Abstract
Notch is a cell-surface receptor that controls cell fate decisions and is regulated by O-glycans attached to epidermal growth factor-like (EGF) repeats in its extracellular domain. Protein O-fucosyltransferase 1 (Pofut1) modifies EGF repeats with O-fucose and is essential for Notch signaling. Constitutive activation of Notch signaling has been associated with a variety of human malignancies. Therefore, tools for inhibiting Notch activity are being developed as cancer therapeutics. Towards this end, we screened L-fucose analogs for their effects on Notch signaling. Two analogs, 6-alkynyl and 6-alkenyl fucose, were substrates of Pofut1 and were incorporated directly into Notch EGF repeats in cells. Both analogs were potent inhibitors of binding to and activation of Notch1 by Notch ligands Dll1 and Dll4, but not by Jag1. Mutagenesis and modeling studies suggest that incorporation of the analogs into EGF8 of Notch1 markedly reduces the ability of Delta ligands to bind and activate Notch1.
Introduction
The Notch signaling pathway is highly conserved across all metazoa and plays important roles in cell fate determination during development and in adult tissue homeostasis1,2. In mammals there are four Notch receptor homologs (Notch1–4) and five Notch ligands: Jagged (Jag) 1, 2 and Delta-like (Dll) 1, 3 and 42. Notch signaling is initiated upon binding of Notch ligand to Notch receptor leading to a cascade of events that ultimately culminates in the cleavage of the Notch intracellular domain (NICD) by γ-secretase. NICD then translocates to the nucleus, where it regulates transcription of target genes3.
Given the critical role that Notch plays in tissue development and maintenance, it is not surprising that dysregulation of Notch signaling leads to several human disorders. Excessive Notch signaling has been implicated in a wide range of human cancers including cervical, renal, lung, hepatocellular, hematologic, and neurologic malignancies4–6. Initial efforts to inhibit Notch signaling in treatment of these cancers has focused largely on the development of γ-secretase inhibitors (GSIs), small molecules that prevent NICD cleavage. Unfortunately, this strategy has not been successful to date, largely due to dose limiting side effects7. Alternative anti-Notch therapeutic strategies now in development include use of monoclonal antibodies targeting either specific Notch homologues8 or Notch ligands9–11, with the hope that these may prove more selective and less toxic.
The Notch extracellular domain (NECD) consists of up to 36 tandem epidermal growth factor-like (EGF) repeats including those that interact directly with ligands12,13. EGF repeats with appropriate consensus sequences are post-translationally modified with O-glycans, initiated by O-fucose, O-glucose, or O-GlcNAc14,15. Protein O-fucosyltransferase 1 (Pofut1) is responsible for the transfer of O-fucose to EGF repeats with an O-fucose consensus sequence and is an essential component of Notch signaling in all contexts examined to date16. Members of the Fringe family of enzymes extend the O-fucose modification by adding GlcNAc to further regulate Notch activity17,18.
We hypothesized that inhibition or interference with the normal O-fucosylation process might lead to the development of Notch inhibitors. Recent work has demonstrated that peracetylated fucose derivatives are metabolized to their corresponding GDP-fucose analogs within cells by exploiting a fucose salvage pathway19–21. Previous reports suggest that some of these analogs cause feedback inhibition of the de novo biosynthesis of GDP-fucose, reducing fucosylation of target glycans and proteins and resulting in their altered behavior19,20. In contrast, our group has shown that peracetylated 6-alkynyl fucose, is converted to the corresponding GDP-fucose analog becoming a substrate of Pofut1, which efficiently transfers it to Notch EGF repeats22.
Here, we report the synthesis of a panel of fucose analogs and show that some inhibit Notch activity. Peracetylated versions of selected inhibitory analogs were converted to GDP-fucose analogs within mammalian cells. Fucose analogs incorporated by Pofut1 into Notch EGF repeats disrupted Delta-, but not Jagged-induced Notch signaling. Our data further suggest that fucose analog incorporation caused steric clashes with Delta ligands, but not with Jag1, and was responsible for inhibition of Notch signaling. Finally, these inhibitory fucose analogs were used to block Notch dependent T-cell differentiation. Fucose analogs thus represent a novel tool for the inhibition of Notch signaling.
Results
Fucose analogs inhibit Notch signaling in Zebrafish
We generated a panel of GDP-fucose derivatives (Compounds 1–8, Figure 1a) and corresponding peracetylated fucose analogs (Compounds 9–16, Figure 1b; Supplementary Results, Supplementary Figure 1a) with different substituents at the 6-carbon position of L-fucose (Supplementary Figure 1b). To screen for fucose analogs with an inhibitory effect on Notch signaling, we utilized transgenic Zebrafish Tg(Tp1bglob:eGFP)um14 embryos expressing a Notch reporter transgene (GFP under the control of elements responsive to NICD)23. GFP fluorescence induced by activation of the Notch reporter serves as a sensitive and specific reflection of Notch signaling intensity and was used to monitor Notch signaling at 48 hours post fertilization, a developmental period when activation by Delta ligands predominates in Zebrafish. GDP-fucose analogs were injected into the yolk sac of embryos at the one cell stage, bypassing the fucose salvage pathway. As fertilized eggs begin to develop, they engulf materials from the yolk sac including GDP-fucose analogs and other nutrients. The analogs in our panel had a range of effects on Notch signaling. As expected, untreated and natural GDP-fucose (1) treated embryos expressed relatively high levels of GFP indicating robust Notch signaling (Figure 1c). Inhibition of GDP-fucose biosynthesis by knocking down GDP-mannose-4,6-dehydratase (gmds MO)24 served as a positive control for Notch signaling inhibition due to reduced fucose on Notch (Figure 1c, bottom left panel). GDP-fucose analogs 2 and 5 did not cause any substantial reduction in Notch signaling compared to negative controls. By contrast, compounds 7 and 8 caused a partial reduction in GFP levels, whereas compounds 3, 4 and 6 with the C-6 ethynyl, ethenyl or OH substituents respectively, had the greatest inhibitory effect, almost entirely eliminating the GFP Notch reporter signal (Figure 1c).
Figure 1. Effects of fucose analogs on Notch signaling in Zebrafish embryos.
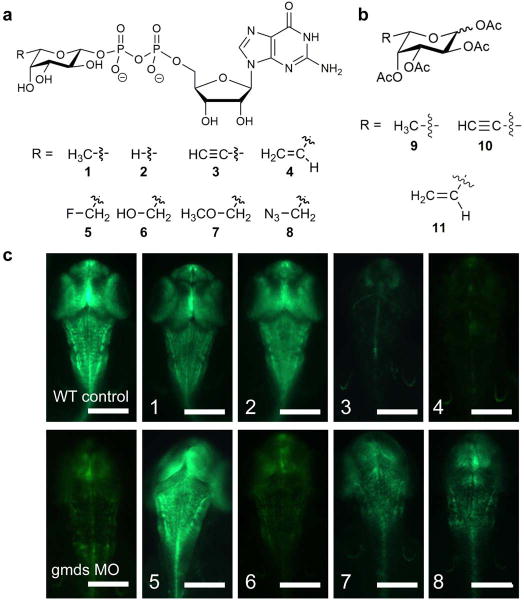
(a) Numbered structures of GDP-fucose analogs screened as potential inhibitors of Notch signaling in Zebrafish. (b) Peracetylated fucose analogs selected for further analysis in cell-based assays. See Supplementary Figure 1a for structures of other peracetylated fucose analogs. (c) Transgenic Zebrafish embryos expressing a GFP fluorescent Notch signaling reporter showed that some injected GDP-fucose analogs, indicated in each panel, reduced Notch signaling. Knock down of GDP-mannose-4,6-dehydratase (gmds MO) to inhibit endogenous GDP-fucose biosynthesis was used as a positive control for the effect of eliminating Notch O-fucosylation and Notch signaling. Scale bar represents 0.5 mm.
Fucose analogs are added to EGF repeats by Pofut1
Based on previous literature describing the importance of fucose for Notch activation16,25, and the effects of sugar analogs in other systems19,20,26–28, we hypothesized that one of two possible mechanisms may explain inhibitory activity of synthetic GDP-fucose analogs on Notch signaling (Figure 1c); either the analogs inhibit the fucosyltransferase activity of Pofut1 resulting in unmodified EGF repeats, or the analogs are transferred by Pofut1, and when incorporated into EGF repeats, interfere with Notch signaling. To address this question, we incubated HEK293T cells expressing EGF1–18 of Notch1 with peracetylated versions of the fucose analogs (Figure 1b, Supplementary Figure 1a), which are more readily taken up by cells than the GDP-fucose analogs in cell culture19,20. The success of this approach requires that a peracetylated analog be taken up by cells, efficiently converted to the corresponding GDP derivative, and transported into the endoplasmic reticulum (Supplementary Figure 1c) for utilization by Pofut119,20. In Zebrafish embryos (Figure 1c), we injected GDP-fucose analogs directly into the yolk, thereby bypassing the need for conversion of the analogs to their corresponding GDP derivatives.
Using mass spectral glycoproteomic methods, we confirmed that HEK293T cells treated with compounds 10 and 11 (the peracetylated versions of 3 and 4, respectively) did not act as inhibitors of Pofut1, but were transferred by Pofut1 onto Notch1 EGF repeats. Extracted ion chromatograms (EICs) were generated to compare the relative amounts of ions corresponding to the fucose analog and fucose-modified glycoforms of a peptide from Notch1 EGF6 that contains an O-fucose consensus sequence (Figure 2, see Supplementary Figure 2 for mass spectra). In the sample treated with peracetylated fucose (9), this peptide is nearly completely modified with fucose (Figure 2a). In the samples treated with compounds 10 and 11, the fucose analogs were incorporated into Notch1 EGF repeats at high stoichiometry (Figure 2b–c). Significantly, there was essentially no signal for unmodified peptide, confirming that the analogs do not inhibit Pofut1 activity. Instead, the analogs nearly completely replaced natural fucose on the peptide. Similar analyses of EGF1–18 produced in cells expressing Lfng demonstrated that EGF repeats modified with analogs 10 and 11 were also efficiently elongated with GlcNAc in cell culture by Lfng (Figure 2d–f), showing that incorporation of these fucose analogs did not interfere with Fringe N-acetylglucosaminyltransferase activity. Elongation to the tri- and tetra-saccharide were also unaffected by fucose analog incorporation (Figure 2d–f). All other peptides with O-fucose modification sites on Notch1 EGF repeats that were identified by mass spectrometry were similarly modified with these fucose analogs (Supplementary Note 1).
Figure 2. Peracetylated fucose analogs are efficiently incorporated into Notch EGF repeats and elongated by Lfng.
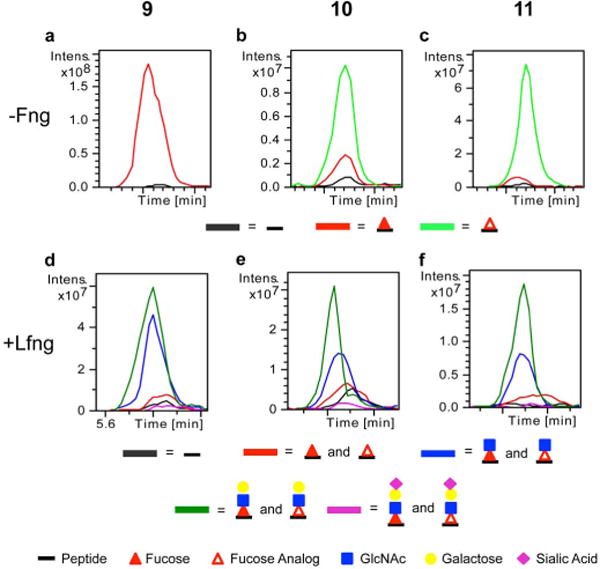
EGF1–18-MycHis from mNotch1 was transfected into HEK293T cells expressing no Fng (a–c) or Lfng (d–f) and the cells were grown in the presence of compounds 9 (a, d), 10 (b, e), or 11 (c, f) for 72 hours. Purified EGF1–18-MycHis was digested and subjected to nano-LC-MS/MS analysis. (a–c) Extracted ion chromatograms (EIC) of the ions corresponding to glycoforms (see key) of this peptide show efficient incorporation of the corresponding compounds. (d–f) Similar experiments in the presence of Lfng demonstrate elongation of fucose analogs with GlcNAc. See Supplementary Figure 2 for corresponding mass spectra and Supplementary Note 1 for m/z ratios used to generate EICs.
None of the other peracetylated fucose analogs corresponding to those evaluated in Figure 1c were incorporated into Notch1 EGF repeats, nor did they inhibit fucose incorporation (Supplementary Figure 3). Compound 6 had a strong inhibitory effect when injected into Zebrafish embryos (Figure 1c), but lack of incorporation of the peracetylated version (Compound 14) suggests that it was not efficiently converted to the corresponding GDP-fucose analog in cells. Compound 13 has been reported to have an inhibitory effect on other fucosyltransferases19,20. This compound was efficiently incorporated into Notch1 EGF repeats but did not inhibit incorporation of fucose, demonstrating it is also a substrate and not an inhibitor of Pofut1 (Supplementary Figure 3). However, it had no significant effect on Notch signaling in a cell-based co-culture reporter assay, consistent with the Zebrafish embryo data using the GDP version of this sugar analog (Figure 1c, compound 5). Compound 17, a previously described inhibitor of fucosyltransferases19, also had no effect, suggesting it does not inhibit Pofut1 activity (Supplementary Figure 3a). Consistent with the lack of incorporation into Notch1 EGF repeats or inhibition of fucose incorporation, we observed no effect of any of these compounds (compounds 12–17) on Notch signaling in a co-culture reporter assay (Supplementary Figure 3b). Thus, we focused on compounds 10 and 11 as potential candidates for development of inhibitors of Notch signaling in vivo.
Compounds 10 and 11 inhibit Dll-induced Notch signaling
To further evaluate the effect that incorporation of inhibitory fucose analogs into Notch EGF repeats has on Notch signaling, we used a cell-based co-culture Notch reporter assay where Dll1 and Jag1 activated Notch1 to nearly identical levels relative to controls (Supplementary Figure 4a). Treatment with peracetylated fucose (9) served as a control and did not inhibit Notch signaling under any of the tested conditions (Figure 3). In contrast, treatment with fucose analogs 10 and 11 reduced Dll1-induced Notch1 signaling to background levels (Figure 3a). Similarly, Dll4-induced Notch1 and Dll1- and Dll4-induced Notch2 signaling were all significantly inhibited by compounds 10 and 11 relative to DMSO and compound 9 treated controls (Figure 3b, d, e). Interestingly, fucose analogs 10 and 11 had no significant effect on Jag1-induced Notch signaling (Figure 3c, f). Thus, analogs 10 and 11 preferentially inhibit Notch activation from Dll ligands. Both of these inhibitors were effective at nanomolar concentrations (Supplementary Figure 4b) and only caused a minimal reduction in cell surface expression of Notch1 (Supplementary Figure 4c). Fucose analogs did not significantly affect the proliferation rate of cells used in these assays over the time period experiments were conducted (Supplementary Figure 4d).
Figure 3. Peracetylated fucose analogs inhibit Dll1- and Dll4- but not Jag1-induced Notch signaling.
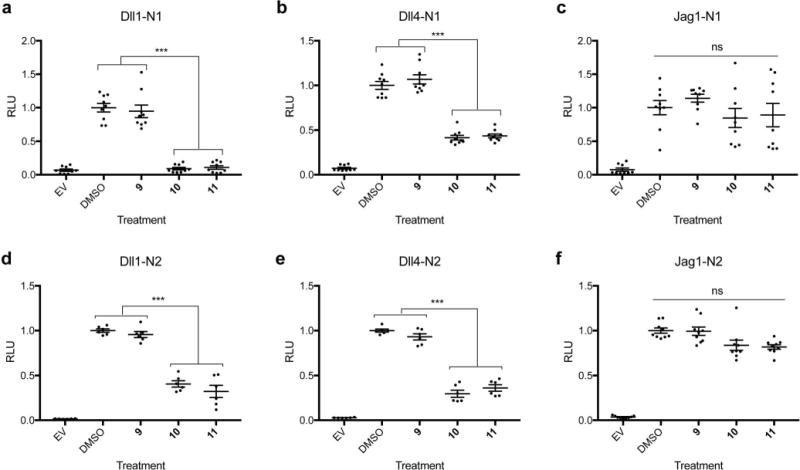
NIH3T3 cells expressing mNotch1 (a–c) or mNotch2 (d–f) were co-cultured with cells expressing Dll1 (a, d), Dll4 (b, e), or Jag1 (c, f) in the presence of peracetylated fucose analogs (compounds 9, 10, 11). Cells transfected with empty vector (EV) were used as a negative control and cells grown in DMSO were a positive control for Notch signaling. All experiments were performed three independent times, and data represents nine biological replicates (n=9). All plots represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, Tukey post-hoc test, adjusted p values.
As reported previously, activation of Notch1 or 2 by Dll1 and Dll4 is enhanced by Lfng, whereas Notch1 or 2 activation by Jag1 is reduced29–31. Since Lfng transfers GlcNAc to both analogs 10 and 11 (Figure 2), we examined whether Lfng had the same effects on Notch activation when Notch was carrying these analogs (Supplementary Figure 5). In the presence of analogs 10 and 11, co-expression of Lfng in Notch receptor-expressing cells enhanced Notch activation by Dll1 or Dll4 (Supplementary Figure 5a, b, d, e), and reduced Notch activation by Jag1 (Supplementary Figure 5c, f), consistent with the effects of Lfng on Notch receptors modified with natural fucose.
Notch ligand-receptor binding correlates with signaling
A flow cytometry based Notch ligand binding assay was used to determine if the decreased Notch signaling caused by fucose analogs 10 and 11 was a result of reduced ligand-receptor binding. Binding experiments with Dll1 and Dll4 correlated well with Notch signaling data. Dll1-Notch1 binding was reduced to background levels after treatment with compounds 10 and 11 (Figure 4a). Dll1-Notch2, Dll4-Notch1 and Dll4-Notch2 binding were all significantly reduced, but to a lesser extent (Figure 4b, d, e). All four Dll-Notch binding interactions were partially rescued by Lfng (Supplementary Figure 6a, b, d, e), similar to the effect of Lfng in Notch signaling assays. These data suggest that reduced ligand-receptor binding is substantially responsible for the effects of inhibitory fucose analogs on Dll-induced Notch signaling.
Figure 4. Compounds 10 and 11 inhibit Notch-Dll ligand binding.
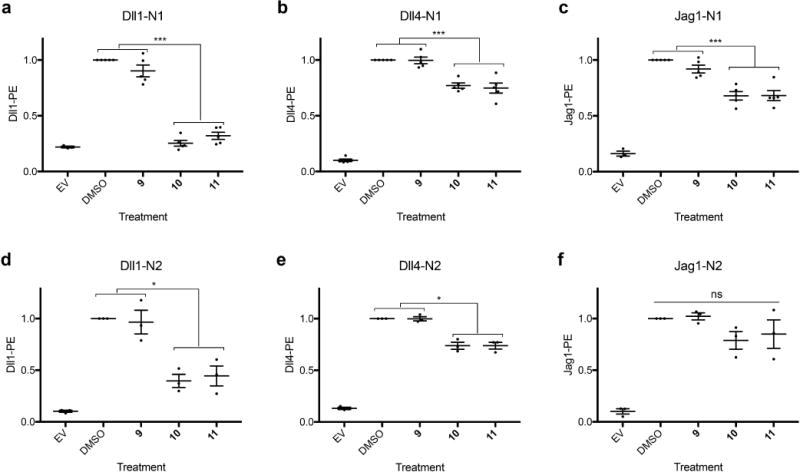
Notch ligand binding experiments were performed to measure the effect of fucose analogs on the ability of mNotch1 (a–c) or mNotch2 (d–f) transfectants to bind soluble Dll1-Fc (a, d), Dll4-Fc (b, e), or Jag1-Fc (c, f) using cell-based flow cytometry assays. EV transfected cells were used as a negative control and cells cultured in the presence of DMSO served as a positive control. Six independent experiments were performed for all samples (n=6). All plots represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, Tukey post-hoc test, adjusted p values.
As we and others have reported, the extent of Jag1 binding levels does not necessarily correlate with signaling intensity30–32. Relative to DMSO and compound 9 treated controls, compounds 10 and 11 caused a modest reduction in Jag1 binding to Notch1 and Notch2 (Figure 4c, f). This reduction in binding did not correlate with a significant effect of these inhibitors on Jag1-induced Notch signaling, although Jag1-induced signaling was generally slightly reduced (Figure 3c, f). As previously reported30–32, Lfng caused a slight increase in Jag1-binding for DMSO controls as well as analog 9, 10 and 11 treated samples (Supplementary Figure 6c, f). These results provide further support for the idea that Jag1-mediated Notch activation is dependent on more than just ligand binding as measured in a flow cytometry binding assay30–32.
Fucose analogs on ligands do not affect Notch activity
In the co-culture Notch reporter assay, both ligand-expressing cells and Notch receptor-expressing cells were exposed to the same treatment conditions, and ligands also contain EGF repeats modified with O-fucose33,34. Thus, we assessed if exposure of the ligand-expressing cells to these conditions reduced their ability to induce Notch signaling and thereby contributed to the inhibitory effects of O-fucose analogs 10 and 11. This was tested in a reverse binding assay, in which cells overexpressing full-length Dll1 grown in the presence of each experimental compound were assessed for their ability to bind a soluble ligand-binding fragment of the Notch1 ECD that was generated with natural fucose. None of the treatment conditions caused any significant change in the ability of Dll1 to bind NECD (Supplementary Figure 7a–b). In addition, we used a plate-coating assay, in which plates were coated with Notch ligands generated with natural fucose. Notch1 reporter cells were plated onto the plate-bound ligands and incubated with fucose analogs. The results showed the same pattern of inhibition of Notch signaling as in the co-culture Notch reporter assay, where cells expressing both Notch ligands and receptors were exposed to fucose analogs (Supplementary Figure 7c). Taken together, these data strongly support the hypothesis that the incorporation of compounds 10 and 11 into EGF repeats of Notch receptors, not into EGF repeats of Notch ligands, is responsible for their inhibitory effect.
Fucose analogs on Notch1 EGF8 inhibit Notch signaling
Recent mutagenesis and structural studies13,31,35 have demonstrated that the O-fucose residues on EGF8 and EGF12 are both directly involved in Notch-ligand interactions. In order to gain further insights into the mechanism by which compounds 10 and 11 inhibit Notch signaling, we generated Notch1 mutants in which the O-fucose modification sites in EGF8, EGF12 or both had been eliminated. We hypothesized that if these fucose analogs interfere with binding to Delta-like ligands when incorporated at either EGF8 or EGF12, then eliminating the O-fucose modification at these sites by mutagenesis might relieve the inhibition. Since treatment with 50 μM analogs completely inhibited Dll1-mediated Notch1 activation (Figure 3a), we used a lower concentration to achieve partial inhibition in these studies (100 nM, Supplementary Figure 4b). As expected, elimination of fucose at these sites caused a substantial reduction in Dll1-induced Notch signaling in natural fucose, while the EGF8/12 double mutant had essentially no activity (Figure 5a). Treatment with 100 nM compounds 10 and 11 caused a further decrease in Notch signaling, indicating that incorporation of the analogs at either of these individual sites was not solely responsible for the inhibitory effects of the fucose analogs. Normalizing Notch activity to the DMSO-treated control for each mutant allowed us to compare the relative decrease caused by each analog with each mutant (Figure 5b). Interestingly, the EGF8 mutant partially rescued inhibition by compounds 10 and 11, but the EGF12 mutant was inhibited as potently as wild type Notch1.
Figure 5. Incorporation of fucose analogs at EGF8 of Notch1 plays an important role in Dll1 and Dll4-mediated Notch activation.
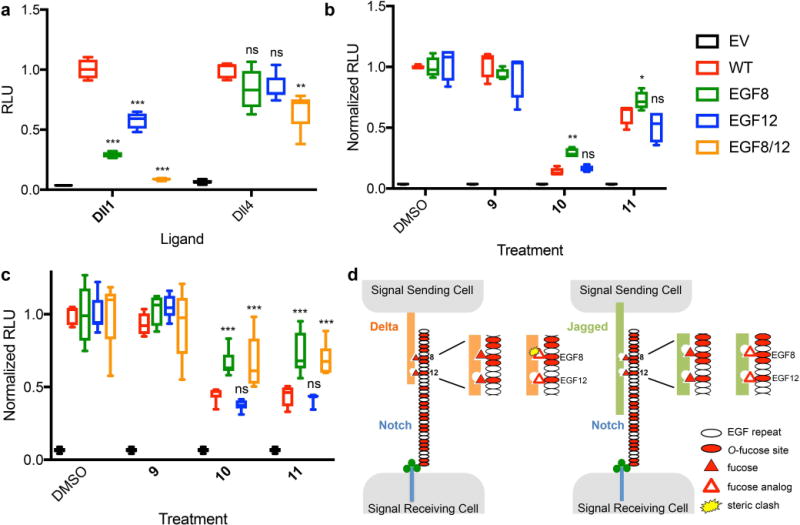
(a) Notch1 activation assays using Dll1-Fc (left) or Dll4-Fc (right) as activating ligands coated on plates showing the effect of mutations at EGF8, 12 or both on Dll1 and Dll4-induced Notch1 signaling. Relative Luciferase Unit (RLU) normalized to wild type (WT) Notch1. (b) Plate coating Notch1 activation assays using Dll1-Fc as activating ligand in the presence of DMSO (control) or 100 nM of the indicated compound. RLU for all constructs (WT and mutants) in DMSO was normalized to 1. (c) Plate coating assays for Dll4-induced signaling using 50 μM fucose analogs. The EGF8/12 double mutant was also tested. RLU for all constructs was normalized to the DMSO control. Box and whisker plots represent six biological replicates (n=6). *p<0.05, **p<0.01, ***p<0.001, Tukey post-hoc test, adjusted p values. (d) Model figure for fucose analog mediated inhibition of Notch1 activation. Elimination of the O-fucose site on EGF8, but not EGF12, partially rescues fucose analog inhibition of Notch1 activation by Dll1 suggesting that binding sites corresponding to O-fucose on EGF12 are tolerant of the additional C6 carbon in compounds 10 and 11. In contrast, the binding site on Dll1 is not tolerant of the additional carbon at EGF8, creating a steric clash, resulting in the observed decreased activation of Notch1. Additional sites contain O-fucose consensus sequences and are modified with O-fucose (red ovals), but contribute little to Notch1-ligand interactions compared to EGF8 and EGF1231.
Similar experiments using Dll4 as the activating ligand also demonstrated that O-fucose on EGF8 and EGF12 are important for optimal Notch1 activation in the absence of analogs (Figure 5a), but the effect was smaller than for Dll1. As with Dll1, elimination of the fucose site on EGF8 partially rescued Notch1 inhibition by compounds 10 and 11 compared with controls (Figure 5c), while mutation of the EGF12 fucose site had no significant effect on the ability of the analogs to inhibit signaling. The EGF8/12 double mutant had a similar effect to the EGF8 single mutant. These data suggest that incorporation of fucose analogs at EGF8 play an important role in mediating the inhibitory effects of fucose analogs 10 and 11, but incorporation of these analogs at some of the other 18 O-fucose sites on Notch1 must also be important.
Since fucose analogs 10 and 11 result in reduced binding to Delta-like ligands (Figure 4), we predict that the presence of the analogs on EGF8 interferes with binding to Delta-like ligands more than to Jag1 (Figure 5d). Although no current structure contains Notch1 EGF8 co-crystallized with a Delta-like ligand, the recent structure of Notch1 EGF8–12 co-crystalized with Jag1 revealed that the O-fucose residues on EGF8 and 12 are directly involved in interactions with Jag135. Modeling of the fucose analogs into this structure revealed no steric clash between Notch1 and Jag1 at either EGF8 or EGF12, consistent with lack of effect of the analogs on Jag1-mediated Notch1 activation (Supplementary Figure 8a). Similar modeling supported the absence of a steric clash between fucose analogs at EGF12 and Dll4, consistent with our mutagenesis data (Supplementary Figure 8b). Confirmation that the presence of the analogs at EGF8 causes a steric clash with Delta-like ligands awaits a co-crystal structure with these ligands.
Compound 10 inhibits T-cell differentiation
To assess the functional consequences of remodeling Notch glycosylation, we investigated the impact of fucose analog 10 on Notch-dependent T-cell differentiation in a co-culture assay. LSK (Lineage−Sca1+ckit+) bone marrow stem cells isolated from mice heterozygous or wild type for all three Fringe enzymes36 were overlaid on OP9-GFP stromal cells alone or OP9-GFP cells expressing Dll1 (OP9-Dll1) or Dll4 (OP9-Dll4) to promote differentiation (Fringe heterozygous mice do not exhibit haploinsufficiency36). Both OP9-Dll1 and OP9-Dll4, but not OP9 alone, promoted differentiation to CD25+ T-cell progenitors of LSK cells grown in the control compound 9 (Figure 6a). However, after treatment with compound 10, neither Dll1 nor Dll4 promoted T-cell differentiation (Figure 6). This same result was obtained when co-cultures were incubated with 1 μM DAPT to inhibit Notch signaling (data not shown).
Figure 6. Fucose analog 10 inhibits the development of T-cell progenitors.
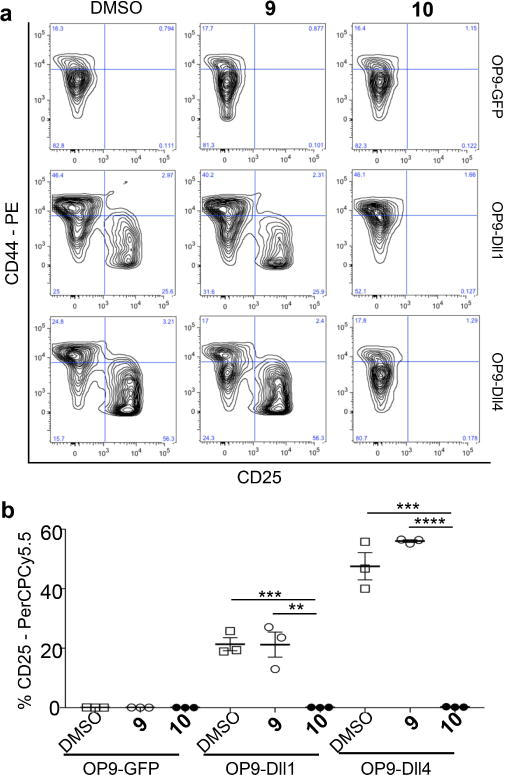
Representative flow cytometric profiles of cells produced from bone marrow LSK cells co-cultured with OP9-GFP, OP9-Dll1 or OP9-Dll4 stromal cells in the presence of DMSO, compound 9 or compound 10 for 8 days. (a) Production of CD25+ T-cell progenitors was evaluated by the expression of CD44 and CD25. (c) Percentage of CD25+ T-cells from mice with a profile typical of panel a (n=3). Mean ± SEM, each symbol represents average data from duplicate wells of LSK cells from one mouse. Data shown are representative of three experiments performed in duplicate. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, unpaired two-tailed Student’s t-test.
Discussion
We demonstrate here that certain fucose analogs serve as inhibitors of Notch signaling, preferentially inhibiting Notch activation from Delta-like ligands. To our knowledge, these are the first examples of small molecule, ligand-specific inhibitors of Notch1 that have been reported. The peracetylated analogs are converted to GDP-fucose analogs intracellularly (Supplementary Figure 1c), utilized as substrates by Pofut1, and incorporated into Notch EGF repeats at high stoichiometries. Further, Lfng can transfer GlcNAc to incorporated fucose analogs and alter their effect on Notch signaling strength. Here, we posit that this type of manipulation of Notch receptor glycosylation will lead to a better understanding of exactly how glycan modifications alter Notch functions. This may then be directly applied toward the development of novel strategies for manipulating Notch signaling in functional processes such as T-cell differentiation or cancer cell proliferation.
Interestingly, the effects of these inhibitory fucose analogs (10 and 11) had different effects on the various Notch-ligand interactions assessed. Among Delta ligands, we saw complete inhibition of Dll1-Notch1 signaling, but only partial inhibition of Dll4-Notch1, Dll1-Notch2, and Dll4-Notch2 signaling. Based on previous reports showing that the Dll1-Notch1 interaction is relatively weaker than the other interactions we examined30,37, our observation suggests that the observed inhibitory effect is more potent in the setting of lower receptor-ligand affinity complexes. A similar hypothesis might also explain why elongation by Lfng, which has been shown to enhance receptor-ligand affinity30, is able to partially rescue the inhibitory effect of these analogs. The added GlcNAc forms stabilizing bonds between Notch ligand and receptor13, which offsets some of the destabilizing effect of the fucose analogs.
Despite somewhat reduced binding affinity in the presence of inhibitory fucose analogs, Jag1-induced Notch signaling was relatively unaffected by these compounds. This could be due to a slightly different signaling mechanism, which does not necessarily correlate with binding affinity and may involve differences in the formation of the catch-bonds recently demonstrated to form between Notch1 and ligands under pulling force35. Dll1-mediated signaling is more sensitive to changes in glycosylation of Notch receptors than Jag1-mediated signaling in several contexts31,38. Here we suggest that variation in steric interactions between receptor and ligand plays a substantial role in controlling these differences and that we can take advantage of these differences to generate ligand specific inhibitors.
Our data also indicate that incorporation of fucose analogs 10 and 11 at EGF8 plays an important role in mediating their inhibitory effects, while EGF12 is less important. This suggests that O-fucose on EGF8 plays an important role at the interface of Dll1 or Dll4 and Notch1 binding. Thus, incorporation of an additional carbon on C6 of fucose at EGF8 is predicted to cause a steric clash with Delta-like, but not Jag ligands (Figure 5d). Consistent with the steric clash model, smaller modifications at C6 (i.e. the fluoro group in compounds 5 or 13) did not alter Notch activity in either Zebrafish or mammalian cell systems, even though this analog was efficiently incorporated into Notch 1 EGF repeats by Pofut1 (Supplementary Figure 3). Larger groups (i.e. compounds 6, 7 and 8) also inhibited Notch activation in the Zebrafish system as would be expected (Figure 1c), but the peracetylated versions of these compounds (compounds 14, 15 and 16) were not incorporated into Notch1 EGF repeats (Supplementary Figure 3), so could not be analyzed further. Structural modeling also suggests that the additional carbons on compounds 10 and 11 incorporated at EGF8 and EGF12 do not cause any steric interference between Notch1 and Jag1 (Supplementary Figure 8a) or Notch1 EGF12 and Dll4 (Supplementary Figure 8b), supporting our steric clash hypothesis (Figure 5d). It is important to note that incorporation at EGF8 is still only partially responsible for the decrease in Delta-like-mediated Notch1 signaling caused by analogs 10 and 11. Others have shown that mutations13 and glycan modifications30,31,39 on Notch EGF repeats outside of the ligand binding domain also affect Notch function. It is possible that the presence of fucose analogs outside of the ligand-binding domain might cause conformational changes in Notch that destabilize Notch-ligand interactions.
The combined data show that peracetylated fucose analogs are taken up by cells, converted to their corresponding GDP-fucose derivatives and incorporated into Notch EGF repeats by Pofut1, and that elongation of fucose by Lfng is not affected by the presence of these groups at the 6-carbon position (Figure 2, Supplementary Note 1). Although compound 6 inhibited Notch signaling in Zebrafish embryos, its peracetylated counterpart (14) was not incorporated into EGF repeats (Supplementary Figure 3a). This was likely due to failure of this compound to be utilized by the fucose salvage pathway. Similarly, peracetylated versions of compounds 2, 7 and 8 (compounds 12, 15 and 16) were not incorporated into Notch EGF repeats in cell culture. Compound 17, an established fucosyltransferase inhibitor, did not affect normal Notch fucosylation. This may be explained by the cePofut1-GDP-fucose co-crystal structure showing that the hydroxyl group on the second position carbon plays a critical role in the binding and transfer of GDP-fucose40.
Previous work from our laboratory demonstrates that fucose analogs such as 10 are poorly incorporated into N-glycans22. Prior work has also shown that altered N-glycan structures on Notch do not affect its ability to be activated by ligand17. Additionally, we have previously shown that changes to O-fucose modifications within EGF repeats behave independently of enzymes responsible for the addition of O-glucose or O-GlcNAc to these to EGF repeats31,41,42. Therefore, we conclude that the incorporation of fucose analogs into Notch EGF repeats directly results in decreased ligand-induced Notch signaling and that the effects of these fucose analogs are unlikely to have been caused by an indirect effect on other glycan modifications within Notch ligands and receptors.
We were able to use the inhibitory fucose analogs to inhibit T-cell differentiation, a functional process with a well-established dependence on Dll-mediated Notch signaling43. This supports the idea that these inhibitors do in fact have a Notch specific effect that can cause functional changes in biological processes. The importance of uncontrolled Notch signaling in the etiology of human malignancy has stimulated intense interest in development of Notch inhibitors as novel cancer therapeutics. While many Notch-mediated cancers rely on ligand independent Notch signaling44, there have been reports suggesting that overexpression of individual Notch activating ligands45,46 and Pofut147–49 also contribute to cancer development. Here, we demonstrate that fucose analogs can be used as inhibitors of Dll-mediated Notch signaling. Similar strategies might be utilized for development of cancer therapeutics in the future.
Online Methods
Zebrafish embryo experiments
All zebrafish (Danio rerio) were maintained following standard procedures and embryos were staged as previously described50. AB and casper fish [roya9;mitfaw2 (AB)]51 were used as wild type fish. The SuH:GFP transgenic line [Tg(TP1bglob:gfp)um13]23 was maintained in an AB background. The towhead (twd)rw685 mutant24 was maintained by intercross. Only healthy Zebrafish larva with normal shape and behavior, as judged by pre-established criteria50, were analyzed. Larvae were assigned randomly to treatment conditions. The gmds Morpholino was used at 2 ng per embryo, as previously described24. GDP-fucose and GDP-fucose analogs were re-suspended in pure water at 20–60 mM and 60 pmol of each analog was injected into embryos at the 1-cell stage. After reaching the desired developmental stages (48 hpf), embryos were mounted to the desired position in 1.5% methyl-cellulose in E3 medium. Images were taken using an Olympus SZ16 fluorescent dissecting microscope and Microfire digital camera (Olympus). Images were taken using the same exposure time and lighting intensity settings to ensure that the GFP signal for all images are comparable. To ensure that each embryo had one allele of GFP-Notch reporter, heterozygous or homozygous [Tg(TP1bglob:gfp)um13] fish were crossed to wild type AB fish. The progeny were collected and injected as described above. At 48 hpf, GFP-positive embryos were collected and each fish was put into one well of a 96-well plate. A group of 60 embryos was treated for each condition. The final number of survived Zebrafish larva ranged from 23 to 48 due to different fertilization rates. Since the treatments of different GDP-fucose analogs created 100% penetrance, no statistical methods were used to determine required sample size. All Zebrafish maintenance and usage was performed with the full compliance of ethical regulation and was approved by the Institutional Animal Care and Use Committee (IACUC) of Albert Einstein College of Medicine.
Expression Constructs
Mouse Notch1 (mNotch1) expression plasmid containing EGF1–18 with C-terminal Myc-His6 tags (pSecTag2, Invitrogen) was described previously52. The plasmid expressing full-length mNotch1 (mN1; pcDNA1-mN1-myc) was generously provided by Dr. Jefferey Nye53. The plasmid expressing full-length mNotch2 (mN2; pTracer-CMV-mN2-Flag) was kindly provided by Dr. Shigeru Chiba54. Fringe plasmids SEAP (EV) and Lfng-AP were previously described17. The TP-1 luciferase reporter construct (Ga981-6) was a gift form Dr. Georg Bornkamm and the gWIZ β-galactosidase construct was from Gene Therapy Systems. A plasmid expressing GFP (pEGFP-N1) was from Clontech. Note that “N1” in this plasmid name refers to a Not1 restriction site following the GFP coding region.
Cell culture
HEK293T and NIH3T3 cells (NIH3T3 CRL-1658) cells were obtained from the American Type Culture Collection (ATCC)(Manassas, VA). These cells were authenticated and tested for mycoplasma contamination by ATCC at the time of purchase. L cells stably expressing Jagged1 (Jag1) or Delta-like 1 (Dll1) were a gift of Dr. Gerry Weinmater (UCLA). MS5 cells stably expressing Delta-like 4 (Dll4) were a gift from Dr. Stephen Blacklow. HEK293T, NIH3T3, L cells, and MS5 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM)(Invitrogen) supplemented with 10% bovine calf serum.
Production of fucose-modified mouse Notch1 EGF 1–18
HEK293T cells were co-transfected with plasmids encoding mouse N1 EGF1–18-MycHis (2 μg) and either SEAP (EV) or Lfng-AP (Lfng) (1 μg) in a 10 cm plate containing 8 mL media using 18 μl of PEI reagent (polyethyleneimine)55 mixed with 300 μl of OPTI-MEM (Invitrogen). Media was changed to OPTI-MEM containing 50 μM of the appropriate fucose analog after 6 hours. Four days later the media was collected and protein was purified using Ni-NTA resin (Qiagen) and eluted with 250 mM imidazole as described previously56.
Glycoproteomic analysis of mouse Notch1 EGF1–18
Affinity-purified protein from each culture condition was reduced, alkylated and digested (in-gel) with trypsin, chymotrypsin or V8 as described previously52,56. The resulting peptides were analyzed by nano LC-MS/MS using Agilent nano-HPLC-CHIP system coupled to a model 6340 Ion Trap mass spectrometer as described52,56. Analysis of glycopeptides modified with some analogs (compounds 12, 15 and 16) was performed on a Thermo-Fischer Q-Exactive Plus coupled to an EasyNano-LC. O-Fucosylated peptides were identified by neutral loss searches and semi-quantitative Extracted Ion Chromatograms (EICs) of selected ions were generated to compare relative amounts of relevant glycoforms of each peptide52,56. EICs were smoothed using a Gaussian algorithm.
Cell-based co-culture Notch reporter assay
NIH3T3 cells (0.5 × 105 cells/well) were seeded in a 24-well tissue culture plate and co-transfected with 0.1 μg of pcDNA1-mN1-myc or pTracer-mN2-Flag, and either 0.05 μg of SEAP (EV) or Lfng-AP (Lfng) plasmid (0.1 μg Lfng-AP was used in Dll1-N2 experiments to better evaluate the effects of Fringe), along with 0.12 μg of TP-1 luciferase Notch signaling reporter construct and 0.06 μg of gWIZ β-galactosidase construct for transfection efficiency normalization using Lipofectamine 2000 (Invitrogen), according to the manufacture’s instructions. After 4 hours, media were changed to media containing the appropriate peracetylated fucose analog (50 μM, unless otherwise specified) or DMSO. Then L cells stably expressing Jag1 or Dll1, or MS5 cells stably expressing Dll4, were overlaid on the NIH3T3 transfectants at a density of 1.5 × 105 cells/well. Cells were lysed after an additional 24 hour culture and luciferase and β-galactosidase assays were performed based on the manufacturer’s instructions (Luciferase Assay system, Promega) as described previously52,57. Three biological replicates were performed in at least two independent experiments (total n≥6, as indicated in figure legends).
Notch ligand coated plate induction assay of Notch signaling
24-well tissue culture plates were coated with Dll1-Fc (R&D Systems, 3970-DL-050), Dll4-Fc (Sino Biological, 10171-H02H-50) or Jag1-Fc (R&D Systems, 599-JG-100) (4 μg/mL per well for each ligand) in PBS for 2 hours at room temperature. NIH3T3 cells (0.5 × 105 cells/well) were plated in ligand-coated wells and incubated overnight. Transfection of control and Notch reporter constructs, media changes, β-galactosidase, and luciferase reporter assays were carried out, as above. Three biological replicates were performed for each condition (n=3).
Cell surface N1 expression
HEK293T cells were co-transfected 1.5 μg of either EV or pcDNA-N1-MycHis, and 0.4 μg of GFP (pEGFP-N1) in a 3.5 cm plate using PEI transfection reagent. After 4 hours in culture, media was changed to media containing 50 μM of the appropriate peracetylated fucose analog or DMSO. At 28–30 h post-transfection, the cells were dissociated with cold PBS pH 7.4 containing 1% bovine serum albumin (BSA) and resuspended in binding buffer (1 mM CaCl2, 1% BSA and 0.05% NaN3 in Hanks’ balanced salt solution pH 7.4 (Gibco)). Cells were incubated with 100 μl of anti-mN1 (ECD) antibody (R&D systems, AF5267) at 10 μg/ml for 1 h at 4 °C. Cells were washed with binding buffer and then incubated with PE-anti-sheep IgG (1:100; Santa Cruz, SC-3757) for 30 min at 4 °C. After two washes with binding buffer, the cells then were analyzed with a FACSCalibur (BD, Bioscience) flow cytometer. The gate was set to collect the GFP positive population of 30,000 events for each sample and analyzed using FlowJo software (ver. 9.4.10). Three independent experiments were performed for each condition (n=3).
Cell-based Notch ligand binding assay
HEK293T cells (8.5 × 105 cells/well) were seeded in a 3.5 cm culture plate and co-transfected with 1.5 μg of pcDNA1-mN1-myc, 0.4 μg of GFP and 0.75 μg of either SEAP (EV) or Lfng-AP (Lfng) using PEI reagent. After 4 hours in culture, media was changed to media containing 50 μM of the appropriate peracetylated fucose analog or DMSO. Cells were dissociated 24–28 hours post-transfection with cold PBS pH 7.4 containing 1% BSA. Cells were then resuspended in binding buffer. Dll1-Fc (R&D Systems, 3970-DL-050), Dll4-Fc (Sino Biological, 10171-H02H-50) or Jag1-Fc (R&D Systems, 599-JG-100) (each at 0.5 μg/mL) were pre-clustered with fluorescent secondary antibody PE-goat anti-mouse IgG (1:100; Invitrogen, P-852) or PE-anti-human IgG (1:100; Jackson Immuno Research, 109155-098) for 30 min at 4 °C. Cells were incubated with clustered ligands at 4 °C. After 1 h, the cells were washed twice with binding buffer and analyzed using a FACSCalibur (BD, Bioscience) flow cytometer. The gate was set to collect 30,000 GFP-positive events for each sample and analyzed using FlowJo software (ver. 9.4.10). Experiments were performed at least three independent times (n≥3, as indicated in figure legends).
For reverse binding assays, HEK293T cells were transfected with 1.5 μg of full-length mDll1 (pTracer) and 0.4 μg of GFP, as above. Cells were dissociated and incubated with 0.5 μg/mL of soluble Notch1-Fc chimera, containing EGF repeats 1–13 (R&D Systems, 5267-TK-050), for 1 hour. Cells were washed in binding buffer and incubated with PE-anti-human IgG (1:100; Jackson Immuno Research, 109155-098). Cells were then washed twice and analyzed, as above. Three independent experiments were performed for each condition (n=3). See Supplementary Figure 9a for typical gating strategy used for these experiments.
Mutagenesis of O-fucosylation sites in mouse Notch1
O-Fucosylation site mutants at EGF repeats 8, 12, or both of the pcDNA1-mN1myc plasmid were provided by Dr. Shinako Kakuda31. Mutations were designed to eliminate the modified residue (threonine to valine) within the O-fucosylation consensus sequence, Cxxxx(S/T)C58. The mutants were confirmed by DNA sequencing.
Purification of LSK cells from Bone Marrow
LSK cells were purified from bone marrow of FVB littermates, 2 males and 2 females, chosen at random. Another experiment was performed with an FVB female heterozygous for each of three Fringe genes (Lfng, Mfng and Rfng)36. Mice were housed in a barrier facility, allowed to eat and drink ad libitum, and used in experiments at 8–10 weeks of age. All experiments were performed with permission from the Albert Einstein Institutional Use and Animal Care Committee. Since only FVB mice were used, no blinding was performed. Briefly, bone marrow cells were prepared by crushing the femur, tibia, hips and vertebrae of FVB mice in cold Flow Buffer (PBS pH 7.4 lacking cations, containing 4% FBS and 100 U/ml penicillin/streptomycin). Cells were incubated in 5 ml RBC lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.2–7.4) for 3 min before adding 40 ml Flow Buffer. After centrifugation at 1200 rpm for 10 min at 4 °C, cells were counted using a Coulter counter. Approximately 1.5 × 108 cells were incubated with 3 μl FcR blocking solution (rat-anti-mouse CD16/CD32) in 250 μl Flow Buffer on ice for 15 min and then depleted of Lineage+ cells using biotin-conjugated antibodies against B220 (1:100; clone RA3-6B2), CD11b (1:500; clone M1/70), Gr-1 (1:500; clone RB6-8C5), CD4 (1:200; clone GK1.5), CD8α (1:200; clone 53-6.7), CD3ε (1:100; clone 145-2C11), Ter119 (1:100; clone TER-119), and CD19 (1:100; clone 6D5) (all biotinylated antibodies were from Biolegend) in a final volume of 300 μl Flow Buffer. After 30 min on ice, cells were washed with 10 ml cold Flow Buffer and the pellet resuspended in 9 ml cold Flow buffer to which 1 ml anti-biotin microbeads (Life Technology) was added. After rotation for 30 min at 4 °C, bead-coated cells were removed by magnetic separation. Lineage-depleted cells were incubated with anti-Sca-1-PE (1:50; Biolegend, clone D7), anti-cKit-APC (1:50; BD Pharmingen, clone–1B8) and streptavidin-PE-Cy7 (1:200; Biolegend) in a final volume of 300 μl Flow Buffer. After 30 min on ice, cells were washed with 10 ml cold Flow buffer, the pellet resuspended in 300 μl Flow Buffer containing 4,6-diamidino-2-phenylindole, dihydrochloride (DAPI; 1:1000). Live, DAPI-negative, Lin−Sca1+cKit+ (LSK) cells were collected by cell sorting using a Aria flow cytometer (BD Biosciences), and analyzed using FlowJo™ (FlowJo, LLC) software. See Supplementary Figure 9b for gating strategy used for these experiments.
LSK cell differentiation assay
OP9-GFP, OP9-Dll1 and OP9-Dll4 cells59 were kindly provided by Cynthia Guidos and were cultured in α-Minimum Essential Medium (MEM, Gibco), supplemented with 10% FBS (Hyclone), 100 U/mL penicillin/streptomycin. OP9-GFP, OP9-Dll1 and OP9-Dll4 cells were plated in 24-well plates (Corning) in 1 ml MEM, supplemented with 20% heat-inactivated FBS (Hyclone), 100 U/mL penicillin/streptomycin and 25 ng/ml Amphotericin B (Gibco), to achieve ~90% confluency after 24 hour. Co-cultures were initiated by overlaying LSK cells (3 × 103 per well) in 1 ml MEM containing 5 ng/mL Flt3-L and 1 ng/mL IL-7 (both from Preprotech, Rocky Hill, NJ). Compound 9 or 10 in DMSO was added to co-cultures to a final concentration of 25 μM and an equal volume of DMSO was added to control wells. Plates were incubated at 37 °C in a humidified atmosphere with 5% CO2. Every 2 days, half the medium was refreshed to maintain the final concentration of DMSO, 9 or 10. On day 8, co-cultures were harvested by forceful pipetting, filtered through a 40 micron cell strainer, and centrifuged at 1200 rpm for 5 min at room temperature. Cell pellets were fixed in 4% paraformaldehyde (PFA) in PBS (pH 7.4, lacking cations) for 15 min at room temperature, and stored at 4 °C. For flow cytometry, fixed cells were washed with 1 ml cold FACS binding buffer (FBB; Hanks’ balanced salt solution (HBSS), 2% BSA, 0.05% sodium azide, pH 7.2–7.4) by centrifugation for 5 min at 1200 rpm. Cells were resuspended in 90 μl FBB containing 1 μl Fc block (rat-anti-mouse CD16/CD32), and incubated for 15 min on ice. Antibody diluted in FBB (10 ml final volume) was added, and the tube was incubated for 30 min at 4 °C. To detect differentiated T-cells, CD44-PE (1:200; clone-IM7, eBioscience) and CD25-PerCPCy5.5 (1:200; clone-PC61.5, eBioscience). Cells were washed twice in 1 ml FBB and transferred to a 5 ml Polystyrene round bottom tube in 250 μl FBB. Immunofluorescence of GFP-negative (non-stromal) cells was analyzed using FACSCalibur (BD Biosciences), and data were analyzed using FlowJo software (FlowJo, LLC). One of four FVB mice gave reduced numbers of CD25+ cells and is not included in the analysis shown in Figure 6. However, when the data were normalized to DMSO control, the results for the 4 mice were qualitatively similar (compound 9 (Ac-GDP-Fuc) averaged 0.97+/−0.15 for Dll1 and 1.11+/−0.11 for Dll4 and compound 10 averaged 0.0+/−0.002 for Dll1 and 0.01+/−0.003 for Dll4. See Supplementary Figure 9c for gating strategy used for these experiments.
Statistical Analysis
For signaling and binding assays two-way ANOVAs were used to assess significance. Tukey’s post-hoc test was used to evaluate differences between individual treatment conditions. Statistically identical conditions were grouped together in graphs. Sidak’s multiple comparisons test was used to evaluate significance between −Fng and +Lfng conditions. Student t-tests were used to assess significance for T-cell differentiation assays. All statistical tests were carried out using Prism 7 software (Graphpad).
Structural modeling of O-fucose analogs
Models of fucose analogs were generated in Maestro (Schrödinger Release 2017-1: Maestro, Schrödinger, LLC, New York, NY, 2017) and superimposed onto the fucose modifications of Notch1 EGF8 and EGF12 in the Notch1-Jag1 complex structure (PDB ID 5UK5). Structural analyses, atomic distance measurements, and figure generation were performed in Pymol (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.).
Data Availability
The data that support the findings of this study are available from the corresponding author(s) upon reasonable request.
Supplementary Material
Acknowledgments
This work was supported by NIH grants RO1GM061126 (R.S.H.), R01GM093282 (P.W.), RO1GM106417 (P. S.), and K99CA204738 (V.C.L). M.S. was partially supported by T32GM00844.
Footnotes
Author Contributions
M.S., P.W., and R.S.H. conceived experiments, analyzed and interpreted data, and wrote the paper. L.U.N. synthesized fucose analogs. M.S., V.K., L.F., H.T., and H.H. designed and performed experiments. V.C.L. and K.C.G. constructed structural models. P.S. and P.W. helped design experiments, interpret data and revise the manuscript.
Competing Financial Interests
The authors declare no competing financial interest.
Main Text References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teodorczyk M, Schmidt MH. Notching on cancer’s door: Notch signaling in brain tumors. Frontiers in oncology. 2015;4:341. doi: 10.3389/fonc.2014.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu G, Wilson G, George J, Qiao L. Modulation of Notch signaling as a therapeutic approach for liver cancer. Current gene therapy. 2015;15:171–181. doi: 10.2174/1566523214666141224100319. [DOI] [PubMed] [Google Scholar]
- 6.Espinoza I, Pochampally R, Xing F, Watabe K, Miele L. Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther. 2013;6:1249–1259. doi: 10.2147/OTT.S36162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin L, Velazquez OC, Liu ZJ. Notch signaling: emerging molecular targets for cancer therapy. Biochemical pharmacology. 2010;80:690–701. doi: 10.1016/j.bcp.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 9.Sagert J, et al. Abstract C158: Tumor-specific inhibition of jagged-dependent notch signaling using a Probody™ therapeutic. Molecular Cancer Therapeutics. 2013;12:C158. doi: 10.1158/1535-7163.targ-13-c158. [DOI] [Google Scholar]
- 10.Tran IT, et al. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. The Journal of clinical investigation. 2013;123:1590–1604. doi: 10.1172/JCI65477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lafkas D, et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature. 2015;528:127–131. doi: 10.1038/nature15715. [DOI] [PubMed] [Google Scholar]
- 12.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 13.Luca VC, et al. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347:847–853. doi: 10.1126/science.1261093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moloney DJ, et al. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. Journal of Biological Chemistry. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura A, et al. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. Journal of Biological Chemistry. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- 16.Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci U S A. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moloney DJ, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 18.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–414. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 19.Rillahan CD, et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol. 2012;8:661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okeley NM, et al. Development of orally active inhibitors of protein and cellular fucosylation. Proceedings of the National Academy of Sciences. 2013;110:5404–5409. doi: 10.1073/pnas.1222263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawa M, et al. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proceedings of the National Academy of Sciences. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Shareffi E, et al. 6-alkynyl fucose is a bioorthogonal analog for O-fucosylation of epidermal growth factor-like repeats and thrombospondin type-1 repeats by protein O-fucosyltransferases 1 and 2. Glycobiology. 2013;23:188–198. doi: 10.1093/glycob/cws140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsons MJ, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mechanisms of development. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohata S, et al. Neuroepithelial cells require fucosylated glycans to guide the migration of vagus motor neuron progenitors in the developing zebrafish hindbrain. Development. 2009;136:1653–1663. doi: 10.1242/dev.033290. [DOI] [PubMed] [Google Scholar]
- 25.Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. Journal of Biological Chemistry. 2003;278:42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann M, von der Lieth CW, Stehling P, Reutter W, Pawlita M. Consequences of a subtle sialic acid modification on the murine polyomavirus receptor. Journal of virology. 1997;71:5922–5931. doi: 10.1128/jvi.71.8.5922-5931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Descheny L, Gainers ME, Walcheck B, Dimitroff CJ. Ameliorating skin-homing receptors on malignant T cells with a fluorosugar analog of N-acetylglucosamine: P-selectin ligand is a more sensitive target than E-selectin ligand. Journal of investigative dermatology. 2006;126:2065–2073. doi: 10.1038/sj.jid.5700364. [DOI] [PubMed] [Google Scholar]
- 28.Gloster TM, et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nature chemical biology. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hicks C, Johnston SH, Collazo A, Vogt TF, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nature cell biology. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 30.Taylor P, et al. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proceedings of the National Academy of Sciences. 2014;111:7290–7295. doi: 10.1073/pnas.1319683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kakuda S, Haltiwanger R. Deciphering the Fringe-mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands. Developmental Cell. 2017;40:193–201. doi: 10.1016/j.devcel.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang LT, et al. Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Molecular biology of the cell. 2005;16:927–942. doi: 10.1091/mbc.E04-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller J, et al. O-fucosylation of the notch ligand mDLL1 by POFUT1 is dispensable for ligand function. PloS one. 2014;9:e88571. doi: 10.1371/journal.pone.0088571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panin VM, et al. Notch Ligands Are Substrates for ProteinO-Fucosyltransferase-1 and Fringe. Journal of Biological Chemistry. 2002;277:29945–29952. doi: 10.1074/jbc.M204445200. [DOI] [PubMed] [Google Scholar]
- 35.Luca VC, et al. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science. 2017;355:1320–1324. doi: 10.1126/science.aaf9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, Kumar V, Wei HX, Qiu J, Stanley P. Lunatic, Manic, and Radical Fringe Each Promote T and B Cell Development. The Journal of Immunology. 2016;196:232–243. doi: 10.4049/jimmunol.1402421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrawes MB, et al. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. Journal of Biological Chemistry. 2013;288:25477–25489. doi: 10.1074/jbc.M113.454850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawaguchi S, et al. O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals. eLife. 2017;6:e24419. doi: 10.7554/eLife.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiruma-Shimizu K, et al. Chemical synthesis, folding, and structural insights into O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor. Journal of the American Chemical Society. 2010;132:14857–14865. doi: 10.1021/ja105216u. [DOI] [PubMed] [Google Scholar]
- 40.Lira-Navarrete E, et al. Structural insights into the mechanism of protein O-fucosylation. PLoS One. 2011;6:e25365. doi: 10.1371/journal.pone.0025365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rana NA, et al. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. Journal of Biological Chemistry. 2011;286:31623–31637. doi: 10.1074/jbc.M111.268243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harvey BM, et al. Mapping sites of O-glycosylation and fringe elongation on Drosophila Notch. Journal of Biological Chemistry jbc. 2016;M116732537 doi: 10.1074/jbc.M116.732537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanley P, Guidos CJ. Regulation of Notch signaling during T‐and B‐cell development by O‐fucose glycans. Immunological reviews. 2009;230:201–215. doi: 10.1111/j.1600-065X.2009.00791.x. [DOI] [PubMed] [Google Scholar]
- 44.Aster JC, Pear WS, Blacklow SC. The Varied Roles of Notch in Cancer. Annual Review of Pathology: Mechanisms of Disease. 2016 doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purow BW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer research. 2005;65:2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 46.Ridgway J, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 47.Sawey ET, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma L, et al. Overexpression of protein O-fucosyltransferase 1 accelerates hepatocellular carcinoma progression via the Notch signaling pathway. Biochemical and biophysical research communications. 2016;473:503–510. doi: 10.1016/j.bbrc.2016.03.062. [DOI] [PubMed] [Google Scholar]
- 49.Yokota S, et al. Protein O-fucosyltransferase 1: a potential diagnostic marker and therapeutic target for human oral cancer. International journal of oncology. 2013;43:1864–1870. doi: 10.3892/ijo.2013.2110. [DOI] [PubMed] [Google Scholar]
- 50.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 51.White RM, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell stem cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rana NA, et al. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J Biol Chem. 2011;286:31623–31637. doi: 10.1074/jbc.M111.268243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nye JS, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 54.Shimizu K, et al. Mouse jagged1 physically interacts with notch2 and other notch receptors assessment by quantitative methods. Journal of Biological Chemistry. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- 55.Thomas M, et al. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5679–5684. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kakuda S, Haltiwanger RS. Analyzing the posttranslational modification status of Notch using mass spectrometry. Methods Mol Biol. 2014;1187:209–221. doi: 10.1007/978-1-4939-1139-4_16. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto S, et al. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science. 2012;338:1229–1232. doi: 10.1126/science.1228745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rana NA, Haltiwanger RS. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr Opin Struct Biol. 2011;21:583–589. doi: 10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van de Walle I, et al. Jagged2 acts as a Delta-like Notch ligand during early hematopoietic cell fate decisions. Blood. 2011;117:4449–4459. doi: 10.1182/blood-2010-06-290049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W, et al. Chemoenzymatic synthesis of GDP-L-fucose and the Lewis X glycan derivatives. Proceedings of the National Academy of Sciences. 2009;106:16096–16101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author(s) upon reasonable request.


