Summary
Invasive cells use small invadopodia to breach basement membrane (BM), a dense matrix that encases tissues. Following the breach, a large protrusion forms to clear a path for tissue entry by poorly understood mechanisms. Using RNAi screening for defects in C. elegans anchor cell (AC) invasion, we found that UNC-6(netrin)/UNC-40(DCC) signaling at the BM breach site directs exocytosis of lysosomes using the exocyst and SNARE SNAP-29 to form a large protrusion that invades vulval tissue. Live-cell imaging revealed that the protrusion is enriched in the matrix metalloprotease ZMP-1 and transiently expands AC volume more than 20%, displacing surrounding BM and vulval epithelium. Photobleaching and genetic perturbations showed that the BM receptor dystroglycan forms a membrane diffusion barrier at the neck of the protrusion that enables protrusion growth. Together these studies define a netrin dependent pathway that builds an invasive protrusion, an isolated lysosome-derived membrane structure specialized to breach tissue barriers.
eTOC Blurb
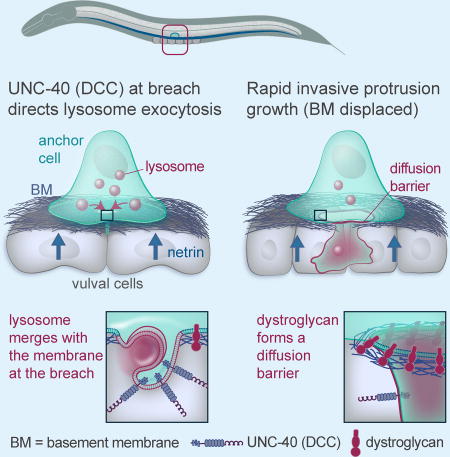
How do invasive cells clear an entry path after basement membrane breach? Naegeli et al. show in C. elegans that directed lysosome exocytosis regulated by a netrin-mediated pathway builds the large invasive protrusion formed after invadopodia. Isolated by a dystroglycan membrane diffusion barrier, the protrusion expands to create tissue openings.
Introduction
The ability of cells to invade and enter tissues is crucial for many developmental and physiological processes, including trophoblast implantation, neural crest migration, and leukocyte trafficking (Lohmer et al., 2014; Madsen and Sahai, 2010). Misregulation of invasion also underlies numerous inflammatory diseases, developmental disorders, and metastasis (Hagedorn and Sherwood, 2011). The first barrier an invading cell encounters is usually a basement membrane (BM), a dense, highly cross-linked extracellular matrix that supports and surrounds tissues (Yurchenco, 2011). Invasive cells use small, matrix metalloprotease-rich (MMP) membrane-associated F-actin structures called invadopodia to create small breaches in BM (Lohmer et al., 2014; Murphy and Courtneidge, 2011). Mechanisms regulating invadopodia formation and turnover have been extensively examined using 2D in vitro assays that mimic flat BM surfaces (Beaty and Condeelis, 2014). The events that follow invadopodium penetration of BM, however, are less clear, as this step in invasion is challenging to visualize in complex vertebrate tissues and difficult to recapitulate with in vitro and ex vivo invasion assays (Beerling et al., 2011; Friedl and Wolf, 2010).
The C. elegans anchor cell (AC) carries out a highly stereotyped cell invasion event that is experimentally accessible to live-cell imaging and genetic perturbations (Sherwood and Sternberg, 2003). The AC uses dynamic, rapidly forming and disassembling invadopodia to breach the BM separating the uterine tissue from the vulval tissue (Hagedorn et al., 2013, 2014; Lohmer et al., 2016). Approximately 10 invadopodia are found at any one time prior to invasion. At the site of the initial invadopodium-mediated BM breach, the netrin receptor UNC-40 (the C. elegans ortholog of vertebrate DCC) becomes enriched, and in response to secreted UNC-6 (netrin) directs the formation of a single large invasive protrusion (Hagedorn et al., 2013). This invasive protrusion shuts down further invadopodia production and dramatically widens the BM opening by degrading and physically displacing the BM as it extends between and contacts the central vulval cells, thus securing uterine-vulval connection (Figure 1A). A few ex vivo invasion studies and tumor tissue sections have also noted the transition of invadopodia to a single large protrusion, as well as the presence of large protrusions that cross BMs in cancer cells (Hotary et al., 2006; Leong et al., 2014; Schoumacher et al., 2010), suggesting this later, poorly understood aspect of the invasion program is conserved.
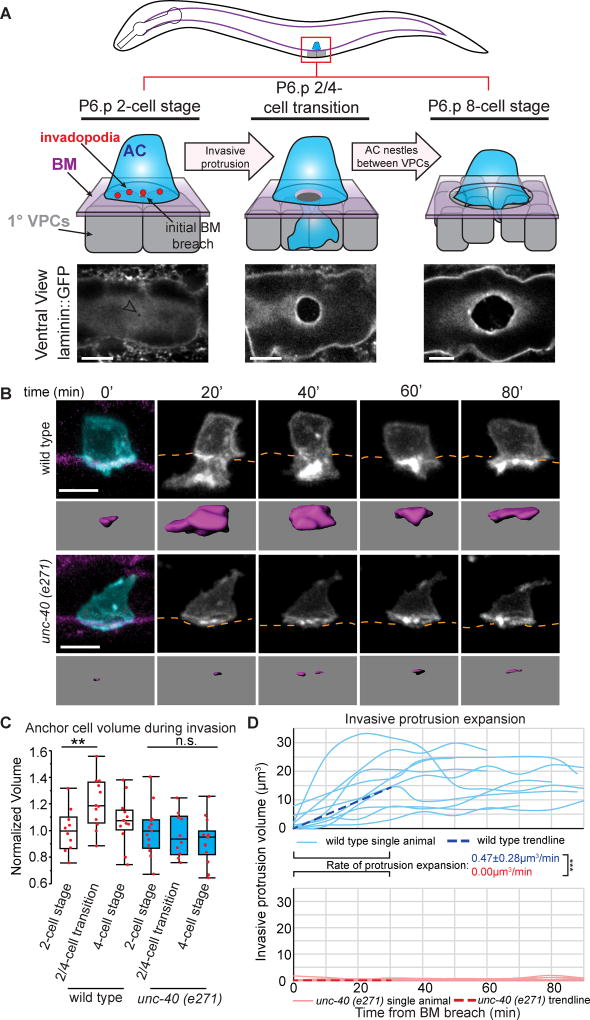
Figure 1. The invasive protrusion locally increases AC area and volume.
(A) AC invasion in C. elegans (top, lateral view schematic; bottom, ventral view of laminin::GFP-labeled BM). Left: During the early L3 larval stage, the AC (blue) sits atop the BM (purple) and the two P6.p VPC descendants (1° VPCs, grey). Actin-rich invadopodia (red) form, and one breaches the BM (arrow top, arrowhead bottom). Center: At the time the underlying P6.p descendants divide, the invasive protrusion expands and the BM hole widens. Right: By completion of next P6.p descendant division, the BM opening has expanded beyond the border of the AC, the invasive protrusion has retracted, and the AC contacts the central P6.p descendants. (B) Time-lapse of AC invasive protrusion formation. AC membrane labeled by cdh-3>GFP::CAAX, blue and greyscale; BM (laminin::mCherry, magenta) position shown by orange dotted lines. Wild type (top) and unc-40 (e271) (bottom) animals. Invasive protrusion isosurfaces (the portion of the AC below the BM, purple). (C) AC volume before protrusion formation (P6.p 2-cell stage), during protrusion expansion (P6.p 2/4-cell transition), and after retraction (P6.p 4-cell stage) for wild type (white) and unc-40 (e271) mutants (blue). Volume is normalized to AC volume of corresponding genotype before invasion – see Methods (n > 10 each category; **p<0.01, n.s. = not significant (p>0.05), One-way ANOVA with Tukey-HSD post-hoc test). (D) AC protrusion volume over time in wild type (blue) and unc-40 (e271) (red) animals from the time of BM breach until time-lapse ended (time points shown every 10 minutes; see Methods). In this and all subsequent figures, protrusion volume growth was compared over the first 30 minutes of protrusion growth, the time of maximum growth in wild type animals. Average expansion rates (dotted lines) ± SD are shown (n=10 animals for each group; ***p<0.001, n.s. = not significant (p>0.05), Student’s t-test). Scale bars = 5µm. See also Figure S1.
BMs and tissues are rigid and densely packed (Halfter et al., 2015), and it is unclear if invasive cells alter their shape, add membrane from internal pools, or harbor membrane within plasma membrane folds to extend large protrusions within the tight confines of tissues to open a path for the invading cell. Interestingly, during invadopodia formation, the membrane-tethered matrix metalloprotease MT1-MMP is dynamically recycled through endosomes and lysosomes to deliver MT1-MMP to invadopodia at the cell surface (Castro-Castro et al., 2016). Whether the invasive protrusion can harness the same membrane compartments to form the large invasive protrusion or uses another membrane source for this distinct invasive structure is not known.
Using quantitative live-cell imaging, genetic analysis, and misexpression studies, we examined the formation of the AC invasive protrusion. We found that UNC-6/UNC-40 interactions localize lysosomes to the site of protrusion formation and that LMP-1 (a lysosomal protein) and ZMP-1 (a membrane-tethered matrix metalloprotease localized to lysosomes) are enriched in the invasive protrusion. The invasive protrusion locally increases the size of the AC by more than 20%, which may contribute strong pushing forces to open a gap in the BM and vulval tissue. Through lysosome perturbation and a focused RNAi screen, we found that lysosome integrity and the exocytic machinery are required for invasive protrusion growth, suggesting lysosomes are exocytosed to form the protrusion. In addition, photobleaching of the invasive protrusion membrane and genetic perturbation studies revealed that it is isolated from the AC body by a diffusion barrier formed by the BM adhesion receptor dystroglycan and that this barrier promotes protrusion growth. Together these results reveal a netrin mediated pathway directs the formation of the invasive protrusion: a distinct lysosome-derived membrane domain that is rapidly exocytosed to breach tissue barriers.
Results
The invasive protrusion forms from localized plasma membrane expansion
Previous studies by our group found that small, rapidly forming (~ 1 minute lifetime) invadopodia breach the BM during AC invasion into the vulval epithelium in C. elegans. Following BM penetration, invadopodia formation ceases, and a large, longer-lived protrusion extends at the site of breach in an UNC-40 (netrin receptor) dependent manner (Hagedorn et al., 2013). UNC-40 is not required for invadopodia formation, indicating that invadopodia and the invasive protrusion are regulated distinctly (Morrissey et al., 2013). We hypothesized two possible mechanisms for expansion of the protrusion: redistribution of the plasma membrane, which would not yield an increase in AC size, or addition of new membrane, which would increase the size of the AC. To begin to distinguish between these two possibilities, we first quantified plasma membrane dynamics during AC invasion using AC-specific expression of GFP tagged with a CAAX prenylation motif (cdh-3>GFP::CAAX), which localizes to plasma membranes. We measured the surface area and volume of the AC throughout the approximately 1.5-hour invasion process (see Methods). The timing of invasive protrusion maturation was assessed using the underlying P6.p vulval precursor cell (VPC) divisions, which adopt a 1° vulval fate and divide in synchrony with AC invasion (Figure 1A). During formation of the invasive protrusion (P6.p 2/4-cell transition), the AC increased 12.7% in surface area and 20.6% in volume (Figures 1B, 1C, and S1A). Protrusion growth occurred during an average 55-minute window (Figure S1B; n=23 animals), with maximum protrusion growth and the fastest invasive protrusions reaching maximal size at 30 minutes (average growth rate of 0.47 µm3/min for 30 min. following BM breach, n=10 animals; Figure 1B, 1D, and Movie S1). AC growth was specifically localized to the single invasive protrusion (Figure S1C). While invadopodia first breach the BM and make a small hole, invasive protrusion expansion was correlated with enlargement of the BM hole (as visualized with the major BM component laminin::GFP; Figure 1A and Figure S1D), supporting the idea that the protrusion actively clears a large BM opening during vulval tissue invasion. Following growth, the invasive protrusion retracted (early P6.p 4-cell stage; n=10 animals observed; Figure 1B and Movie S1), and the AC volume and surface area returned to their approximate original sizes (Figures 1C and S1A). Similar protrusion dynamics were observed in three strains expressing two membrane markers (see Methods), indicating invasive protrusion formation is robust and stereotyped. Notably, ACs from animals lacking the netrin receptor UNC-40 (vertebrate DCC), which is required to form the invasive protrusion (Hagedorn et al., 2013), did not increase in volume or surface area (average growth rate of 0.00 µm3/min; for this and all other experiments growth rates were calculated during the first 30 minutes of protrusion growth after BM breach; Figures 1B–1D and S1A; Movie S1). Together, these results indicate that UNC-40 directs the formation of a large protrusion that locally expands the size of the AC to open a single large gap in the BM. This is in marked contrast to the distinctly different action of invadopodia, which breach BM, are more numerous, smaller, short-lived, and not directed by UNC-40 (Hagedorn et al., 2013, 2014; Lohmer et al., 2016).
Lysosomes contribute membrane to form the invasive protrusion
Lipid membranes can only stretch their surface area 2–3% before rupturing (Mohandas and Evans, 1994). To dynamically increase surface area, cells add membrane from membrane folds or through exocytosis of internal membrane stores (Kay et al., 2008). Previous staining with the outer membrane dye FM1-43 suggested that the AC plasma membrane lacks membrane folds (Hagedorn et al., 2013), and we ruled out folds as the source of protrusion membrane by transmission electron microscopy (Figure S2A). A number of internal membrane sources provide lipids for plasma membrane expansion, including the endoplasmic reticulum, the Golgi apparatus, endosomes, and lysosomes (Dyer et al., 2007; Lecuit and Pilot, 2003; Lecuit and Wieschaus, 2000; Raiborg et al., 2015; Rodríguez et al., 1997). We examined worms expressing fusion proteins that mark the endoplasmic reticulum (CYTB-5.1::GFP), the Golgi apparatus (AMAN-2::GFP), early endosomes (mCherry::RAB-5), late endosomes (mCherry::RAB-7), recycling endosomes (mCherry::RAB-11), and lysosomes (LMP-1::GFP and GFP::CUP-5) (Sato et al., 2014). We hypothesized that if one of these membrane sources builds the protrusion, it should localize to the BM breach site. Notably, LMP-1::GFP and CUP-5::GFP-positive vesicles were polarized strongly along the AC’s invasive membrane prior to invasive protrusion formation (Figures 2A and S2B–C). Further, both lysosome markers were enriched within the invasive protrusion during its growth (n=20/20 animals; Figures 2A and S2D).
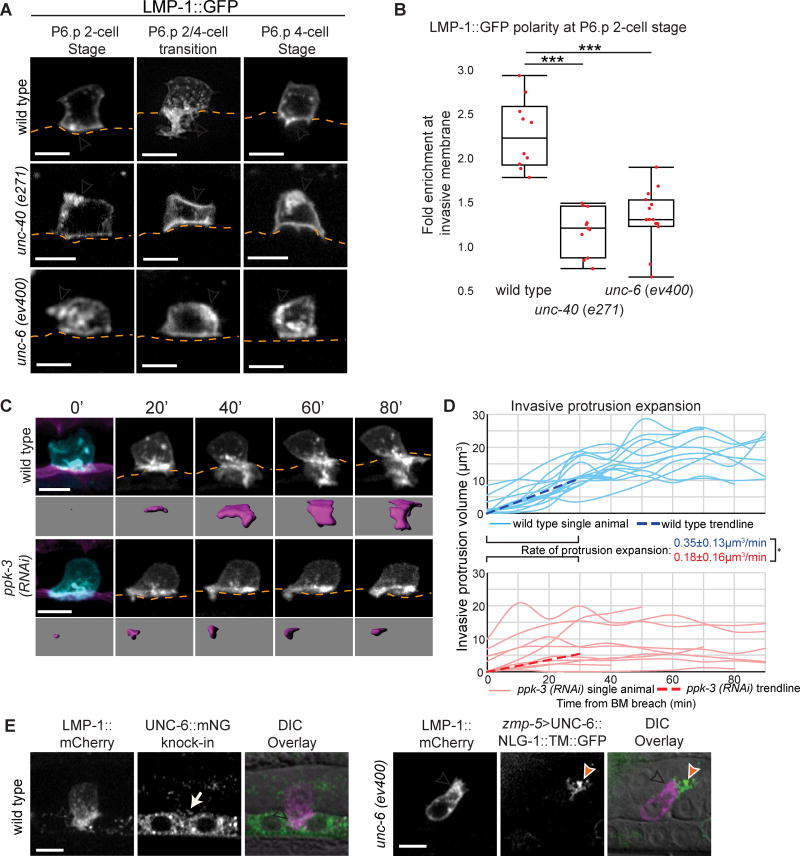
Figure 2. UNC-6 (netrin) polarizes lysosomes to build the invasive protrusion.
(A) Lysosomes (LMP-1::GFP) in the ACs of wild type (top), unc-40 (e271) (middle), and unc-6 (ev400) (bottom) animals before protrusion formation (P6.p 2-cell stage), during protrusion expansion (P6.p 2/4-cell transition), and after retraction (P6.p 4-cell stage). Arrowheads indicate lysosome localization. BM position, orange. (B) Polarity of LMP-1::GFP at the AC invasive membrane before BM breach. (n≥11 animals per genotype; ***p<0.001, One-way ANOVA with Tukey HSD post-hoc test). (C) Time-lapse of AC invasive protrusion formation in a uterine-specific RNAi background. AC membrane labeled with cdh-3>mCherry::PLCδPH, blue and greyscale, purple isosurface below; BM (laminin::GFP, magenta) position shown by orange dotted lines. (D) AC protrusion volume over time in control (blue) and ppk-3 (RNAi) (red) animals. Average expansion rates (dotted lines) ± SD for the first 30 minutes of protrusion growth (n=14 animals for wild type, n=10 animals for ppk-3 (RNAi); *p<0.05, Student's t-test). (E) Lysosomes (LMP-1::mCherry, magenta) polarize within the invasive protrusion (arrowhead) toward UNC-6::mNG (green, unc-6 (cp190)), which is expressed in the VPCs and localizes in extracellular punctae (arrow). An unc-6 (ev400) mutant with membrane-tethered UNC-6 (zmp-5>unc-6::nlg-1::TM::GFP) expressed in a dorsal uterine cell (orange arrowhead). The AC extends a small protrusion with the lysosome marker LMP-1::mCherry (white arrowhead) polarized toward the ectopic UNC-6::GFP. Scale bars = 5µm. See also Figure S2.
Lysosomes are known to play roles in invasion via secretion or delivery of membrane associated proteases (Castro-Castro et al., 2016; Kirkegaard and Jäättelä, 2009), but they have never been investigated as potential membrane sources for building invasive structures in cells. To test if the invasive protrusion is formed from lysosomal membrane, we used RNAi knockdown of the phosphatidylinositol phosphate kinase ppk-3, which regulates maturation and integrity of lysosomes (Nicot et al., 2006). Because the null allele of ppk-3 is lethal (Nicot et al., 2006), we performed these experiments using a uterine-specific RNAi strain (see Methods). Importantly, knockdown of ppk-3 decreased the total amount of LMP-1-positive vesicles within the AC but not early endosomes (RAB-5 marked) or late endosomes (RAB-7 marked) (Figure S2E), indicating a specific loss of lysosomes in the AC. Knockdown of ppk-3 in the uterine cells strongly perturbed AC invasion (Table S1) and invasive protrusion formation (Figure 2C). As other uterine cells do not contribute to AC invasion (Sherwood and Sternberg, 2003), these data indicate that ppk-3 function is specifically required in the AC for invasion. Quantification of invasive protrusion growth revealed that loss of ppk-3 resulted in a significantly reduced rate of invasive protrusion growth (0.18 µm3/min versus 0.35 µm3/min in control animals; Figures 2C and 2D; Movie S2), and the volume of the AC did not increase (Figure S2F), consistent with an absence of membrane addition. Taken together, these observations suggest that lysosomes contribute membrane to form the invasive protrusion.
Netrin/DCC interactions direct lysosome polarity
The netrin receptor UNC-40 (vertebrate DCC) localizes to the site of BM breach and organizes invasive protrusion formation in response to UNC-6 (netrin). If lysosomes are a local membrane source for the protrusion, we hypothesized that the UNC-6/UNC-40 interaction may polarize them. Consistent with this notion, LMP-1-labeled lysosomes were no longer polarized at the invasive membrane prior to or during the normal time of invasion in unc-40 (e271) and unc-6 (ev400) mutants (Figures 2A and 2B; polarity was calculated as enrichment of lysosomes at the invasive cell membrane compared to density at the apical membrane; see Methods).
To examine how endogenous UNC-6 acts to polarize lysosomes to form the protrusion, we used CRISPR/Cas9 to fuse the coding sequence of fluorescent protein mNeonGreen (mNG) to the unc-6 genomic locus (unc-6::mNG knock-in) (Shaner et al., 2013). UNC-6::mNG was detected at high levels in the P6.p vulval precursor cell and its descendants prior to and during protrusion formation, as well as in extracellular punctae around the AC and its invasive protrusion (n=12/12 animals, Figure 2E). These results suggest that a short-range UNC-6 signal is present on the ventral side of the AC to polarize lysosomes to the site of protrusion formation.
To directly test whether UNC-6 is sufficient to polarize lysosomes, we expressed a membrane-tethered form of UNC-6 in the dorsal uterine cells (zmp-5>UNC-6::NLG-1::TM::GFP) in unc-6 (ev400) null animals (Figure 2E) (Wang et al., 2014). In unc-6 null mutants alone, lysosomes are mispolarized (Figure 2A), no protrusion forms, and AC invasion fails (Ziel et al., 2009). Dorsal presentation of UNC-6, however, robustly directed lysosomes (marked with LMP-1::GFP) toward ectopic UNC-6 (n=15/15 animals) and in some cases the AC extended a small LMP-1-enriched protrusion toward the ectopic UNC-6 (n=6/15 animals; Figure 2E). These observations suggest that localized UNC-6 directs the polarized addition of lysosomal membrane to form the invasive protrusion.
The t-SNARE SNAP-29 promotes invasive protrusion formation
Localized exocytosis of the lysosomes that concentrate at the breach site might add membrane to promote invasive protrusion formation. To test this idea, we performed a RNAi screen of 116 genes encoding proteins involved in membrane trafficking, looking for perturbations in AC invasion (Sato et al., 2014). RNAi-mediated knockdown of 22 genes yielded defects in AC invasion (Table S2). In addition, loss of the C. elegans vertebrate Rab11 orthologs rab-11.1 and rab-11.2, which primarily regulate recycling endosomes (Grant and Donaldson, 2009; Stenmark, 2009), disrupted invasive protrusion retraction (Figure S3A). Both whole body and uterine specific targeting of rab-11.1 and rab-11.2 disrupted retraction at the P6.p 4-to-6 cell stage transition (in 6/10 and 17/62 whole body RNAi treated animals, and 8/22 and 2/15 uterine specific RNAi treated animals the ACs invasive protrusion was not retracted, respectively, versus 1/22 ACs in wild type animals), suggesting that membrane recycling is important for protrusion withdrawal. As depletion of the SNAP-25-family t-SNARE snap-29 produced the strongest defect in invasion (Table S2), we characterized the function of SNAP-29 during protrusion formation.
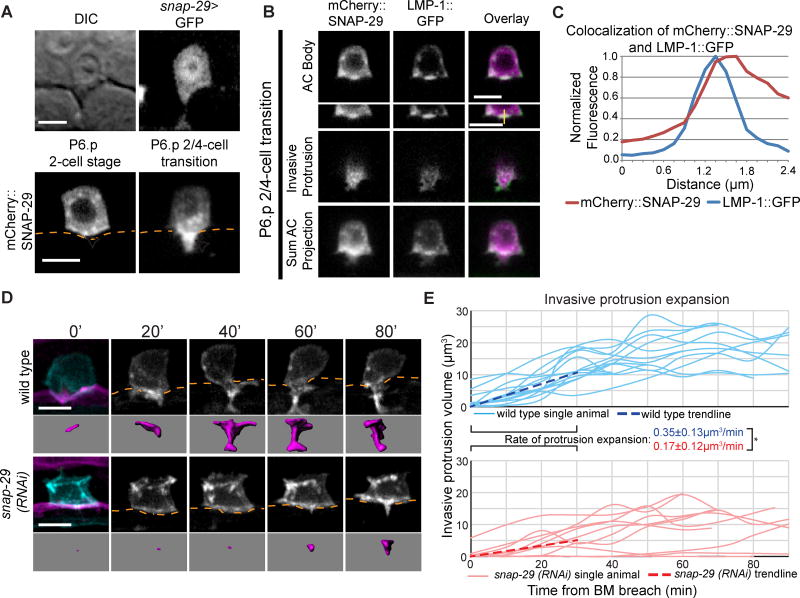
The vertebrate orthologue of snap-29 (SNAP25) promotes exocytosis and plasma membrane expansion during axon branch formation through interactions with the vertebrate UNC-40 ortholog DCC (Winkle et al., 2014) and thus might form a conserved polarized membrane addition circuit. Supporting this possibility, UNC-40 contains the intracellular P3 domain that has been implicated in regulating interactions with vertebrate SNAP25 during exocytosis (Winkle et al., 2014; Ziel and Sherwood, 2010). Uterine-specific RNAi knockdown of snap-29 disrupted AC invasion to the same extent as whole body treatment (Table S1), suggesting that SNAP-29 functions in the AC. A snap-29>GFP transcriptional reporter was expressed in the AC and mCherry::SNAP-29 was polarized to the invasive membrane before invasion and concentrated in the invasive protrusion (Figures 3A and S3B). The lysosome marker LMP-1::GFP and mCherry::SNAP-29 colocalized at the invasive surface of the AC and in the invasive protrusion (Figures 3B and 3C), suggesting that SNAP-29 may mediate lysosome exocytosis to form the invasive protrusion. Consistent with this idea, uterine-specific knockdown of snap-29 resulted in a reduction of invasive protrusion growth rate and protrusion volume (Figures 3D and 3E; Movie S3), and AC size did not increase (Figure S3C). Loss of unc-40 did not alter SNAP-29 polarity, indicating that SNAP-29 is enriched at the invasive membrane independent of UNC-40 (Figure S3D). Importantly, knockdown of snap-29 also did not affect UNC-40 polarization at the invasive membrane (Figures S3E and S3F) or lysosome polarization (Figure S3G), indicating that snap-29 does not alter the invasive protrusion through misregulation of UNC-40 or lysosome localization. Taken together these data suggest that SNAP-29 directs exocytosis of lysosomes to form the invasive protrusion.
Figure 3. The t-SNARE SNAP-29 facilitates invasive protrusion expansion.
(A) A snap-29 transcriptional reporter (snap-29>GFP; top right) expressed in the AC. An AC expressed translational reporter (cdh-3>mCherry::SNAP-29; bottom) is polarized (arrowheads) to the invasive membrane before invasion (left) and within the protrusion (right); BM position, orange. (B) Colocalization of mCherry::SNAP-29 and LMP-1::GFP during protrusion formation. 0.5µm confocal z-slices of the AC body (top,) and invasive protrusion (middle); sum projection of the AC (bottom). Yellow line (top inset, right) indicates point of fluorescence measurement shown in (C). (C) Fluorescence measurement shows colocalization of peak SNAP-29 (red) and LMP-1 (blue) signal. (D) Time-lapse of AC protrusion formation in a uterine specific RNAi strain. AC membrane labeled by cdh-3>mCherry::PLCδPH, blue and greyscale, purple isosurface below; BM (laminin::GFP, magenta) position shown by orange dotted lines. (E) AC protrusion volume over time for wild type (blue) and snap-29 (RNAi) (red) animals. Average expansion rates (dotted lines) ± SD for first 30 minutes of protrusion growth (n=14 animals for control, n=10 animals for snap-29 (RNAi); *p<0.05, Student's t-test). Wild type controls were the same as in Figure 2D. Scale bars = 5µm. See also Figure S3.
The exocyst complex is required for generating the invasive protrusion
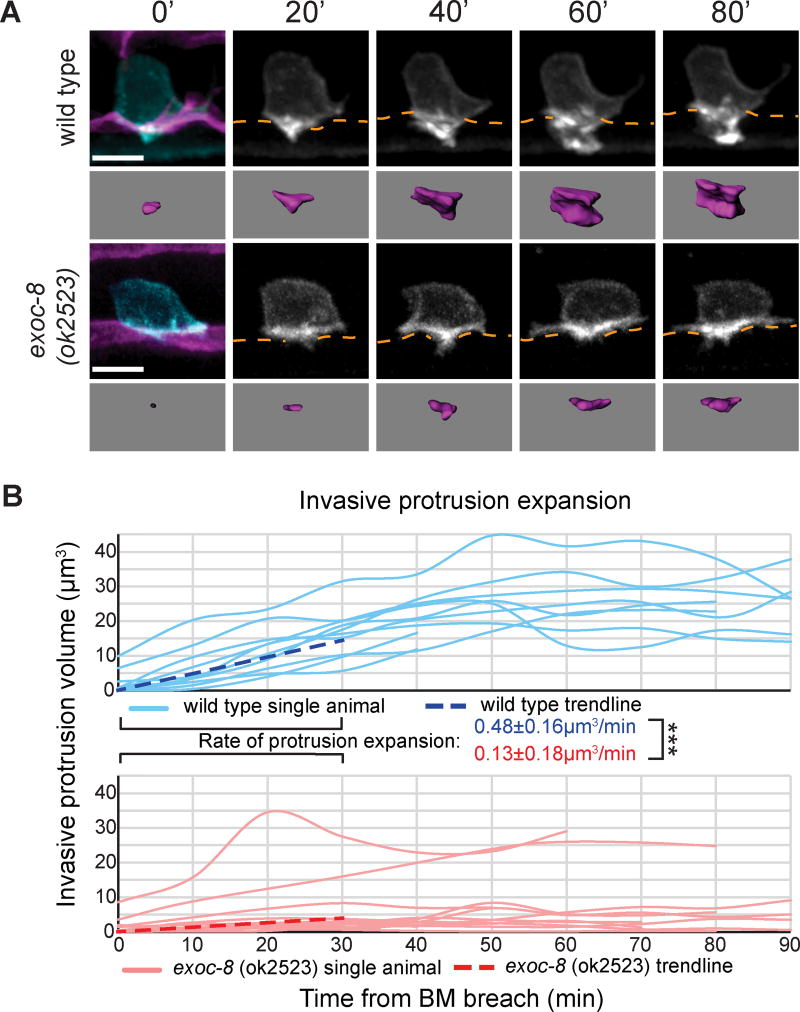
Three components of the exocyst complex were also identified in our screen (Table S2). The exocyst complex regulates transport, tethering, and docking of vesicles prior to SNARE function at the plasma membrane (He and Guo, 2009), and is composed of eight subunits, encoded in C. elegans by the genes sec-3, sec-5, sec-6, sec-8, sec-10, sec-15, exoc-7, and exoc-8 (Jiu et al., 2012). Uterine-specific RNAi knockdown of exocyst components yielded defects in AC invasion (Table S1), suggesting that the complex acts in the AC. Endogenously-tagged SEC-5::YFP was expressed in the AC, but it and other exocyst components were not polarized (Figure S4A) (Armenti et al., 2014). We assayed protrusion formation in a null mutant of exoc-8 (ok2523), which is a regulatory subunit not required for animal viability (Jiu et al., 2012). ACs in exoc-8 mutants breached the BM but had dramatically diminished rates of protrusion growth and volume (Figures 4A and 4B; Movie S4). Consistent with a lack of exocytosis, the ACs in exoc-8 mutants also did not increase in size (Figure S4B). Notably, UNC-40::GFP and LMP-1::mCherry polarities were normal in exoc-8 (ok2523) mutants (Figures S4C and S4D), indicating that the exocyst complex is not necessary for UNC-40 or lysosome localization. Together, these data provide evidence that invasive protrusion formation requires exocytosis of lysosomes, which is mediated by the exocyst complex and the SNAP-25 family t-SNARE SNAP-29.
Figure 4. The exocyst complex promotes invasive protrusion formation.
(A) Time-lapse of AC protrusion formation. AC membrane labeled by cdh-3>mCherry::PLCδPH, blue and greyscale, purple isosurface below; BM (laminin::GFP, magenta) position shown by orange dotted lines. (B) AC protrusion volume over time for wild type (blue) and exoc-8 (ok2523) (red). Average expansion rates (dotted lines) ± SD for first 30 minutes of protrusion growth (n=10 animals for wild type, n=13 animals for exoc-8 (ok2523); ***p<0.001, Student's t-test). Asterisks indicate statistical significance – see Methods. Scale bars = 5µm. See also Figure S4.
The invasive protrusion is an isolated membrane domain
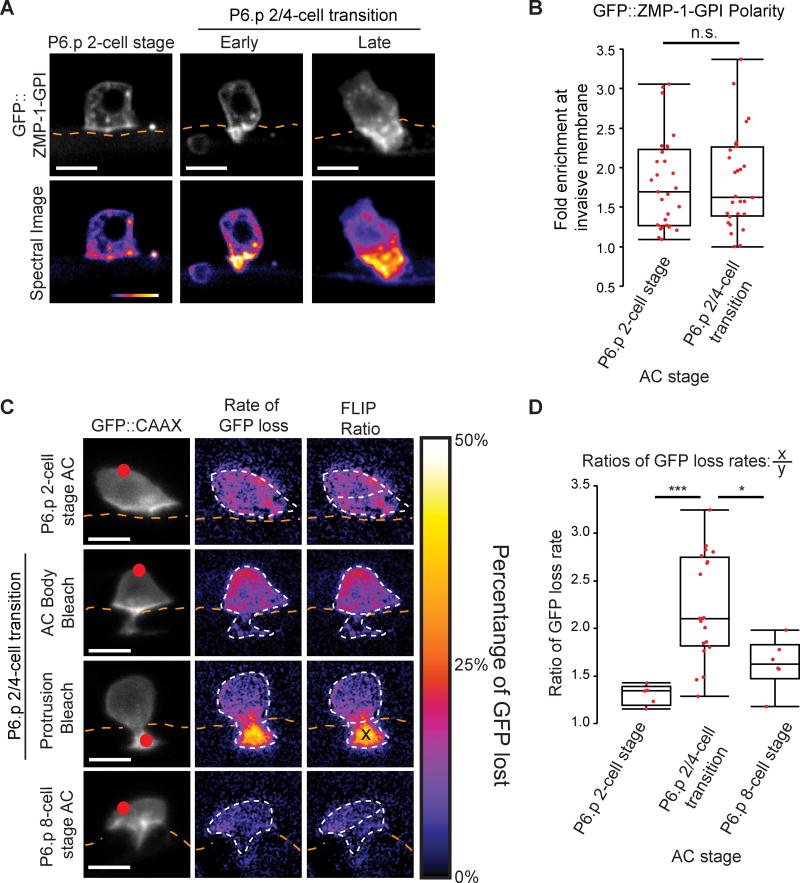
We noted that LMP-1::GFP, a transmembrane protein and marker of lysosomes, was concentrated along the cell surface of the invasive protrusion (Figure 2A). This localization raised the possibility that the invasive protrusion membrane is isolated from the rest of the AC and may be a specialized invasive structure. To further test this idea, we examined the localization of the GPI-anchored membrane associated MMP ZMP-1, which promotes BM removal during AC invasion (Sherwood et al., 2005). A ZMP-1 localization reporter (zmp-1>GFP::zmp-1-GPI) co-localized with the lysosome marker LMP-1 prior to AC invasion (Figures S5A and S5B, n=5 animals) and was strongly enriched in the invasive protrusion (Figures 5A and 5B, n=22/22 animals). These observations support the idea that the invasive protrusion is a specialized structure and that it might be a distinct membrane domain.
Figure 5. The invasive protrusion is a segregated membrane domain.
(A) A reporter of ZMP-1 localization (zmp-1>GFP::ZMP-1-GPI) (top) and corresponding spectral representation of fluorescence intensity (bottom) shows that ZMP-1 becomes highly concentrated in the invasive protrusion during its formation. (B) Quantification of GFP::ZMP-1-GPI polarity before invasion (P6.p 2-cell stage, n=29 animals) and in the invasive protrusion (P6.p 2/4-cell transition, n=28 animals; n.s. = not significant (p>0.05), Student's t-test). (C) Fluorescence loss in photobleaching (FLIP) of AC membrane reporter (cdh-3>GFP::CAAX). Red dot indicates target of photobleaching (left). A smoothed spectral intensity map of percentage of GFP lost (center). The white dotted lines outline the AC. FLIP ratios (right) were determined by dividing the average loss in the region of photobleaching (X) by the average loss in the region of the AC not targeted (Y). (D) FLIP ratios before invasion (P6.p 2-cell stage, n=5 animals), during protrusion formation (targeting the invasive protrusion or the AC body, P6.p 2/4-cell transition, n=20 animals), and after AC invasion (P6.p 8-cell stage, n=6 animals) (*p<0.05, ***p<0.001, One-way ANOVA with Tukey HSD post-hoc test). Scale bars = 5µm. See also Figure S5.
If the invasive protrusion is a distinct membrane domain, fluorescence loss in photobleaching (FLIP) would show that photobleached GFP::CAAX from the AC cell body does not diffuse into the protrusion (and vice versa). We performed FLIP in ACs prior to protrusion formation (P6.p 2-cell stage), during protrusion formation (P6.p 2/4-cell transition) and after retraction (P6.p 8-cell stage) by photobleaching a 1µm-diameter region of AC membrane and measuring the rate of GFP loss. When targeting the body of the AC, we observed a significantly higher rate of GFP signal loss in the body than in the protrusion (Figures 5C and 5D). Reciprocal FLIP of the protrusion revealed a similar rate of GFP signal loss localized to the protrusion (Figures 5C and 5D). In contrast, in ACs that had not yet formed a protrusion and in ACs after protrusion retraction, we found close rates of GFP signal loss throughout the AC (Figures 5C and 5D). We conclude that the invasive protrusion membrane harbors unique proteins and is a separate compartment that is isolated from the rest of the AC.
An expanding ring of dystroglycan and F-actin localizes to the neck of the protrusion
Our FLIP results suggested that a membrane diffusion barrier may separate the invasive protrusion from the AC body. Membrane diffusion barriers impede lipid and protein movement in structures such as tight junctions in epithelial cells and the axon hillock in neurons, where cytoskeletal and transmembrane proteins are concentrated (Trimble and Grinstein, 2015). Because the neck of the invasive protrusion, which contacts the BM, appeared to be the boundary between the invasive protrusion and AC body (Figure 5C), we hypothesized that a BM adhesion receptor might concentrate at the neck of the protrusion and form a diffusion barrier.
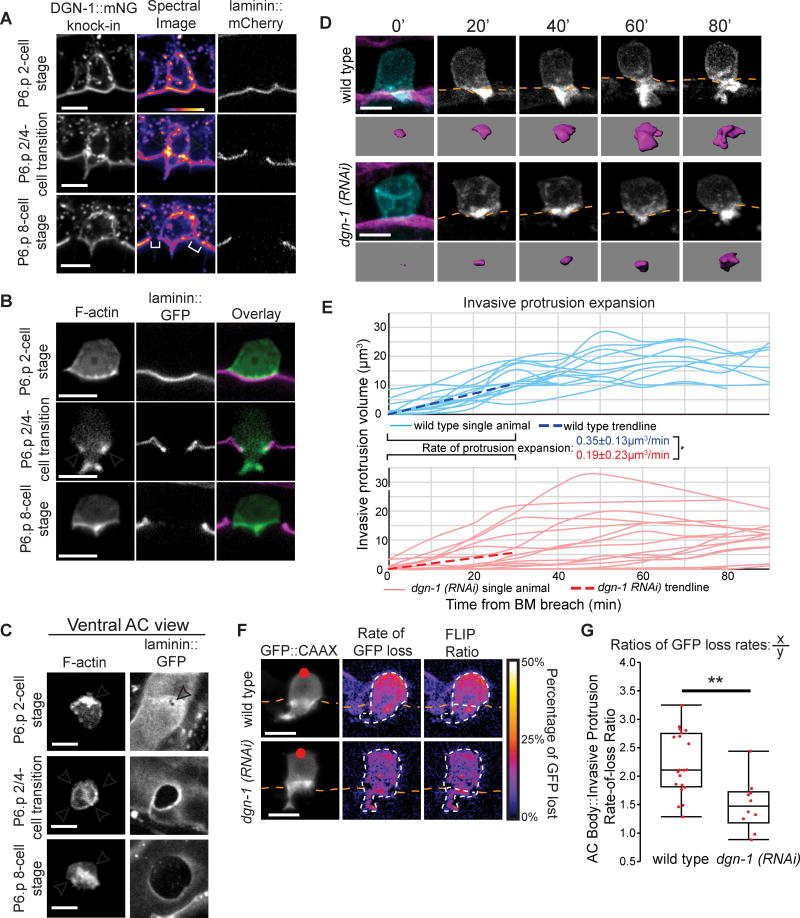
The two major classes of adhesion receptors that link BM to the cytoskeleton are integrin family members and the receptor dystroglycan (Bello et al., 2015; Yurchenco, 2011). The INA-1/PAT-3 heterodimer is the only integrin expressed in the AC at the time of invasion (Hagedorn et al., 2009), where it is required for invadopodia formation (Hagedorn et al., 2013). A functional PAT-3::GFP fusion protein, however, was not concentrated at the neck of the invasive protrusion (Figure S6A; n=19/19 animals). To examine dystroglycan localization, we used CRISPR/Cas9 to fuse the coding sequence of mNG (mNeonGreen) to the dgn-1 genomic locus (dgn-1::mNG knock-in). DGN-1::mNG was present at high levels in the AC prior to and throughout invasion. Importantly, DGN-1::mNG was enriched at the AC-BM interface in an expanding ring at the neck of the invasive protrusion during its growth (Figure 6A; n=26/27 animals). Following retraction, when the AC loses contact with the BM and moves between the central vulval cells, DGN-1::mNG enrichment on the basal sides of the AC was lost (Figure 6A; n=15/15 animals). As actin networks also contribute to diffusion barriers (Trimble and Grinstein, 2015), we next examined the localization of F-actin. Notably, a similar localization pattern at the neck of the protrusion was observed for F-actin (Figures 6B and 6C; n=24/24 and 15/15 animals, respectively). These observations indicate that dystroglycan and F-actin form a ring at the neck of the invasive protrusion, which may function as a membrane diffusion barrier that isolates the protrusion from the AC body.
Figure 6. The BM receptor dystroglycan (DGN-1) forms a membrane diffusion barrier that promotes protrusion formation.
(A) ACs expressing dgn-1::mNG (dgn-1 qy18) (left) and spectral representations of fluorescence intensity (center), with BM (laminin::GFP, right). DGN-1::mNG protein localizes to the invasive membrane before invasion (top), concentrates at the AC-BM interface (arrowheads) at the neck of the protrusion (middle), and is lost at this location (bottom) as the BM moves beyond the AC (brackets). The protrusion also retracts at this time. (B) F-actin (cdh-3>mCherry::moesinABD) similarly concentrates at the neck of the invasive protrusion at the AC-BM interface (arrowheads) during protrusion formation. (C) Ventral view of the AC-BM interface shows Factin localization at the site of initial BM breach (top, arrowhead), in a ring (arrowheads) at the AC-BM contact site as the BM hole expands (middle), which is then lost (arrowheads) after the BM gap extends beyond the AC (bottom). (D) Time-lapse of AC protrusion formation in a uterine-specific RNAi strain with the AC membrane labeled by cdh-3>mCherry::PLCδPH, blue and greyscale, purple isosurface below; BM (laminin::GFP, magenta) position shown by orange dotted lines. (E) AC protrusion volume over time for wild type (blue) and dgn-1 (RNAi) (red). Average expansion rates (dotted lines) ± SD for first 30 minutes of protrusion growth (n=14 animals for wild type, n=16 animals for dgn-1 (RNAi); *p<0.05, Student's t-test). Wild type controls were the same as in Figure 2D. (F) Fluorescence loss in photobleaching (FLIP) of AC membrane reporter (cdh-3>GFP::CAAX). Red dot indicates target of photobleaching (left). A smoothed spectral intensity map of percentage of GFP lost (center). The white dotted lines outline the AC. FLIP ratios (right) were determined by dividing the average loss in the region of photobleaching (X) by the average loss in the region of the AC not targeted (Y). (G) FLIP ratios for wild type and dgn-1 (RNAi). Controls were shared with the 2/4-cell transition data in Figure 5D (n=20 animals for wild type, n=10 animals for dgn-1 (RNAi); **p<0.01, Student's t-test). Scale bars = 5µm. See also Figure S6.
Dystroglycan acts as a diffusion barrier and promotes protrusion growth
To test whether dystroglycan functions as a diffusion barrier, we performed time-lapse analysis of invasive protrusion formation following uterine-specific RNAi knockdown of dgn-1. Knockdown of dgn-1 (an average 75% reduction of DGN-1::mNG fluorescence within the AC; Figure S6B) resulted in a dramatic reduction in invasive growth rate and volume (Figures 6D and 6E; Movie S5) and the overall size of the AC failed to increase (Figure S6C). We performed FLIP analyses on 10 animals after dgn-1 knockdown, and these animals showed an increased rate of GFP diffusion between the small invasive protrusions and the AC body compared to controls (Figures 6F and 6G), indicating a breakdown of the diffusion barrier. We also examined ZMP-1 reporter localization after dystroglycan knockdown. The ZMP-1 reporter was still enriched in the reduced protrusions (Figures S6D and S6E). However, as the ZMP-1 reporter localizes to lysosomes, which are polarized by UNC-6/UNC-40 interactions at the invasive membrane (Figures 2A and 2B), it is possible that UNC-40 is sufficient to concentrate ZMP-1 within the reduced protrusions. RNAi knockdown of dgn-1 did not significantly impact the localization of F-actin to the AC-BM interface at the necks of the smaller invasive protrusions (Figure S6F), suggesting that a DGN-1-independent mechanism regulates actin localization. Notably, loss of INA-1/PAT-3 (integrin), either through a viable loss of function ina-1 allele (gm39) or a dominant negative form of pat-3 expressed in the AC (Hagedorn et al., 2009), did not alter invasive protrusion formation or disrupt the membrane diffusion barrier (Figures S6G–I). Thus, integrin is neither localized to or functionally involved in the AC protrusion diffusion barrier. These observations suggest that a diffusion barrier set up by dystroglycan-BM interactions at the neck of the AC promotes invasive protrusion growth by maintaining it as a distinct membrane domain.
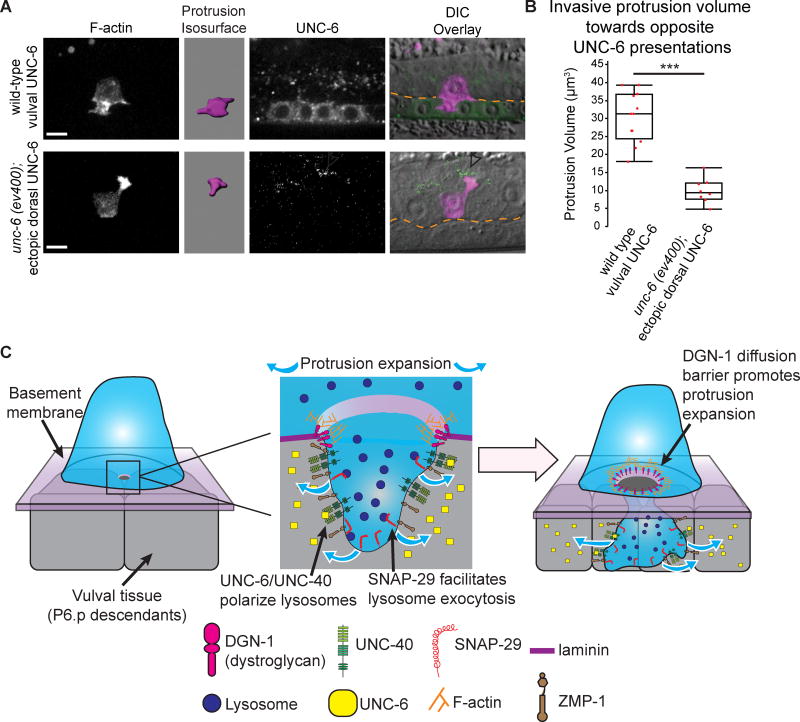
We further tested whether BM contact with the neck of the invasive protrusion is necessary for the polarized addition of a large amount of membrane. The invasive protrusions that formed in response to ectopic UNC-6 (Figure 2E) do not breach or otherwise interact with a BM. As they rarely formed protrusions, we wanted to more carefully examine protrusions by using a marker for F-actin (cdh-3>moeABD::mCherry) in the AC. Consistent with the requirement for a BM-dependent diffusion barrier, most ACs failed to form a protrusion upon contacting ectopic UNC-6 (n=18/26 ACs observed). The few protrusions that did form were much smaller than normal invasive protrusions (Figures 7A and 7B). Taken together, our observations indicate that the AC invades into the vulval tissue by forming a membrane diffusion barrier at the AC-BM interface, which allows localized exocytosis of lysosomes to promote expansion of a large, MMP-enriched protrusion that breaches tissue boundaries.
Figure 7. The BM is a scaffold for protrusion growth.
(A) F-actin (cdh-3>mCherry::moesinABD; left) marks the invasive protrusion (purple isosurface), UNC-6 (center), and overlay on DIC (right, BM position, orange). Endogenous UNC-6 (top) and dorsal uterine cell membrane tethered UNC-6 (bottom, arrowhead). Scale bars = 5µm. (B) Maximum protrusion volume (n=12 animals for wild type, n=8 animals for unc-6 (ev400) with ectopic dorsal UNC-6; ***p<0.001, Student's t-test). (C) Following invadopodial breach, the netrin receptor UNC-40 localizes to the breach site where it is activated by UNC-6 (netrin), which is secreted from in the central vulval cells (P6.p and its descendants). UNC-40 polarizes lysosomes, which add membrane to form the invasive protrusion through SNAP-29-mediated exocytosis. During protrusion growth GPI-anchored ZMP-1 concentrates in the invasive protrusion, and the BM receptor dystroglycan and F-actin localize to a ring at the ACBM interface, forming a membrane diffusion barrier. This diffusion barrier allows focused membrane addition and expansion of the protrusion that clears an opening in the BM and vulval tissue allowing the AC to enter the vulval tissue.
Discussion
The importance of invadopodia in breaching BM and the mechanisms regulating their formation are well-established (Castro-Castro et al., 2016; Genot and Gligorijevic, 2014; Lohmer et al., 2014; Murphy and Courtneidge, 2011). In contrast, little is known about the molecular and cellular processes that regulate large protrusion formation at sites of invadopodial breach and in particular the mechanisms by which membrane is added to these protrusions within the tight confines of densely packed tissues. Using C. elegans AC invasion into the vulval tissue as a model, we found that in response to UNC-6 (netrin) secreted by the vulval tissue, the netrin receptor UNC-40 (DCC) directs lysosomes to the site of BM breach. Our results indicate that UNC-40 acts through the targeting t-SNARE SNAP-29 to trigger exocytosis of lysosomes to form an MMP-enriched invasive protrusion that dramatically increases in volume to create a large opening in the BM and underlying vulval tissue. We show that the BM receptor dystroglycan clusters at the neck of the protrusion (the AC-BM interface) and forms a membrane diffusion barrier that enables protrusion growth. We further discovered that the protrusion is short-lived: as it degraded and displaced the BM, the dystroglycan-BM diffusion barrier was lost, and the protrusion retracted. Together these results define a netrin directed pathway that builds an invasive protrusion, a transient lysosome-derived membrane structure that is specialized to breach tissue barriers (Figure 7C).
By using quantitative live-cell imaging during the stereotyped process of AC invasion, we discovered that invasive protrusion formation rapidly increases the AC’s surface area approximately 15%, far beyond the 2–3% that a cell can stretch (Mohandas and Evans, 1994). We show that UNC-40/UNC-6 interactions polarize lysosomes, organelles that contribute lipids for membrane repair and neurite outgrowth (Arantes and Andrews, 2006; Reddy et al., 2001), to the breach site. As the invasive protrusion formed, it became enriched with the lysosomal protein LMP-1, and disruption of lysosomes inhibited protrusion formation, suggesting that lysosomes are used to build the protrusion. Finally, through a membrane trafficking RNAi screen we found that both the target SNARE SNAP-29 and the exocyst complex, which together mediate the docking, tethering, and fusion of exocytic vesicles (He and Guo, 2009), are expressed in the AC and promote invasive protrusion formation. Given that no protrusion formed in the absence of UNC-40 or UNC-6, our results suggest that the UNC-40/UNC-6 interaction acts through the t-SNARE SNAP-29 and the exocyst complex to facilitate exocytosis of lysosomes to form the invasive protrusion. In addition to membrane addition, lysosome exocytosis might contribute cathepsin proteases that could promote BM removal (Kirkegaard and Jäättelä, 2009). However, there are at least 43 genes encoding cathepsins in C. elegans (Xu et al., 2014) and extensive genetic and RNAi screens have not identified a role for cathepsins during AC invasion (Lohmer et al., 2016; Matus et al., 2010). Thus, if cathepsins do promote invasion they may function redundantly. Netrin-mediated membrane addition may be a common mechanism to construct invasive protrusions, as the vertebrate netrin-1 ligand is overexpressed in numerous metastatic cancers and stimulates invasion in pancreatic, breast, colorectal, malignant melanoma, hepatocellular, and multiple brain tumors examined in vitro and in vivo (Ylivinkka et al., 2016).
The mechanisms that promote AC invasive protrusion formation have marked similarities to those guiding axon outgrowth and branching. The vertebrate UNC-40 receptor DCC interacts with the vertebrate homolog of SNAP-29 (the t-SNARE SNAP-25) to mediate exocytosis of membrane for axon outgrowth and branching (Winkle et al., 2014). Biochemical evidence suggests that this is directed in part through the intracellular P3 domain of DCC, a domain shared with UNC-40 (Winkle et al., 2014; Ziel and Sherwood, 2010). Notably, DCC/SNAP-25 exocytosis is partially dependent on VAMP-7, a lysosomal V-SNARE (Winkle et al., 2014), suggesting a similar lysosomal membrane source is also used in neurons. Further, the exocyst complex is required for axon outgrowth (Tojima and Kamiguchi, 2015). Thus, the circuitry that directs membrane addition during axon outgrowth and AC invasion appears to be shared. This connection might reflect parallel functions of invasive protrusions and extending axons, as axon growth cones must also invade tissues during establishment of proper synaptic connections (Santiago-Medina et al., 2015).
Diffusion barriers are thought to help form and maintain specialized membrane domains in cells (Trimble and Grinstein, 2015). Several barriers have been described in plasma membranes, including the initial segment of axons, the phagocytic cup of macrophages, and the neck of budding yeast cells (Freeman et al., 2016; Nakada et al., 2003; Takizawa et al., 2000). Although the mechanisms that establish these barriers are poorly understood, it is thought that densely packed transmembrane and cytoskeletal proteins can act as molecular fences that contribute to barrier formation (Nakada et al., 2003; Trimble and Grinstein, 2015). We have discovered that the dystroglycan transmembrane BM receptor and F-actin concentrate in a ring around the neck of the protrusion at the AC-BM interface. Fluorescence loss in photobleaching (FLIP) and knockdown studies revealed that dystroglycan restricts the diffusion of an inner leaflet-anchored GFP protein between the protrusion and AC body, suggesting that dystroglycan forms a barrier that helps isolate the invasive protrusion. Loss of dystroglycan did not perturb the F-actin ring. The F-actin ring might help localize dystroglycan, as dystroglycan can be stabilized at specific cellular domains by anchoring to the cytoskeleton (Nakaya et al., 2013). F-actin has also been implicated as possibly stabilizing an integrin receptor diffusion barrier during phagocytosis (Freeman et al., 2016). Rings of matrix adhesion proteins surround invadopodia in cancer cells, invasive podosomes in dendritic cells, and podosomes during sprouting angiogenesis (Branch et al., 2012; Gawden-Bone et al., 2010; Seano et al., 2014), suggesting that diffusion barriers might be a common mechanism to form protrusions that invade tissues.
Our results suggest that the primary function of the dystroglycan-BM diffusion barrier is to allow or maintain focused growth of the invasive protrusion. RNAi-mediated reduction of dystroglycan or the absence of the dystroglycan-BM barrier led to a loss of the protrusion or dramatic reduction in protrusion size. Disassembly of the diffusion barrier may also trigger protrusion withdrawal; when the BM gap eventually widens beyond contact with the protrusion, the dystroglycan-BM barrier at the neck of the protrusion breaks down, and the protrusion retracts. The diffusion barrier at the neck of the invasive protrusion might have functional similarities with the barrier organized by a ring of septins at the neck of budding yeast, where this barrier restricts growth to the new bud by focusing vesicle trafficking and membrane addition (Caudron and Barral, 2009). Dystroglycan’s role in the AC diffusion barrier may be to physically restrict exocytosed membrane to allow for localized membrane expansion or to locally restrict exocytosis to facilitate deposition of new membrane only within the invasive protrusion. Because of the loss or dramatic reduction of the protrusion after dystroglycan knockdown, it was difficult to determine if the dystroglycan diffusion barrier also maintains the lysosome-localized, membrane-bound protease ZMP-1. Given that lysosomes are also polarized by UNC-6/UNC-40 activity, and UNC-40 localizes to the invasive protrusion (Hagedorn et al., 2013), multiple mechanisms might function to maintain the unique molecular identity of the invasive protrusion.
Recent studies have shown that many mammalian cell types transiently expand in volume 10–30% during mitosis (Son et al., 2015; Zlotek-Zlotkiewicz et al., 2015). This expansion is not accompanied by an increase in mass, and thus dividing cells briefly decrease in density. A transient density decrease may also occur in the AC. It has been suggested that osmotic swelling might create strong pushing forces to allow mitotic cells to round in dense tissues (Stewart et al., 2011; Zlotek-Zlotkiewicz et al., 2015). Similarly, we suspect that focused membrane addition allows the invasive protrusion to penetrate initially small, confined gaps in tissues. Subsequent osmotic swelling coupled with the concentration of proteases and further membrane addition would then allow these gaps to be progressively widened by the expanding protrusion to clear a path for the invading cell. This idea is consistent with optical highlighting experiments demonstrating that the AC’s invasive protrusion both degrades and physically displaces BM during invasion (Hagedorn et al., 2013).
Work examining cell migration and invasion in confined extracellular matrix and cellular in vitro and in vivo environments has revealed that cells harbor a remarkable repertoire of strategies to move through restrictive barriers, including shifts between amoeboid and mesenchymal migration, the ability to withstand nuclear envelope rupture, and even osmotic and nuclear-piston driven cell movement (Paul et al., 2017). Because of the challenges of examining cell size during the dynamic process of tissue invasion, however, it was not known if invasive cells can dynamically modulate membrane area and cell volume to enter tissues. Our work here establishes that an invasive cell can actively target membrane addition to the site of invasion and increase its volume locally to generate a large protrusion that promotes invasion through dense tissues. Given observations that cancer cells use single large protrusions to cross BMs into tissues (Hotary et al., 2006; Leong et al., 2014; Schoumacher et al., 2010), we expect this may be a common strategy for invading cells to breach tissue barriers.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David R. Sherwood (david.sherwood@duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were maintained on standard NGM media and fed E. coli OP50. For RNAi experiments, wild type controls were fed E. coli HT115(DE3) containing L4440 (see RNAi experiments, STAR Methods). Unless otherwise noted, all strains were maintained at 20°C. All animals scored were hermaphrodites during the L3 stage when the anchor cell (AC) invades. AC invasion was precisely staged in reference to VPC divisions and gonad development as previously described (Sherwood et al., 2005). Briefly, the AC is positioned over the central P6.p vulval precursor cell cell prior to invasion in the early L3 larval stage. During the mid L3 stage the P6.p cell divides once (P6.p 2-cell stage). At the late P6.p 2-cell stage the AC initiates BM breach (near the time with the distal tip cells of the gonad arm begin migrating dorsally). At the P6.p 2-to-4-cell transition (when the P6.p daughters divide) the AC protrusion forms and clears an opening in the basement membrane and extends around and between the P6.p vulval precursor cell descendants. At the early P6.p 4-cell stage (mid-to-late L3 stage), the protrusion retracts back into the AC (Figure 1A).
METHOD DETAILS
Construction of genomically-edited strains
Endogenous tagging of the C-terminus of unc-6 with the mNG::3xFLAG sequence was accomplished using CRISPR/Cas9-mediated genome editing using a homologous repair template containing 500bp homology arms, the mNG::3XFLAG, and a selection markers as described previously (Dickinson et al., 2015). For unc-6 we used a guide RNA (sgRNA) with a targeting sequence of 5’- TATCTGTGTGACGTAATCTCTGG-3’. GFP was similarly knocked into the dgn-1 locus by CRISPR/Cas9-triggered homologous recombination using a homologous repair template with a selectable marker, GFP, and 1.7kb homology arms at the HindIII site 7 amino acids upstream of the stop codon (Johnson et al., 2006) using two sgRNA targeting sequences, 5’- GATGAAGCATGTcCGAGACGCGG-3’ (antisense) and 5’-GCCAGCAACTCTCCGCGTCTCGG-3’ (sense). Silent mutations were introduced into the homology arms using site directed mutagenesis. See Table S3 for homology arm oligonucleotide primer sequences. For construction of both genome edited strains, the sgRNA targeting sequences were cloned into the pDD162 Cas9-sgRNA expression vector for C. elegans. The homologous repair template and Cas9-sgRNA plasmids were coinjected into the gonad of young adult N2 worms. Animals that were recombinant were identified in the F3 offspring of injected animals based on the presence of selectable markers (dominant-negative sqt-1 rol phenotype and hygromycin resistance). Following strain isolation, the selectable markers were removed from the genome through Cre-Lox recombination and proper genome editing was confirmed by amplification and sequencing of the edited region.
Construction of fusion proteins
GFP::CAAX was amplified from pSA129 and then cloned into pBlueScript containing 1.5kb AC-regulatory region of the C. elegans cdh-3 promoter at SalI and SacI sites (see Table S3 for oligonucleotide sequences) (Sherwood et al., 2005).
The AC-specific endoplasmic reticulum marker cdh-3>cytb-5.1::GFP was constructed by PCR fusion. A 3.0kb fragment of the full length cytb-5.1 sequence, GFP, and the let-858 3’UTR from pHD189 was fused with a 2.2kb fragment of the cdh-3 promoter region amplified from pPD107.94 (see Table S3 for oligonucleotide sequences).
The AC-specific Golgi apparatus marker cdh-3>aman-2::GFP was constructed by PCR fusion. A 2.3kb fragment of aman-2, GFP, and the let-858 3’UTR from pHD93 was fused with a 2.2kb fragment of the cdh-3 promoter region amplified from pPD107.94 (see Table S3 for oligonucleotide sequences).
Fusion of the lin-29 promoter (5.4kb, see Table S3) to the snap-29 open reading frame starting from the ATG translation start site amplified from C. elegans genomic DNA (1.3kb) and mCherry from a modified pBlueScript (0.9kb) was accomplished by Gibson assembly using pBlueScript as a backbone (3.6kb).
Exocyst components (exoc-8, exoc-7, sec-15, and sec-5) fused to GFP previously (Zou et al., 2015) were amplified from their corresponding plasmids and fused to 1.7kb of the cdh-3 promoter amplified from pBlueScript by PCR fusion (see Table S3 for oligonucleotide sequences).
A 0.9kb fragment of LMP-1 encompassing the complete open reading frame from the ATG translation start site was amplified using genomic DNA (see Table S3 for oligonucleotide sequences) was cloned into pBlueScript containing zmp-1>mCherry at AgeI and PacI sites to generate zmp-1>LMP-1::mCherry.
The snap-29>GFP transcriptional reporter was generated from 1.4kb of sequence 5’ of the snap-29 transcription start site expressed as an extrachromosomal array (see Table S3 for oligonucleotide sequences).
The zmp-1>GFP::ZMP-1-GPI construct was generated by amplifying the zmp-1 GPI membrane targeting sequence (an 84 base pair fragment encompassing the final 28 amino acids of zmp-1; see Table S3 for oligonucleotide sequences) from genomic DNA and cloning this fragment into pBlueScript containing GFP using EcoRV and NotI sites. The resulting GFP::GPI sequence was then amplified with oligonucleotides for GFP::GPI (forward) and the unc-54 3’ UTR (reverse) and joined to the 2.7kb of sequence 5’ of the zmp-1 transcription start site and the zmp-1 signal sequence (first 66 amino acids) by PCR fusion.
All constructs were injected into the syncytial gonads of young adult unc-119 (ed4) hermaphrodites along with 50ng/µL unc-119 rescue DNA, 50ng/µL pBSSk(−), and 50ng/µL EcoRI-digested salmon sperm DNA. F1 animals were selected for recovery of wild type animal movement (rescue of the unc-119 phenotype), and stable lines were selected based on transmission of stable extrachromosomal transgenes into the F2 generation. Integrated lines were generated by gamma irradiation as previously described (Sherwood et al., 2005). Briefly, approximately 50 young adult hermaphrodites carrying the extrachromosomal transgenes were irradiated with 3800rad of γ irradiation from Cesium-137 of and rescued to plates to produce progeny. F1 animals (5 per parent) exhibiting rescue of the unc-119 phenotype were singled to NGM plates. Plates that showed 100% rescue of the unc-119 phenotype in the F2 were considered stable integrants, evaluated for transgene expression, and then backcrossed into strain N2.
Microscopy and image acquisition
All time-lapse and polarity images were acquired using an EM-CCD or Orca-R2 camera (Hamamatsu Photonics) and a spinning disk confocal microscope (CSU-10, Yokogawa) mounted on an upright AxioImager microscope (Carl Zeiss) with a Plan-APOCHROMAT 100×/1.4 oil differential interference contrast objective controlled by µmanager software (version 1.4) (Edelstein et al., 2010). Time-lapse acquisition was performed as described previously (Kelley et al., 2017). Synchronized L3 hermaphrodites were anesthetized in 0.2% tricine and 0.02% levamisole in M9 for 20 minutes and then transferred to 5% noble agar pads. The cover slip was sealed with VALAP and worms were imaged at 23°C for two hours. For protrusion analysis, confocal stacks of marked ACs with protrusions (23 optical slices, each at 0.5µm thickness) were acquired every five minutes to avoid photobleaching. Some time-lapses were acquired every minute to construct movies with better temporal resolution.
For single-timepoint AC snapshots and all scoring of AC invasion, worms were anesthetized on 5% noble agar pads with 0.01M sodium azide. Fluorescence images of the AC were acquired as confocal z-stacks with 0.5µm optical slices spanning the entire cell.
Images of ACs on rab-11.1 (RNAi) and wild type controls (Figure S3A) were acquired on a Zeiss AxioImager A1 microscope with a 100× plan-apochromat objective and Zeiss AxioCam MRm CCD camera controlled by Zeiss Axiovision software (Zeiss Microimaging).
RNAi experiments
RNAi was delivered by feeding worms E. coli feeding strain HT115(DE3) expressing double-stranded RNA (Fire et al., 1998). Bacteria harboring an empty RNAi vector (L4440) was used as a negative control for all RNAi experiments. RNAi clones targeting C. elegans genes originated from the C. elegans ORF-RNAi Collection V1.1 (Open Biosystems) and an RNAi library constructed by the Ahringer lab (Fraser et al., 2000). Transcription of RNAi vector expression was induced with 1mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and cultures were plated on plates containing NGM and topical application of 5µL each of 30mg/mL carbenicillin and 1M IPTG. Synchronized L1-arrested larvae were plated on the RNAi-expressing HT115(DE3) and grown for 40–48 hours at 16°C before scoring. RNAi that produced phenotypes were sequenced to verify correct insert.
For uterine-specific RNAi experiments, the strain NK1316 was utilized (Haerty et al., 2008; Hagedorn et al., 2009). The strain harbors mutations in rrf-3 (pk1426), an RNA-directed RNA polymerase whose loss sensitizes the worms to RNAi, as well as rde-1 (ne219), an argonaute protein required for RNAi (Hagedorn et al., 2009). Expression of rde-1 in the somatic uterine cells (fos-1a>rde-1) specifically restores RNAi in the uterine cells.
Electron microscopy
Transmission electron micrographs were acquired in serial sections as described previously (Hall et al., 2012; Morrissey et al., 2014). L3 worms were anesthetized with 1% phenyl-isopropanol in fixative buffer and then fixed in 2.5% glutaldehyde, 1.5% paraformaldehyde in 0.1M Na-cacodylate buffer. Worms were cut open in the head or tail region and incubated for 2.5 hours. Samples were then washed three times in 0.1M Na-cacodylate buffer, stained in 1% UAC in 0.1M Na-acetate buffer, and then washed once in Na-acetate buffer and twice in Na-cacodylate buffer. Worms were then embedded in 3% agarose and dehydrated in ethanol. Samples were then embedded in Embed812 resin and propylene oxide solutions followed by embedding in 100% Embed812 resin for two days. Samples were then incubated at 60°C for 2 days. Longitudinal sections were cut across the gonad to allow for evaluation of the tissue to establish precise developmental stage and to identify the anchor cell. Images were then acquired on a Philips CM12 transmission electron microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image processing and AC protrusion volume
Acquisition of images is described above in “Microscopy and Image Acquisition”. Acquired confocal z-stack images were processed with Fiji (ImageJ 1.51f) and Photoshop (CC 2015; Adobe) (Schindelin et al., 2012). Time-lapse analysis and 3D reconstructions of protrusion formation and growth were built from confocal z-stacks, analyzed, and exported using Imaris 7.6.5 (Bitplane, Inc). Importantly, in some instances out-of-focus fluorescence from intense membrane signal above the protrusion was captured in slices. This signal was clearly distinguished and excluded from the relevant slices (and thus volume analysis), but this out-of-focus fluorescence in some instances appeared to broaden and obscure the edges of the protrusion in 3D Imaris renderings. Supplementary movies were exported at 20 frames per second as .mp4 files using the 3D reconstructions of time-lapse data in the Imaris 7.6.5 Animation module with key frames designated every 20 minutes.
Invasive protrusion volume and total AC volume were calculated as previously described by our laboratory (Kelley et al., 2017). In time-lapses, AC protrusion volume was calculated by hand-tracing the protrusion (the region of the AC extending below the BM) in each slice to generate an isosurface for each time point in Imaris 7.6.5. All rates of invasive protrusion expansion were calculated over the first thirty minutes of time-lapse image series following BM breach (images collected every five minutes, see above), as this was the time period of maximal growth of the invasive protrusion. The difference in protrusion volume at the time of initial breach was subtracted from the size of the protrusion thirty minutes after breach; this difference was then divided by the time that transpired (30 minutes) to obtain a rate of expansion for each animal observed.
For surface area and volume measurements of the entire AC, isosurfaces were generated in Imaris by manual thresholds of fluorescence intensity using 0.300µm surface detail. The volume of the invasive protrusion in these comparisons (Figure S1C) was then calculated by using the Imaris isosurface slicer tool to cut the AC isosurface at the AC-BM junction, thus separating the AC from the invasive protrusion. In comparisons across stages of invasion (P6.p 2-cell, P6.p 2/4-cell transition, and P6.p 4-cell stages), AC surface area or volume was normalized to average AC surface area or volume for ACs of the corresponding genotype before invasion (P6.p 2-cell stage). In dominant negative integrin (zmp-1>HA-βtail) animals, AC invasion is delayed, so data were collected from ACs that breached the BM at the P6.p 8-cell stage before the BM hole expanded beyond the AC.
Three strains were used for invasive protrusion analysis: NK881 (AC labeled with qyIs166 (cdh-3>GFP::CAAX), Figure 1), NK361 (AC labeled with qyIs24 (cdh-3>mCherry::PLCδPH, Figure 4), and NK1316 (uterine-specific RNAi line with the AC labeled with qyIs24 (cdh-3>mCherry::PLCδPH, Figures 2, 3, and 6). We found that strains NK881 and NK361 showed slightly faster rates of protrusion formation (0.47 and 0.48um3/min) while the uterine specific line was a little slower (0.35um3/min), although the rate was not statistically different (p>0.05, Student's t-test).
Analysis of AC polarity
AC polarity measurements were generated from background-subtracted sum projections of the AC using Fiji. Polarity was measured using a 5-pixel wide line tracing the invasive membrane (P6.p 2-cell stage) or invasive protrusion membrane (P6.p 2/4-cell transition) from centermost image slices. This value was then divided by the mean intensity of a 5-pixel wide line tracing the apical membrane of the AC in its centermost slice to determine fold enrichment of fluorescently tagged proteins at the invasive cell membrane. ImageJ macros which semi-automate this method of polarity measurement is available upon request.
Colocalization analysis
Colocalization analysis was performed in Fiji as described previously by our laboratory (Wang et al., 2014). The centermost slice of the AC was selected from confocal z-stacks with signal from two fluorophores. A 3-pixel wide line was drawn through the AC membrane and the absolute intensity of each fluorophore was measured every 0.15µm along this line (reported by Fiji as Integrated Density). The intensity profile for each fluorophore was then normalized to the maximum measured intensity of that fluorophore along the line.
Fluorescence loss in photobleaching (FLIP)
Images from fluorescence loss in photobleaching (FLIP) experiments were collected using a point scanning confocal microscope (Zeiss 780; Carl Zeiss) with a GaAsP high QE 32 channel spectral array detector with a 63×/1.4 oil Plan-Apochromat objective (Carl Zeiss) controlled by Zen software (version 2010; Carl Zeiss). Synchronized L3 hermaphrodites were anesthetized and prepared for imaging as described for time-lapse acquisition above. The FLIP region of interest (ROI) was a 25-pixel (approximately 1µm) diameter circle placed on the perimeter of the AC along either the apical membrane of invasive protrusion membrane. Four cycles of z-stack images were acquired at 18 second intervals at with 1.0µm slice thickness for 14 slices centered on the AC. Each cycle of acquisition was followed by a cycle of photobleaching of the FLIP ROI. Bleaching was performed at 100% laser power with 25 iterations.
All quantifications for FLIP experiments were performed in Fiji. Relative GFP loss was calculated by subtracting the pixel values of a sum projection of the AC taken at the fourth acquisition cycle (three cycles of bleaching performed every 18 seconds, then a fourth acquisition performed 54 seconds from acquisition start) from the corresponding pixel values of a sum AC projection taken at the first acquisition cycle (before any photobleaching). The resulting difference was then divided by the initial pixel intensity from the first acquisition cycle to generate a relative rate of loss. Regions for FLIP ratios were established by drawing a 3-pixel wide line across the AC to separate the two halves (for P6.p 2-cell and 8-cell controls) or by drawing a 3-pixel wide line across the AC at the neck of the invasive protrusion. The average rate of loss of the two AC regions was calculated from the relative rates of loss of the pixels in each region. FLIP ratios display the quotient of the average of the region with the photobleaching ROI (Region X) divided by the average of the region without the photobleaching ROI (Region Y). Spectral images for display purposes were generated by adding a background value of 100 to each pixel of the first acquisition cycle to eliminate background noise before the calculation of relative loss and applying the Fire lookup table in Fiji with constrained pixel ranges from 0.00 to 0.50. The resulting image was then smoothed for display. An ImageJ macro which automates this process is available upon request.
Calculation of basement membrane hole opening rates
Rates of BM hole expansion were quantified as published previously by our laboratory (Kelley et al., 2017). Time-lapse image series of AC protrusion formation acquired as described above were reconstructed in 3D using Imaris 7.6.5 (Bitplane, Inc.) The 3D-rendered BM was rotated 90° about the anterior-posterior axis of the worm to view the ventral surface of the animal. The signal from the BM fluorescence channel only was then exported as a .TIFF series. This series was opened in Fiji and BM hole size was then analyzed at five minute intervals through the time-lapse series by manually thresholding the BM hole and measuring the area of the thresholded region.
Blinding and unbiasing of data
For polarity and fluorophore intensity measurements, data sets were randomized using an ImageJ macro (courtesy of Martin Hoehne) to blind analysis. For samples in which blind analysis was not possible, randomly-selected samples were chosen for re-analysis to confirm precision of measurements.
Statistical analysis and data presentation
Statistical analyses were performed using JMP version 12.0 (SAS Institute). For all figure legends, asterisks indicate statistical significance as follows: n.s. = not significant (p>0.05); * p<0.05; **p<0.01; ***p<0.001. All comparisons of means were accomplished using a two-tailed unpaired Student’s t-test or One-way ANOVA with Tukey HSD post-hoc test as appropriate, and the specific test used and all sample sizes are indicated in the corresponding figure legend. For comparisons of parametric samples in which statistically significant differences were observed, sample sizes and distributions were validated by assaying normality of variance using a Shapiro-Wilk’s normality test. All data sets displayed normal variance with the exception of protrusion size in the wild type animals shown in Figure S1G (Figure S1G). Figures and graphs were constructed using Excel 2010 (Microsoft) and Illustrator (CC 2015; Adobe).
Supplementary Material
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>GFP::CAAX) and a unc-40 mutant animal (right), in which the AC breaches the BM but does not extend a protrusion. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies are 90 min. and were constructed from projections of 0.5µm z-sections. Time points in the wild type (left) were acquired in 1-min. intervals and in the unc-40 mutant (right) in 2-min. intervals. The video corresponds to the animals shown in Figure 1B.
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>mCherry::PLCδPH) and ppk-3 (RNAi) (right) treated animal, in which the AC breaches the BM, but fails to form a large protrusion. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies were constructed from projections of 0.5µm z-sections. Movies are 90 min., with time points acquired at 2-min. intervals. The video corresponds to the animals shown in Figure 2C.
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>mCherry::PLCδPH) and snap-29 (RNAi) (right) treated animal, in which the AC breaches the BM, but the protrusion forms at a reduced rate. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies were constructed from projections of 0.5µm z-sections. Movies are 120 min. The wild type (left) time-lapse was acquired in 5-min. intervals and the snap-29 (RNAi) in 2-min. intervals. The video corresponds to the animals shown in Figure 3D.
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>mCherry::PLCδPH) and exoc-8 (ok2523) mutant (right), in which the AC breaches the BM, but the AC fails to form a large protrusion. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies were constructed from projections of 0.5µm z-sections. Movies are 90 min. The wild type (left) time-lapse was acquired in 2-min. intervals and the exoc-8 mutant in 1-min. intervals. The video corresponds with the animals shown in Figure 4A.
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>mCherry::PLCδPH) and a dgn-1 (RNAi) (right) treated animal, in which the AC breaches the BM, but the AC does not form a large invasive protrusion. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies were constructed from projections of 0.5µm z-sections. Movies are 90 min. and were acquired in 5-min. intervals. The video corresponds to the animals shown in Figure 6D.
Highlights.
Netrin receptor UNC-40 directs lysosome exocytosis to form an invasive protrusion
UNC-40 mediates lysosome exocytosis through the t-SNARE SNAP-29 and the exocyst
The membrane anchored MMP ZMP-1 localizes to the invasive protrusion
A dystroglycan membrane diffusion barrier isolates the invasive protrusion
Acknowledgments
We thank Hanna Fares for organelle markers; Sam Johnson of the Duke University LMCF for imaging advice; Wei Zou for advice; and the University of Minnesota Caenorhabditis Genetics Center and National BioResource Project for strains used in this research. We thank Agnieszka Kawska (info@illuscientia) for her work on the graphical abstract. E. H. is supported by postdoctoral fellowship 129351-PF-16-024-01-CSM from the American Cancer Society. K. G. is supported by postdoctoral fellowship GM121015-01 from the National Institutes of Health. A. M. P. was supported by postdoctoral fellowship F32 GM115151 from the National Institutes of Health. This work was supported by NIGMS R01 GM083071 to B. G. and NIGMS R01 GM079320 and NIGMS R35 MIRA GM118049 to D. R. S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, K.M.N. and D.R.S; Methodology, K.M.N., Q.C.; Formal Analysis, K.M.N., A.G.; Investigation, K.M.N., E.H., Z.W., D.P.K., M.M.; Resources, K.M.N., Z.W., D.P.K., A.M.P., L.C.K., M.M., Q.C.; Writing – Original Draft, K.M.N., E.H., K.L.G., and D.R.S.; Writing – Revised Draft, K.M.N. and D.R.S.; Supervision, D.R.S.; Funding Acquisition, E.H., K.L.G., A.M.P., B.G. and D.R.S.
References
- Arantes RME, Andrews NW. A role for synaptotagmin VII-regulated exocytosis of lysosomes in neurite outgrowth from primary sympathetic neurons. J Neurosci. 2006;26:4630–4637. doi: 10.1523/JNEUROSCI.0009-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenti ST, Lohmer LL, Sherwood DR, Nance J. Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development. 2014;141:4640–4647. doi: 10.1242/dev.115048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello V, Moreau N, Sirour C, Hidalgo M, Buisson N, Darribère T. The dystroglycan: nestled in an adhesome during embryonic development. Dev Biol. 2015;401:132–142. doi: 10.1016/j.ydbio.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Branch KM, Hoshino D, Weaver AM. Adhesion rings surround invadopodia and promote maturation. Biology Open. 2012;1:711–722. doi: 10.1242/bio.20121867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Castro A, Marchesin V, Monteiro P, Lodillinsky C, Rossé C, Chavrier P. Cellular and Molecular Mechanisms of MT1-MMP-Dependent Cancer Cell Invasion. Annu Rev Cell Dev Biol. 2016;32:555–576. doi: 10.1146/annurev-cellbio-111315-125227. [DOI] [PubMed] [Google Scholar]
- Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 2015;200:1035–1049. doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer N, Rebollo E, Domínguez P, Elkhatib N, Chavrier P, Daviet L, González C, González-Gaitán M. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development. 2007;134:4437–4447. doi: 10.1242/dev.010983. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using µManager. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Freeman SA, Goyette J, Furuya W, Woods EC, Bertozzi CR, Bergmeier W, Hinz B, van der Merwe PA, Das R, Grinstein S. Integrins Form an Expanding Diffusional Barrier that Coordinates Phagocytosis. Cell. 2016;164:128–140. doi: 10.1016/j.cell.2015.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawden-Bone C, Zhou Z, King E, Prescott A, Watts C, Lucocq J. Dendritic cell podosomes are protrusive and invade the extracellular matrix using metalloproteinase MMP-14. J Cell Sci. 2010;123:1427–1437. doi: 10.1242/jcs.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Artieri C, Khezri N, Singh RS, Gupta BP. Comparative analysis of function and interaction of transcription factors in nematodes: extensive conservation of orthology coupled to rapid sequence evolution. BMC Genomics. 2008;9:399. doi: 10.1186/1471-2164-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn EJ, Sherwood DR. Cell invasion through basement membrane: the anchor cell breaches the barrier. Curr Opin Cell Biol. 2011;23:589–596. doi: 10.1016/j.ceb.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn EJ, Yashiro H, Ziel JW, Ihara S, Wang Z, Sherwood DR. Integrin acts upstream of netrin signaling to regulate formation of the anchor cell’s invasive membrane in C. elegans. Dev Cell. 2009;17:187–198. doi: 10.1016/j.devcel.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn EJ, Ziel JW, Morrissey MA, Linden LM, Wang Z, Chi Q, Johnson SA, Sherwood DR. The netrin receptor DCC focuses invadopodia-driven basement membrane transmigration in vivo. J Cell Biol. 2013;201:903–913. doi: 10.1083/jcb.201301091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn EJ, Kelley LC, Naegeli KM, Wang Z, Chi Q, Sherwood DR. ADF/cofilin promotes invadopodial membrane recycling during cell invasion in vivo. J Cell Biol. 2014;204:1209–1218. doi: 10.1083/jcb.201312098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Hartwieg E, Nguyen KCQ. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 2012;107:93–149. doi: 10.1016/B978-0-12-394620-1.00004-7. [DOI] [PubMed] [Google Scholar]
- He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary K, Li X-Y, Allen E, Stevens SL, Weiss SJ. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 2006;20:2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiu Y, Jin C, Liu Y, Holmberg CI, Jäntti J. Exocyst subunits Exo70 and Exo84 cooperate with small GTPases to regulate behavior and endocytic trafficking in C. elegans. PLoS ONE. 2012;7:e32077. doi: 10.1371/journal.pone.0032077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RP, Kang SH, Kramer JM. C. elegans dystroglycan DGN-1 functions in epithelia and neurons, but not muscle, and independently of dystrophin. Development. 2006;133:1911–1921. doi: 10.1242/dev.02363. [DOI] [PubMed] [Google Scholar]
- Kay RR, Langridge P, Traynor D, Hoeller O. Changing directions in the study of chemotaxis. Nat Rev Mol Cell Biol. 2008;9:455–463. doi: 10.1038/nrm2419. [DOI] [PubMed] [Google Scholar]
- Kelley LC, Wang Z, Hagedorn EJ, Wang L, Shen W, Lei S, Johnson SA, Sherwood DR. Live-cell confocal microscopy and quantitative 4D image analysis of anchor-cell invasion through the basement membrane in Caenorhabditis elegans. Nat Protoc. 2017;12:2081–2096. doi: 10.1038/nprot.2017.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard T, Jäättelä M. Lysosomal involvement in cell death and cancer. Biochim Biophys Acta. 2009;1793:746–754. doi: 10.1016/j.bbamcr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Pilot F. Developmental control of cell morphogenesis: a focus on membrane growth. Nat Cell Biol. 2003;5:103–108. doi: 10.1038/ncb0203-103. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Wieschaus E. Polarized insertion of new membrane from a cytoplasmic reservoir during cleavage of the Drosophila embryo. J Cell Biol. 2000;150:849–860. doi: 10.1083/jcb.150.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong HS, Robertson AE, Stoletov K, Leith SJ, Chin CA, Chien AE, Hague MN, Ablack A, Carmine-Simmen K, McPherson VA, et al. Invadopodia are required for cancer cell extravasation and are a therapeutic target for metastasis. Cell Rep. 2014;8:1558–1570. doi: 10.1016/j.celrep.2014.07.050. [DOI] [PubMed] [Google Scholar]
- Lohmer LL, Kelley LC, Hagedorn EJ, Sherwood DR. Invadopodia and basement membrane invasion in vivo. Cell Adh Migr. 2014;8:246–255. doi: 10.4161/cam.28406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmer LL, Clay MR, Naegeli KM, Chi Q, Ziel JW, Hagedorn EJ, Park JE, Jayadev R, Sherwood DR. A Sensitized Screen for Genes Promoting Invadopodia Function In Vivo: CDC-42 and Rab GDI-1 Direct Distinct Aspects of Invadopodia Formation. PLoS Genet. 2016;12:e1005786. doi: 10.1371/journal.pgen.1005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus DQ, Li X-Y, Durbin S, Agarwal D, Chi Q, Weiss SJ, Sherwood DR. In vivo identification of regulators of cell invasion across basement membranes. Sci Signal. 2010;3:ra35. doi: 10.1126/scisignal.2000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas N, Evans E. Mechanical properties of the red cell membrane in relation to molecular structure and genetic defects. Annu Rev Biophys Biomol Struct. 1994;23:787–818. doi: 10.1146/annurev.bb.23.060194.004035. [DOI] [PubMed] [Google Scholar]
- Morrissey MA, Hagedorn EJ, Sherwood DR. Cell invasion through basement membrane: The netrin receptor DCC guides the way. Worm. 2013;2:e26169. doi: 10.4161/worm.26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey MA, Keeley DP, Hagedorn EJ, McClatchey STH, Chi Q, Hall DH, Sherwood DR. B-LINK: a hemicentin, plakin, and integrin-dependent adhesion system that links tissues by connecting adjacent basement membranes. Dev Cell. 2014;31:319–331. doi: 10.1016/j.devcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DA, Courtneidge SA. The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada C, Ritchie K, Oba Y, Nakamura M, Hotta Y, Iino R, Kasai RS, Yamaguchi K, Fujiwara T, Kusumi A. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat Cell Biol. 2003;5:626–632. doi: 10.1038/ncb1009. [DOI] [PubMed] [Google Scholar]
- Nakaya Y, Sukowati EW, Sheng G. Epiblast integrity requires CLASP and Dystroglycan-mediated microtubule anchoring to the basal cortex. J Cell Biol. 2013;202:637–651. doi: 10.1083/jcb.201302075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot A-S, Fares H, Payrastre B, Chisholm AD, Labouesse M, Laporte J. The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3062–3074. doi: 10.1091/mbc.E05-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CD, Mistriotis P, Konstantopoulos K. Cancer cell motility: lessons from migration in confined spaces. Nat Rev Cancer. 2017;17:131–140. doi: 10.1038/nrc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, Vietri M, Nisi V, Bucci C, Brech A, et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–169. doi: 10.1016/s0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- Rodríguez A, Webster P, Ortego J, Andrews NW. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J Cell Biol. 1997;137:93–104. doi: 10.1083/jcb.137.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Medina M, Gregus KA, Nichol RH, O’Toole SM, Gomez TM. Regulation of ECM degradation and axon guidance by growth cone invadosomes. Development. 2015;142:486–496. doi: 10.1242/dev.108266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Norris A, Sato M, Grant BD. C. elegans as a model for membrane traffic. WormBook. 2014:1–47. doi: 10.1895/wormbook.1.77.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189:541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seano G, Chiaverina G, Gagliardi PA, di Blasio L, Puliafito A, Bouvard C, Sessa R, Tarone G, Sorokin L, Helley D, et al. Endothelial podosome rosettes regulate vascular branching in tumour angiogenesis. Nat Cell Biol. 2014;16:1. doi: 10.1038/ncb3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood DR, Sternberg PW. Anchor cell invasion into the vulval epithelium in C. elegans. Dev Cell. 2003;5:21–31. doi: 10.1016/s1534-5807(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, Butler JA, Kramer JM, Sternberg PW. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell. 2005;121:951–962. doi: 10.1016/j.cell.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Son S, Kang JH, Oh S, Kirschner MW, Mitchison TJ, Manalis S. Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. J Cell Biol. 2015;211:757–763. doi: 10.1083/jcb.201505058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature. 2011;469:226–230. doi: 10.1038/nature09642. [DOI] [PubMed] [Google Scholar]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Tojima T, Kamiguchi H. Exocytic and endocytic membrane trafficking in axon development. Dev Growth Differ. 2015;57:291–304. doi: 10.1111/dgd.12218. [DOI] [PubMed] [Google Scholar]
- Trimble WS, Grinstein S. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J Cell Biol. 2015;208:259–271. doi: 10.1083/jcb.201410071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Linden LM, Naegeli KM, Ziel JW, Chi Q, Hagedorn EJ, Savage NS, Sherwood DR. UNC-6 (netrin) stabilizes oscillatory clustering of the UNC-40 (DCC) receptor to orient polarity. J Cell Biol. 2014;206:619–633. doi: 10.1083/jcb.201405026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkle CC, McClain LM, Valtschanoff JG, Park CS, Maglione C, Gupton SL. A novel Netrin-1-sensitive mechanism promotes local SNARE-mediated exocytosis during axon branching. J Cell Biol. 2014;205:217–232. doi: 10.1083/jcb.201311003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Liu Y, Zhao L, Gan Q, Wang X, Yang C. The lysosomal cathepsin protease CPL-1 plays a leading role in phagosomal degradation of apoptotic cells in Caenorhabditis elegans. Mol Biol Cell. 2014;25:2071–2083. doi: 10.1091/mbc.E14-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylivinkka I, Keski-Oja J, Hyytiäinen M. Netrin-1: A regulator of cancer cell motility? Eur J Cell Biol. 2016;95:513–520. doi: 10.1016/j.ejcb.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel JW, Sherwood DR. Roles for netrin signaling outside of axon guidance: a view from the worm. Dev Dyn. 2010;239:1296–1305. doi: 10.1002/dvdy.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel JW, Hagedorn EJ, Audhya A, Sherwood DR. UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nat Cell Biol. 2009;11:183–189. doi: 10.1038/ncb1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotek-Zlotkiewicz E, Monnier S, Cappello G, Le Berre M, Piel M. Optical volume and mass measurements show that mammalian cells swell during mitosis. J Cell Biol. 2015;211:765–774. doi: 10.1083/jcb.201505056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Yadav S, DeVault L, Nung Jan Y, Sherwood DR. RAB-10-Dependent Membrane Transport Is Required for Dendrite Arborization. PLoS Genet. 2015;11:e1005484. doi: 10.1371/journal.pgen.1005484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>GFP::CAAX) and a unc-40 mutant animal (right), in which the AC breaches the BM but does not extend a protrusion. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies are 90 min. and were constructed from projections of 0.5µm z-sections. Time points in the wild type (left) were acquired in 1-min. intervals and in the unc-40 mutant (right) in 2-min. intervals. The video corresponds to the animals shown in Figure 1B.
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>mCherry::PLCδPH) and ppk-3 (RNAi) (right) treated animal, in which the AC breaches the BM, but fails to form a large protrusion. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies were constructed from projections of 0.5µm z-sections. Movies are 90 min., with time points acquired at 2-min. intervals. The video corresponds to the animals shown in Figure 2C.
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>mCherry::PLCδPH) and snap-29 (RNAi) (right) treated animal, in which the AC breaches the BM, but the protrusion forms at a reduced rate. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies were constructed from projections of 0.5µm z-sections. Movies are 120 min. The wild type (left) time-lapse was acquired in 5-min. intervals and the snap-29 (RNAi) in 2-min. intervals. The video corresponds to the animals shown in Figure 3D.
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>mCherry::PLCδPH) and exoc-8 (ok2523) mutant (right), in which the AC breaches the BM, but the AC fails to form a large protrusion. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies were constructed from projections of 0.5µm z-sections. Movies are 90 min. The wild type (left) time-lapse was acquired in 2-min. intervals and the exoc-8 mutant in 1-min. intervals. The video corresponds with the animals shown in Figure 4A.
Time-lapses show protrusion extension through the BM (laminin::mCherry, purple, shown in first and last frames) in a wild type (left, membrane marker cdh-3>mCherry::PLCδPH) and a dgn-1 (RNAi) (right) treated animal, in which the AC breaches the BM, but the AC does not form a large invasive protrusion. Images were acquired using a spinning disc confocal microscope (CSU-10 scan head; Yokogawa, mounted on a Zeiss AxioImager compound microscope). Movies were constructed from projections of 0.5µm z-sections. Movies are 90 min. and were acquired in 5-min. intervals. The video corresponds to the animals shown in Figure 6D.