rectangle). (p) Box-and-whiskers plot showing peak current amplitudes (pA) for CoChR-GFP and CoChR-KA2(1–150)-GFP (soCoChR-GFP) in cultured hippocampal neurons illuminated with blue light (480 nm, 34.84 mW/mW2, 5 ms; n = 13 neurons from 3 cultures and 13 neurons from 3 cultures for CoChR-GFP and CoChR-KA2(1–150)-GFP respectively). Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values respectively. Not significant; Wilcoxon rank sum test; see Supplementary Table 3 for full statistics. (q) Box-and-whiskers plot showing photocurrent decay time (τoff, in ms) for CoChR-GFP and CoChR-KA2(1–150)-GFP (soCoChR-GFP) in cultured hippocampal neurons illuminated with blue light (480 nm, 34.84 mW/mW2, 5 ms; n = 13 neurons from 3 cultures and 13 neurons from 3 cultures for CoChR-GFP and CoChR-KA2(1–150)-GFP respectively). Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values respectively. ***P < 0.0001; Wilcoxon rank sum test; see Supplementary Table 3 for full statistics.
Results
Creation of a high performance somatic opsin
We screened for soma-targeting sequences that could localize opsins to neuronal cell bodies. We first searched the literature for proteins that express somatically. We chose 9 such proteins (summarized, with further info added as described below, in Supplementary Table 1) for consideration: myelin proteolipid proteins srPLP and DM2020, the potassium channel KV2.121, sodium channels NaV1.2 and NaV1.622, the adhesion molecule L1 with the soma-retention causing mutation R184Q23, the dynein adaptor protein Bicaudal-D (BicD) truncated after 50 codons (out of 782; this truncation impairs transport of FMRP out of the soma)24,25, the adaptor protein AnkyrinG26, and the kainate receptor subunit KA218.
In some studies, neuron somatic localization had been explored further by fusing the proteins to reporters – namely, NaV1.2, NaV1.6, L1-R184Q and AnkyrinG were fused to fluorescent proteins (FPs)22,23,26, KA2 to a myc-tag18, and KV2.1 to an HA-tag21. For some of the above soma-restricting proteins, fragments were shown to be sufficient to cause soma targeting of a reporter (Supplementary Table 1). For each of NaV1.2 and NaV1.6, a 27–amino acid segment within the intracellular loop between transmembrane domains II and III, termed NaV1.2(II-III) and NaV1.6(II-III) respectively, was sufficient22; for KV2.1 a 65 amino acid segment at the C-terminus sufficed21,27; for AnkyrinG the N-terminal 837 amino acid segment of the protein sufficed28; and for BicD the N-terminal 50 amino-acid segment located at locus r5 in the Drosophila BiCD gene sufficed25.
We fused GFP to full length clones of the 4 soma-targeting proteins described above for which no sufficient fragment was reported (srPLP, DM20, L1-R84Q, and KA2), as well as to the reported fragments for the other proteins (Supplementary Table 1). We transfected these GFP fusions into cultured hippocampal neurons, and visually observed that 2 of the 9 sequences tested appeared to target GFP primarily to the cell body (Supplementary Table 1).
We chose two proteins for further consideration, KA2 and NaV1.2(II/III)-GFP. Since KA2 is a 979 amino acid protein, and thus unwieldy from a viral packaging standpoint, we divided KA2 into three parts. This resulted in fragments (listed from N terminal to C terminal) of length 360 amino acids (containing one transmembrane domain29), 360 amino acids (containing three transmembrane domains), and 259 amino acids (containing one transmembrane domain and an arginine-rich ER retention sequence30). GFP fused to the first fragment was somatic, but GFP fused to the latter two were not (Supplementary Table 1). We tried the N-terminal 150 amino acids of fragment 1, which we termed KA2(1–150), and found that this targeted GFP to the cell body (see Supplementary Fig. S1 for comparison of the fluorescence between KA2-GFP, KA2(1–150)-GFP and GFP and Supplementary Table 2 for statistical analysis of this comparison), but KA2(1–75) did not. As for NaV1.2(II/III), an earlier study using this to target ChR2-YFP31 to the axon hillock of neurons revealed photocurrents smaller than those of ChR2-YFP31, as well as alterations of neuron excitability32, so we retained this motif for comparison purposes (Supplementary Fig. S2 for images of CoChR-NaV1.2(II/III)-GFP and Supplementary Table 3 for statistics), but did not pursue it further.
Key to efficient use of 2P light power for multicell activation is the usage of an opsin with high currents. Accordingly, we examined opsins that exhibited very high photocurrents but whose performance in intact mammalian brain circuitry had not yet been explored. We fused KA2(1–150) to the C-terminus of CoChR, a powerful opsin with perhaps the largest photocurrent described to date19, at least when assessed in cultured neurons. We examined the resulting localization in cultured neurons, finding that unlike wild-type CoChR-GFP (Fig. 1c–f), CoChR-KA2(1–150)-GFP (called soma-targeted CoChR or soCoChR for short) appeared primarily at and near the cell body (Fig. 1g–j). Quantitatively, at 50 µm from the edge of the cell body, CoChR-KA2(1–150)-GFP fluorescence was 4.67±1.29% of the average brightness across the cell body, in contrast to 35.56 ± 8.33% for the wild-type opsin (mean ± standard error; n = 9 neurites taken from 7 cells from 3 cultures for the KA2 fusion, n = 7 neurites taken from 5 cells from 2 cultures for the wild-type, Fig. 1m,n). We found that for most distances between 0 to 100 µm from the edge of the cell body, in CoChR-KA2(1–150)-GFP expressing cells the neurite fluorescence was significantly lower than in CoChR-GFP expressing cells (n = 9 neurites from 7 cells from 3 cultures for CoChR-KA2(1–150)-GFP; n = 7 neurites from 5 cells from 2 cultures for CoChR-GFP; see Supplementary Table 3 for Bonferroni-corrected Kolmogorov-Smirnov tests run for each bin).
As for Kv2.1, for which a motif has been previously used by multiple groups to target ChR2 to the cell body27,33, we found that the expression of CoChR-GFP-Kv2.1motif was not purely at the cell body (Supplementary Figure S3). Quantitatively, at 50µm from the edge of the cell body, CoChR-GFP-Kv2.1motif fluorescence was 33.60 +/− 4.75% of the average brightness across the cell body, whereas this ratio was 35.56 ± 8.33% for wild-type CoChR (mean ± standard error; n = 5 neurites taken from 5 cells from 2 cultures for the Kv2.1motif fusion, n = 7 neurites taken from 5 cells from 2 cultures for the wild-type, Supplementary Figure S3). Statistically, the brightness difference between CoChR-GFP and CoChR-GFP-Kv2.1 was not significant along neurites (n = 5 neurites from 3 cells from 2 cultures for CoChR-GFP-Kv2.1motif; n = 7 neurites from 5 cells from 2 cultures for CoChR-GFP; see Supplementary Table 3 for Bonferroni-corrected Kolmogorov-Smirnov tests run for each bin). Thus we chose not to further pursue CoChR-GFP-Kv2.1motif.
CoChR- GFP and CoChR-KA2(1–150)-GFP could both mediate action potentials in expressing neurons in response to blue light pulses (Fig. 1k,l). Photocurrent measurements (Fig. 1o,p; 5 ms light pulses at 480nm, 34.84 mW/mm2) revealed that CoChR-GFP and CoChR-KA2(1–150)-GFP had average photocurrents of 1029 ± 217 pA and 720pA ± 156 pA respectively (mean ± standard error of the mean, n = 13 and 13 cells each from 3 and 3 cultures for CoChR-GFP and CoChR-KA2(1–150)-GFP respectively; see Supplementary Table 3 for full statistics). As previously reported19, CoChR-GFP under 1P widefield activation has a long photocurrent decay time (Fig. 1q; τoff = 150 ± 18 ms, value is mean ± standard error of the mean). This value was shorter for CoChR-KA2(1–150)-GFP (52 ± 6 ms; mean ± standard error of the mean; n = 13 neurons from 3 cultures, 13 neurons from 3 cultures for CoChR-GFP and CoChR-KA2(1–150)-GFP respectively; Fig. 1q; see Supplementary Table 3 for full statistics), perhaps because the patch pipette is measuring just the currents near the cell body, and not the currents propagating in from distant neurites that would be driven when a wild-type opsin is used.
Using CoChR- and soCoChR-expressing cultured neurons, we measured the action spectra of these molecules (Supplementary Fig. S4a), the spike probability (Supplementary Fig. S4b) and photocurrent (Supplementary Fig. S4c) mediated by these molecules as a function of irradiance, and the spike fidelity mediated by these molecules as a function of the stimulation frequency (Supplementary Fig. S4d). We also examined basic cell health parameters. The passive membrane properties for neurons containing CoChR-GFP (n = 10 cells from 3 cultures) vs. CoChR-KA2(1–150)-GFP (n = 10 cells from 3 cultures) were not significantly different (P > 0.05, Kruskal-Wallis Test for these membrane properties: resting potential, membrane capacitance, holding current and membrane resistance; see Supplementary Fig. S5 for comparison between soma-targeted vs. non-targeted CoChR).
Somatic CoChR enables single cell optogenetic control using 1P photostimulation
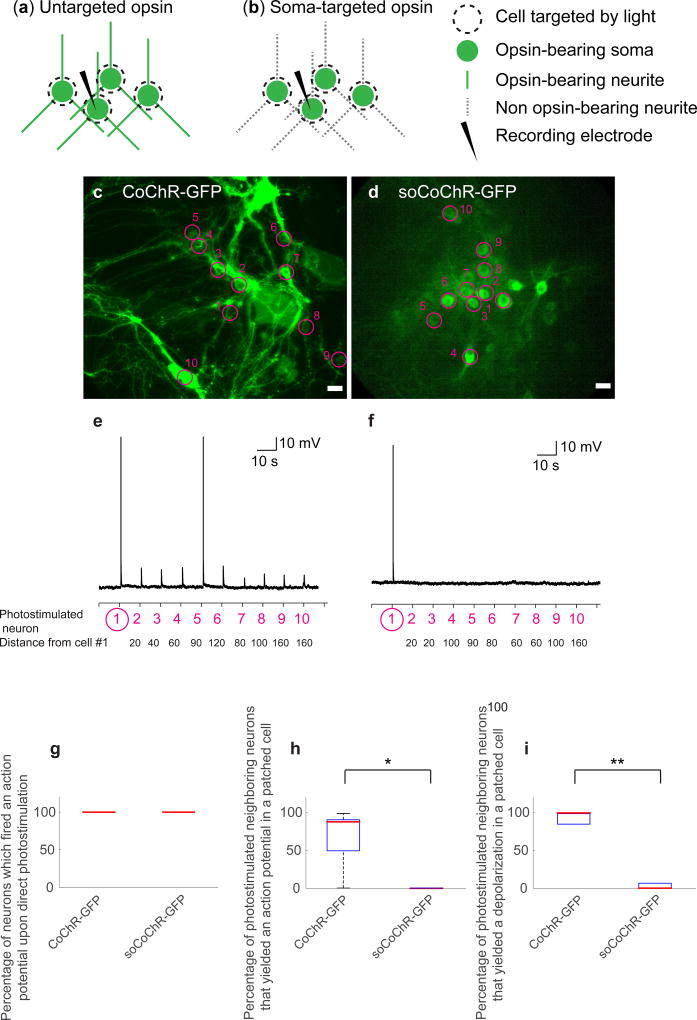
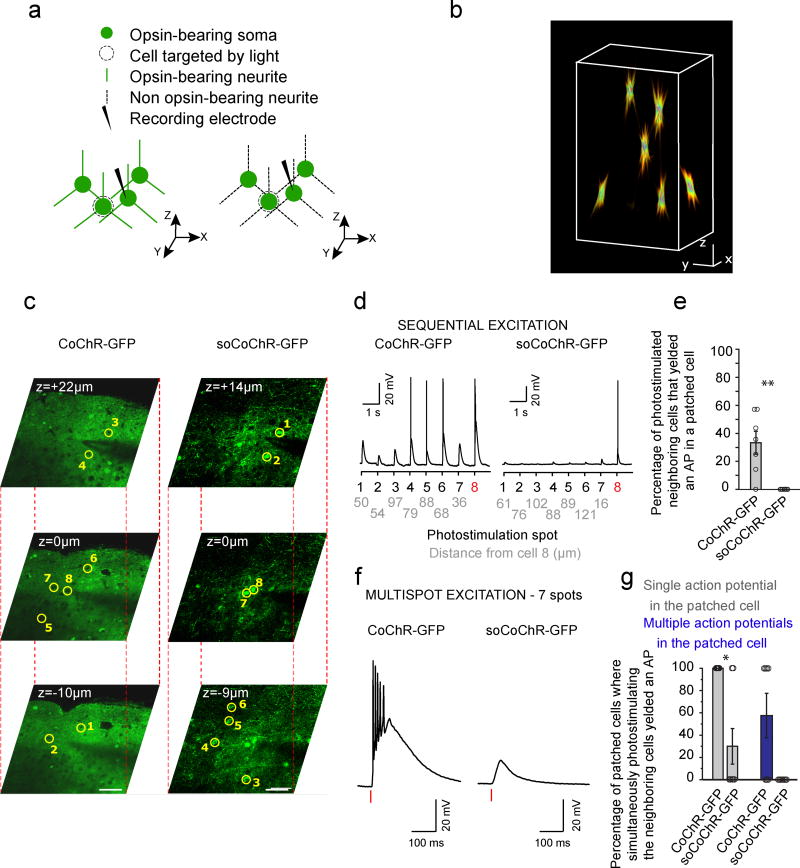
To test whether soCoChR-GFP could mediate single-photon (1P) stimulation of cultured neurons without stimulating nearby cells, we patched a single cell under widefield fluorescence microscopy, and used a digital micromirror device (DMD) to photo-stimulate the patched cell and its neighbors with 20 microns diameter spots (Fig. 2a,b for schematic of the experiment), under synaptic blockade. We found that for light of this power, and for both CoChR-GFP or soCoChR-GFP expressing cells (n = 5 for both opsins; average distance of photostimulated cell from patched cell = 76.89 ± 6.98 µm and 74.22 ± 6.50 µm respectively; Fig. 2c,d for images of opsin-expressing neurons; see also Supplementary Table 3), illuminating the cell body of the patched cell always yielded an AP in that cell. Furthermore, while patching a CoChR-GFP expressing cell and photostimulating the somata of neighboring non-patched CoChR-GFP expressing cells, we recorded an AP when photostimulating 62.0 ± 16.3% of the neighboring cells (Fig. 2e,h; mean ± s.e.m.; n = 5 patched cells from 4 cultures); in contrast, while patching a soCoChR cell, photostimulating the neighboring cells' somata never resulted in an AP in the patched cell (P < 0.01, Wilcoxon rank sum test; Fig. 2f–h; n = 5 patched cells from 5 cultures). Analyzing the data further, we found depolarizations of > 0.5 mV above baseline (defined as the average voltage over the 100 ms preceding the voltage change, see Online Methods) in patched cells within CoChR-GFP-expressing networks when 96.0 ± 2.4% of the nearby cells were stimulated (Fig. 2e,i; mean ± s.e.m.), in contrast to 4.0 ± 4.0% of the nearby-cell stimulations for soCoChR-expressing neurons (P < 0.001, Wilcoxon rank sum test; Fig. 2f,i; mean ± s.e.m.).
Figure 2. Zero-spike crosstalk single cell optogenetic control of cultured neurons with one-photon illumination.
(a,b) Schematic of the experiment. Cultured hippocampal neurons two weeks after transduction with AAV8-Syn-CoChR-GFP (untargeted CoChR, a) or AAV8-Syn-soCoChR-GFP (somatic CoChR, b). One cell in the field of view was patched (current clamp). The patched cell along with 9 neighboring cells were photo-stimulated using a digital micromirror device (DMD) consecutively. (c) Cultured hippocampal neurons expressing CoChR-GFP; regions of stimulation are highlighted by magenta circles. Numbers denote the order of stimulation. Scale bar: 20 µm. (d) As in c, but for soCoChR-GFP. (e) Representative current-clamp recording for a cultured hippocampal neuron expressing CoChR-GFP. Cells were photostimulated sequentially using the DMD (470 nm, 40.7 mW/mm2, 1 ms light pulse). The order of the photo-stimulation, as indicated by the numbers in c, is given on the x-axis. The patched cell is numbered 1. The distance of cells 2–9 from the patched cell is given below each cell number, in microns. (f) As in e, but for soCoChR-GFP. (g) Box-and-whiskers plot showing the percentage of cultured hippocampal neurons which fired an action potential (AP) upon direct photostimulation, using light pulses as above (n = 5 cells from 4 cultures for CoChR-GFP; n = 5 cells from 5 cultures for soCoChR-GFP). Red line denotes the median. (h) Box-and-whiskers plot showing, averaged across all patched neurons, the percentage of photostimulated neighboring cells that yielded an AP in each patched cell (n = 5 cells from 4 cultures for CoChR-GFP; n = 5 cells from 5 cultures for soCoChR-GFP). Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values respectively. *P=0.0476, Wilcoxon rank sum test; Supplementary Table 3 for full statistics. (i) Box-and-whiskers plot showing, averaged across all patched neurons, the percentage of photostimulated neighboring cells that yielded a depolarization in each patched cell (n = 5 cells, in 4 cultures for CoChR-GFP; n = 5 cells, in 5 cultures for soCoChR-GFP). Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. **P=0.0079, Wilcoxon rank sum test; Supplementary Table 3 for full statistics.
Two-photon holographic control of soCoChR-expressing neurons in brain slice
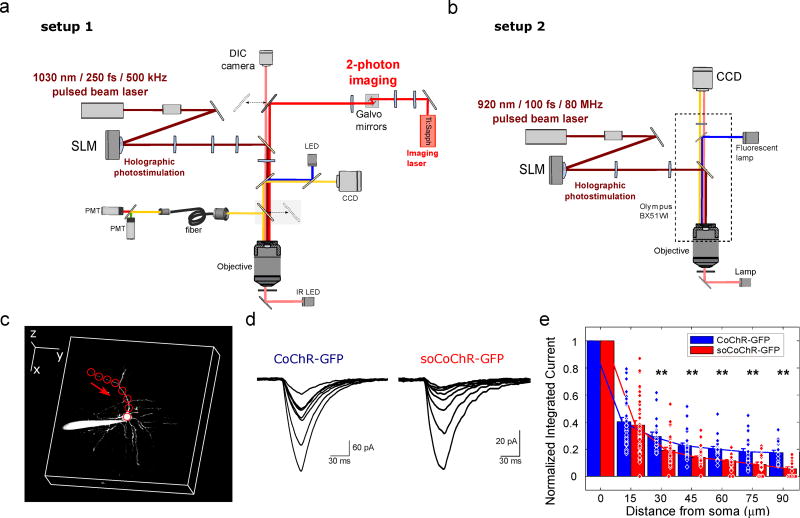
We next sought to assess the potential for soCoChR to mediate single cell optogenetic control in intact mouse brain slices. The strong scattering undergone by visible light in tissue will induce a broadening of the excitation spot and therefore spurious activation of distant neurons by both out of focus and scattered photons34,35. To avoid this possibility, we used 2P activation, and to optimize temporal resolution we used parallel holographic light illumination. We implemented holographic 2P activation in two different microscopes (Fig. 3a,b and Supplementary Fig. S6). In setup 1, a custom made 2P scanning system was combined with a 2P CGH setup, using a low repetition amplified fiber laser (λ = 1030 nm; exit average power = 10 W; repetition rate = 500 kHz; pulse duration = 250 fs) as excitation source. In setup 2, a 1P epifluorescence microscope was combined with a 2P CGH system using as the excitation source a conventional mode locked Ti:Sapphire fs laser (λ = 920 nm; exit average power = 1.6 W; repetition rate = 80 MHz; pulse duration = 100 fs). Setup 1 was designed to achieve simultaneous activation of multiple cells, since the high output power and low repetition rate of the laser source enabled parallel illumination with high-energy pulses, while keeping average power low. Setup 2, in contrast, had lower output power and used conventional and commonly available laser sources. The tunable laser source used in setup 2 enabled us to tune the excitation wavelength to the peak of the 2P spectrum (Supplementary Fig. S7), while the laser source used in setup 1 has a fixed excitation wavelength at 1030 nm, which corresponds to ~60% of the excitation peak (Supplementary Fig. S7).
Figure 3. Photocurrents of neurites expressing soCoChR-GFP are significantly smaller than in neurites expressing CoChR-GFP, in neurons virally expressing these opsins in mouse cortical brain slice.
(a–b) Schematic representation of the two experimental setups used for holographic illumination. In setup 1 (a), holographic photostimulation was achieved using an amplified fiber laser and was coupled with a two-photon (2P) scanning imaging system. In setup 2 (b) holographic photostimulation was achieved using a conventional Ti:Sapphire pulsed laser and was coupled with a widefield epifluorescence imaging system. Detailed descriptions of the setups are reported in Methods section and Supplementary Figure 6. (c) Schematic of 2P holographic stimulation along one neurite, showing Alexa 594 fluorescence (obtained via 2P scanning at 780 nm) from dye injected into a patched CoChR-GFP expressing neuron. The Alexa 594 fluorescence was used to guide holographic spot placement (red circles) to different points along a neurite, at different distances from the soma. Red arrow indicates the order of photostimulation. The bright emission to the left of the cell represents the patch pipette filled with Alexa 594 (scale bars: 50 µm along all three axes, image acquired with setup 1). (d) Representative whole-cell currents recorded from a CoChR-GFP expressing neuron (left) and a soCoChR-GFP expressing neuron (right; some 50 Hz electrical noise is apparent in the traces on the right), when illuminated with a power density corresponding to the spiking threshold power density (18 µW/µm2 and 101 µW/µm2 for the CoChR-GFP and the soCoChR-GFP expressing cells respectively; λ = 1030 nm, using setup 1). (e) Bar plot of the integral of the elicited photocurrent, normalized to that obtained with the spot at the soma, as a function of distance from the soma, for CoChR-GFP (blue bars) and soCoChR-GFP (red bars) expressing neurons (spacing between spots ~10 µm, λ = 1030 nm or 920 nm for setup 1 or setup 2, respectively; data were pooled across both setups). Bars reports mean ± s.e.m. Dots denote values for single neurites. For each cell, the photostimulation was done at the power density threshold determined for that cell (average powers: 29±10 µW/µm2 for CoChR-GFP expressing cells; 90±60 µW/µm2 for soCoChR-GFP expressing cells; power values referred to setup 1, see Methods). The normalized current integral was significantly higher in CoChR-GFP relative to soCoChR-GFP expressing cells for distances of 30 µm or more from the soma. ** P<0.0017 for distances ≥ 30 µm; Kolmogorov-Smirnov (KS) test with Bonferroni correction (n=16 neurites from 8 CoChR-GFP cells from 7 mice; n=27 neurites from 16 soCoChR-GFP cells from 13 mice; see Supplementary Table 4 for full statistics for Fig. 3). Data obtained using both setup 1 and setup 2.
We quantified the spatial confinement of soCoChR vs CoChR by measuring the photostimulated current integral while steering a 10 µm diameter holographic spot away from the soma of a patched neuron in ~10 µm steps along a neurite of a mouse cortical brain slice, with synaptic receptor blockers (NBQX, gabazine, strychnine, APV) applied (Fig. 3c,d). 2P photo-stimulation power density was set, for each cell, at the minimum value necessary to reliably (in all three of three consecutive trials) trigger one AP using a 30 ms photo-stimulation pulse illuminating the whole soma (threshold power for CoChR-GFP: 28 ± 10 µW/µm2 and 92 ± 93 using setups 1 and 2 respectively, n=7 cells from 5 mice in setup 1 and n=10 cells from 8 mice in setup 2; threshold power for soCoChR-GFP: 83 ± 39 µW/µm2 and 261 ± 190 µW/µm2 using setups 1 and 2 respectively, n=4 cells from 4 mice in setup 1 and n=9 cells from 6 mice in setup 2). The difference in power required for the two opsins is possibly due to a difference in expression efficiency for the two opsins in vivo. We cannot exclude that with the axial resolution used in the experiment (20–40 µm; Supplementary Fig. S8) the surrounding neurites contribute to the observed current, this effect being more pronounced for the non-soma targeted opsin. For each illumination position along each neurite, we normalized the measured current integral to the corresponding value measured at the soma (Fig. 3e). The normalized current integral decayed more sharply along neurites expressing soCoChR-GFP than along neurites expressing CoChR-GFP (P < 0.05 at sites 30 µm from soma and beyond; P < 0.01 at sites 45 µm from the soma and beyond; n = 16 neurites from 8 CoChR-GFP cells from 7 mice; n = 27 neurites from 16 soCoChR-GFP cells from 13 mice). We note that the sharper current decay was observed even though for soCoChR-GFP we used higher photostimulation powers than for CoChR-GFP. Absolute values of currents corresponding to Fig. 3e are reported and analyzed in Supplementary Fig. S10 and Supplementary Table 4 (the decay of the fluorescence reporter (GFP) along the neurites is shown in Supplementary Fig. S9). Thus, photo-evoked currents fall off more rapidly down soCoChR-bearing neurites than those bearing CoChR, a crucial condition to support neuronal stimulation with single cell resolution.
Millisecond-timescale activation of neurons in brain slices
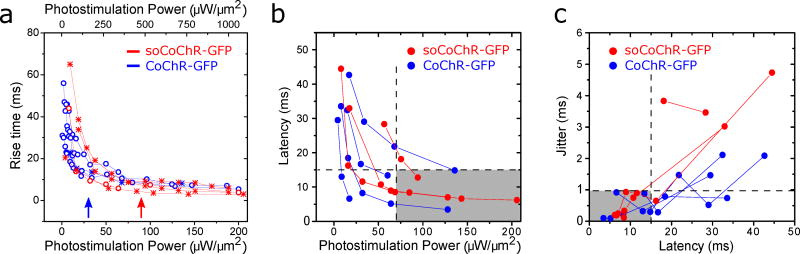
We next searched for illumination conditions that enabled the triggering of APs with <1 millisecond temporal jitter in cells expressing soCoChR-GFP vs CoChR-GFP. We measured the rise time (τon) and decay time (τoff) of photocurrents generated by a holographic spot covering the cell body of an opsin-expressing neuron while we increased illumination power. For both CoChR-GFP and soCoChR-GFP expressing neurons, τon decreased with increasing illumination power (Fig. 4a), reaching an asymptotic value (see methods) of 5.7 ± 2.6 ms in CoChR-GFP cells (n = 4 cells from 4 mice) and 4.1 ± 2.5 ms in soCoChR-GFP cells (n = 4 cells from 4 mice). τoff values were independent of the illumination power and equal to 37±13 ms in CoChR-GFP expressing cells (n=4 cells out of 4 mice) and 33.3±5.6 ms in soCoChR-GFP expressing cells (n = 4 cells out of 4 mice). Notably, these numbers were similar, in contrast to what was seen for 1P widefield illumination in cultured neurons (Fig. 1q), suggesting that restricting light illumination to the soma effectively equalized the decay times of photo-evoked currents for targeted vs non-targeted opsins.
Figure 4. 2P power necessary to enable millisecond control of cortical neuron activation in mouse brain slice.
(a) Rise time of soCoChR-GFP (red) and CoChR-GFP (blue)-mediated photocurrents, measured in mouse brain slice cortical neurons, as a function of 2P stimulation power (n = 4–12 photostimulation powers per cell, 6 cells from 6 mice for CoChR-GFP; n = 2–11 photostimulation powers per cell, for 5 cells from 5 mice for soCoChR-GFP). Data collected from setup 1 are plotted with circles, and correspond to the power scale on the bottom x-axis; data collected from setup 2 are plotted with asterisks, and correspond to the top x-axis. The length of the top x-axis was rescaled with respect to the bottom x-axis by a power conversion factor k = 5.3 (see Methods). Lines connect data acquired from the same neuron. Blue and red arrows indicate average threshold powers relative to setup 1 for CoChR-GFP and soCoChR-GFP expressing neurons respectively (for CoChR-GFP: 28 ± 10 µW/µm2, n = 7 cells from 5 mice; for soCoChR-GFP: 83 ± 39 µW/µm, n = 4 cells from 4 mice). (b) The AP latency, defined as the time from the onset of 2P stimulation to the peak of the AP, plotted vs. 2P stimulation power in setup 1 units (n = 3–5 powers per cell, for 4 cells from 3 mice for CoChR-GFP, blue dots; n = 3–5 powers per cell, for 3 cells from 3 mice for soCoChR-GFP, red dots). Lines connect data acquired from the same neuron. Horizontal dashed line denotes 15 ms latency, and vertical dashed line denotes 70 µW/µm2 stimulation power, for comparison to panel c. Setups 1 and 2 were used interchangeably; power values used on setup 2 were scaled to equivalent power values for setup 1 (see Methods). See (c) for description of the gray area. (c) AP temporal jitter (measured as standard deviation of the AP latency across a series of 5 photostimulations for a given cell) plotted as a function of AP latency (n = 3–5 powers per cell, for 4 cells from 3 mice for CoChR-GFP, blue dots; n = 3–5 powers per cell, for 3 cells from 3 mice for soCoChR-GFP, red dots). Lines connect data acquired from the same neuron. Setups 1 and 2 were used interchangeably. Jitter below 1 ms (horizontal dashed line) was ensured when the latency was kept below 15 ms (vertical dashed line), corresponding to a photostimulation power higher than approximately 70 µW/µm2. The gray areas in both (b) and (c) mark the region at which such conditions were satisfied.
As a next step in understanding the temporal properties of 2P CGH excitation of soCoChR-bearing neurons, we assessed how AP latency (defined as the time interval between the onset of photoactivation and the peak of the AP) and AP jitter (defined as the standard deviation of the aforementioned latency) depended on the illumination power, for both wild-type CoChR and soCoChR. Increasing the photo-stimulation power density to values above 70 µW/µm2 on setup 1 (or 360 µW/µm2 on setup 2) enabled for both CoChR-GFP and soCoChR-GFP-expressing neurons a spike latency below 15 ms (Fig. 4b), and a spike jitter below 1 ms (Fig. 4c).
Single cell activation of soCoChR-bearing neurons in brain slices using 3D holography
We assessed whether soCoChR-bearing neurons in combination with 2P holographic stimulation could enable millisecond-precision single-cell optogenetics in brain slices. We performed crosstalk experiments as before, patching an opsin-expressing neuron and attempting to activate both the patched neuron and nearby neurons (experiments done using setup 1 and schematically represented in Fig. 5a). We patched an opsin-expressing neuron, with synaptic transmission blocked, and acquired a 2P image stack to reconstruct the 3D volume (~200×200×80 µm3) surrounding the patched cell. 3D holographic illumination allowed to sequentially or simultaneously target multiple locations in such volume (Fig. 5b). We randomly selected 5 to 8 neighboring neurons in this volume (Fig. 5c). We then sequentially illuminated these cells with a 10–14 µm holographic spot while recording from the patched cell. We used excitation powers above 70 µW/µm2 that, accordingly to Fig. 4, assure < 15 ms latency and <1 millisecond jitter. In each sequence, for both CoChR-GFP or soCoChR-GFP, the patched cell responded with at least one AP when the holographic spot was placed on its cell body. For CoChR-GFP expressing cells, in 33±8% of the cells nearby to a patched cell (n = 7 patched cells from 6 mice, Fig. 5d,e), illumination evoked an AP in the patched cell, whereas for soCoChR-GFP expressing cells, illumination of neighboring cells one at a time never evoked an AP in the patched cell (P<0.01; n = 7 patched cells from 7 mice; Fig. 5d,e). Thus, soCoChR used in combination with 2P-CGH illumination could ensure zero-spike crosstalk, single cell stimulation of neurons in intact brain circuitry and <1 millisecond jitter of the spikes.
Figure 5. Soma-targeted CoChR enables single cell control with 3D photoactivation in brain slices.
(a) Schematic of 3D holographic activation. One cell expressing CoChR-GFP or soCoChR-GFP was patched, then neighboring cells at different z-planes were illuminated with 10–14 µm diameter holographic spots. (b) Reconstruction of multi-spot 3D holographic pattern obtained by measuring induced fluorescence from a thin layer of rhodamine 6G (spin coated on a glass coverslip), imaged through a second microscope objective; spot diameter: 10 µm. Scale bars: 35 µm along all three axes. (c) 2P images (imaging λ = 920 nm) from a 3D z-stack used to draw 10–14 µm holographic spots (λ = 1030 nm, setup 1) on neighboring cells expressing CoChR-GFP (left, yellow circles numbered 1–7) or soCoChR-GFP (right, yellow circles numbered 1–7) and on the patched cell (yellow circle, numbered 8), inside a volume of approximately 200×200×70 µm (scale bars: 50 µm). (d) Whole-cell recording of a CoChR-GFP expressing cell (left) and a soCoChR-GFP expressing cell (right) while sequentially positioning the holographic spot on neighboring cells one at a time (represented in panel (c); λ = 1030 nm, setup 1, photostimulation power: 100 µW/µm2 for CoChR-GFP and for soCoChR, 30 ms duration). The distance of cells 1–7 from the patched cell 8 is given below each cell number, in gray. (e) Bar plot showing the percentage of neighboring cells that, when stimulated, yielded an AP in a given patched cell (and averaged across all patched cells; n = 7 cells from 6 mice for CoChR-GFP; n = 7 cells from 7 mice for soCoChR-GFP. Values are mean ± s.e.m.). Dots denote values for single cells. λ = 1030 nm, setup 1, average photostimulation power: CoChR-GFP 142±24 µW/µm2, soCoChR-GFP: 173±26 µW/µm2, 30 ms duration. **P < 0.01, Wilcoxon rank sum test (P=0.004). (f) Whole-cell recording of a CoChR-GFP expressing cell (left) and a soCoChR-GFP cell (right; both cells presented in d) during simultaneous photostimulation of the neighboring cells represented in c, without a stimulation spot on the soma of the patched cell. Photostimulation power density for each spot was equal to the one used in d (λ = 1030 nm, setup 1, photostimulation power: 100 µW/µm2 for CoChR-GFP and for soCoChR-GFP, 30 ms duration, red dash under traces). (g) Bar plot showing the percentage of simultaneous photostimulations of neighboring cells that yielded APs in a given patched cell (averaged across all patched cells). Grey bars indicate the generation of 1 AP; blue bars indicate the generation of more than one AP (n = 7 cells expressing CoChR-GFP from 6 mice; n = 7 cells expressing soCoChR-GFP from 7 mice). Values are mean ± s.e.m. Dots denote values for single cells. λ = 1030 nm, setup 1, average photostimulation power: 142±24 µW/µm2 and 173±26 µW/µm2 for CoChR-GFP expressing cells and for soCoChR-GFP expressing cells respectively; 30 ms duration. For generation of 1AP: *P < 0.05; χ2-test (χ2 = 3.97, P = 0.046, df=1). For generation of more than one AP, χ2 = 3.15, P = 0.08, df=1. Average distances between patched cells and neighboring cells are reported in Supplementary Figure S12 and Supplementary Table 4.
Although individually illuminating neighboring soCoChR-GFP expressing cells did not evoke APs in the patched cell, the illumination still induced a depolarization of 2.6 ± 1 mV in the patched cell (Fig. 5d; n =7 cells from 7 mice). Accordingly, we assessed whether simultaneous stimulation of multiple neighboring cells could push a patched neuron to fire one or more APs, by exciting between 5 and 8 randomly selected neighboring cells. We generated multiple spots, each with power density equal to the one used for the corresponding experiment with sequential cell stimulation (142±24 µW/µm2 and 173±26 µW/µm2 for CoChR-GFP and soCoChR-GFP expressing cells respectively; see also Methods and Supplementary Fig. S11). For CoChR-GFP, in 57% of the cells we observed multiple APs (n = 7 patched cells from 6 mice; Fig. 5f,g). In contrast, for soCoChR-GFP we never observed multiple APs (n = 7 patched cells from 7 mice; Fig. 5f,g). However, single APs were observed in 100% and 33% of patched cells, for CoChR-GFP and soCoChR-GFP respectively (n = 7 cells from 6 mice and 10 cells from 9 mice, respectively) in the simultaneous excitation condition.
These experiments were done at a power just below saturation (Supplementary Fig. S8d). This enabled us to assure a high enough channel recruitment without too much broadening of the axial resolution (Supplementary Fig. S8d) and consequently helped minimize artifactual out of focus neurite activations.
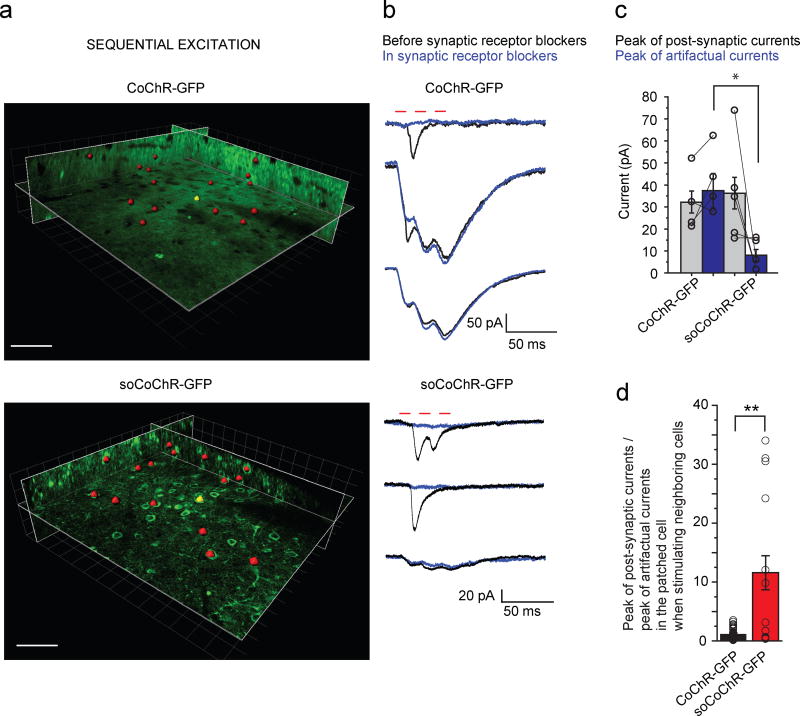
Finally, we performed local functional connectivity experiments in cortical slices from layer II/III of mouse primary visual cortex. To this end, we patched a neuron expressing CoChR-GFP or soCoChR-GFP and we selected 15–25 neighboring opsin-positive cells in a volume of about 200×200×70 µm3 centered around the patched cell (Figs. 6a,b; Supplementary Fig. S13). We then sequentially stimulated the neighboring cells (three 10 ms laser pulses at 50 Hz) while recording the currents in the patched cell (Fig. 6a,b), and repeated the entire sequence three times. To trigger APs with short (<10 ms) latency and ms jitter in the presynaptic cells we applied laser powers in the range of 130 ± 30 µW/µm2, for CoChR-GFP, and 180 ± 20 µW/µm2, for soCoChR-GFP (Fig. 4b,c). In both preparations, we repeated the same experiment in presence of synaptic receptor blockers (NBQX, gabazine, strychnine, APV), so that we could isolate the synaptic vs. artifactual components.
Figure 6. 2P holographic stimulation of soCoChR enables mapping of functional connectivity in brain slices.
(a) Schematic of the connectivity experiment: a 2P image (λ = 920 nm, setup 1) of the volume around a patched opsin positive cell (yellow spot), expressing CoChR-GFP (top) or soCoChR-GFP (bottom), is used to sequentially position a 10–14 µm holographic spot (red spots) on opsin expressing nearby cells and to calculate the corresponding phase masks. Scale bar: 40 µm. (b) Representative currents recorded in whole-cell configuration from a CoChR-GFP (top) or soCoChR-GFP (bottom) expressing cell while sequentially positioning the holographic spot on neighboring cells. Red bars represent the photostimulation periods for each cell (three light pulses at 50 Hz of 10 ms duration). Photostimulation power: 120 µW/µm2 for CoChR-GFP; 150 µW/µm2 for soCoChR-GFP; λ = 1030 nm, setup 1. Black traces: currents recorded before the perfusion of receptor blockers (average of three trials on each spot position). Blue traces: artifactual currents recorded after the perfusion of receptor blockers (average of three trials on each spot position). (c) Bar plots showing the peak amplitudes of currents sensitive to receptor blockers (PSCs; gray bars), and the peak amplitudes of artifactual currents recorded after the perfusion of receptor blockers (IART, blue bars) (n = 4 CoChR-GFP cells from 4 mice; n = 5 soCoChR-GFP cells from 5 mice). Currents were recorded in whole-cell configuration from cells expressing CoChR-GFP or soCoChR-GFP while sequentially positioning the holographic spot on neighboring cells one at a time (λ = 920 or 1030 nm, setup 1 or 2; photostimulation power (relative to setup 1): 130 ± 30 mW/µm2 for CoChR-GFP, and 180 ± 20 mW/µm2 for soCoChR-GFP; three light pulses at 50 Hz of 10 ms duration). For mean peak current comparison: *P < 0.05; Wilcoxon rank sum test (P=0.015). (d) Bar plots showing the ratio PSC / IART (n = 54 out of 78 photostimulated neighboring cells triggering PSCs in the patched cell, from 4 CoChR-GFP cells from 4 mice; n = 15 out of 109 photostimulated neighboring cells triggering PSCs in the patched cell from 5 soCoChR-GFP cells from 5 mice). Currents were recorded in whole-cell configuration from cells expressing CoChR-GFP or soCoChR-GFP while sequentially positioning the holographic spot on neighboring cells one at a time (λ = 920 or 1030 nm, setup 1 or 2; photostimulation power (relative to setup 1): 130 ± 30 mW/µm2 for CoChR-GFP, and 180 ± 20 mW/µm2 for soCoChR-GFP; three light pulses at 50 Hz of 10 ms duration). For comparison of ratio PSC / IART: **P < 0.01; Wilcoxon rank sum test (P = 0.003).
When performing the experiments in soCoChR-GFP expressing cells we found that 13 ± 7% of photostimulated neighboring cells evoked currents, ITOT, in the patched cell. The application of synaptic receptor blockers left a residual current (8 ± 6 pA; Fig. 6c) that corresponded to the artifactual component (IART), which came from the direct stimulation of dendrites or axons of the patched cell crossing the photostimulation volume. The component sensitive to receptor blockers (ITOT−IART) was thus an estimate of the true postsynaptic current (PSCs) (36 ± 16 pA; Fig. 6c).
In the case of CoChR-GFP expressing cells, 71±14% of photostimulated neighboring cells evoked currents, ITOT, in the patched cell with an artifactual component, IART, of 37 ± 15 pA, which was of about the same order as the PSCs (32 ± 10 pA). These results revealed for CoChR the presence of a strong artifactual contribution. Overall, the ratio between the real PSCs and the artifactual peak currents was significantly higher in soCoChR-GFP expressing cells compared to the case of CoChR-GFP expressing cells (11 ± 10 in soCoChR-GFP; 1.4 ± 1.2 in CoChR-GFP; Fig. 6d).
Discussion
We here demonstrate that optical activation of single cells in dense mammalian neural circuitry, with <1 millisecond temporal jitter, is possible without causing stray spiking in neighboring neurons. We achieved this combined temporally and spatially precise optogenetics through molecular engineering – creating a soma-targeted version of the powerful opsin CoChR19, or soCoChR for short, as well as optimal 2P-holographic control of individual neurons. We screened potential trafficking sequences from 9 different soma-localized molecules, first testing entire molecules as well as fragments with GFP, then CoChR. We discovered that the first 150 amino acids on the N-terminus of the kainate receptor subunit 2 enabled efficient targeting of CoChR to the soma, restricting CoChR expression to the first 20–50 µm of dendrites and axons, without alteration of cellular function. We showed that we could get zero-spike crosstalk 2P-excitation of individual cells, i.e. without driving APs in neighbors, but activating many neighbors at once could cause a nearby neuron to be excited to the point of spiking. This thus provides an avenue for future investigation.
We performed local functional connectivity experiments in cortical slices and showed that soma-targeted opsins enabled identification and measurement of downstream postsynaptic currents onto defined cells. It is important to note that although the use of soma-targeted CoChR enabled significant reduction in the artifactual component arising from the direct stimulation of postsynaptic cell processes, this contribution was not completely eliminated. Further improved soma-targeting strategies combined with reduced axial confinement of the excitation volume, e.g. by combining CGH12 with temporal focusing (Supplementary Fig. S8f,g) will enable further reductions in this artifactual component.
Optogenetic analysis of synaptic connectivity has been demonstrated before in intact mammalian circuits36,37. To avoid artifactual components in the PSCs, previous studies have used expression of opsins in the presynaptic cell only, thereby ensuring that the postsynaptic cells do not contain an opsin38. Alternative strategies have used sparse labeling of the ChR2 expressing neurons39 or selective opsin expression in a specific subset of neurons40. Pioneering experiments have used ChR2 targeted to the soma through Kv2.130,36 together with 1P36 and 2P33 activation. In the latter case, the authors were able to show for the first time the use of somatic opsins to map neuronal connectivity. However, these first experiments required long photostimulation pulses (150ms) and used relatively high excitation power (~180 mW/cell)33 which corresponded to a jitter of 17ms, thus limiting the maximum number of targets (2 to 5) that could be simultaneously reached and the precision in the temporal control of spiking generation.
Here we demonstrated that using somatic CoChR enables control of neuronal firing using short illumination pulses (10–30 ms) with temporal jitter short enough (<1ms) to control precise neural codes. For soCoChR-GFP-expressing neurons we demonstrated that APs could be generated reliably by using either a conventional mode locked fs Ti:Sapphire laser or an amplified fiber laser. The first approach is of easier implementation, since such laser sources are common in 2P microscopy. However, we found that reaching the AP threshold with soCoChR at a depth of about 50 µm required an excitation power of about 30 mW/cell. This value, considering an available exit power after the objective of ~ 200 mW (at 920 nm), will limit to 6–7 cells the maximum number of simultaneously achievable targets. Amplified fiber lasers enable higher 2P absorption compared to typical mode-locked Ti:Sapphire laser oscillators due to lower pulse repetition rates of the femtosecond laser (the two-photon excited signal , with f the repetition rate, τ the pulse width and Pavg the average beam power41). This feature enabled reducing to ~80 µW/µm2 (corresponding to about 9 mW/cell) the AP spiking threshold and therefore, considering the several-watt exit power of amplified fiber lasers (corresponding in our setup to ~2 W after the objective) would make it possible to simultaneously photostimulate up to 200 cells. Performances similar to the one achieved with targeted CoChR and 2P holographic illumination might be reached by other opsins when fused to the KA2(1–150) motif, e.g. Jaws for neuronal inhibition42.
Single cell optogenetics may prove powerful for mapping the connectivity of neurons within functional networks, a topic of great interest in the understanding of how individual cells work together in networks to implement neural computation. Past attempts to map connectivity, either by multi-neuron simultaneous whole cell patch clamp43, by glutamate uncaging onto populations of cells44, by examining spontaneous activity in neural networks45, or by laser scanning photoactivation of populations of neurons equipped with optogenetic actuators46 have revealed many tantalizing and non-random features of mammalian neural circuits, with consequences for understanding how individual neurons and networks compute, generate behavior, and contribute to brain pathology. With zero-spike crosstalk single cell optogenetics, it will be possible to patch one neuron and photostimulate many neighboring cells, measuring synaptic strength as well as synaptic release kinetics, and perhaps also to image neural activity network-wide in response to each neuron within the network being excited in turn. Thus, the technologies we report here will be useful for bridging the structural and functional domains of the field of connectomics, important for ultimately realizing its impact on the understanding of behavior and disease.
Online Methods
Primary neuron culture, transfection and transduction
All procedures involving animals were in accordance with the National Institutes of Health Guide for the care and use of laboratory animals and approved by the Massachusetts Institute of Technology Animal Care and Use Committee. Hippocampal neuron cultures were prepared from postnatal day 0 or day 1 Swiss Webster (Taconic or Charles River) mice as previously described19.
For neuronal expression of GFP fusions with opsins or GFP fusions with trafficking sequences during the screen for soma targeting sequences, we performed transfection at 4 days in vitro (DIV) with a commercial calcium phosphate kit (Invitrogen). We added an additional washing step with acidic MEM buffer (pH 6.8 – 6.9) after calcium phosphate precipitate incubation to completely re-suspend residual precipitates47. We used 1µg of DNA. Neurons were imaged 14–18 DIV (10–14 days post-transfection).
For neuronal expression of mCherry we transduced the already transfected neurons 10–14 DIV, with AAV8-Syn-mCherry-WPRE virus by adding 1µl of the virus (titer = 1.4×1013 infectious units per mL).
For neuronal expression of either CoChR-GFP or soCoChR-GFP in culture (as seen in Fig. 2), hippocampal cultures were transduced 3–4 DIV with either AAV8-Syn-CoChR-GFP-WPRE or AAV8-Syn-soCoChR-GFP-WPRE viruses (at a titer of 4.7×1012 and 4.0 ×1012 respectively) and imaged 14 days later.
AAV particles were produced by the University of North Carolina Chapel Hill Vector Core.
Whole-cell electrophysiology in vitro
Current and voltage clamp recordings of cultured neurons
Whole cell patch clamp recordings in culture were made using Axopatch 200B or Multiclamp 700B amplifiers, a Digidata 1440 digitizer, and a PC running pClamp (Molecular Devices). For in vitro current-clamp recordings, neurons were patched 14–18 DIV (10–14 days post-transfection) to allow for neurons to mature to the point of being capable of spiking. Neurons were bathed in room temperature Tyrode containing 125 mM NaCl, 2 mM KCl, 3 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 30 mM glucose and the receptor blockers 0.01 mM NBQX and 0.01 mM gabazine. The Tyrode pH was adjusted to 7.3 with NaOH and the osmolarity was adjusted to 300 mOsm with sucrose. For in vitro voltage-clamp recordings, neurons were patched 19–21 DIV (17–20 days post-transfection) under similar conditions as for the current-clamp recordings, except the Tyrode also contained 1 µM tetrodotoxin (TTX, Tocris Bioscience). No all-trans-retinal was supplemented for any cultured neuron recordings.
For recordings, borosilicate glass pipettes (Warner Instruments) with an outer diameter of 1.2 mm and a wall thickness of 0.255 mm were pulled to a resistance of 5–10 MΩ with a P-97 Flaming/Brown micropipette puller (Sutter Instruments) and filled with a solution containing 125 mM K-gluconate, 8 mM NaCl, 0.1 mM CaCl2, 0.6 mM MgCl2, 1 mM EGTA, 10 mM HEPES, 4 mM Mg-ATP, and 0.4 mM Na-GTP. The pipette solution pH was adjusted to 7.3 with KOH and the osmolarity was adjusted to 298 mOsm with sucrose. For voltage clamp experiments, cells were clamped at −65 mV. For current clamp experiments, access resistance was monitored throughout recording. Data was analyzed using Clampfit (Molecular Devices) and custom MATLAB scripts (Mathworks, Inc.).
Current clamp recordings of cultured neurons during digital micromirror device (DMD) photostimulation experiments
Whole cell patch clamp recordings were made using Axopatch 200B or Multiclamp 700B amplifiers, a Digidata 1440 digitizer, and a PC running pCLAMP (Molecular Devices). In each experiment (Fig. 2), we patched (current clamp) cell #1. Then, starting from cell #1, we photostimulated cell #1 and 9 other cells using the DMD to focus the light. Photostimulations were made 10 seconds apart (470 nm; 40.7 mW/mm2) and the responses for the photostimulation were recorded in pCLAMP.
Molecular Cloning and virus production
All opsin genes were synthesized (Genscript) with mammalian codon optimization and subcloned as previously described19,48. For screening in cultured neurons, all genes were subcloned into the FCK lentiviral backbone under a CaMKII promoter and with a C-terminal GFP fusion. For soCoChR-GFP, the first 450 bp were cloned 3’ to CoChR, with no further trafficking sequences added. For CoChR-NaV1.2 (II/III)-GFP, the KGC49 followed by ER250 trafficking sequences from the potassium channel Kir2.1, with the resulting molecule named CoChR-KGC-NaV1.2(II/III)-ER2-GFP (see51 for earlier details). For virus production, the genes were cloned into the pAAV plasmid, using a synapsin promoter. AAV (serotype 8) was produced by the UNC vector core.
Photostimulation experiments in vitro
Whole field neural stimulation
Neuron voltage clamp photo-stimulation experiments were done with a LED mounted on a microscope for widefield illumination (Leica 3000B), with a nominal wavelength at 480 nm (X-Cite XLED1, Excelitas Tecnologies). We filtered the LED light with the 472/30 nm BrightLine single-band bandpass filter (Semrock). Light was triggered by pClamp (Molecular Devices). Light power was measured as 34.84 mW/mm2, through a Leica HCX APO L 40× objective (air, NA=0.6). For each trace recorded, a 10 ms current injection was given (to make sure a neuron could spike), followed by a 1 ms light pulse (480nm; 34.84 mW/mm2) 5 seconds later.
Stimulation of cell bodies with a DMD
Stimulation of neural cell bodies was performed with a Leica 6000 B widefield microscope, mounted with a Mosaic DMD system (Andor) and a Zyla 5.5 sCMOS camera (Andor). The experiments were performed with an LED with nominal wavelength at 470 nm (Thorlabs, M470L2) and power of 40.7 mW/mm2. Neurons were imaged and stimulated through a 1 MP-2262-59022XR-360T GFP/mCherry filter set (dual GFP/mCherry, Andor). For the photostimulation experiment, an image of the green fluorescence was first acquired, and cell bodies were then identified by eye based on their donut shaped fluorescence. A cell was chosen randomly to be patched (to avoid sample bias, we patched the neuron which was closest to the middle of the field of view), with the only requirement being that it was in a dense area. This cell was referred to as #1. Thereafter, 9 circles, with a diameter of 20 µm were defined around the cell bodies of interest using Metamorph software (Molecular Devices) with distances ranging between 10–200 µm from cell #1. Then, starting from cell #1, we photostimulated cell #1 and 9 other cells sequentially. Stimulations were made 10 s apart.
Single photon characterization of CoChR and soCoChR
Action spectrum data for CoChR and soCoChR were taken and analyzed as previously described19. Analysis of spike probability as a function of irradiance for CoChR and soCoChR expressing cells, photocurrent as a function of irradiance for CoChR and soCoChR expressing cells, and light driven spike fidelity for CoChR and soCoChR expressing cells were all conducted as previously described19.
Imaging in vitro
GFP-fusions with trafficking sequences, opsin-GFP with trafficking sequences, and cytosolic mCherry expressed in cultured neurons were imaged with a LED mounted (X-Cite XLED1, Excelitas Technologies) on a microscope for widefield illumination (Leica 3000B), through either a Leica HCX APO L 40× objective (air, NA=0.6) or a Leica HCX APO L 20× objective (air, NA=0.5). Imaging was performed with a Hamamatsu Orca Flash 4.0 camera under identical illumination conditions throughout: a 480 nm LED using GFP-3035D filter cube (Semrock) for GFP fluorescence (34.84 mW/mm2) and a 540 nm LED with 543 nm ± 11 nm excitation filter (Semrock) for mCherry fluorescence. Images were taken with an exposure time of 300 ms.
Cultured neurons expressing CoChR-GFP, KA2-GFP or KA2(1–150)-GFP were imaged using similar parameters: fluorescence was excited with a 480 nm LED filtered by a 472/30 nm BrightLine single-band bandpass filter (Semrock) and focused on the sample through a Leica HCX APO L 20× objective (air, NA = 0.6), with a power of 25.19 mW/mm2. Images were acquired with a Hamamatsu Orca Flash 4.0 with an exposure time of 300 ms.
Viral injections and whole-cell electrophysiology in brain slices
All experimental procedures were conducted in accordance with guidelines from the European Union and institutional guidelines on the care and use of laboratory animals (council directive 86/609 of the European Economic Community) that were approved by the Paris Descartes Ethics Committee for Animal Research (registered number CEEA34.EV.118.12).
Stereotactic injections of the viral vectors AAV8-Syn-CoChR-GFP, AAV8-Syn -CoChR-KGC-NaV1.2(II/III)-GFP-ER2 and AAV8-Syn-soCoChR-GFP were performed in 4 week old male Swiss mice (Janvier Labs). For connectivity experiments the promoters Syn, CaMKII or CAG were used. Animals were housed from 3 to 5 per cage with a light dark cycle of 12+12 hours. Mice were anesthetized with ketamine (80 mg/Kg)-xylazine (5 mg/Kg) solution and a small craniotomy (0.7 mm) was made on the skull overlying V1 cortex. Injection of 1–1.5 µl solution containing the viral vector was made with a cannula at about 80–100nl/min at 200–250 µm below the dural surface. The skin was sutured and the mouse recovered from anesthesia.
Brain slices of V1 cortex were prepared from mice 7–15 weeks after viral injection. Mice were deeply anesthetized with isoflurane (5% in air), decapitated, and the brain rapidly removed. Sagittal slices of 300 µm were obtained (VT1200S Leica Biosystems, Germany) in room temperature or ice-cold solution containing the following (in mM): 85 NaCl, 2.5 KCl, 0.5 CaCl2, 4 MgCl2, 65 sucrose, 25 glucose, 0.5 ascorbic acid. Slices were transferred in a recovery chamber held at 35° for 45 minutes, in a bath containing 20% sucrose solution and 80% ACSF containing the following (in mM): 125 NaCl, 2.5 KCl, 26 NaHCO3, 1.25 NaH2PO4, 1 MgCl2, 1.5 CaCl2, 25 glucose, 0.5 ascorbic acid. All solutions were aerated with 95% O2 and 5% CO2 to a final pH of 7.4.
Slices were placed in a recording chamber under the microscope objective, and were patched while monitoring IR transmitted light images acquired at approximately video rate. Cells were patched at 40–70 µm depth and clamped at −70 mV in voltage-clamp configuration and membrane potential was kept at −70 mV with currents injections ranging from −5 to −35 pA in current-clamp configuration. Cell type was established based on morphology and AP firing properties. Electrophysiology data were acquired with a Multiclamp 700B amplifier, a Digidata 1322A digitizer (MolecularDevices) or a National Instrument device, and a PC running pClamp10 software (MolecularDevices) or Neuromatic software running on the IgorPro interface (Wavemetrics). Voltage and current clamp recordings were filtered at 6–10 kHz and sampled at 20–50 kHz. On the curves displayed in Fig. 3 a smoothing between 150–250 data points was applied, and in Fig. 5 a smoothing of 10 data points was applied.
The following receptor blockers were added to the artificial cerebro-spinal fluid (ACSF) solution in all experiments to block any synaptic effect: strychnine, picrotoxin/gabazine and NBQX (1–5 µM each; from Abcam or Tocris). Borosilicate glass pipettes (outer diameter 1.5 mm and inner diameter 0.86 mm) were pulled with a micropipette puller (Sutter Instruments) and filled with a solution containing the following (mM): 130 K-gluconate, 7 KCl, 4 MgATP, 0.3 mM NaGTP, 10 Na-phosphocreatine, and 10 mM HEPES (pH adjusted to 7.28 with KOH; osmolarity 280 mOsm). Pipette resistance in the bath was 5–7 MΩ. Data were acquired with a Multiclamp 700B amplifier, a Digidata 1322A digitizer (Molecular Devices) or a National Instrument device, and a PC running pClamp10 software (Molecular Devices) or Neuromatic software running on the IgorPro interface (Wavemetrics).
Imaging and photostimulation in brain slices
Holographic photostimulation was implemented in two different setups, with two different photostimulation laser sources and imaging systems. Setups 1 and 2 were used to acquire the data presented in Figs. 3, 4 and 6 whereas data presented in Fig. 5 was acquired solely with setup 1.
Setup 1
Setup 1 consists of a homemade system, built around a commercial upright microscope (SliceScope, Scientifica) in which the holographic photostimulation path was combined with 3 imaging pathways: a two photon (2P) raster-scanning pathway, a one photon (1P) widefield epi-fluorescence pathway, and an infrared (IR) transmitted light imaging pathway (see detailed schematic in Supplementary Fig. S6). 2P imaging was performed with a pulsed Ti-Sapphire laser source (Coherent Chameleon Vision II, pulse width 140 fs, tuning range 680 nm – 1080 nm). The beam was raster scanned across the sample via a pair of XY galvanometric mirrors (3mm aperture, 6215H series, Cambridge Technology) imaged at the back aperture of the microscope objective (40× W APO NIR, Nikon) through an afocal telescope (scan lens: f = 100mm, Thorlabs #AC508-300-B; tube lens: f = 300mm, Thorlabs #AC508-100-B). Fluorescence was collected with a fiber-coupled detection scheme52, based on a large diameter collector lens (f=75mm, Thorlabs #LB1309-A) and a 5mm diameter liquid light guide (LLG, Series 300, Lumatec customized with a f = 14.5mm doublet lens glued at the fiber entrance by Till Photonics, and an anti-reflective coating provided at the fiber exit). The exit of the fiber was projected onto two GaAsP photomultiplier tubes (H10770-40 SEL, Hamamatsu H10770-40 SEL, active area 5mm) by a set of three matching aspheric lenses (f = 23.5mm, Melles Griot #LAG-32.5-23.5-C). Fluorescence light was filtered by an infrared-light blocking filter (FF01-750sp, Semrock), spectrally split into two channels by a dichroic mirror (FF555-Di03, Semrock) and further filtered through two emission filters (FF01-510/84 and FF02-617/73, Semrock). The power of the 2P imaging laser was controlled through a liquid crystal variable phase retarder (LRC-200-IR1, Meadowlark Optics) and a polarizer cube (BB-050-IR1, Meadowlark Optics).
1P imaging was based on illumination provided by a LED source (M470L2, Thorlabs), whose emission spectra was filtered by a bandwidth excitation filter (FF01-452/45, Semrock). To provide homogenous widefield illumination, the emission was directed though a diffuser (DG10-1500, Thorlabs) and, successively, through an achromatic lens (f = 30mm, #LA1805 Thorlabs). 1P induced fluorescence, after being separated from the excitation light using a dichroic mirror (FF510-Di02, Semrock), was collected through a tube lens (f = 200mm), filtered by a band-pass filter (FF01-609/181, Semrock) and detected by a CCD camera (Orca-05G, Hamamatsu). Fluorescence induced either by 2P raster scanning or 1P widefield illumination was collected by PMTs or a CCD respectively; in the detection pathway we had a switchable dichroic mirror (FF705-Di01, 70×50mm custom size, Semrock).
The transmitted IR oblique illumination imaging path was based on an IR-LED source (M780L2, Thorlabs), coupled with a DODT-contrast tube (DODT tube, Scientifica) and a condenser. Light transmitted through the sample was collected with an IR CCD (IR-1000, DAGE-MIT).
2P photoactivation consisted of arbitrary illumination patterns obtained with computer generated holography, based on phase modulation of the laser wave front via the use of a spatial light modulator (SLM).
The laser source used consisted of a femtosecond pulsed beam delivered by a fiber laser source (pulse width 250 fs, repetition rate 500 kHz, pulse energy 20 µJ, λ = 1030 nm; Satsuma, Amplitude Systems). The beam was enlarged by a telescope and reflected off the SLM (LCOS-SLM, X10468-07 Hamamatsu Photonics). The beam was then projected on the back focal plane of the objective with a double afocal telescope (f = 500mm Thorlabs #AC508-500-B, f = 300mm, Thorlabs #AC508-300-B, f = 200mm, Thorlabs #AC508-200-B and f = 300mm Thorlabs #AC508-300-B). The SLM was driven by custom-designed software (Wave Front Designer53) based on a Gerchberg and Saxton (GS) iterative algorithm54 which converts an arbitrary intensity pattern on the sample plane to a specific phase profile to be addressed at the SLM plane.
As originally described for low55 and, more recently, high NA objectives12, adding lens-phase modulations to 2D-phase holograms enables remote axial displacement and 3D positioning of bidimensional light patterns. This allowed us to target neural processes, following their path in x-y-z (Fig. 3) and to address multiple targets, sequentially or simultaneously, on multiple planes (Fig. 5 and 6).
The zeroth order of diffraction was eliminated by introducing a single or a double cylindrical lens56. 2P imaging scanning and 2P photoactivation beams were combined through a large dichroic mirror (Chroma T970dcspxr, 50mm×70mm custom sized, Chroma).
Setup 2
An analogous holographic photostimulation path was coupled with widefield epifluorescence imaging on a second system, here denoted as setup 2 (see Supplementary Fig. S6).
This system was built around an Olympus BX51WI upright microscope, capable of widefield epifluorescence imaging using illumination with an arc lamp, (OptoSource Illuminator, Cairn Research, coupled with a monochromator, Optoscan Monochromator, Cairn Research), and an Orca Flash 4.0 Hamamatsu CCD camera for epifluorescence imaging. The native infrared differential-interference contrast (DIC) path of the Olympus microscope allowed DIC imaging on the CCD.
The holographic photoactivation laser source consisted of a conventional pulsed Ti:Sapphire laser, used at 920 nm (pulse width: 100 fs, repetition rate: 80 MHz, Mai-Tai, Deep-See, Spectra Physics).
The holographic path was analogous to the one described for setup 1: a beam expander enlarged the beam in front of the spatial light modulator (LCOS-SLM X10468-02), whose plane was projected at the back focal plane of a 40×-NA 0.8 objective (LUM PLAN FI/IR, Olympus) by an afocal telescope (f=750mm, Thorlabs #AC508-750-B and f=500mm Thorlabs #AC508-500-B). The holographic beam was coupled to the optical axis of the microscope by a dichroic mirror (FF670, SDi01, 25×36 mm, Semrock). Photostimulation light pulses were generated by a Pockels cell (350-80, Conoptics).
Power conversion between Setup 1 and 2
The differences between the illumination conditions (wavelength, pulse width and repetition rate) used in setups 1 and 2 required finding a proper criterion to compare the experiments performed on the two setups. To do so, we empirically estimated a scaling conversion factor k between the power of the two systems, P1 on setup 1 and P2 on setup 2, based on measurements of the rise times (τon) of photo-induced currents (see Fig. 4): we take as the conversion factor k the ratio between the powers that in each setup induced a photocurrent with the same rise time. We fit the dependence of the rise times on the excitation power (n=4 cells measured in setup 1, among which 3 expressed CoChr-GFP and 1 soCoChR-GFP; n=5 cells measured on setup 2, among which 2 expressed CoChr-GFP and 3 soCoChR-GFP), using the expression:
| (1) |
where Pi is the photo-stimulation power density (with i= 1 or 2 indicating setup 1 or setup 2, respectively) and c1, c2 and c3 are three free fitting parameters. For each cell, the experimental dependence of the rise time on the illumination power included at least 5 power values.
The conversion factor k = P2/P1 was obtained as the factor that minimized the mean square difference between the values of the curve τon,1(P1) and those of the curve with the rescaled power values τon,2(kP1). The so-obtained value was k = P2/P1 ≈ 5.3, meaning that powers used in setup 1 were about 5 times lower than those used in setup 2.
Diffraction efficiency corrections
Holographic generated light patterns suffer from a position dependent diffraction efficiency12,57,58, meaning that the intensity of a holographic spot decreases the more is moved away from the center of the field of view or away from the nominal focal plane of the objective.
Therefore, generation of illumination patterns with uniform intensity, crucial for experiments in Fig. 3, 5 and 6, required the implementation of corrective algorithms to compensate for this effect.
In the case of sequential targeting of different locations, homogenization of light distribution was achieved by generating, together with the excitation holographic spots of surface Aspot, an additional correction spot at the edge of the field of view, of surface Acorr. By adjusting the size of the correction spot we could modulate the light intensity in the excitation spot while moving the latter across the excitation field thus achieving constant light illumination (see Supplementary Fig. S11). More precisely, at first we characterized experimentally the position dependent diffraction efficiency, DE(x, y, z); then, given a sequence of N illumination spots of coordinates (xi, yi, zi), we set the surface of the correction spot for each illumination site as , where is the lower diffraction efficiency value over the N illumination sites. The correction spot was blocked at an intermediate image plane after the Spatial Light Modulator, preventing it from reaching the sample.
For simultaneous illumination of multiple spots we used, as described in refs. Conti et al.59 and Hernandez et al.12, graded input patterns to calculate with the iterative GS algorithm the corresponding phase mask (see Supplementary Fig. S11). Briefly, each illumination spot, the i-th in the ensemble of N, drawn in the input image for the GS algorithm, was weighted with a subunitary coefficient wi, given by . This enabled us to direct more power to spots corresponding to low-diffraction efficiency regions while sending less power to the spots closer to the center of the excitation field and/or the objective focal plane.
Finally, we equalized the excitation power densities used for single spot sequential stimulation and simultaneous multiple spot stimulation by adjusting the exit laser power.
Holographic photostimulation along neurites
Patched cells in slices were loaded with Alexa594 hydrazide dye (15–20 µM; Invitrogen) to visualize cell morphology. We imaged the patched neuron loaded with Alexa594 with a 2P scanning imaging system (imaging laser at 780 nm60, setup 1) or widefield illumination (at 570 nm, setup 2). A z-stack of the fluorescence emission was acquired in order to reconstruct the 3D morphology of the neurites (in a range of typically ±40 µm) and to target, with holographic illumination, specific x-y-z positions along neurites.
The photo-stimulation protocol consisted of sequentially displacing 10 µm diameter holographic stimulation spots from the distal end of the process towards the soma, with a step-size of approximately 10 µm. 2P stimulation consisted of 30 ms pulses with an illumination power density corresponding to the power threshold density required to trigger one AP when stimulating the whole soma. Practically, the power threshold density was obtained by progressively increasing the excitation power up to a value that allowed to reliably (in three consecutive trials) trigger one AP each time.
Measurement of opsin and photo-induced AP kinetic parameters
Neurons expressing CoChR-GFP or soCoChR-GFP were patched and photo-stimulated with holographic spots whose diameter was between 10 and 15 µm – i.e., covering the whole soma. The kinetics of the response was monitored in both current-clamp and voltage-clamp configurations. The photo-stimulation consisted of pulses of holographic illumination with a duration varying from 300 ms at low power, down to a few ms at high power. The reported rise time τon corresponds to the time constant of a mono-exponential fit of the ascending part of the photo-induced current.
To estimate the asymptotic value of the rise time, we fit the rise time vs. power curve of each cell with expression (1) and the coefficient c3 was taken as the asymptotic value of the rise time for each cell, and then averaged across cells. We considered in the average only cells in which the rise time vs. power curve had a sufficient number of experimental points (≥5) to assure the reliability of the fit.
The AP latency was defined as the time delay between the onset of 2P stimulation and the peak of the photostimulated AP. The plotted values were obtained by averaging AP latencies across 5 photostimulations (separated by 30 s). The duration of the photostimulation pulse was initially set for 30 ms for low stimulation power (22±19 µW/µm2). For high photostimulation power (107±29 µW/µm2), we decreased the duration of photo-stimulation to below 30 ms, in order to minimize the generation of multiple spikes. The reported jitter was calculated as the standard deviation of the AP latency computed across 5 consequent photostimulations. The values were extracted from the same data used to obtain the latency values.
Multiple cell-soma holographic photostimulation in slices
Opsin-expressing cells were visualized with a 2P scanning imaging system (imaging laser at λ = 920 nm in setup 1) and patched. Thereafter, a z-stack of the GFP fluorescence emission in the volume around the patched cell was acquired in order to identify neighboring positive cells. The scanning imaging lasers, although inducing some photocurrents (I=23±13 pA; imaging power P=14±2 mW, scanning frequency 0.74 Hz; n=3 soCoChR expressing cells from 3 mice) didn’t induced APs during imaging. In slices expressing soCoChR-GFP, cells were clearly distinguishable, since their somata were predominantly fluorescent with minimal neuropil fluorescence. In contrast, slices expressing CoChR-GFP presented a diffuse and homogeneous green fluorescence, in which cells could be recognizable as dark spots (spots with lower fluorescence compared to the background).
In the volume around the patched cells, we randomly identified and selected an ensemble of neighboring fluorescent cells that we targeted with 10–14 µm holographic spots. Depending on the field of view and cell distribution, we selected 5 to 8 cells. In order to avoid axial resolution broadening due to the axial overlapping of multiple spots, for cells separated by an axial distance smaller than the spot FWHM (see Supplementary Fig. S8) we choose cells that were laterally shifted with respect to one another. These cells, in the vicinity of the patched cell, were sequentially stimulated with a holographic spot (30 ms illuminations, 5 s apart) and the generated membrane depolarizations recorded. The last cell to be photostimulated for each stimulation sequence was the patched cell. Receptor blockers were added to the ACSF solution in all experiments to block any synaptic effects (strychnine, picrotoxin/gabazine and NBQX, 1–5 µM; Abcam or Tocris).
For sequential and simultaneous illumination experiments, the same power density was delivered to each spot in the pattern, thanks to our diffraction efficiency compensating algorithm (see Methods above). Since the axial separation between the patched cell and targeted neighboring cells was < 30 µm, we estimated the effects of scattering on the relative intensity between spots on the patched soma and on neighboring cells were less than 10%61 and were therefore negligible. For higher spatial separations, a uniform light distribution was obtained by adding further corrections to the intensity of each illumination spot according to axial position.
3D holographic pattern reconstruction and calibration
To reconstruct the 3D holographic illumination pattern of Fig. 5b and Supplementary Figs. S8 and S11, we used a dual microscope configuration as previously described9,12,53,62. Briefly, below the main upper objective we placed a second objective (NA 1.2, water-immersion, 60×). While the upper objective focused the holographic pattern on a thin layer of rhodamine 6G (spin coated and dried on a glass coverslip), the second objective collected the fluorescence generated by the rhodamine layer that was then recorded on a CCD camera. By changing the vertical position of the upper objective, we reconstructed the whole x-y-z distribution of the holographic excitation volume.
Daily calibration of the holographic illumination system consisted in verifying the spatial alignment between the patterns of illumination and the imaging system. In setup 2 (combining 2P-CGH with widefield 1P imaging) we used the holographic patterns to excite a thin fluorescent layer and we checked the position of the pattern in the field by imaging the evoked fluorescence on the camera. In setup 1 (combining 2P-CGH with 2P scanning imaging) we used the holographic pattern to bleach a thick fluorescent plate (Chroma - Part No. 92001) and then we re-imaged the bleached pattern with the 2P scanning laser. In this way, we obtained the exact positioning of the holographic patterns in the coordinates of the imaging system. By iteratively adjusting the coordinate system of the holographic patterns (specifically an x-y-z stretch and translation, together with a rotation in the x-y plane), we could achieve a precise overlap with the imaging coordinate system. During experiments, this calibration allowed us to use fluorescent images as a reference to place with micrometric precision photostimulation spots on the structures of interest.
Image analysis – determining the fluorescence brightness from the soma to the neurites
Images for this analysis were taken 14–18 DIV (10–14 days post-transfection) from cultured neurons. The image analysis was performed in ImageJ. For each neuron we first defined the boundaries of the soma. To that end, we drew a 20 µm diameter circle near the soma, inside which there was no apparent fluorescence from the soma or from neurites. We defined the average fluorescence in this circle as background fluorescence.
We considered pixels with fluorescence intensity of at least 10% above background levels as part of the soma and processes, and we defined the boundary between soma and processes by the cell morphology. Then, we drew a polygon along the defined soma boundary and measured the average fluorescence inside of it, and subtracted the previously calculated background value. The resulting value was considered soma fluorescence. To measure fluorescence intensities along the neurites, we defined 1µm2 rectangles along the neurite that were up to 100 µm away from soma at increments of 10 µm. The distance between each rectangle and the soma was measured along the neurites (note well – this did not correspond to the minimal linear distance from the soma, since neurites were curved). We then defined the background value as described above for the soma. We made sure that the pixel intensity values at the boundaries of the rectangle were at least 10% above background levels, to be considered inside the neurite. We averaged the fluorescence intensity in each rectangle, then subtracted the background, then divided it by the average soma fluorescence and plotted the resulting ratios with respect to their distances along the neurites. The ratios for each distance were averaged across neurites and data was plotted (using Matlab) as average and standard error of the mean (for CoChR-GFP, n = 5 neurites from 5 cells from 2 cultures; for soCoChR-GFP, n= 5 neurites from 5 cells from 3 cultures; for CoChR-NaV1.2 (II/III)-GFP, n= 5 neurites from 5 cells from 4 cultures). For the Kolmogorov-Smirnov (KS) test verified that the dependent variable (fluorescence) was at least ordinal, that the two samples (cells expressing CoChR vs cells expressing soCoChR) were independent and that in both cases cells were chosen randomly. Image analysis in brain slices was performed the same way; however, due to the dense labeling of cells we needed to determine the continuity of any given measured cell. Since only patched cells were labeled with Alexa 594 we verified cell continuity using the red channel, and measured the fluorescence in the green (GFP) channels. This was performed for cells expressing CoChR vs. soCoChR.
Sample size determination
Because the experiments here were aimed at developing a technology, rather than testing a formal scientific hypothesis, the animal numbers in the paper were forecasted based upon past neuroengineering projects we have done, using the experience we have attained since starting the Synthetic Neurobiology group at MIT, similar to Klapoetke et al19. This follows guidelines recommended by the NIH63 regarding “experiments based on the success or failure of a desired goal.”
Data analysis
Analyzing current amplitudes and current decay time following 1P photostimulation experiments
For current amplitude analysis, during photostimulation experiments, voltage clamp traces were recorded and the amplitudes of the current were taken directly from Clampfit (Molecular Devices) and copied into an Excel spreadsheet. For each patched cell, there was one current value. We averaged the photocurrent amplitude values across cells for each opsin targeting variant, and calculated the standard error of the mean (n = 13 neurons from 3 cultures and 13 neurons from 3 cultures for CoChR-GFP and soCoChR-GFP respectively). We then performed a two-sided Wilcoxon rank sum test (in Matlab). Full statistics are in Supplementary Table 3. For the Wilcoxon rank sum test, we verified that we were comparing two independent categorical groups (the first group with cells expressing CoChR, and the second group with cells expressing soCoChR), that the dependent variable (percentage) was ordinal, and that observations of dependent variables were independent (because these were different experiments, on different neurons).
For τoff analysis, during photostimulation experiments, voltage clamp traces were recorded and their current decay times (τoff) were extracted from mono-exponential fits in Clampfit (Molecular Devices). For each patched cell, there is one τoff value. We averaged the τoff values across cells for each opsin targeting variant, and calculated the standard error of the mean (n = 10 neurons from 3 cultures and 13 neurons from 3 cultures for CoChR-GFP and soCoChR-GFP). We then performed a two-sided Wilcoxon rank sum test (in Matlab). Full statistics are in Supplementary Table 3. For the Wilcoxon rank sum test, we verified that we were comparing two independent categorical groups (first group with cells expressing CoChR, second group with cells expressing soCoChR), that the dependent variable (percentage) was ordinal, and that the observations of dependent variables were independent (because these were different experiments, on different neurons).
Analysis procedure for DMD experiments
To find the percentage of patched cells spiking when directly photostimulated, we defined the patched cell as having an AP when its membrane voltage depolarized by at least 35 mV upon photostimulation. For each of the 5 cells patched, we recorded the data as ‘1’ if the patched cell yielded an AP when the patched cell was photostimulated, and ‘0’ if the patched cell did not yield an AP when the patched cell was photostimulated. We found the percentage of APs in patched cells resulting from direct stimulation, by summing all ‘1’s and dividing the sum by the number of patched cells (n = 5 cells from 4 cultures for CoChR-GFP; n = 5 cells from 5 cultures for CoChR-KA2(1–150)-GFP)).
To find the percentage of photostimulated neighboring somata yielding an AP in the patched cell, we defined the patched cell as having an AP when its membrane voltage was depolarized by at least 35 mV upon photostimulation (baseline was calculated as the average of the voltage values over the 100ms period which preceded the event). For each of the 5 cells patched, 9 cells neighboring the patched cell were photostimulated. We then counted the number of neighboring cells whose stimulation led to an AP in the patched cell, and divided it by the number of photostimulated neighboring cells. This value was then averaged across the 5 patched cells (n = 5 cells from 4 cultures for CoChR-GFP; n = 5 cells from 5 cultures for soCoChR-GFP)). We then made a bar-plot showing the average percentage and standard error of the mean for CoChR-GFP and soCoChR-GFP. We compared this percentage between CoChR-GFP and soCoChR-GFP via a two-sided Wilcoxon rank sum test (Matlab). Before performing the Wilcoxon rank sum test, we verified that we were comparing two independent categorical groups (first group with cells expressing CoChR, second group with cells expressing soCoChR), that the dependent variable (percentage) was ordinal, and that the observations of dependent variables were independent (because these were different experiments, on different neurons).
To find the percentage of photostimulated neighboring somata yielding a depolarization in the patched cell, we defined the patched cell as having a depolarization when its membrane voltage increased from baseline by more than 0.5 mV upon photostimulation (baseline was calculated as the average of the voltage values over the 100ms period which preceded the event). For each of the 5 cells patched, 9 cells neighboring the patched cell were photostimulated. We then counted the number of neighboring cells whose stimulation led to a depolarization in the patched cell, and divided it by the number of photostimulated neighboring cells. The value was averaged across the 5 patched cells (n = 5 cells from 4 cultures for CoChR-GFP; n = 5 cells from 5 cultures for soCoChR-GFP)). We then made a bar-plot showing the average percentage and standard error of the mean for CoChR-GFP and soCoChR -GFP. We compared this percentage between CoChR-GFP and soCoChR-GFP by a two-sided Wilcoxon rank sum test (Matlab).
Measurement of 2P excitation spectrum
The wavelength dependence of the 2P excitation of CoChR, reported in Supplementary Figure S7, was measured on cultured CHO cells stimulated using the tunable laser in setup 1. In order to cover the entire cell body, the optical path of the imaging laser was modified to underfill the objective back aperture and generate a large 2P Gaussian excitation (FWHM=38 µm) at the sample plane. Wavelength scanning was performed using photostimulation pulses of 200 ms at constant photon flux (2.4±0.3 1026 photons/s·m2, spatially averaged over the Gaussian spot surface over the investigated wavelength range, assuming constant laser pulse width. From the photocurrent curves, we extracted the wavelength dependence of the peak currents and of the exponential rise and decay times, which we used to deduce the normalized excitation cross-section spectrum (Supplementary Figure S7b) using equation (13) in Chaigneau et al.10
Modeling of the power dependence of the axial resolution of 2P holographic photostimulation
The non-linear opsin response to the illumination power can affect the effective axial resolution of photostimulation. In order to evaluate and model this effect, we firstly recorded the response of a CoChR expressing CHO cell to different 2P holographic illumination powers (shown in the inset of Supplementary Fig. S8d). The use of cultured CHO cells allowed, in contrast to neurons, avoiding any influence of excitation of neurites around the soma. As expected, the photoinduced peak currents at high excitation power showed saturation as power increased. In a first approximation, the curve of the normalized peak current near saturation could be fitted with an exponential curve: I(P) = 1 − e−P/P′, with P′ ≈ 0.06 mW/µm2 (λ=1030 nm, setup 1). Secondly, we extracted the normalized intensity axial profile p(z) of the excitation holographic spot from a Gaussian fit of the corresponding axial fluorescence profile (Supplementary Fig. S8b,c). Finally we modeled the axial profile of the photocurrent by using the expression I(z) = 1 − e−P0·p(z)/P′, with P0 the nominal power density (per unit surface) at z=0. The results of the model are reported and compared with experimental results in Supplementary Fig. S8d,e.
A further optimization of the model would include a more precise fit of the quadratic behavior of the photocurrent at lower power, far below saturation, and would also consider the convolution of the illumination profile with the exact morphology of the neuron cell body (here considered simply as a thin disk) and the spatial distribution of its axon and dendritic tree.
Analyzing photocurrent integrals under photostimulation along neurites
For each photostimulation location, 1 to 3 trials were acquired and the resulting traces were averaged. The current integral over time was calculated by integrating the current response recorded at the soma in voltage-clamp configuration from the onset of the photostimulation until the time of recovery to the initial resting-value of the current. In the calculation of the integral, an offset in the current curve was introduced, corresponding to the integral of the holding current at rest.
The distance from the soma was estimated based on the coordinates of the photostimulation spots placed along neurites. This sequence was manually defined during the experiment and consisted of a series of 10µm spots starting from the center of the soma and moving away from it along a neurite with a step of approximately 10 µm up to a maximum distance of 110 µm from the soma (while taking into account the curvature of neurites). Post-experimentally, the distance between each pair of consecutive spots di was defined as the cartesian distance between the center of the consecutive spots (taking into account the x, y and z coordinate of each). The distance to the soma from the i-th spot was then calculated as . The photostimulation sequence was then performed starting from the most peripheral spot on the neurites to the spot on the soma.
Current integrals over time at various neurite locations were normalized to the current integral recorded at the soma. The current integral values were then binned according to their distance from soma in intervals of 15 µm. Values were averaged across neurites (1 to 3 neurites per cell were investigated) and data plotted as mean ± s.e.m, in Fig. 3. To estimate the statistical significance of the difference between CoChR-GFP and soCoChR-GFP photocurrents, we performed a two-sample Bonferroni-corrected Kolmogorov-Smirnov test for each binning interval. The test was two-sided. For the Kolmogorov-Smirnov (KS) test we verified that the dependent variable (photocurrent integral) was at least ordinal, that the two samples (cells expressing CoChR vs. cells expressing soCoChR) were independent and that both samples were random (neurons and neurites to work on were chosen randomly).
Results of the test are reported in Supplementary Table 4. Data were analyzed using pClamp10 software (MolecularDevices), IgorPro and MATLAB (Mathworks, Inc.).
Analyzing single and multiple soma holographic stimulations in brain slices
For each patched cell we counted the number of neighboring cells whose stimulation led to an AP in the patched cell, and divided this number by the number of photostimulated neighboring cells, and then averaged this ratio across all patched cells. Data are presented as mean ± s.e.m in Fig. 5. Significance was tested with the non-parametric two-sided Wilcoxon rank sum test (Matlab) for continuous variable (percentage) in Fig. 5e, and the non-parametric χ2–test for discrete values in Fig. 5g. For the Wilcoxon rank sum test, we verified that we were comparing two independent categorical groups (cells expressing CoChR and soCoChR), that the dependent variable (percentage) was ordinal, and that observations of dependent variables were independent (since obtained on different experiments and different neurons). For the χ2–test, we verified that variables were mutually exclusive and that the studied groups were independent (cells expressing CoChR and soCoChR).
For simultaneous stimulation experiments we calculated the percentage of patched cells firing a single AP or multiple APs when their neighbors were simultaneously stimulated. Illumination patterns were composed of 5 to 8 holographic spots. The results were averaged across the patched cells.
Connectivity experiments were performed by sequentially photostimulating presynaptic cells. The whole photostimulation sequence was repeated three times. The postsynaptic traces from the three trials were averaged for analysis and for the traces reported in the figures. Whenever stimulation of presynaptic cell elicited a current (ITOT) in the patched cell, we defined the artifactual component, IART, as the residual current left after application of synaptic blockers. The PSC component was then calculated as PSC=ITOT−IART. The ratio PSC/IART was used to compare connectivity experiments performed using soCoChR-GFP or CoChR-GFP.Data are presented as mean ± s.e.m. in Fig. 6. Data were analyzed using pClamp10 software (MolecularDevices), IgorPro and R-studio.
Excluding data points
For recording in slices, data points (singular trials) were excluded if the recording was unstable, meaning the baseline potential changed by more than 5 mV during the recording. Cells were excluded if a gigaohm seal was lost or a high leak current appeared.
For recording in culture, data points (cells) were excluded if we could not establish a gigaohm seal.
Statistics
For comparison of GFP brightness vs position along the neurite between constructs, two-sample Kolmogorov-Smirnov (K-S) tests with Bonferroni correction were performed. For comparing spike probability vs. irradiance, Bonferroni-corrected Wilcoxon rank sum tests were used. For comparisons of normalized integrated photocurrent along neurites, two-sample Kolmogorov-Smirnov tests with Bonferroni correction were used. For comparing of the average distances between patched cells and photostimulated cells, Wilcoxon rank sum tests were performed. For comparing single and multiple spike probabilities, the χ2 test was used. Wilcoxon rank sum tests were used for comparison of PSC and IART currents and for comparison of the PSC / IART ratios. Data collection and analysis were not performed blind to the conditions of the experiments. Animals and cultures were randomly assigned to the different experimental groups. Information about the experimental design and statistics are given in the Life Sciences Reporting Summary. Normality and equal variances were not assumed, therefore all the tests were non parametric tests (K-S test, χ2 test, Wilcoxon rank sum test).
Supplementary Material
Figure S1. An N-terminal domain of kainate receptor subunit 2 is retained in the soma. (a) A hippocampal neuron expressing GFP. (b) A hippocampal neuron expressing KA2-GFP. (c) A hippocampal neuron expressing KA2(1–150)-GFP. Scale bars for a–c: 50 µm. (d, e, f) Box-and-whiskers plots of GFP brightness versus position along a neurite, normalized to GFP brightness at the soma for GFP (n=5 neurites from 3 cells), KA2-GFP (n=5 neurites from 3 cells) and KA2(1–150)-GFP (n=5 neurites from 3 cells) respectively. Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values respectively. See Supplementary Table 2 for Bonferroni-corrected Kolmogorov-Smirnov tests for Figure S1.
Figure S2. CoChR-NaV1.2(II–III)-GFP expression and photostimulation in cultured hippocampal neurons. (a–d) Images are presented for cultured hippocampal neurons expressing CoChR-NaV1.2(II–III)-GFP, along with the countermarker mCherry. (a) A hippocampal neuron in culture expressing CoChR-NaV1.2(II–III)-GFP and mCherry, seen in the GFP channel (scale bar: 100 µm). (b) Zoomed-in image from the yellow rectangle of a. (c) The neuron of b, seen in the mCherry channel (magenta), along with other nearby neurons. (d) Merge of b and c (scale bar for b–d: 20 µm). (e) Whole cell current clamp recording of a cultured hippocampal neuron expressing CoChR-NaV1.2(II–III)-GFP, under current injection (10 ms duration; gray rectangle) and optical stimulation (480nm, 34.84mW/mm2, 1 ms duration; blue rectangle). Rectangles not to scale. (f) Box-and-whiskers plot of GFP brightness versus position along a neurite, normalized to GFP brightness at the soma, extracted from neurites of cultured hippocampal neurons expressing CoChR-NaV1.2(II–III)-GFP (n = 5 neurites taken from 5 cells from 4 cultures). Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values respectively.
Figure S3. CoChR-GFP-Kv2.1motif expression is somatodendritic. (Ai, Aii) Hippocampal neurons expressing CoChR-GFP-Kv2.1motif. (B, C) Box-and-whisker plots of brightness versus position along a neurite, normalized to brightness at the soma for CoChR-GFP-Kv2.1motif (B, n = 5 neurites from 5 cells from 2 cultures) and CoChR-GFP (C, n = 7 neurites from 5 cells from 2 cultures). Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Red line denotes the median. Top and bottom whiskers indicate the highest and lowest values respectively. n.s., not significant, comparing B vs. C; see Supplementary Table 3 for Bonferroni-corrected Kolmogorov-Smirnov tests for Figure S3. Scale bar: 20µm.
Figure S4. In vitro single photon characterization of CoChR and soCoChR. (a) Action spectra for CoChR (n = 9 cells) and soCoChR (n = 10 cells) measured in cultured neurons. Plotted data are mean ± s.e.m. (b) Blue light driven spike probability as a function of irradiance for CoChR (magenta) and soCoChR (black) expressing cells. Plotted data are mean ± s.e.m (n = 6 cells for each opsin). All blue light spiking protocols used 2 ms width light pulses at 480 nm. (c) Blue light driven photocurrents as a function of irradiance for CoChR (magenta) and soCoChR (black) expressing cells. Plotted data are mean ± s.e.m (n = 7 cells for each opsin). All blue light protocols used 2 ms pulse width at 480 nm. (d) Blue light driven spike fidelity for CoChR (magenta) and soCoChR (black) expressing cells. All blue light spiking protocols used a train of 40 pulses, 2 ms pulse width, at 480 nm, at 5 mW/mm2. Plotted data are mean ± s.e.m (n = 7 – 10 cells for each opsin). See Supplementary Table 3 for full statistics for Figure S4.
Figure S5. Membrane properties of neurons expressing somatic vs. untargeted molecules.
Cultured hippocampal neurons expressing CoChR-GFP (n = 10 cells from 3 cultures), KA2(1–150)-GFP (n = 10 cells from 3 cultures), and soCoChR-GFP (n = 10 cells from 3 cultures) were patched 14 days following AAV transduction. (a) Resting potential. We performed a Kruskal-Wallis Test (not significant, P = 0.7549, chi-square = 0.5624). (b) Holding current. Kruskal-Wallis Test, not significant, P = 0.9875, chi-square = 0.0252. (c) Membrane capacitance. Kruskal-Wallis Test, not significant, P = 0.9817, chi-square = 0.0369. (d) Membrane resistance. Kruskal-Wallis Test, not significant, P = 0.9205, chi-square = 0.1656. For all panels, red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values.
Figure S6. Detailed schematics of the holographic microscopes, for both setup 1 and setup 2. PC, Polarizer Cube; L/2, Half Wave Plate; M, Mirror; L, Lens; SLM, Spatial Light Modulator; CL, Cylindrical Lens; D, Dichroic; GM, Galvanometric Mirrors; F, Filter; OBJ, Objective; C, Condenser; PoC, Pockels Cell. See detailed description in Methods section.
Figure S7. 2P action spectrum and normalized excitation cross-section of CoChR. (a) Normalized peak current of CoChR as a function of wavelength, measured in cultured CHO cells. Photostimulation was performed using a 2P Gaussian spot (FWHM=38 µm) at different wavelengths, at constant photon flux (2.4±0.3 1026 photons/s·m2) over the investigated wavelength range. Plotted data are mean ± s.e.m., with raw data in open circles, n=5 cells. (b) Normalized 2P excitation cross section, extracted from the photocurrent kinetics (see Methods). Plotted data are mean ± s.e.m., with raw data in open circles, n=5 cells.
Figure S8. Axial resolution of 2P holographic spots. Top (a) and lateral (b) view of the fluorescence emission generated by a 10 µm diameter holographic spot on a thin fluorescent layer. The fluorescence is collected by a second bottom objective and relayed to a CCD camera; the z-stack is acquired by scanning the vertical position of the upper objective. Scale bar, 10 µm. Measured performed on setup 1 (λ=1030 nm). (c) Normalized axial intensity profile from the z-stack of panel b. (d) Simulated axial dependence of the photocurrent’s peak at different excitation powers using the excitation spot of panels a,b. The black curve represents a Gaussian fit of the spot’s axial profile shown in panel c (FWHM=17.5 µm). Colored curves represent the simulated axial dependence of the photocurrent for different excitation powers, taking into account the experimental photocurrent dependence on the excitation power shown in the inset (see Methods for more details on the model used) Inset: Black dots represent measured normalized photocurrent from a CoChR expressing CHO cell (λ=1030 nm, setup 1), fitted by an exponential (red curve). (e) Experimental (violet dots; n=2 CoChR expressing cells) and simulated (red line) axial dependence of photocurrent’s peak for the 10 µm diameter holographic spot of panels a,b using an excitation power of 80 µW/µm2. The blue curve is a Gaussian fit of the experimental data (FWHM=43 µm). The black curve represents the Gaussian fit of the spot’s axial profile shown in panel c. (f) Lateral (x,z) view of the fluorescence emission generated by a 10 µm diameter holographic temporally focused spot on a thin fluorescent layer. Scale bar, 10 µm. Measurement performed on setup 2, modified by adding a diffraction grating for temporal focusing. (g) Experimental (violet dots) axial dependence of the photocurrent’s peak and Gaussian fit (blue line) generated by using the temporally focused spot shown in (f) (n=1 CoChR expressing cell; n=1 soCoChR expressing cell). The black curve represents the optical axial resolution of the temporally focused spot of panel f.
Figure S9. Fluorescence plots along neurites, for cells expressing CoChR-GFP or soCoChR-GFP in slices. (a–b) Box-and-whiskers plots of GFP intensity versus position along a neurite, normalized to GFP intensity at the soma, extracted from 2P images (setup 1) of neurites of (a) CoChR-GFP (n = 9 neurites taken from 4 cells from 3 slices from 3 mice) and (b) CoChR-KA2(1–150)-GFP expressing cells (soCoChR-GFP; n = 9 neurites taken from 6 cells from 2 slices from 2 mice) in acute slices. Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values, respectively. For CoChR the GFP fluorescence decays slower than the corresponding current (Fig. 3e); this is probably due to a contribution to the fluorescence from the dense distribution of out of focus processes from neighbouring cells.
Figure S10. Absolute currents at different distances away from the soma for CoChR- and soCoChR-expressing cells in slices. A bar plot showing the peak currents induced by illumination of neurites at different distances from the soma (corresponding to experiments reported in Fig. 3e), for CoChR-GFP- (blue bars) and soCoChR-GFP (red bars) -expressing neurons. Bars reports are mean ± s.e.m.; dots denote values for single neurites.
Figure S11. Multi-spot holographic illumination and diffraction efficiency correction. (a) Max projection of the fluorescence generated by a sequence of 15 µm diameter holographic spots placed on a square grid. The generation of a variable size (from 0 to 15 µm) correction holographic spot (not shown) is applied to correct for the position-dependent diffraction efficiency (see Methods). Scale bar: 30 µm. (b) Vertical and horizontal intensity profiles corresponding to panel a with and without the correction of diffraction efficiency, averaged over the 5 columns and lines of the matrix, respectively. (c) Fluorescence generated by the simultaneous projection of 7 randomly placed holographic spots with equal intensity, corrected for diffraction efficiency. Scale bar: 30 µm. (d) Graded input pattern used to calculate the hologram generating the illumination pattern in panel c (see Methods). Scale bar: 30 µm. (e) Axial intensity profile of the fluorescence induced by a 10 µm holographic spot displaced over different axial positions relative to the microscope objective focal plane without diffraction efficiency correction (black curves) and with the correction (red curve) using an external correction spot (not shown). (f) Superposition of the axial intensity profiles of panel e (with correction of diffraction efficiency), after being centered at z=0. The black curve represents the average profile with FWHM= 18.5 µm. (g) Axial intensity profile of the multispot holographic pattern reported in Figure 5b. The black curve represents the average profile with FWHM=17.5 µm. (h) Lateral view of the 3D fluorescence pattern generated by 3 holographic spots (diameter 10 µm) of equal intensity at axial positions of z = −120 µm; 0; +120µm. Scale bar: 30 µm. (i) Graded input pattern used to calculate the hologram generating the illumination pattern in panel h. As in panel d, different input intensities were assigned to each spot to compensate for the diffraction efficiency dependence on the vertical position and to obtain the same output power density on each spot. (j) Superposition of the axial intensity profiles of the spots of panel h. The black curve represents the average profile with FWHM=18 µm.
Figure S12. Average distances between the soma of the patched cell and the somata of neighboring photostimulated cells. This boxplot corresponds to experiments reported on Fig. 5e,g. Blue and red symbols represent average distances between the several photostimulated neighboring cells (n=5 to 8 cells) and the patched cells (7 patched cells expressing CoChR-GFP from 6 mice, blue symbols; 7 patched cells expressing soCoChR-GFP from 7 mice, red symbols). Horizontal dashed lines represent the average distance over all experiments (n=7 patched CoChR-GFP expressing cells, n=7 patched soCoChR-GFP expressing cells). The height of the rectangles represents two standard deviations of the distribution of the average distances over different experiments. The differences in the distances in the two groups were not significant under a Wilcoxon rank sum test. Statistics are reported in Supplementary Table 4.
Figure S13. Mapping of functional connectivity in brain slices. (a) Currents recorded in whole-cell configuration from a CoChR-GFP expressing cell (same patched cell shown in Fig. 6b) while sequentially positioning the holographic spot on neighboring cells (λ = 920 nm, setup 2, photostimulation power: 120 µW/µm2; three light pulses at 50 Hz of 10 ms duration). Currents corresponding to neighboring cells numbered 3, 5 and 18 (green numbers) are shown in Fig. 6). The distances between each neighboring cell and the patched cell are reported next to each curve (grey numbers). Each trace is the average of three trials. Currents were recorded before the perfusion of synaptic receptor blockers (ITOT) and after the perfusion of synaptic receptor blockers (IART). The current component sensitive to application of synaptic receptor blockers was: PSCs = ITOT − IART. Black traces: ITOT where the ratio PSCs/IART > 1. Gray traces: ITOT where the ratio PSCs/IART <1. Red traces: ITOT where no PSCs were observed (ITOT = IART). Blue traces: IART. (b) Currents recorded in whole-cell configuration from a soCoChR-GFP expressing cell while sequentially positioning the holographic spot on neighboring cells (λ = 1030 nm, setup 1, photostimulation power: 120 µW/µm2; three light pulses at 50 Hz of 10 ms duration). Currents corresponding to neighboring cells numbered 6, 11 and 22 are shown in Fig. 6). The distances between each neighboring cell and the patched cell are reported next to each curve (grey numbers). Color code as in panel (a).
Acknowledgments
We thank Marta Gajowa for participating in preliminary opsin screening, Fabio Simony and Marta Gajowa for help with viral injections, Christophe Tourain for technical support in building the holographic system, Vincent de Sars for software development, and Jouffret Cécile for her help with CHO cell culture and transfection.
VE thanks the Agence Nationale de la Recherche (ANR-10-INBS-04-01, France-BioImaging Infrastructure network; ANR-14-CE13-0016, Holohub), the National Institutes of Health (NIH 1-U01-NS090501-01), the FRC and the Rotary Club through the program « Espoir en Tete and the Getty Lab. OAS thanks the Simons Foundation for the Social Brain Fellowship and the ISEF (International Sephardic Educational Foundation) for the ISEF postdoctoral fellowship. VE and EB thank the Human Frontiers Science Program (Grant RGP0015/2016) for financial support. EB additionally acknowledges, for funding, John Doerr, the Open Philanthropy Project, the HHMI-Simons Faculty Scholars Program, NIH R44EB021054, the MIT Media Lab, NIH 1R24MH106075, NIH 2R01DA029639, NIH 1R01NS087950, NIH 1R01MH103910, NIH Director's Pioneer Award 1DP1NS087724, and NIH 1R01GM104948.
Footnotes
Data Availability Statement
All relevant data are available from the authors. Sequences are available in this manuscript under Supplementary information. Clones will be made available through the nonprofit DNA distributor Addgene, as well as the UNC and UPenn Viral cores.
Author contributions
O. A. S. designed, screened and tested soma targeted opsins. O.A.S, C.L. and K.P performed and analyzed 1P experiments in cultured cells. V. Z. and D. T. performed and analyzed 2P experiments in cultured cells and brain slices. V.Z. performed and optimized virus injection and implemented electrophysiological recording on the 2P rigs. E.R. and E.P. designed and built up setup 1, D. T. designed and built up setup 2, and optimized multicell stimulation and designed calibration procedures. O.A. S, E. S. B., D. T., V. Z. and V. E. interpreted data, designed experiments, and wrote the paper with contributions from all authors. E. S. B. and V. E. conceived and supervised the project.
Competing Financial Interests Statement
O.A.S., E.S.B. and C.L. are inventors on pending patents covering the described work.
Contributor Information
Or A. Shemesh, Media Laboratory, Massachusetts Institute of Technology (MIT), Cambridge, Massachusetts, USA; Department of Biological Engineering, MIT, Cambridge, Massachusetts, USA; Center for Neurobiological Engineering, MIT, Cambridge, Massachusetts, USA; Department of Brain and Cognitive Sciences, MIT, Cambridge, Massachusetts, USA; McGovern Institute for Brain Research, MIT, Cambridge, Massachusetts, USA.
Dimitrii Tanese, Neurophotonics Laboratory, Wave front engineering microscopy group, CNRS UMR8250, Université Paris Descartes, 45, rue des Saints Pères, 75270 Paris Cedex 06, France.
Valeria Zampini, Neurophotonics Laboratory, Wave front engineering microscopy group, CNRS UMR8250, Université Paris Descartes, 45, rue des Saints Pères, 75270 Paris Cedex 06, France.
Changyang Linghu, Media Laboratory, Massachusetts Institute of Technology (MIT), Cambridge, Massachusetts, USA; Department of Biological Engineering, MIT, Cambridge, Massachusetts, USA; Center for Neurobiological Engineering, MIT, Cambridge, Massachusetts, USA; Department of Brain and Cognitive Sciences, MIT, Cambridge, Massachusetts, USA; McGovern Institute for Brain Research, MIT, Cambridge, Massachusetts, USA.
Kiryl Piatkevich, Media Laboratory, Massachusetts Institute of Technology (MIT), Cambridge, Massachusetts, USA; Department of Biological Engineering, MIT, Cambridge, Massachusetts, USA; Center for Neurobiological Engineering, MIT, Cambridge, Massachusetts, USA; Department of Brain and Cognitive Sciences, MIT, Cambridge, Massachusetts, USA; McGovern Institute for Brain Research, MIT, Cambridge, Massachusetts, USA.
Emiliano Ronzitti, Neurophotonics Laboratory, Wave front engineering microscopy group, CNRS UMR8250, Université Paris Descartes, 45, rue des Saints Pères, 75270 Paris Cedex 06, France.
Eirini Papagiakoumou, Neurophotonics Laboratory, Wave front engineering microscopy group, CNRS UMR8250, Université Paris Descartes, 45, rue des Saints Pères, 75270 Paris Cedex 06, France; Institut national de la santé et de la recherche médicale (Inserm), France.
Edward S. Boyden, Media Laboratory, Massachusetts Institute of Technology (MIT), Cambridge, Massachusetts, USA; Department of Biological Engineering, MIT, Cambridge, Massachusetts, USA; Center for Neurobiological Engineering, MIT, Cambridge, Massachusetts, USA; Department of Brain and Cognitive Sciences, MIT, Cambridge, Massachusetts, USA; McGovern Institute for Brain Research, MIT, Cambridge, Massachusetts, USA.
Valentina Emiliani, Neurophotonics Laboratory, Wave front engineering microscopy group, CNRS UMR8250, Université Paris Descartes, 45, rue des Saints Pères, 75270 Paris Cedex 06, France.
References
- 1.Boyden ES. A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biol. Rep. 2011;3:11. doi: 10.3410/B3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 2010;13:1433–40. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li C-YT, Poo M-M, Dan Y. Burst spiking of a single cortical neuron modifies global brain state. Science. 2009;324:643–6. doi: 10.1126/science.1169957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houweling AR, Brecht M. Behavioural report of single neuron stimulation in somatosensory cortex. Nature. 2008;451:65–68. doi: 10.1038/nature06447. [DOI] [PubMed] [Google Scholar]
- 5.Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15025–30. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaziri A, Emiliani V. Reshaping the optical dimension in optogenetics. Current Opinion in Neurobiology. 2012;22:128–137. doi: 10.1016/j.conb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Papagiakoumou E, et al. Scanless two-photon excitation of channelrhodopsin-2. Nat. Methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papagiakoumou E, De Sars V, Oron D, Emiliani V. Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt. Express. 2008;16:22039–22047. doi: 10.1364/oe.16.022039. [DOI] [PubMed] [Google Scholar]
- 9.Bègue A, et al. Two-photon excitation in scattering media by spatiotemporally shaped beams and their application in optogenetic stimulation. Biomed. Opt. Express. 2013;4:2869–79. doi: 10.1364/BOE.4.002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaigneau E, et al. Two-Photon Holographic Stimulation of ReaChR. Front. Cell. Neurosci. 2016;10:234. doi: 10.3389/fncel.2016.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ronzitti E, et al. Sub-millisecond optogenetic control of neuronal firing with two-photon holographic photoactivation of Chronos. J. Neurosci. 2017 doi: 10.1523/JNEUROSCI.1246-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez O, et al. Three-dimensional spatiotemporal focusing of holographic patterns. Nat. Commun. 2016;7:11928. doi: 10.1038/ncomms11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papagiakoumou E, et al. Functional patterned multiphoton excitation deep inside scattering tissue. Nat. Photonics. 2013;7:274–278. [Google Scholar]
- 14.Packer AM, et al. Two-photon optogenetics of dendritic spines and neural circuits. Nat. Methods. 2012;9:1202–U103. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anselmi F, Ventalon C, Begue A, Ogden D, Emiliani V. Three-dimensional imaging and photostimulation by remote-focusing and holographic light patterning. Proc Natl Acad Sci U S A. 2011;108:19504–19509. doi: 10.1073/pnas.1109111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dal Maschio M, Donovan JC, Helmbrecht TO, Baier H. Linking Neurons to Network Function and Behavior by Two-Photon Holographic Optogenetics and Volumetric Imaging. Neuron. 2017;94:774–789.e5. doi: 10.1016/j.neuron.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 17.Nicholson C, Syková E. Extracellular space structure revealed by diffusion analysis. Trends Neurosci. 1998;21:207–215. doi: 10.1016/s0166-2236(98)01261-2. [DOI] [PubMed] [Google Scholar]
- 18.Valluru L, et al. Ligand binding is a critical requirement for plasma membrane expression of heteromeric kainate receptors. J. Biol. Chem. 2005;280:6085–93. doi: 10.1074/jbc.M411549200. [DOI] [PubMed] [Google Scholar]
- 19.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11:338–46. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs EC, Bongarzone ER, Campagnoni CW, Kampf K, Campagnoni AT. Soma-restricted products of the myelin proteolipid gene are expressed primarily in neurons in the developing mouse nervous system. Dev. Neurosci. 2003;25:96–104. doi: 10.1159/000072259. [DOI] [PubMed] [Google Scholar]
- 21.Lim ST, Antonucci DE, Scannevin RH, Trimmer JS. A Novel Targeting Signal for Proximal Clustering of the Kv2.1 K+ Channel in Hippocampal Neurons. Neuron. 2000;25:385–397. doi: 10.1016/s0896-6273(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 22.Garrido JJ, et al. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science. 2003;300:2091–4. doi: 10.1126/science.1085167. [DOI] [PubMed] [Google Scholar]
- 23.Schäfer MKE, et al. L1 syndrome mutations impair neuronal L1 function at different levels by divergent mechanisms. Neurobiol. Dis. 2010;40:222–37. doi: 10.1016/j.nbd.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 24.Bianco A, Dienstbier M, Salter HK, Gatto G, Bullock SL. Bicaudal-D regulates fragile X mental retardation protein levels, motility, and function during neuronal morphogenesis. Curr. Biol. 2010;20:1487–92. doi: 10.1016/j.cub.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran B, Bopp R, Suter B. Null alleles reveal novel requirements for Bic-D during Drosophila oogenesis and zygotic development. Development. 1994;120:1233–42. doi: 10.1242/dev.120.5.1233. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Bennett V. Restriction of 480/270-kD ankyrin G to axon proximal segments requires multiple ankyrin G-specific domains. J. Cell Biol. 1998;142:1571–81. doi: 10.1083/jcb.142.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Ivanova E, Zhang Y, Pan Z-H. rAAV-mediated subcellular targeting of optogenetic tools in retinal ganglion cells in vivo. PLoS One. 2013;8:e66332. doi: 10.1371/journal.pone.0066332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberg KP, Pham A, Werblin FS. Differential targeting of optical neuromodulators to ganglion cell soma and dendrites allows dynamic control of center-surround antagonism. Neuron. 2011;69:713–20. doi: 10.1016/j.neuron.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014;43 doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren Z, et al. Multiple Trafficking Signals Regulate Kainate Receptor KA2 Subunit Surface Expression. J. Neurosci. 2003;23:6608–6616. doi: 10.1523/JNEUROSCI.23-16-06608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grubb MS, Burrone J. Channelrhodopsin-2 localised to the axon initial segment. PLoS One. 2010;5:e13761. doi: 10.1371/journal.pone.0013761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Feng J, Wu C, Lu Q, Pan Z-H. Targeted Expression of Channelrhodopsin-2 to the Axon Initial Segment Alters the Temporal Firing Properties of Retinal Ganglion Cells. PLoS One. 2015;10:e0142052. doi: 10.1371/journal.pone.0142052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker CA, Elyada YM, Parra A, Bolton MM. Cellular resolution circuit mapping with temporal-focused excitation of soma-targeted channelrhodopsin. Elife. 2016;5 doi: 10.7554/eLife.14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 35.Szabo V, Ventalon C, De Sars V, Bradley J, Emiliani V. Spatially Selective Holographic Photoactivation and Functional Fluorescence Imaging in Freely Behaving Mice with a Fiberscope. Neuron. 2014;84:1157–1169. doi: 10.1016/j.neuron.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Brill J, Mattis J, Deisseroth K, Huguenard JR. LSPS/Optogenetics to Improve Synaptic Connectivity Mapping: Unmasking the Role of Basket Cell-Mediated Feedforward Inhibition. eNeuro. 2016;3 doi: 10.1523/ENEURO.0142-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohara K, et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat. Neurosci. 2014;17:269–79. doi: 10.1038/nn.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Little JP, Carter AG. Subcellular Synaptic Connectivity of Layer 2 Pyramidal Neurons in the Medial Prefrontal Cortex. J. Neurosci. 2012;32 doi: 10.1523/JNEUROSCI.1616-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pala A, et al. In Vivo Measurement of Cell-Type-Specific Synaptic Connectivity and Synaptic Transmission in Layer 2/3 Mouse Barrel Cortex. Neuron. 2015;85:68–75. doi: 10.1016/j.neuron.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–5. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu C, Webb WW. Measurement of two-photon excitation cross sections of molecular fluorophores with data from 690 to 1050 nm. J. Opt. Soc. Am. B. 1996;13:481. [Google Scholar]
- 42.Chuong AS, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat. Neurosci. 2014;17:1123–1129. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song S, Sjöström PJ, Reigl M, Nelson S, Chklovskii DB. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. 2005;3:e68. doi: 10.1371/journal.pbio.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura Y, Dantzker JLM, Callaway EM. Excitatory cortical neurons form fine-scale functional networks. Nature. 2005;433:868–873. doi: 10.1038/nature03252. [DOI] [PubMed] [Google Scholar]
- 45.Ikegaya Y, et al. Synfire chains and cortical songs: temporal modules of cortical activity. Science. 2004;304:559–64. doi: 10.1126/science.1093173. [DOI] [PubMed] [Google Scholar]
- 46.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 2007;10:663–8. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 47.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat. Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 48.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma D, et al. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–9. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 50.Hofherr A, Fakler B, Klöcker N. Selective Golgi export of Kir2.1 controls the stoichiometry of functional Kir2.x channel heteromers. J. Cell Sci. 2005;118:1935–43. doi: 10.1242/jcs.02322. [DOI] [PubMed] [Google Scholar]
- 51.Chuong AS, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat. Neurosci. 2014;17:1123–9. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ducros M, et al. Efficient large core fiber-based detection for multi-channel two-photon fluorescence microscopy and spectral unmixing. J. Neurosci. Methods. 2011;198:172–80. doi: 10.1016/j.jneumeth.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 53.Lutz C, et al. Holographic photolysis of caged neurotransmitters. Nat. Methods. 2008;5:821–7. doi: 10.1038/nmeth.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerchberg R, Saxton W. A practical algorithm for the determination of the phase from image and diffraction plane pictures. Opt. 1972;35 [Google Scholar]
- 55.Haist T, Schönleber M, Tiziani HJ. Computer-generated holograms from 3D-objects written on twisted-nematic liquid crystal displays. Opt. Commun. 1997;140:299–308. [Google Scholar]
- 56.Hernandez O, Guillon M, Papagiakoumou E, Emiliani V. Zero-order suppression for two-photon holographic excitation. Opt. Lett. 2014;39:5953–6. doi: 10.1364/OL.39.005953. [DOI] [PubMed] [Google Scholar]
- 57.Golan L, Reutsky I, Farah N, Shoham S. Design and characteristics of holographic neural photo-stimulation systems. J. Neural Eng. 2009;6:66004. doi: 10.1088/1741-2560/6/6/066004. [DOI] [PubMed] [Google Scholar]
- 58.Yang S, et al. Three-dimensional holographic photostimulation of the dendritic arbor. J. Neural Eng. 2011;8:46002. doi: 10.1088/1741-2560/8/4/046002. [DOI] [PubMed] [Google Scholar]
- 59.Conti R, Assayag O, de Sars V, Guillon M, Emiliani V. Computer Generated Holography with Intensity-Graded Patterns. Front. Cell. Neurosci. 2016;10 doi: 10.3389/fncel.2016.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mütze J, et al. Excitation spectra and brightness optimization of two-photon excited probes. Biophys. J. 2012;102:934–44. doi: 10.1016/j.bpj.2011.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horton NG, et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photonics. 2013;7:205–209. doi: 10.1038/nphoton.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papagiakoumou E, de Sars V, Oron D, Emiliani V. Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt. Express. 2008;16:22039–47. doi: 10.1364/oe.16.022039. [DOI] [PubMed] [Google Scholar]
- 63.Dell RB, Holleran S, Ramakrishnan R. Sample Size Determination. ILAR J. 2002;43:207–213. doi: 10.1093/ilar.43.4.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. An N-terminal domain of kainate receptor subunit 2 is retained in the soma. (a) A hippocampal neuron expressing GFP. (b) A hippocampal neuron expressing KA2-GFP. (c) A hippocampal neuron expressing KA2(1–150)-GFP. Scale bars for a–c: 50 µm. (d, e, f) Box-and-whiskers plots of GFP brightness versus position along a neurite, normalized to GFP brightness at the soma for GFP (n=5 neurites from 3 cells), KA2-GFP (n=5 neurites from 3 cells) and KA2(1–150)-GFP (n=5 neurites from 3 cells) respectively. Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values respectively. See Supplementary Table 2 for Bonferroni-corrected Kolmogorov-Smirnov tests for Figure S1.
Figure S2. CoChR-NaV1.2(II–III)-GFP expression and photostimulation in cultured hippocampal neurons. (a–d) Images are presented for cultured hippocampal neurons expressing CoChR-NaV1.2(II–III)-GFP, along with the countermarker mCherry. (a) A hippocampal neuron in culture expressing CoChR-NaV1.2(II–III)-GFP and mCherry, seen in the GFP channel (scale bar: 100 µm). (b) Zoomed-in image from the yellow rectangle of a. (c) The neuron of b, seen in the mCherry channel (magenta), along with other nearby neurons. (d) Merge of b and c (scale bar for b–d: 20 µm). (e) Whole cell current clamp recording of a cultured hippocampal neuron expressing CoChR-NaV1.2(II–III)-GFP, under current injection (10 ms duration; gray rectangle) and optical stimulation (480nm, 34.84mW/mm2, 1 ms duration; blue rectangle). Rectangles not to scale. (f) Box-and-whiskers plot of GFP brightness versus position along a neurite, normalized to GFP brightness at the soma, extracted from neurites of cultured hippocampal neurons expressing CoChR-NaV1.2(II–III)-GFP (n = 5 neurites taken from 5 cells from 4 cultures). Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values respectively.
Figure S3. CoChR-GFP-Kv2.1motif expression is somatodendritic. (Ai, Aii) Hippocampal neurons expressing CoChR-GFP-Kv2.1motif. (B, C) Box-and-whisker plots of brightness versus position along a neurite, normalized to brightness at the soma for CoChR-GFP-Kv2.1motif (B, n = 5 neurites from 5 cells from 2 cultures) and CoChR-GFP (C, n = 7 neurites from 5 cells from 2 cultures). Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Red line denotes the median. Top and bottom whiskers indicate the highest and lowest values respectively. n.s., not significant, comparing B vs. C; see Supplementary Table 3 for Bonferroni-corrected Kolmogorov-Smirnov tests for Figure S3. Scale bar: 20µm.
Figure S4. In vitro single photon characterization of CoChR and soCoChR. (a) Action spectra for CoChR (n = 9 cells) and soCoChR (n = 10 cells) measured in cultured neurons. Plotted data are mean ± s.e.m. (b) Blue light driven spike probability as a function of irradiance for CoChR (magenta) and soCoChR (black) expressing cells. Plotted data are mean ± s.e.m (n = 6 cells for each opsin). All blue light spiking protocols used 2 ms width light pulses at 480 nm. (c) Blue light driven photocurrents as a function of irradiance for CoChR (magenta) and soCoChR (black) expressing cells. Plotted data are mean ± s.e.m (n = 7 cells for each opsin). All blue light protocols used 2 ms pulse width at 480 nm. (d) Blue light driven spike fidelity for CoChR (magenta) and soCoChR (black) expressing cells. All blue light spiking protocols used a train of 40 pulses, 2 ms pulse width, at 480 nm, at 5 mW/mm2. Plotted data are mean ± s.e.m (n = 7 – 10 cells for each opsin). See Supplementary Table 3 for full statistics for Figure S4.
Figure S5. Membrane properties of neurons expressing somatic vs. untargeted molecules.
Cultured hippocampal neurons expressing CoChR-GFP (n = 10 cells from 3 cultures), KA2(1–150)-GFP (n = 10 cells from 3 cultures), and soCoChR-GFP (n = 10 cells from 3 cultures) were patched 14 days following AAV transduction. (a) Resting potential. We performed a Kruskal-Wallis Test (not significant, P = 0.7549, chi-square = 0.5624). (b) Holding current. Kruskal-Wallis Test, not significant, P = 0.9875, chi-square = 0.0252. (c) Membrane capacitance. Kruskal-Wallis Test, not significant, P = 0.9817, chi-square = 0.0369. (d) Membrane resistance. Kruskal-Wallis Test, not significant, P = 0.9205, chi-square = 0.1656. For all panels, red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values.
Figure S6. Detailed schematics of the holographic microscopes, for both setup 1 and setup 2. PC, Polarizer Cube; L/2, Half Wave Plate; M, Mirror; L, Lens; SLM, Spatial Light Modulator; CL, Cylindrical Lens; D, Dichroic; GM, Galvanometric Mirrors; F, Filter; OBJ, Objective; C, Condenser; PoC, Pockels Cell. See detailed description in Methods section.
Figure S7. 2P action spectrum and normalized excitation cross-section of CoChR. (a) Normalized peak current of CoChR as a function of wavelength, measured in cultured CHO cells. Photostimulation was performed using a 2P Gaussian spot (FWHM=38 µm) at different wavelengths, at constant photon flux (2.4±0.3 1026 photons/s·m2) over the investigated wavelength range. Plotted data are mean ± s.e.m., with raw data in open circles, n=5 cells. (b) Normalized 2P excitation cross section, extracted from the photocurrent kinetics (see Methods). Plotted data are mean ± s.e.m., with raw data in open circles, n=5 cells.
Figure S8. Axial resolution of 2P holographic spots. Top (a) and lateral (b) view of the fluorescence emission generated by a 10 µm diameter holographic spot on a thin fluorescent layer. The fluorescence is collected by a second bottom objective and relayed to a CCD camera; the z-stack is acquired by scanning the vertical position of the upper objective. Scale bar, 10 µm. Measured performed on setup 1 (λ=1030 nm). (c) Normalized axial intensity profile from the z-stack of panel b. (d) Simulated axial dependence of the photocurrent’s peak at different excitation powers using the excitation spot of panels a,b. The black curve represents a Gaussian fit of the spot’s axial profile shown in panel c (FWHM=17.5 µm). Colored curves represent the simulated axial dependence of the photocurrent for different excitation powers, taking into account the experimental photocurrent dependence on the excitation power shown in the inset (see Methods for more details on the model used) Inset: Black dots represent measured normalized photocurrent from a CoChR expressing CHO cell (λ=1030 nm, setup 1), fitted by an exponential (red curve). (e) Experimental (violet dots; n=2 CoChR expressing cells) and simulated (red line) axial dependence of photocurrent’s peak for the 10 µm diameter holographic spot of panels a,b using an excitation power of 80 µW/µm2. The blue curve is a Gaussian fit of the experimental data (FWHM=43 µm). The black curve represents the Gaussian fit of the spot’s axial profile shown in panel c. (f) Lateral (x,z) view of the fluorescence emission generated by a 10 µm diameter holographic temporally focused spot on a thin fluorescent layer. Scale bar, 10 µm. Measurement performed on setup 2, modified by adding a diffraction grating for temporal focusing. (g) Experimental (violet dots) axial dependence of the photocurrent’s peak and Gaussian fit (blue line) generated by using the temporally focused spot shown in (f) (n=1 CoChR expressing cell; n=1 soCoChR expressing cell). The black curve represents the optical axial resolution of the temporally focused spot of panel f.
Figure S9. Fluorescence plots along neurites, for cells expressing CoChR-GFP or soCoChR-GFP in slices. (a–b) Box-and-whiskers plots of GFP intensity versus position along a neurite, normalized to GFP intensity at the soma, extracted from 2P images (setup 1) of neurites of (a) CoChR-GFP (n = 9 neurites taken from 4 cells from 3 slices from 3 mice) and (b) CoChR-KA2(1–150)-GFP expressing cells (soCoChR-GFP; n = 9 neurites taken from 6 cells from 2 slices from 2 mice) in acute slices. Red line denotes the median. Top and bottom edges of the box indicate the 75th and 25th percentiles, respectively. Top and bottom whiskers indicate the highest and lowest values, respectively. For CoChR the GFP fluorescence decays slower than the corresponding current (Fig. 3e); this is probably due to a contribution to the fluorescence from the dense distribution of out of focus processes from neighbouring cells.
Figure S10. Absolute currents at different distances away from the soma for CoChR- and soCoChR-expressing cells in slices. A bar plot showing the peak currents induced by illumination of neurites at different distances from the soma (corresponding to experiments reported in Fig. 3e), for CoChR-GFP- (blue bars) and soCoChR-GFP (red bars) -expressing neurons. Bars reports are mean ± s.e.m.; dots denote values for single neurites.
Figure S11. Multi-spot holographic illumination and diffraction efficiency correction. (a) Max projection of the fluorescence generated by a sequence of 15 µm diameter holographic spots placed on a square grid. The generation of a variable size (from 0 to 15 µm) correction holographic spot (not shown) is applied to correct for the position-dependent diffraction efficiency (see Methods). Scale bar: 30 µm. (b) Vertical and horizontal intensity profiles corresponding to panel a with and without the correction of diffraction efficiency, averaged over the 5 columns and lines of the matrix, respectively. (c) Fluorescence generated by the simultaneous projection of 7 randomly placed holographic spots with equal intensity, corrected for diffraction efficiency. Scale bar: 30 µm. (d) Graded input pattern used to calculate the hologram generating the illumination pattern in panel c (see Methods). Scale bar: 30 µm. (e) Axial intensity profile of the fluorescence induced by a 10 µm holographic spot displaced over different axial positions relative to the microscope objective focal plane without diffraction efficiency correction (black curves) and with the correction (red curve) using an external correction spot (not shown). (f) Superposition of the axial intensity profiles of panel e (with correction of diffraction efficiency), after being centered at z=0. The black curve represents the average profile with FWHM= 18.5 µm. (g) Axial intensity profile of the multispot holographic pattern reported in Figure 5b. The black curve represents the average profile with FWHM=17.5 µm. (h) Lateral view of the 3D fluorescence pattern generated by 3 holographic spots (diameter 10 µm) of equal intensity at axial positions of z = −120 µm; 0; +120µm. Scale bar: 30 µm. (i) Graded input pattern used to calculate the hologram generating the illumination pattern in panel h. As in panel d, different input intensities were assigned to each spot to compensate for the diffraction efficiency dependence on the vertical position and to obtain the same output power density on each spot. (j) Superposition of the axial intensity profiles of the spots of panel h. The black curve represents the average profile with FWHM=18 µm.
Figure S12. Average distances between the soma of the patched cell and the somata of neighboring photostimulated cells. This boxplot corresponds to experiments reported on Fig. 5e,g. Blue and red symbols represent average distances between the several photostimulated neighboring cells (n=5 to 8 cells) and the patched cells (7 patched cells expressing CoChR-GFP from 6 mice, blue symbols; 7 patched cells expressing soCoChR-GFP from 7 mice, red symbols). Horizontal dashed lines represent the average distance over all experiments (n=7 patched CoChR-GFP expressing cells, n=7 patched soCoChR-GFP expressing cells). The height of the rectangles represents two standard deviations of the distribution of the average distances over different experiments. The differences in the distances in the two groups were not significant under a Wilcoxon rank sum test. Statistics are reported in Supplementary Table 4.
Figure S13. Mapping of functional connectivity in brain slices. (a) Currents recorded in whole-cell configuration from a CoChR-GFP expressing cell (same patched cell shown in Fig. 6b) while sequentially positioning the holographic spot on neighboring cells (λ = 920 nm, setup 2, photostimulation power: 120 µW/µm2; three light pulses at 50 Hz of 10 ms duration). Currents corresponding to neighboring cells numbered 3, 5 and 18 (green numbers) are shown in Fig. 6). The distances between each neighboring cell and the patched cell are reported next to each curve (grey numbers). Each trace is the average of three trials. Currents were recorded before the perfusion of synaptic receptor blockers (ITOT) and after the perfusion of synaptic receptor blockers (IART). The current component sensitive to application of synaptic receptor blockers was: PSCs = ITOT − IART. Black traces: ITOT where the ratio PSCs/IART > 1. Gray traces: ITOT where the ratio PSCs/IART <1. Red traces: ITOT where no PSCs were observed (ITOT = IART). Blue traces: IART. (b) Currents recorded in whole-cell configuration from a soCoChR-GFP expressing cell while sequentially positioning the holographic spot on neighboring cells (λ = 1030 nm, setup 1, photostimulation power: 120 µW/µm2; three light pulses at 50 Hz of 10 ms duration). Currents corresponding to neighboring cells numbered 6, 11 and 22 are shown in Fig. 6). The distances between each neighboring cell and the patched cell are reported next to each curve (grey numbers). Color code as in panel (a).