Abstract
Protein complexes exhibit great diversity in protein membership, post-translational modifications and noncovalent cofactors, enabling them to function as the actuators of many important biological processes. The exposition of these molecular features with current methods lacks either throughput or molecular specificity, ultimately limiting the use of protein complexes as direct analytical targets in a wide range of applications. Here, we apply native proteomics, enabled by a multistage tandem mass spectrometry approach, to characterize 125 intact endogenous complexes and 217 distinct proteoforms derived from mouse heart and human cancer cell lines in discovery mode. The native conditions preserved soluble protein–protein interactions, high-stoichiometry noncovalent cofactors, covalent modifications to cysteines, and, remarkably, superoxide ligands bound to the metal cofactor of superoxide dismutase 2. The data enable precise compositional analysis of protein complexes as they exist in the cell and demonstrate a new approach that uses mass spectrometry as a bridge to structural biology.
Introduction
Protein complexes play critical roles in an array of biological processes, including cancer1, senescence2, and cell cycle arrest3. Given the vital functions of these macromolecular assemblies, their identification and structural characterization is foundational to our understanding of normal and disease biology. Thus, examination of cellular products at progressively higher levels of complexity, from peptides4, 5 to proteins6–8 to protein complexes9–11, paints an increasingly complete picture of the actuators of biological systems.
Current methodologies for analysis of protein complexes fall into two groups: those that provide detailed characterization of a single target and those that provide low-level information on a large number of interactions. Examples of the former include X-ray crystallography12, 13, fluorescence imaging14, cryo-electron microscopy15, 16, and co-immunoprecipitation17, which are lower-throughput techniques that can require large amounts of purified and optimized samples. In contrast, the latter category includes the use of tandem mass spectrometry (MS) techniques in a peptide-based “bottom-up” methodology18. While bottom-up approaches can provide extensive maps of physical interactions19–21, they often fail to determine stoichiometry or the complete modification states of complex subunits.
In this study, we describe the first large-scale study utilizing our new proteomic platform22–25 that occupies the space between existing approaches to gain high molecular detail from relatively small amounts of unpurified endogenous protein complexes. We applied this pipeline to human and mouse samples and identified previously unreported protein–protein interactions, enzyme metal cofactors, and post-translational modifications. The detailed characterization and increased level of coverage achieved with native proteomics represents a powerful new middle ground for both targeted and discovery proteomics.
Results
An Approach to Native Proteomics
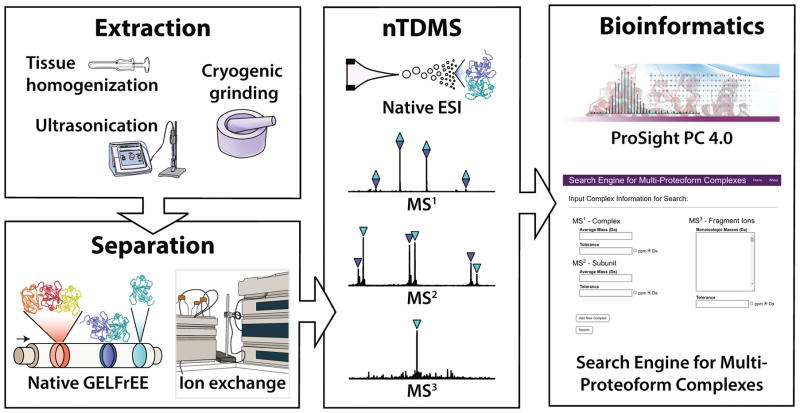
Native proteomics aims to directly identify and characterize proteoforms and multi-proteoform complexes (MPCs)25. The latter is a term defined recently25 as the distinct set of protein complexes formed by the association of different proteoforms (monomeric proteins and their co- and post-translational modifications (PTMs)26) derived from the same or different genes. Complete elucidation of MPCs requires a “top-down” (TD) approach that begins with the lysis of samples with methods that maintain many endogenous non-covalent interactions (Fig. 1). Extracted soluble proteins and complexes are not chemically reduced and are subsequently subjected to one-dimensional separation using either ion exchange (IEX)27 or native GELFrEE (nGF) chromatography23, 24. Fractions are then subjected to detergent removal, desalting, or both, prior to analysis using a three-tiered tandem MS approach to native top-down mass spectrometry (nTDMS) to controllably disassemble protein complexes in the gas phase22. Lastly, the data extracted from the nTDMS experiment are searched with ProsightPC 4.0 or an online search engine for MPCs (SEMPC, http://complexsearch.kelleher.northwestern.edu)25, 28 to identify, characterize, and score MPCs. Native proteomics is therefore a “discovery mode” experiment, as the identity of the complex is not known prior to analysis.
Figure 1. Native proteomics implemented in untargeted mode.
Samples are produced via native cell lysis or tissue homogenization methods and are fractionated with either native GELFrEE or ion exchange chromatography. The resulting fractions are analyzed using a three-tiered approach to native top-down mass spectrometry (nTDMS) which maintains many of the fragile noncovalent interactions prior to controlled fragmentation of protein complexes into subunits and their backbone fragment ions. Finally, the nTDMS data are analyzed with ProSight PC 4.0 and a Search Engine for Multi-Proteoform Complexes to identify and characterize the complexes and proteoforms formed endogenously within the cell or tissue samples.
In native proteomics, the nTDMS step provides the key to extensive molecular characterization of MPCs. Briefly, and using the human nucleoside diphosphate kinase (NDPK) heterohexamer as an illustrative example, a single charge state of one intact multi-proteoform complex is isolated (Supplementary Fig. 1a), and activated to eject one or multiple subunits (Supplementary Fig. 1b). Next, activation is performed prior to isolation and each of the ejected monomers is isolated and fragmented individually (Supplementary Fig. 1c). The simultaneous activation of all ions in the source formally disconnects ejected monomers from their complexes. However, with validation of stoichiometry and measurement of the overall mass, confident assignments are made in nearly all cases. For NDPK, we confidently identified both ejected monomers as NDPKA (P15531) and NDPKB (P22392); the observed distribution of three intact MPCs was consistent with hexamers containing ratios of between 3:3 and 1:5 of the alpha and beta subunits, respectively (Supplementary Fig. 1a). In identifying and characterizing three related MPCs in a single experiment, a feat that is difficult or impossible using other approaches, this example demonstrates that stoichiometry and precise subunit composition can be quickly determined using a native top-down approach to protein complex analysis.
Untargeted Characterization of Mammalian Complexes
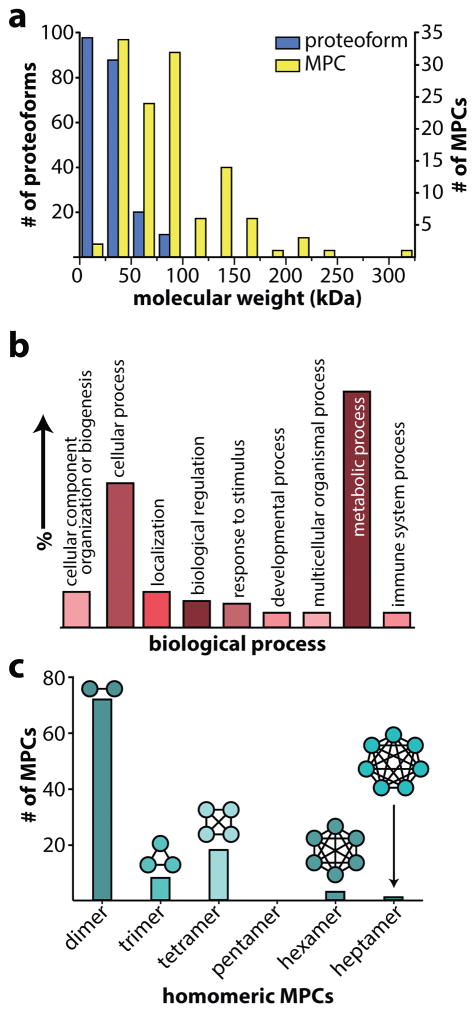
Next, we applied native proteomics to identify a variety of primary metabolic enzymes from CD-1 mouse hearts and characterize a wider range of MPCs from four human cancer cell lines (see Methods for more details). From ~600 fractions individually analyzed over the course of this study, we identified with high confidence 164 unique gene products (proteins); 81 of these formed 125 unique MPCs (Supplementary Data Set 1) and the remaining 83 were characterized as monomeric proteoforms (Supplementary Data Set 2). Overall, the intact masses of the identified proteins and complexes span from 5 to 316 kDa (Fig. 2a), demonstrating the applicability of the method to species far greater in mass than those reported by many previous denaturing top-down studies6, 7. Further increases in experimental capabilities will increase the mass range of identified MPCs (native MS of a 9 MDa complex has been reported29), but the identification of these larger complexes also presents substantial challenges25. Here, many of the proteins identified are involved in cellular metabolism, including coenzyme metabolic processes, generation of precursor metabolites and energy, and primary metabolic processes (Fig. 2b). Thus, we extensively characterized the abundant enzymes driving primary metabolism, particularly those with high energy needs such as the cancer cell lines and heart tissue.
Figure 2. A summary of results from performing native proteomics.
(a) The molecular weight distribution of proteoforms and their assemblies characterized in this study. (b) The distribution of biological processes of the identified gene products. (c) The observed distribution of stoichiometries of identified homomeric multi-proteoform complexes (MPCs).
Native proteomics can directly determine protein complex stoichiometry, which is known to modulate enzyme activity30 and is critical to our understanding of many disease states1. In our study, the majority of MPCs identified were homomeric and ranged from homodimers to a homoheptamer (Fig. 2c). Two homomeric complexes that we identified were annotated differently in UniProt: the 10 kDa heat shock protein (P61640), which we identified as a homoheptamer and is annotated as a homohexamer, and the 3-hydroxyisobutyrate dehydrogenase (Q99L13) homotetramer that is annotated as a homodimer. Furthermore, we identified three homodimers (Supplementary Data Set 1) that were not predicted to form homomeric interactions of any stoichiometry, including the heart isozyme of murine fatty acid binding protein (FABPH, P11404). Serum levels of human FABPH (86% sequence identity) have been used to diagnose myocardial infarction31; the dimerization of this biomarker may be of future relevance in clinical research. In addition to homomeric complexes, we also identified several heteromeric MPCs (Supplementary Data Set 1), including two novel heterodimeric interactions: peptidyl-prolyl cis-trans isomerase A (PPIA, P62937) binding to destrin (P60981) or to cofilin (P23528) (Supplementary Fig. 2). The ability of PPIA to catalyze the cis-trans isomerization of proline residues may serve to regulate the action of destrin and cofilin on actin fibrils. Thus, native proteomics can be used to both count and characterize known subunits with precision and discover new protein complexes. The results are complementary to those from bottom-up crosslinking studies32, as they provide the stoichiometry, as well as where complexes “start” and “stop” that is often difficult to determine from large-scale interaction networks. Moreover, unlike crystallography and cryo-electron microscopy, it does not require extensive purification, preparation, and potentially crystallization to determine MPC stoichiometry.
Characterization of Endogenous Metalloenzymes
In addition to protein–protein interactions, we were able to characterize metal cofactors binding to both proteoforms and MPCs. There are 1,378 metal cation cofactors annotated on the 20,197 curated human proteins in SwissProt (~7%), with many more likely unreported. Here, we observed and characterized 28 metal binding events, involving Zn2+, Cu1+, Mg2+, Mn2+, Ca2+, Fe3+, and one 3Fe4S cluster, including three previously unreported (Supplementary Table 1). With one exception, the bound metals corresponded to the proteins’ reported biological cofactors, including Mn2+, which according to the Irving-Williams series is the easiest ion to replace in vitro33. Thus, the metal binding we observed is consistent with that found in the cellular environment. In the case of the one observed discrepancy, between the Mg2+ reported from in vitro studies34, and the Zn2+ cofactor we observed on isopentenyl-diphosphate isomerase (Q13907, Supplementary Fig. 3), we posit the native proteomics result to be more reflective of the in vivo system. Many enzyme cofactors depend critically on their local cellular environment; by eschewing traditional overexpression-based techniques in bacterial systems, native proteomics provides direct information on endogenously formed interactions requiring no prior perturbation of the tissue, cellular or genetic context.
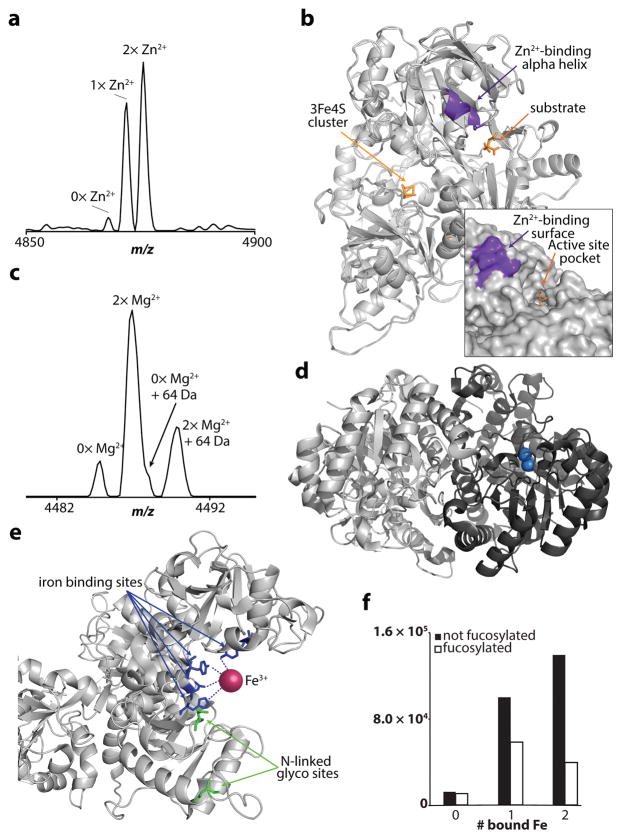
Of the 28 systems characterized with metal cations, two, the murine aconitate hydratase (aconitase, Q99KI0) monomer and the human alpha enolase (P06733) dimer, best exemplify the ability of native proteomics to characterize untargeted metalloproteomic systems. We observed the citric acid cycle enzyme aconitase (82.8 kDa) with an additional mass consistent with a 3Fe4S cluster and between zero and two copies of a Zn2+ cofactor (Fig. 3a). Tandem MS provided confident protein identification and partial localization of the Zn2+, to a region near the active site but far from the 3Fe4S cluster as predicted by the crystal structure (Fig. 3b and Supplementary Fig. 4). The addition of Zn2+ has been shown previously to inhibit the enzymatic turnover of aconitase, but its binding was not localized35. The zinc binding in conjunction with the observation of the catalytically inactive 3Fe4S cluster (as opposed to the active 4Fe4S cluster36) provide compelling evidence that we observed the catalytically inactive form of the aconitase. In addition, we characterized the Mg2+-binding motif of the alpha enolase dimer. Mass spectra of the intact dimer and ejected monomer each showed evidence of zero and one copy of the previously reported di-Mg2+ cofactor (Fig. 3c), while the ESI-produced native monomer showed no evidence of the cofactor (Supplementary Fig. 5). Thus, the holoenzyme was only present in the dimeric complex, indicating the ability of the di-Mg2+ to stabilize the dimer. However, the most abundant dimer peak consisted of one apo- and one holo-subunit (Fig. 3d), and we did not observe the two holo-subunits per dimer reported previously37. By maintaining the fragile noncovalent interactions in the system, subunit stoichiometry and metal binding information are obtained simultaneously, which provides a complete view of new MPC associations and composition.
Figure 3. Direct readout of metal cofactors by native proteomics.
(a) An intact mass spectrum of murine aconitase showing the distribution of Zn2+ binding in addition to a mass shift consistent to within 0.3 Da with the 2+ charge state of its 3Fe4S cluster. (b) The crystal structure of porcine aconitase (98% sequence identity, PDB: 5acn) with the tandem MS-localized Zn2+ binding site indicated in purple. (c) The intact mass spectrum of the enolase dimer bound to zero and one copy (but not two) of its di-Mg2+ cofactors. (d) A crystal structure of the enolase dimer indicating the di-Mg2+ binding pocket that is only occupied on one monomer. (e) One of two iron-binding pockets of human serotransferrin (72% sequence identity) is located near two probable sites of N-linked glycosylation. (f) The normalized relative abundance of species with zero, one, and two bound Fe3+ cofactors indicates a 52% increase in the efficiency of binding two irons with a non-fucosylated glycan. See Supplementary Figure 9 for more details.
Elucidation of Sequence Polymorphisms and Processing
Amino-acid and spliced isoform variants can modulate protein subcellular localization38 and drive specific disease states39. By ejecting subunits and then fragmenting them, we can directly characterize this variation from the proteins that participate in protein complexes. We observed and characterized two mouse hemoglobin subunit beta-1 chains (P02088), which differed by three amino acids and corresponded to products of the S- and D-alleles (Supplementary Fig. 6a, b). We also observed each as a heterodimer bound to hemoglobin alpha chain (P01942), which had a major species corresponding to the S69N and G79A variant, and a minor (~20% abundance) form with just the S69N variant (Supplementary Fig. 6c, d). In addition, we characterized many primary sequence processing events that are critical for protein transport and function. Of the identified proteoforms, 152 of the 217 were observed with the initiator methionine removed (–Metini), and an additional 35 were observed with their signal peptide cleaved, providing strong evidence for the subcellular location of the protein. Many of these events closely corresponded to annotations in UniProt, though a few did not. For example, two forms of the nipsnap homologue 3A monomer (Q9UFN0) were identified with the first 25 and 27 residues cleaved, indicating a potentially novel signal or mitochondrial transit peptide (Supplementary Fig. 7a, b). Likewise, the NAD(P)H-hydrate epimerase (Q8NCW5) homodimer was characterized with each monomer consisting of residues 53–288, instead of residues 48–288 as is predicted following mitochondrial transit peptide removal (Supplementary Fig. 7c). Thus, the pipeline can be used for precise characterization of a gene product’s isoform and subcellular localization as well as to provide de novo information on unannotated processed protein products.
Identification of Post-Translational Modifications
The fact that PTMs contribute to the proteoform complexity of intact proteins in denaturing top-down proteomics is well established40. Native proteomics also characterizes these modifications, but performs better with larger and more heterogeneous proteoform distributions due to the reduced number of charge states in the spectrum41. At the monomer or subunit level, we identified and characterized 116 examples of N-terminal acetylation, two geranylgeranylations and C-terminal carboxymethylations on the RHOA and CDC42 monomers, part of a known enzymatic processing event that allows the Rho-family proteins to anchor to the cellular membrane42. Additionally, up to four pyridoxal phosphate modifications were observed bound to lysines on the dimeric murine cytosolic aspartate aminotransferase (P05201, Supplementary Fig. 8), and a single one bound to human phosphoserine aminotransferase (Q9Y617). In each of these cases the pyridoxal phosphate modifications activate the enzyme’s binding pocket43. However, for the former, only one modification per subunit was annotated prior to this study. Native proteomics is also useful for untargeted characterization of large glycoproteins. For example, the ~74 kDa serotransferrin from mouse (Q921I1) was found to have its single glycosylation site occupied with a biantennary, sialated glycan that was ~30% fucosylated (Supplementary Fig. 9). However, while native proteomics offers high detail on glycan abundance, it does not provide the structure and complete molecular detail of these glycosylated proteoforms. In addition, it was observed bound to between zero and two copies of its Fe3+ cofactor, with the non-fucosylated species showing a 52% increase in normalized di-iron binding when compared to the fucosylated form (Fig. 3e, f). According to the crystal structure of human serotransferrin, we hypothesize that the steric vicinity of N-linked glycans in the C-terminal lobe can hinder the binding of one of the irons. Glycoproteins have proven to be especially challenging to identify with traditional top-down and bottom-up proteomics methods and are often chemically modified or cleaved; here we show the simultaneous identification and characterization of a large intact glycoprotein and its metal cofactor with no prior glycan derivatization.
Beyond monomer-level proteoform characterization, native proteomics also provides MPC-level information of PTM occupancy on intact complexes that is not available from denaturing top-down and bottom-up methodologies25. In many cases, intact complex PTM stoichiometry did not appear to depend on PTM occupancy of the subunits, a “random” association that then produces a distribution of MPC masses with relative abundances that follow a multinomial distribution (Supplementary Fig. 10). However, this was not always observed, as exemplified by the triosephosphate isomerase (TPI, P60174) dimer. In this case, we characterized three major MPCs, each with roughly equal abundance (Supplementary Fig. 11a). Isolation of each of the MPCs individually, monomer ejection (Supplementary Fig. 11b), and fragmentation of each of the three subunit proteoforms (Supplementary Figs. 11c and 12, Supplementary Data Set 2) indicated that they were made up of the –Metini, the N-terminally acetylated, and the –Metini and phosphorylated forms of isoform 2 of TPI. However, the intact MPCs did not follow a multinomial distribution; the phosphorylated and acetylated forms were not observed dimerizing with themselves or each other (Supplementary Fig. 11d). The lack of observable interactions between these forms indicates that their modification states inhibit dimerization, consistent with a proteoform-specific “stoichiometric control.” While the N-terminal acetylation is distal to the dimerization domain, consistent with an allosteric interaction, the phosphorylation, as currently annotated, lies near the dimer interface, which may directly interfere with dimerization (Supplementary Fig. 11e). Differential modification of proteins, especially phosphorylation, has long been thought to modulate protein complexation. However, the precise and simultaneous measurement of subunit modification and complex stoichiometry, difficult or impossible with other methodologies, now allows these situations of proteoform-specific stoichiometric control to be read out quickly with minimal perturbation.
Post-translational modifications of cysteine residues are rarely annotated, with only four cysteine persulfide and eight cysteine glutathionylations annotated on the entire validated UniProt human proteome. However, many of the non-terminal modifications characterized here were, in fact, located on cysteines. We observed 15 gene products with cysteine modifications (Supplementary Table 2), forming 22 proteoforms, with many combining to form MPCs. Only four of these characterized cysteine modifications were annotated in UniProt. As an example, the dimeric creatine kinase M from mouse heart (P07310) was observed with four monomer proteoforms corresponding to: the unmodified form, a cysteine persulfide modification, a cysteine persulfide and a homocysteinylation, and a glutathionylation (Supplementary Fig. 13). The high stoichiometry and heterogeneity of cysteine modifications in creatine kinase, all previously unknown, strongly support previous evidence that the modification state of this cysteine modulates enzyme function44. On the other hand, the Parkinson’s disease-related dimeric protein deglycase DJ-1 (Q99497) has been reported to undergo cysteine modification as a response to cellular oxidative stress45. Ejected from the dimer, we observed the unmodified form, a form with the active cysteine modified to a sulfinic acid, and a form with the cysteine sulfated (Supplementary Fig. 14). The prevalence of these cysteine modifications across a variety of proteins imparts an enhanced ability for cells to handle and respond quickly to cellular stress, likely through redox-sensitive regulatory mechanisms. Furthermore, these findings also allude to previously unreported and ubiquitous cysteine modifications in a variety of cellular proteins that are essential to normal function.
Of the 746 annotated internal acetylations and succinylations across the 164 proteins identified in this study, none were observed. In an attempt to increase acetylation, we treated HEK293T cells with Trichostatin A (TSA), a non-specific deacetylase inhibitor46, prior to lysis and analysis (Supplementary Fig. 15a). Even mitochondrial malate dehydrogenase, an abundant homodimer that has been previously reported as tri-acetylated after TSA treatment47 was only observed as the unmodified form (Supplementary Fig. 15b, c). The dynamic range of our measurement was ~800, indicating that no acetylated forms were present at greater than ~0.1% abundance. Further, N-terminal acetylations were observed on over half of the proteoforms reported here, strongly indicating that acetylated proteoforms can be maintained in native proteomics. The sub-stoichiometric abundances of many acetylated and phosphorylated proteoforms can be attributed in part to the increased sensitivity and dynamic range of the bottom-up techniques used to identify these sites; a previous study of global internal acetylation in yeast determined that ~1% of sites were occupied at >1% stoichiometry48. In some cases, modifications at extremely low stoichiometry may drive signaling cascades and other highly sensitive biological mechanisms. However, they may also represent stochastic cellular processes that have little effect on the action of the protein, or they may also be caused by chemical modification during sample preparation49. In contrast to bottom-up studies, the ability of native proteomics to measure the relative stoichiometry and dynamics of these modifications should help clarify which post-translational modifications play critical roles in the functioning of cells and tissues.
Observation of Protein-bound Metals and Small Molecules
In addition to post-translational modifications, we also characterized several fragile small-molecule binding events. Three of the Ras-superfamily small guanosine triphosphate (GTP)ase proteins were observed bound to guanosine diphosphate (GDP), the dephosphorylated form of their substrate. The first, monomeric GTP-binding nuclear protein Ran (P62826), was observed bound to GDP with ~40% of the ions also bound to either Zn2+ or Cu1+ (Supplementary Fig. 16); GDP binds predominantly to the cytosolic form of the protein and affects nucleocytoplasmic transport50, while the metal binding has not been previously reported. The other two, RHOA (P61586) and CDC42 (P60953), are both in the Rho family of small GTPases, and were both observed in heterodimeric form bound to RHOGDI1 (P52565, Supplementary Fig. 17). Each complex was bound to GDP and Mg2+, two cofactors reported to drive formation of the heterodimeric complex and allow the GTPase to migrate through the cytosol to various locations on the plasma membrane50. Noncovalent enzyme cofactors are almost universally lost upon denaturation of an endogenous sample during preparation for denaturing top-down or bottom-up analysis; native proteomics provides a promising new method for characterization of the thousands of human protein cofactors, in addition to the potential to identify unknown protein-small molecule interactions.
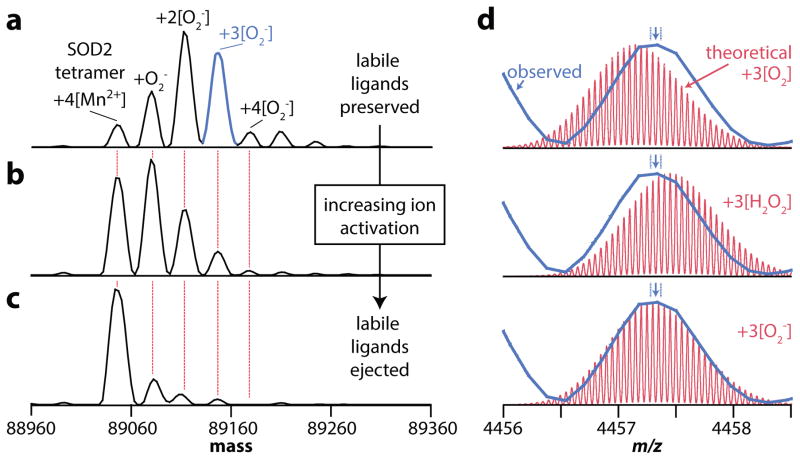
However, the identification of labile small molecules was not limited to cofactors. The intact mass spectrum of the tetrameric human mitochondrial superoxide dismutase (SOD2, P04179) exhibited five peaks differing in mass by 33 ± 1 Da; the lowest-mass peak is consistent with the tetramer and its four Mn2+ cofactors (Fig 4a). Gentle activation of the ions rapidly dissociated these peaks, converting them into the ~89 kDa holo-tetramer (Fig 4b, c; isotopically resolved in Supplementary Fig. 18). Mapping the isotopic distributions of the holo-enzyme with three copies of either its substrate (superoxide) or products (oxygen and hydrogen peroxide) over the observed data provided strong evidence that the additional peaks were consistent with multiple copies of the superoxide substrate (Fig 4d). This example demonstrates the ability of native proteomics to identify an enzyme as well as its stoichiometry, cofactors, and even substrate in a single experiment and in discovery mode. Capturing these extremely fragile interactions could provide critical information about enzymatic mechanisms in a large variety of protein systems.
Figure 4. Detection of labile superoxide bound to mitochondrial superoxide dismutase.
(a–c) The intact mass spectrum of the tetrameric human mitochondrial superoxide dismutase bound to between zero and four copies of its superoxide substrate in addition to four copies of its Mn2+ cofactor. Slightly increasing the amount of ion activation in the electrospray source region, from 5 V (a) to 20 V (b) to 30 V (c), progressively ejected more of the weakly bound substrates while retaining the intact tetramer and all four of the Mn2+ cofactors. (d) A comparison of the observed fourth-highest mass peak from a (in blue) with theoretical isotopic distributions for the intact tetramer, its four Mn2+ cofactors, and three copies of either O2, H2O2, and O2−. The arrow above each peak indicates the centroid, with the dotted lines corresponding to shifts of ±10 ppm; thus, the results strongly indicate the presence of superoxide and not either of the products. The difference in mass between O2− and O2 is due to the difference in formal charge, changing the number of protons on the ions required to produce a net charge state of 20+.
Discussion
As demonstrated throughout, an integrated platform for native proteomics for the untargeted characterization of abundant complexes in mammalian proteomes is now feasible, providing a new type of compositional information about endogenous complexes and their molecular associations. We applied native proteomics to mouse hearts and a variety of human cancer cell lines; the methodology is applicable to diverse tissues, fluids or cell-based sources. Additional modifications, already demonstrated in the literature, could further expand the proteome coverage of native proteomics to membrane and lower abundance complexes. Further, the current platform requires substantially more sample than traditional denaturing bottom-up, crosslinking experiments,32 or denaturing top-down proteomics7, but future application of ‘online’ native separations with automated tandem MS will dramatically improve these methodological requirements.
Leveraging a greatly expanded ability to tackle large molecular weight proteins and their complexes, we provide a first glimpse at the possibility of direct measurement of protein–protein and protein–ligand interactions. The general applicability of the methodology to these noncovalent interactions will catalyze new techniques for determination of drug targets and metalloenzyme composition and function. However, characterization of proteoforms and their complexes is not limited to noncovalent interactions, with this wave of results contributing to the view that even complex arrays of covalent modifications such as those involving N-glycosylation are tractable in a top-down approach to molecular analysis. Yet native proteomics goes beyond the characterization of protein modification determined by traditional proteomics techniques, allowing the modifications to be placed within the context of the intact molecular machinery connecting tightly with the function of endogenous proteins.
Online Methods
Preparation of mouse hearts
CD-1 mouse hearts were purchased from BioreclamationIVT. For the mitochondrial and cytosolic preparations, 20 mouse hearts were diced with a razor blade over ice and diluted to 100 mg/mL with lysis buffer (250 mM sucrose, 10 mM Tris-HCl, 0.1 mM EGTA, pH 7.4) and frozen. Good results were generally obtained from ~100 mg of heart tissue (roughly one mouse heart). The heart tissue was homogenized using a glass-teflon homogenizer, and subcellular fractionation was performed as described previously23 to produce separate mitochondrial and cytosolic subcellular fractions. Heart samples that were not subcellularly fractionated were diced with a razor blade, and then cryo-ground in liquid nitrogen with a mortar and pestle. The ground tissue was then resuspended in lysis buffer and HALT protease inhibitor (Thermo Fisher Scientific) and frozen prior to analysis.
Preparation of cell lines
Human cell lines were obtained from ATCC or DSMZ and cultured according to manufacturer recommendations. In general, successful results were obtained from ~50 million cells. All reagents for cell culture were obtained from ThermoFisher, unless otherwise noted. Lymphocyte cell lines included Ramos (Burkitt Lymphoma, B cell), Hg-3 (Chronic lymphocytic leukemia, B cell), and Jurkat (Acute lymphoblastic leukemia, T cell); all grown in RPMI-1640 supplemented with 10% fetal bovine serum, 10 mM HEPES, and 1 mM sodium pyruvate. In addition to lymphocyte cell lines, human embryonic kidney cells (HEK-293T) cells were grown in DMEM supplemented with 10% fetal bovine serum, 50 U/mL penicillin, and 50 μg/mL streptomycin. All cells were grown in 150 cm2 flasks and harvested while cells were in log phase growth. Lymphocytes were grown in suspension, whereas HEK-293T cells were adherent and were lifted using 0.05% Trypsin-EDTA solution. HEK-293T were treated with 20 μM Trichostatin A (TSA, Sigma) and 5 mM nicotinamide (NAM, Sigma) for 18 hours, similar to previously established methods.46 Cells were pelleted by centrifugation, washed using cold PBS, re-pelleted, and cell pellets were stored at −80° C until processed. Mycoplasma testing was performed quarterly for all cell lines used in this study using either the LookOut Mycoplasma Kit (Sigma) or the MycoProbe Mycoplasma Detection Kit (R&D systems). Cell lines were authenticated using the GenePrint® 10 System (Promega, Chicago, IL), which uses short tandem repeat (STR) analysis.
Cells were lysed using a detergent-free hypotonic lysis buffer consisting of 15 mM Tris-HCl (pH 7), 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 250 mM sucrose (all from Sigma), 1x HALT protease and phosphatase inhibitor (ThermoFisher), and 10 mM sodium butyrate (Sigma). Cell pellets were resuspended using 10:1 volume of lysis buffer to approximate volume of cell pellet. Cells were allowed to swell for 1–2 hours in a constant temperature of 4° C. Mechanical disruption was used to complete the lysis, using either a probe sonicator or the gentleMACS dissociator. For sonication, cells were lysed using the following parameters: 33% amplitude, 2x-30 sec pulse. For dissociation, cells were lysed using the “E” program (most disruptive setting). Cellular debris and intact organelles were pelleted by centrifugation (10,000 × g for 5 minutes at 4° C) and the supernatant, which contained primarily soluble cytosolic proteins, was transferred to a clean, siliconized microcentrifuge tube. The lysate was either fractionated immediately or stored at −80° C until fractionated.
Native GELFrEE
Mouse heart samples were fractionated by native-GELFrEE as described previously23, 24. Briefly, 0.5–1 mg of protein extract was mixed with 3 volumes of solubilization buffer (50 mM imidazole, 500 mM aminocaproic acid, 1 mM EDTA, pH 7.0) and incubated on ice for 30 min. The samples were cleaned with solubilization buffer (3 times 300 μL) and concentrated using a 30 kDa molecular weight cutoff spin filter (Millipore). The final volume of 100 μL of sample was mixed with 30 μL of 70% glycerol and 0.1% Ponceau S for loading. Fractionation was performed in a 10 cm long 1–10 or 1–12% T gradient gel using a 6 mm ID tube via imidazole-based clear native GELFrEE buffer system. A voltage range of 200–1000 V was applied for protein separation, and 150 μL fractions were collected manually.
Ion exchange chromatography (IEX)
Prior to loading, samples were buffer exchanged into 10 mM ammonium acetate using a 10, 30, or 100 kDa molecular weight cutoff filter (Millipore), depending on the targeted molecular weight range. Samples were filtered using a 0.45 μm syringe filter, loaded on a 500 uL sample loop and injected on an integrated CATWAX column (PolyLC) using an Agilent series 1100 (degasser, pumps, column compartment, UV module) and 1200 (fraction collector, fraction chiller) high-performance liquid chromatography setup. The CATWAX columns used contained equal amounts of PolyCAT A and PolyWAX LP, however two variations of columns were used: (1) 12 μm particle and 1500 Å pore and (2) 5 μm particle and 1000 Å pore. Samples were run on exploratory and optimized gradients of buffer A (10 mM ammonium acetate, pH 7), and buffer B (1 M ammonium acetate, pH 7) with a flow rate of 0.25 mL/min to 0.5 mL/min using the Agilent ChemStation control software. To maximize the identification of new complexes, the IEX gradients were varied, but generally had a duration of 3 hours. Fractions were collected by time every 90 seconds, and did not depend on the elution profile. The separation was monitored by UV absorbance at 280 nm.
Antibody Blotting
For targeted studies of mitochondrial malate dehydrogenase, the lysates were separated using ion exchange and a dot blot was performed to localize fractions containing malate dehydrogenase. For this dot blot, 2 μL of each fraction was spotted onto a nitrocellulose membrane. For blocking, the membrane was incubated in 5% (w/v) dry skim milk in tris-buffered saline with 0.02% Tween-20 (TBST) for 30 minutes. The primary antibody was anti-human MDH2 rabbit monoclonal (ab181873; Abcam, Cambridge, UK) and was diluted 1:10,000 in 5% (w/v) skim milk in TBST; the membrane was incubated in primary antibody for 2 hours at room temperature with agitation. Next, the membrane was washed for 3 × 5 min washes with TBST. The secondary antibody was goat anti-rabbit IgG H&L horseradish peroxidase conjugated (ab97051; Abcam) and was diluted 1:20,000 in 5% (w/v) skim milk in TBST; the membrane was incubated in secondary antibody for 30 minutes at room temperature with agitation. The membrane was washed for 5 × 3 min washes with TBST. The membrane was then treated with SuperSignal West Pico Chemiluminescent Substrate (Thermo) for 5 minutes in the dark with agitation. The immunoblot was imaged using the Bio-Rad GelDoc™ XR+ and was automatically processed via the Image Lab™ software (Bio-Rad, Hercules, CA).
For measurement of global acetylation levels, the anti-acetyl blot was performed using the Anti acetylated-lysine (Ac-K2-100) multiMab Rabbit monoclonal antibody mix, which was purchased from Cell Signaling (#9814S).
Sample preparation for native TDMS
Fractions from native GELFrEE were first treated to remove detergent with 1–2 applications of a HiPPR detergent removal column (Thermo Fisher). Native GELFrEE and ion exchange fractions were exchanged into 100–200 mM ammonium acetate and concentrated using a 30 kDa molecular weight cutoff spin filter (Millipore).
Native Top-Down MS analysis
Samples were introduced into the mass spectrometer with a custom nano electrospray source as described previously25 using 0.8 to 1.6 kV spray voltage. In total, approximately 600 hundred samples were analyzed, with some being regenerated and interrogated more than once. All samples were analyzed with a Q-Exactive HF (Thermo) mass spectrometer modified as described previously22. To analyze protein complexes, we used a three-tiered approach22. Briefly, the intact complex mass spectrum was measured (MS1) using only moderate source energy to reduce salt adduction. One or more of the intact complex charge states was then quadrupole-isolated, and the monomer was ejected in the HCD cell (MS2). Finally, the monomer was ejected in the source, quadrupole-isolated, and fragmented for identification and characterization (MS3). In some cases, when the monomers were too tightly bound, the MS3 spectrum was obtained by increasing the activation energy of the intact complex in the MS2 step. A range of resolving powers were used (7,500 – 960,000), however, the MS3 spectrum was always collected at 60,000 or above to provide ample resolution for fragment ion deconvolution. Intact protein monomers were quadrupole-isolated and fragmented in the HCD cell using equivalent parameters to those for MS2 and MS3 described above. The acquisition parameters in the mass spectrometer (mainly the pressure and energy of collisions in the source and HCD cell) were varied and optimized manually for each proteoform and MPC.
Data analysis and processing
Low resolution MS1 and MS2 spectra were deconvoluted manually, with MagTran software51, or with Protein Deconvolution 4.0 (Thermo). All MS1 and MS2 mass value refer to average mass, when isotopic resolution was achieved the centroid average of the isotopic distribution was calculated with mMass software52. High resolution MS3 spectra were deconvoluted with Xtract software (Thermo); in some cases artefactual peaks were removed manually. Additional fragment ion analysis was performed with mMass.
Many of the acquired spectra proved to be challenging to analyze with traditional data analysis software packages and had to be curated manually. Specifically, the Xtract deconvolution software would often produce large numbers of spurious neutral masses corresponding to singly charged fragment ions, which had to be corrected by manual deletion or addition of peaks. Therefore, many of the metrics usually reported with high-throughput proteomic reports (p-score, E-value, FDR, etc.) must be considered with caution. However, the raw data and fragment maps are publicly accessible (vide infra), and each identification was scrutinized to determine whether sufficient data was present to report the identification.
Masses derived from MS2 and MS3 data (MS1 and MS2 for monomers) were entered into the Prosight PC 4.0 software (Thermo) and searched against a database consisting of either UniProtKB human or mouse proteins, shotgun-annotated to account for protein modifications. Annotated modifications mentioned in the main text refer to the July 6, 2016 release of UniProtKB. Protein complex stoichiometries were then determined manually and with the SEMPC, which is based on the CORUM database53 and information from UniProtKB to characterize and score an MPC with low or high molecular specificity depending on the underlying nTDMS data quality25.
Data Availability
The mass spectra .RAW data files and proteoform fragment maps that support the findings of this study are publicly available online at www.proteomexchange.org under accession number PXD005313.
Supplementary Material
Acknowledgments
Funding for this project was provided by the W. M. Keck foundation (DT061512) and Northwestern University. This research was also supported by the Paul G. Allen Family Foundation (Grant Award 11715). The authors would also like to acknowledge helpful discussion with Adam Catherman, Ryan Fellers, Joseph Greer, and Bryan Early. Helpful assistance from Thermo Fisher Scientific was provided by Mikhail Belov, Stevan Horning, and Alexander Makarov. OSS and PFD are supported by US National Science Foundation graduate research fellowships (2014171659 and 2015210477, respectively). LHFDV is supported under CNPq research grant 400301/2014-8 from the Brazilian government. LFS was supported by the Chemistry of Life Processes Predoctoral training program at Northwestern University. Additional support for the maintenance of the SEMPC from the National Resource for Translational and Developmental Proteomics (GM108569) is acknowledged.
Footnotes
Author Contributions
Data acquisition and analysis was performed by OSS with assistance from NAH, LF, and RDM and from all authors. NAH, RDM, LF, LHFDV and PFD prepared samples with help from OSS and LFS. OSS wrote the manuscript with critical insights from PDC and additional assistance from all co-authors. NLK and PDC conceived of and oversaw the project.
Competing financial interests
NLK serves as a paid consultant to Thermo Fisher Scientific, whose instruments were utilized in this work.
References
- 1.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 2.Alcorta DA, et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunaief JL, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 4.Hummon AB, et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science. 2006;314:647–649. doi: 10.1126/science.1124128. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 6.Tran JC, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catherman AD, et al. Large-scale top-down proteomics of the human proteome: membrane proteins, mitochondria, and senescence. Mol Cell Proteomics. 2013;12:3465–3473. doi: 10.1074/mcp.M113.030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu S, et al. Top-Down Characterization of the Post-Translationally Modified Intact Periplasmic Proteome from the Bacterium Novosphingobium aromaticivorans. Int J Proteomics. 2013;2013:279590. doi: 10.1155/2013/279590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou M, et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Wongkongkathep P, Van Orden SL, Ogorzalek Loo RR, Loo JA. Revealing ligand binding sites and quantifying subunit variants of noncovalent protein complexes in a single native top-down FTICR MS experiment. J Am Soc Mass Spectrom. 2014;25:2060–2068. doi: 10.1007/s13361-014-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 12.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 13.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachir AI, et al. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr Biol. 2014;24:1845–1853. doi: 10.1016/j.cub.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du J, Lu W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 17.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- 18.Link AJ, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotech. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 19.Havugimana PC, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huttlin EL, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rees JS, et al. In vivo analysis of proteomes and interactomes using Parallel Affinity Capture (iPAC) coupled to mass spectrometry. Mol Cell Proteomics. 2011;10:M110002386. doi: 10.1074/mcp.M110.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belov ME, et al. From protein complexes to subunit backbone fragments: a multi-stage approach to native mass spectrometry. Anal Chem. 2013;85:11163–11173. doi: 10.1021/ac4029328. [DOI] [PubMed] [Google Scholar]
- 23.Skinner OS, et al. Native GELFrEE: a new separation technique for biomolecular assemblies. Anal Chem. 2015;87:3032–3038. doi: 10.1021/ac504678d. [DOI] [PubMed] [Google Scholar]
- 24.Melani RD, et al. CN-GELFrEE - Clear Native Gel-eluted Liquid Fraction Entrapment Electrophoresis. J Vis Exp. 2016:53597. doi: 10.3791/53597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skinner OS, et al. An informatic framework for decoding protein complexes by top-down mass spectrometry. Nat Methods. 2016;13:237–240. doi: 10.1038/nmeth.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith LM, Kelleher NL Consortium for Top Down, P. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havugimana PC, Wong P, Emili A. Improved proteomic discovery by sample pre-fractionation using dual-column ion-exchange high performance liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:54–61. doi: 10.1016/j.jchromb.2006.10.075. [DOI] [PubMed] [Google Scholar]
- 28.Skinner OS, Schachner LF, Kelleher NL. The Search Engine for Multi-Proteoform Complexes: An Online Tool for the Identification and Stoichiometry Determination of Protein Complexes. Curr Protoc Bioinformatics. 2016;56:13 30 11–13 30 11. doi: 10.1002/cpbi.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Waterbeemd M, et al. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat Methods. 2017;14:283–286. doi: 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- 30.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 31.McMahon CG, et al. Diagnostic accuracy of heart-type fatty acid-binding protein for the early diagnosis of acute myocardial infarction. Am J Emerg Med. 2012;30:267–274. doi: 10.1016/j.ajem.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Liu F, Rijkers DT, Post H, Heck AJ. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods. 2015;12:1179–1184. doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- 33.Foster AW, Osman D, Robinson NJ. Metal preferences and metallation. J Biol Chem. 2014;289:28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng W, et al. The crystal structure of human isopentenyl diphosphate isomerase at 1.7 A resolution reveals its catalytic mechanism in isoprenoid biosynthesis. J Mol Biol. 2007;366:1447–1458. doi: 10.1016/j.jmb.2006.12.055. [DOI] [PubMed] [Google Scholar]
- 35.Costello LC, Liu Y, Franklin RB, Kennedy MC. Zinc inhibition of mitochondrial aconitase and its importance in citrate metabolism of prostate epithelial cells. J Biol Chem. 1997;272:28875–28881. doi: 10.1074/jbc.272.46.28875. [DOI] [PubMed] [Google Scholar]
- 36.Robbins AH, Stout CD. Structure of activated aconitase: formation of the [4Fe-4S] cluster in the crystal. Proc Natl Acad Sci U S A. 1989;86:3639–3643. doi: 10.1073/pnas.86.10.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang HJ, Jung SK, Kim SJ, Chung SJ. Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme. Acta Crystallogr D Biol Crystallogr. 2008;64:651–657. doi: 10.1107/S0907444908008561. [DOI] [PubMed] [Google Scholar]
- 38.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 39.Oyer JA, et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28:198–201. doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durbin KR, et al. Quantitation and Identification of Thousands of Human Proteoforms below 30 kDa. J Proteome Res. 2016;15:976–982. doi: 10.1021/acs.jproteome.5b00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Barendregt A, Kamerling JP, Heck AJ. Analyzing protein micro-heterogeneity in chicken ovalbumin by high-resolution native mass spectrometry exposes qualitatively and semi-quantitatively 59 proteoforms. Anal Chem. 2013;85:12037–12045. doi: 10.1021/ac403057y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki T, Takai Y. The Rho small G protein family-Rho GDI system as a temporal and spatial determinant for cytoskeletal control. Biochem Biophys Res Commun. 1998;245:641–645. doi: 10.1006/bbrc.1998.8253. [DOI] [PubMed] [Google Scholar]
- 43.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 44.Reddy S, Jones AD, Cross CE, Wong PS, Van Der Vliet A. Inactivation of creatine kinase by S-glutathionylation of the active-site cysteine residue. Biochem J. 2000;347(Pt 3):821–827. [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson MA. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid Redox Signal. 2011;15:111–122. doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vigushin DM, et al. Trichostatin A Is a Histone Deacetylase Inhibitor with Potent Antitumor Activity against Breast Cancer in Vivo. Clinical Cancer Research. 2001;7:971–976. [PubMed] [Google Scholar]
- 47.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinert BT, et al. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skinner OS, Kelleher NL. Illuminating the dark matter of shotgun proteomics. Nat Biotechnol. 2015;33:717–718. doi: 10.1038/nbt.3287. [DOI] [PubMed] [Google Scholar]
- 50.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J Am Soc Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 52.Strohalm M, Kavan D, Novak P, Volny M, Havlicek V. mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal Chem. 2010;82:4648–4651. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- 53.Ruepp A, et al. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2010;38:D497–501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectra .RAW data files and proteoform fragment maps that support the findings of this study are publicly available online at www.proteomexchange.org under accession number PXD005313.