Abstract
The relationship of the gastrointestinal microbiome to health and disease is of major research interest, including the effects of the gut microbiota on age related conditions. Here we report on the outcome of a project to collect stool samples on a large number of community dwelling elderly men using the OMNIgene-GUT stool/feces collection kit (OMR-200, DNA Genotek, Ottawa, Canada). Among 1328 men who were eligible for stool collection, 982 (74%) agreed to participate and 951 submitted samples. The collection process was reported to be acceptable, almost all samples obtained were adequate, the process of sample handling by mail was uniformly successful. The DNA obtained provided excellent results in microbiome analyses, yielding an abundance of species and a diversity of taxa as would be predicted. Our results suggest that population studies of older participants involving remote stool sample collection are feasible. These approaches would allow large scale research projects of the association of the gut microbiota with important clinical outcomes.
Keywords: Microbiome, Aging, Men, Gastrointestinal, Genotyping, Stool
Abbreviations: CMMR, Alkek Center for Metagenomics and Microbiome Research; DNA, Deoxyribonucleic Acid; DXA, Dual Energy X-ray Absorptiometry; FFQ, Food Frequency Questionnaire; HRpQCT, High Resolution Peripheral Quantitative Computed Tomography; MrOS, Osteoporotic Fractures in Men Study; NCATS, National Center for Advancing Translational Sciences; NIA, National Institute on Aging; NIAMS, National Institute of Arthritis and Musculoskeletal and Skin Diseases; OTU, Operational Taxonomic Units; rDNA, Recombinant Deoxyribonucleic Acid; SILVA database, a comprehensive on-line resource for quality checked and aligned ribosomal RNA sequence data; UPARSE, cluster algorithm; USEARCH, search and cluster algorithm
1. Introduction
The human microbiome is the composition of microorganisms (e.g. bacteria, virus, fungi, and parasites) and microbial products that inhabits the human body. The microbes in a healthy human adult are estimated to at least equal the number of human cells [1]. It has been long known that bacteria are involved in certain body processes, such as digesting food and producing vitamins, but the microbiome may have a much broader impact on our health than was previously realized. The community of microbes in an individual may influence the susceptibility to certain infectious diseases, as well as contribute to disorders such as obesity [2], diabetes [3], and some chronic illnesses of the gastrointestinal system such as Crohn's disease and irritable bowel syndrome [4].
The influence of the gastrointestinal microbiome on health and disease is of major research interest. Stool specimens offer the most accessible means of assessing the gut microbial community, and the number of reports concerning the association of stool microbiome with physiological and medical conditions is increasing quickly. Nevertheless, there are few data concerning practical methods for collecting stool specimens for microbiome analyses. This is particularly true in the context of large, population-based studies that demand successful recruitment approaches, acceptable specimen collection methods, and successful transport and processing of large numbers of samples. These issues may be especially challenging in the study of elderly populations in which remote sample collection is imperative.
Here we report the results of an effort to collect stool samples from a large number of older men enrolled in the Osteoporotic Fractures in Men Study (MrOS). Using convenient stool collection methods, we developed a successful approach for remote sample acquisition and preservation that could be applied to similar population-based research.
2. Materials and methods
2.1. Subjects and overall MrOS study protocol
MrOS study is a prospective study of 5994 older men, recruited at six clinical U.S. sites between 2000 and 2002. The cohort and recruitment methods have been previously described [5]. At baseline, participants were at least 65 years of age, able to consent, walked without assistance of another person, and did not have bi-lateral hip replacement or any condition that in the judgment of the site investigator that would likely impair participation in the study [6]. The Institutional Review Boards at all sites reviewed and approved the study and all participants provided informed consent.
Since its inception and baseline examination, participants have returned for at least 4 full in-clinic examinations, completed tri-annual postcards concerning outcomes of interest, and completed two extensive interim questionnaires concerning lifestyle and medical issues. The rate of ongoing participation in the MrOS study has been excellent. Of surviving participants, 99.4% had completed all study visits and 88.0% completed all tri-annual postcards used for regular follow-up.
In May 2014, men began returning for a fourth clinic visit that was comprehensive and complex. Participants completed a health history questionnaire at home, obtained objective activity monitoring by wearing accelerometry equipment for 7 days, and completed a variety of measures during a research clinic visit at one of the six participating institutions. Clinic measures included whole body, hip and spine dual energy x-ray absorptiometry (DXA), high resolution peripheral quantitative computed tomography (HRpQCT), measures of physical performance (short physical performance battery; 400 m walk, force plate for lower extremity power), measures of height and weight, blood and urine specimen collection, questionnaires and interviews. Participants were asked to rate their overall health in relationships to their similarly-aged peers (excellent, good, fair, poor). The visit took approximately 4–5 h to complete. The goal was to obtain these measures on 1950 men (70% of the surviving cohort).
2.2. Stool collection protocol
In March–April 2015, all six institutions began the collection of stool specimens for microbiome analyses. To recruit men for stool sample collection, study staff asked subjects at the research clinic visit if they would agree to provide a stool specimen to study the microbiome. For those who agreed, staff demonstrated the process with the collection materials in hand. Participants then took home a collection kit that included a toilet hat, instructions for collecting/mailing the specimen, a collection tube, exam gloves, alcohol wipes, a postage-paid return mailing envelope, and biohazard mailing bag. A short questionnaire about the collection time and date was included in the collection kit. A dietary recall questionnaire (Block 98.2 food frequency questionnaire (FFQ); modified for MrOS to capture the most frequently consumed sources of calcium, vitamin D, and other selected nutrients influencing risk of osteoporosis and prostate cancer in US men (Nutritionquest, Berkeley CA)) was completed by the participant at home and returned with the stool specimen.
Participants at all 6 MrOS sites mailed the samples directly to the Portland site for initial processing. Immediately upon receipt, study staff opened the packages to check for stool sample adequacy and to ensure the paperwork (time and date of collection) had been completed. Samples were then stored in a −80 °C freezer. If after two weeks an expected stool sample had not arrived at the central lab, the participant's study site was notified and a follow up phone call was made to ensure a sample was collected.
The participants from the Portland site completed an additional short questionnaire regarding the tolerability of the collection, time required and any perceived complications. This was mailed to the clinic with the specimen.
2.3. Kit for stool sample collection and preservation
The OMNIgene-GUT stool/feces collection kit (OMR-200, DNA Genotek, Ottawa, Canada) was used to collect stool samples. The OMNIgene-GUT collection kit is designed for the self-collection of a consistent volume of stool and preservation of microbial DNA. The tube includes a non-toxic stabilizing reagent and mixing apparatus and is safe for home use; personal protective equipment (e.g. gloves, eyewear, etc.) is not required. After the sample is collected and the tube is capped, the user vigorously shakes the tube for 30 s to homogenize and liquefy the sample. At that point the stool DNA is preserved for at least 60 days at ambient temperature [7], [8]. The collection process has been successfully used in other clinical studies [9], [10].
2.4. Bacterial genotyping
Six hundred specimens from unique participants were sent to the Alkek Center for Metagenomics and Microbiome Research (CMMR) at Baylor College of Medicine in Houston, Texas for microbiome analysis. Samples were arrayed in boxes and shipped on dry ice over-night with an accompanying sample manifest that included de-identified sample IDs and box positions. Upon delivery, samples were reconciled with the provided manifest and stored at −80 °C until further processing. For bacterial genomic DNA extraction, samples were thawed at room temperature to re-liquefy the samples and 200 μL of stool suspension were transferred to the extraction deep-well plate. For samples where the fecal material was too thick to pipette, an equivalent volume was transferred using a sterile and disposable spatula. DNA extraction was carried out in the Hamilton STARlet platform following the standard MoBio PowerMag Soil DNA extraction protocol. Extracted DNA was subjected to 16S (v4) rDNA amplification using primers 515F and 806R containing Illumina adapters and a single-end barcode allowing pooling and direct sequencing of PCR products [11]. Amplicons were visualized via gel electrophoresis and quantified via automated Quant-iT PicoGeen assay. Quantified amplicons were normalized and pooled according at DNA mass of 100 ng per sample, and the resulting amplicon pool was cleaned using the ChargeSwitch PCR Clean-up Kit (Invitrogen). The amplicon pool was sequenced on two lanes of an Illumina MiSeq reagent kit v2 (2 × 250 bp) and resulting sequences were demultiplexed based on the unique molecular barcodes, and reads were merged using USEARCH v7.0.1090 [12], allowing zero mismatches and a minimum overlap of 50 bases. Merged reads were trimmed at first base with Q5. In addition, a quality filter was applied to the resulting merged reads and reads containing above 0.05 expected errors were discarded.
The analytic pipeline for 16S rDNA analysis leverages custom analytic packages and pipelines developed at the CMMR to provide summary statistics and quality control measurements for each sequencing run, as well as multi-run reports and data-merging capabilities for validating built-in controls (known and blank) and characterizing microbial communities across large numbers of samples or sample groups.
16Sv4 rDNA gene sequences were clustered into Operational Taxonomic Units (OTUs) at a similarity cutoff value of 97% using the UPARSE algorithm [13]. OTUs were mapped to an optimized version of the SILVA Database [14] containing only the 16Sv4 region to determine taxonomies. Abundances were recovered by mapping the demultiplexed reads to the UPARSE OTUs. A custom script constructed a rarefied OTU table from the output files generated in the previous two steps for downstream analyses of alpha-diversity, beta-diversity [15] and phylogenetic trends.
3. Results
The mean age of the men at the time of their visit was 85.0 ± 4.5 years. After the stool collection process began 1328 men completed Visit 4 and 982 (74%) agreed to collect a stool specimen. Three hundred and forty-six (26%) men refused or were deemed ineligible to collect stool samples. There were a few significant differences between those who agreed and those who refused, but the differences were very small. The men who agreed were slightly younger, more active, stronger, and more often reported good or excellent health than those who declined, but were otherwise similar (see Table 1). The reasons stool collection was refused included a belief the collection process was unpleasant or discomfortable with collecting a stool sample, unwillingness to add another collection measure to an already lengthy research encounter, and health problems that made it difficult or impossible to collect a sample. In some instances, clinic staff decided against asking for stool collection because of cognitive or physical limitations, including vision impairment and a lack of comprehension of the stool collection directions when the kit was presented. There was variation among the clinic sites in the proportion of men who agreed to participate. The reasons for this variation were unclear although may have been related to both staff and participant characteristics.
Table 1.
Characteristics of men who agreed to participate in the stool sample study compared to men who did not agree to participate in the study, among men who were asked to participate (n = 1328). Results are reported in mean (±SD) or n (%).
| Agreed to participate (n = 982) | Did not agree to participate (n = 346) | p-valuea | |
|---|---|---|---|
| Age | 84.2 (±4.0) | 85.9 (±4.9) | <0.001 |
| BMI | 27.0 (±3.8) | 26.8 (±3.7) | 0.53 |
| Race | 0.64 | ||
| White | 875 (89.1) | 310 (89.6) | |
| African American | 30 (3.1) | 15 (4.3) | |
| Asian | 41 (4.2) | 10 (2.9) | |
| Hispanic | 22 (2.2) | 7 (2.0) | |
| Other | 14 (1.4) | 4 (1.2) | |
| Site | <0.001 | ||
| Birmingham | 120 (12.2) | 72 (20.8) | |
| Minneapolis | 164 (16.7) | 92 (26.6) | |
| Palo Alto | 137 (14.0) | 60 (17.3) | |
| Pittsburgh | 142 (14.5) | 44 (12.7) | |
| Portland | 177 (18.0) | 66 (19.1) | |
| San Diego | 242 (19.1) | 12 (3.5) | |
| Self-reported health | 0.05 | ||
| V poor/poor/fair | 108 (11.0) | 51 (14.7) | |
| Good/excellent | 873 (89.0) | 287 (83.0) | |
| PASE score | 119.4 (±65.4) | 86.3 (±64.1) | <0.001 |
| Grip strength | 35.6 (±7.7) | 33.9 (±8.5) | 0.001 |
| Unable | 28 (2.9) | 28 (8.1) | <0.001 |
Comparisons were made by chi-square tests for categorical variables or ANOVA for continuous variables.
Of the 982 men who said they would collect a sample, 951 completed the collection. Only 20 of the 951 (2%) samples were unacceptable (samples that were completely dry or the collection tube was improperly sealed). Of those 20, a second stool sample was obtained in all but six.
At the time of the stool collection, men at the Portland site were asked to complete a short questionnaire about the collection process. Of the 174 Portland men who successfully collected a stool sample, 157 (90%) completed the questionnaire. Almost all of those men (97%) reported the instructions were easy to follow. On a scale that rated the difficulty of collection (0 = least, 10 = most) the average was 2.2, and the average time to collect a sample was 9 min. Most Portland men (87%) said they would participate in stool collection again (Table 2).
Table 2.
Stool collection acceptability questionnaire.
| Portland only questionnaire | Volunteer again (y/n) | Easy instructions (y/n) | How easy/difficult (0–10) | Average time to collect (min) |
|---|---|---|---|---|
| Collected stool = 174 | ||||
| Completed the questionnaire = 157 (90%) | Yes = 136 (87%) | Yes = 152 (97%) | 2.2 | 9 |
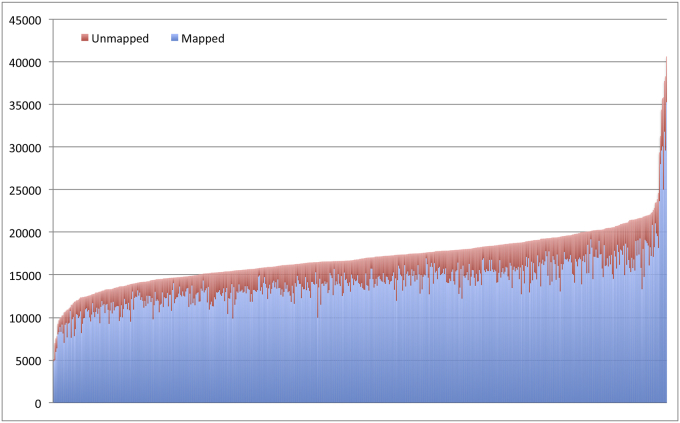
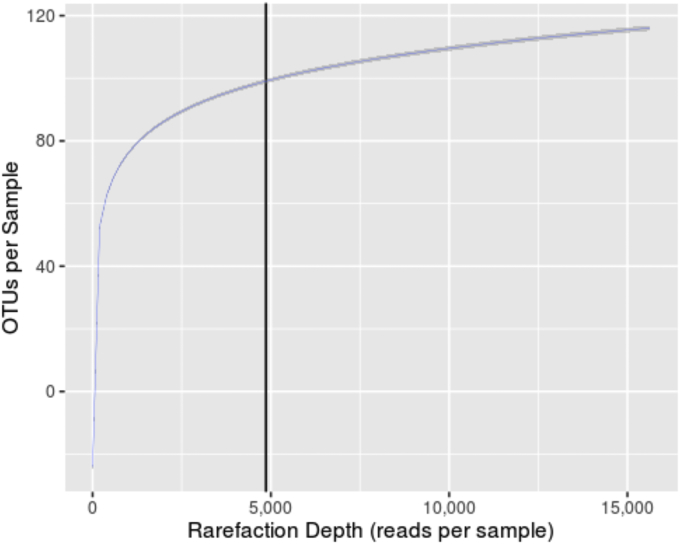
The stool samples were uniformly found to be of sufficient quality to support microbiome analyses. A total of 10,243,047 16Sv4 rDNA reads were successfully analyzed and merged following the parameters described (Methods), of which 82.9% were mapped to the SILVA (v123) database [14]. Merged-raw and merged-mapped reads were evenly distributed acrossed the dataset (Fig. 1). For merged-raw reads, the minimum number reads obtained for a sample was 5115 and the maximum was 40,629. The mean amount of merged-raw reads obtained was 17,072 and the median 16,880. For merged-mapped reads, the minimum number reads obtained for a sample was 4863 and the maximum was 35,263. The mean amount of merged-raw reads obtained was 14,156 and the median 14,034. For overall diversity and taxonomic analyses, the dataset was uniformly normalized to 4863 mapped reads per sample (Fig. 2). This resulted in the inclusion of all 600 samples in the analysis.
Fig. 1.
Distribution of merged reads mapping to the SILVA (v123) database at 97% identity. 600 samples sorted by total merged reads (mapped + unmapped).
Fig. 2.
Average amount of Operational Taxonomic Units (OTUs) identified at the rarefaction depth chosen (vertical line) for this dataset.
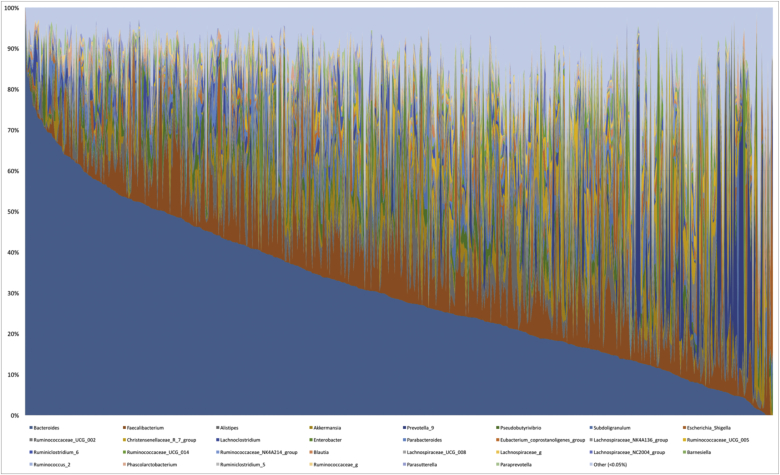
A total of 1040 bacterial Operational Taxonomic Units (OTUs) were found in the dataset broadly ranging in relative abundance. These OTUs were mapped to 275 distinct taxa of which 238 were resolved to the genus level. Table 3 describes the mean and median relative abundance of the top 30 genera identified in the dataset. Genera of the Bacteroides and Firmicutes phyla dominated in the microbiomes identified followed by Proteobacteria and Verrucomicrobia. The most common genus in the dataset was Bacteroides followed by Faecalibacterium, Alistipes, Akkermansia, and Prevotella.
Table 3.
Genera representing 0.5% or more in average relative abundance across the entire dataset (n = 600). Operational taxonomic units (OTUs) that could not be resolved to the genus level are represented by the next level of taxonomic resolution available.
| Genera | Mean | Median | Std. Deviation | Range |
|---|---|---|---|---|
| Bacteroides | 31.19% | 28.38% | 18.84% | 0.02%–97.68% |
| Faecalibacterium | 8.50% | 7.18% | 7.23% | 0.00%–48.55% |
| Alistipes | 4.54% | 3.59% | 4.40% | 0.00%–32.74% |
| Akkermansia | 3.75% | 0.99% | 7.14% | 0.00%–66.23% |
| Pervotella_9 | 3.54% | 0.00% | 12.10% | 0.00%–82.13% |
| Pseudobutyrivibrio | 3.03% | 2.47% | 2.49% | 0.00%–18.40% |
| Subdoligranulum | 2.69% | 1.57% | 3.72% | 0.00%–57.39% |
| Escherichia_Shigella | 2.58% | 0.04% | 7.24% | 0.00%–72.69% |
| Ruminococcaceae_UCG_002 | 2.46% | 1.76% | 2.68% | 0.00%–17.23% |
| Christensenellaceae_R_7_group | 2.28% | 0.61% | 3.73% | 0.00%–25.83% |
| Lachnoclostridium | 1.99% | 1.34% | 2.35% | 0.00%–33.33% |
| Enterobacter | 1.88% | 0.02% | 6.82% | 0.00%–70.55% |
| Parabacteroides | 1.88% | 1.21% | 2.51% | 0.00%–25.97% |
| Eubacterium_coprostanoligenes_group | 1.51% | 0.65% | 1.96% | 0.00%–17.05% |
| Lachnospiraceae_NK4A136_group | 1.32% | 0.74% | 1.73% | 0.00%–16.57% |
| Ruminococcaceae_UCG_005 | 1.27% | 0.48% | 1.90% | 0.00%–15.40% |
| Ruminiclostridium_6 | 1.24% | 0.28% | 2.27% | 0.00%–19.31% |
| Ruminococcaceae_UCG_014 | 0.89% | 0.23% | 1.64% | 0.00%–11.76% |
| Ruminococcaceae_NK4A214_group | 0.89% | 0.44% | 1.33% | 0.00%–11.19% |
| Blautia | 0.85% | 0.53% | 1.43% | 0.00%–21.14% |
| Lachnospiraceae_UCG_008 | 0.85% | 0.43% | 1.95% | 0.00%–27.04% |
| Lachnospiraceae_g | 0.83% | 0.56% | 0.95% | 0.00%–6.35% |
| Lachnospiraceae_NC2004_group | 0.77% | 0.04% | 1.63% | 0.00%–14.70% |
| Barnesiella | 0.76% | 0.02% | 1.25% | 0.00%–9.25% |
| Ruminococcus_2 | 0.72% | 0.29% | 1.45% | 0.00%–20.58% |
| Phascolarctobacterium | 0.65% | 0.29% | 1.25% | 0.00%–15.32% |
| Ruminiclostridium_5 | 0.62% | 0.37% | 0.74% | 0.00%–5.26% |
| Ruminococcaceae_g | 0.61% | 0.45% | 0.60% | 0.00%–5.45% |
| Parasutterella | 0.57% | 0.06% | 1.03% | 0.00%–9.99% |
| Paraprevotella | 0.56% | 0.00% | 1.46% | 0.00%–9.54% |
A total of 275 genera were identified in the dataset broadly ranging in relative abundance. Bacteroides was found to be the genera in most abundance (mean 31.2%, range: 0.02%–97.68%), followed by Faecalibacterium, Alistipes, Akkermansia, and Prevotella (Fig. 3).
Fig. 3.
Distribution of the relative abundance of top 30 genera identified order by abundance of Bacteroides.
4. Discussion
The collection of stool samples from a large number of community dwelling, elderly people could present challenges in terms of recruitment, study staff resource management and sample collection. We developed a process that was very feasible, and demonstrated that it is quite acceptable to participants in an established epidemiological cohort. The quality of the DNA derived from samples collected using these methods was excellent and will support analyses of associations of taxonomic classes with clinical metadata collected from MrOS participants. Similar protocol designs should be useful in other studies involving large populations.
Of 1328 men who were asked to provide stool samples, 74% agreed. Our experience suggests that the most successful approach for optimizing acceptance involved face-to-face discussions with the participant, including clear instructions and demonstrations about the sample collection process. The DNA Genotek OMNIgene collection kit was accepted well by the participants who were surveyed, and the process of sample transfer by mail was uniformly successful.
There are alternative methods for obtaining stool samples, including sample collection at a central collection clinic with immediate processing. This approach is more controlled, could reduce bias inherent in home collections, and there might be specific workers to help with the collection process. But, of course, it costs more. Another option is at-home sample collection, potentially with sample freezing, and shipping to the lab by the participant. Because of inconvenience, these methods are a barrier to participation for many research volunteers. Since the OMNIgene collection method involves rapid stabilization of nucleotides without freezing, the approach we describe offers a more acceptable alternative. As well, it may yield a lower cost compared to the shipment of frozen samples, particularly if transport involves dry ice that is expensive and can be difficult for research participants to handle properly [7].
The DNA obtained by these methods appeared to be well preserved for genotyping. 16S rRNA gene sequence-based taxonomic signatures were uniformly good and the species abundance and diversity of taxa in our cohort were as predicted. These results are similar to those of other recent publications. Mathay et al. [16] reported that stool samples stabilized with OMNIgene reagents yielded DNA suitable for metagenomic analyses. Choo et al. [17] compared a small number of samples collected and stabilized with the OMNIgene kit to parallel samples frozen immediately after collection or freshly extracted without storage. The stabilized samples were found to be superior to those either frozen or immediately processed in terms of amounts of DNA recovered and equivalent in terms of the results of whole genome sequencing. Song et al. reported similar results [8], and Anderson et al. [18] found that fecal samples collected with the OMNIgene system were of similar or superior quality for sequencing DNA and RNA. Overall, collection and stabilization using this approach appears to provide high value samples for nucleic acid analyses.
Some potential limitations of our findings should be considered. Almost 90% of our participants were white men, which may limit the generalizability of our approach. Although we don't anticipate large differences in women or in other racial/ethnic groups, similar studies in other cohorts would be useful. Also, almost 90% of participants reported good/excellent health compared to their peers. The usefulness of these results in a more impaired or frail population may be different.
In summary, we describe methods for successfully collecting stool samples for analysis of the gut microbiota in a large cohort of community dwelling older men. The collection process was well accepted and yielded samples very suitable for analysis. This approach should be very useful for similar large-scale, population-based microbiome research.
Disclosure statement
All authors state that they have no actual or potential conflict of interest including any financial, personal or other relationship with other people or organizations within three years of beginning the work submitted that could have inappropriately influenced their work.
Funding
The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the National Institutes on Aging (NIA), the National Institutes of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Center for Advancing Translational Sciences (NCATS), and NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. OMNIgene-GUT stool/feces collection kits were provided by DNA Genotek, Ottawa, Canada.
Contributor Information
Melanie Abrahamson, Email: abrahamm@ohsu.edu.
Elizabeth Hooker, Email: hookere@ohsu.edu.
Nadim J. Ajami, Email: Nadim.Ajami@bcm.edu.
Joseph F. Petrosino, Email: Joseph.Petrosino@bcm.edu.
Eric S. Orwoll, Email: orwoll@ohsu.edu.
References
- 1.Sender R., Fuchs S., Milo R. Are we really vastly Outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164(3):337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Nehra V. Gut Microbiota: Modulation of host Physiology in obesity. Physiol. (Bethesda) 2016;31(5):327–335. doi: 10.1152/physiol.00005.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paun A., Danska J.S. Modulation of type 1 and type 2 diabetes risk by the intestinal microbiome. Pediatr. Diabetes. 2016 doi: 10.1111/pedi.12424. [DOI] [PubMed] [Google Scholar]
- 4.Baylor College of Medicine . 2016. The Human Microbiome Project.https://www.bcm.edu/departments/molecular-virology-and-microbiology/research/the-human-microbiome-project Available from: [Google Scholar]
- 5.Orwoll E. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study–a large observational study of the determinants of fracture in older men. Contemp. Clin. Trials. 2005;26(5):569–585. doi: 10.1016/j.cct.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Blank J.B. Overview of recruitment for the osteoporotic fractures in men study (MrOS) Contemp. Clin. Trials. 2005;26(5):557–568. doi: 10.1016/j.cct.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Hill C.J. Effect of room temperature transport vials on DNA quality and phylogenetic composition of faecal microbiota of elderly adults and infants. Microbiome. 2016;4(1):19. doi: 10.1186/s40168-016-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song S.J., Amir A., Metcalf J.L., Amato K.R., Xu Z.Z., Humphrey G., Knight R. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems. 2016;1(3) doi: 10.1128/mSystems.00021-16. e00021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeevi D. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell C.M. The effect of prebiotic supplementation with inulin on cardiometabolic health: rationale, design, and methods of a controlled feeding efficacy trial in adults at risk of type 2 diabetes. Contemp. Clin. Trials. 2015;45(Pt B):328–337. doi: 10.1016/j.cct.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso J.G. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 13.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 14.Quast C. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathay C. Method optimization for fecal sample collection and fecal DNA extraction. Biopreserv. Biobank. 2015;13(2):79–93. doi: 10.1089/bio.2014.0031. [DOI] [PubMed] [Google Scholar]
- 17.Choo J.M., Leong L.E., Rogers G.B. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015;5:16350. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson E.L. A robust ambient temperature collection and stabilization strategy: enabling worldwide functional studies of the human microbiome. Sci. Rep. 2016;6:31731. doi: 10.1038/srep31731. [DOI] [PMC free article] [PubMed] [Google Scholar]