Abstract
Gene by environment (GxE) interaction studies have investigated the influence of a number of candidate genes and variants for major depressive disorder (MDD) on the association between childhood trauma and MDD. Most of these studies are hypothesis driven and investigate only a limited number of SNPs in relevant pathways using differing methodological approaches. Here (1) we identified 27 genes and 268 SNPs previously associated with MDD or with GxE interaction in MDD and (2) analyzed their impact on GxE in MDD using a common approach in 3944 subjects of European ancestry from the Psychiatric Genomics Consortium who had completed the Childhood Trauma Questionnaire. (3) We subsequently used the genome-wide SNP data for a genome-wide case-control GxE model and GxE case-only analyses testing for an enrichment of associated SNPs. No genome-wide significant hits and no consistency among the signals of the different analytic approaches could be observed. This is the largest study for systematic GxE interaction analysis in MDD in subjects of European ancestry to date. Most of the known candidate genes/variants could not be supported. Thus, their impact on GxE interaction in MDD may be questionable. Our results underscore the need for larger samples, more extensive assessment of environmental exposures, and greater efforts to investigate new methodological approaches in GxE models for MDD.
INTRODUCTION
Major depressive disorder (MDD) is known to be substantially heritable but also has a huge number of well-established environmental and lifestyle factors that contribute to the disease risk (Cerdá, 2010). Although the proportion of variance attributable to genome-wide SNPs (SNP heritability) for MDD has been estimated to be about 21% to 32% (Lee et al., 2013; Lubke et al., 2012), there are only a few genome-wide significant hits for MDD or depressive symptoms that have been detected to date (Hek et al., 2013; Okbay et al., 2016; CONVERGE, 2015; Hyde et al., 2016) and their biological impact on depression is largely unknown. Many factors could explain the lack of success so far including limited sample size, the high symptom heterogeneity for MDD as well as the strong contribution of environmental and, lifestyle factors and life events (Flint and Kendler, 2014), and it has been hypothesized that genetic factors need the presence of environmental triggers to exhibit an effect on the individual (Caspi et al., 2003; Dun et al., 2015).
Early childhood trauma (CT) is the most frequently investigated environmental factor, which shows a high impact on major depression and many other psychiatric disorders (Mandelli et al., 2015). Previous studies have suggested interaction effects between CT and individual variants from several genes including the highly studied serotonin-transporter-linked polymorphic region (5-HTTLPR) (Caspi et al., 2003; Van der Auwera et al., 2014; Karg et al., 2011), but also for BDNF, TPH2, FKBP5, DRD2 and many other genes from candidate pathways (Mandelli and Serretti, 2013; Grabe et al., 2011; Appel et al., 2011). These candidate gene approaches in gene-environment (GxE) interaction analyses select single variants in specific genes belonging to plausible disease-related pathways. Although this approach seems sensible, there are many challenges in this work. Many factors impact on their interpretation, not least non-significant results are less likely to be taken forward for publication generating a reporting bias (Duncan and Keller, 2011). Comparisons between studies are difficult because of the different environmental exposures considered such as stressful life events, abuse and neglect subtypes, social support or living in rural/urban areas, different methods of assessment (questionnaires vs. interviews), and the different quantification of exposures (binary, categorical or continuous) (Mandeli and Serretti, 2013; Dunn et al., 2011). Likewise, the phenotype definition varies between a binary lifetime major depression variable to a dimensional score of current depression. Moreover, there are no common guidelines regarding how to perform GxE analyses in MDD (type of regression model (linear or logistic model), mode of action (multiplicative or additive) or assumed genetic effect (additive, allelic, dominant or recessive model)). Until now, only two GxE interaction analyses for depressive symptoms have been performed on a genome-wide level. One was published by Dunn et al. (2016) in a sample of African American and Hispanic women. In N=7179 African American women one genome-wide significant hit was found near CEP350, a centrosomal protein which has never been associated with a psychiatric phenotype before. Another study in a Japanese population (N=320) reported the genome-wide hit rs10510057 near RGS10 (Otowa et al., 2016), but given the small sample size this result should be regarded with caution. Both studies performed a linear regression GxE analysis assessing the p-value of the interaction term with a dimensional depression score as the outcome, and stressful life events during the past 12 months as the environmental exposure. Whether these findings can be replicated in a population of European ancestry, although the environmental exposure is different, needs to be elucidated.
Here, we focus on the most important known risk exposure for MDD, early childhood trauma, and combine all cohorts from the Psychiatric Genomics Consortium (PGC) with available CT and MDD data to perform gene-environment (GxE) interaction. At the outset, despite being the largest European ancestry study to date, we recognized that our study was likely under-powered to detect GxE effects on a genome-wide level, so we sought to reduce the multiple testing burden by screening the literature and identified genes and SNPs previously implicated in MDD or GxE in MDD. Our aims were 1) to analyze candidate variants, 2) to compare different methodological approaches, and 3) to analyze the genome-wide summary statistics of the GxE analyses for an enrichment of significant findings. We hypothesize that candidate genes/SNPs for a GxE interaction in MDD that have been proposed in the past should at least show a nominally significant association (p<0.05) in our analyses.
MATERIALS AND METHODS
Participants
The Psychiatric Genomics Consortium (PGC) collates genome-wide genotypic and phenotypic data for MDD (PGC steering committee, 2009). Subjects were recruited from the PGC wave 2 for MDD, with phenotypic and genetic data of 16823 MDD cases and 25632 controls from 24 different cohorts with individuals from European ancestry. All cases were diagnosed according to DSM-IV lifetime MDD using structured diagnostic instruments from direct interviews by trained interviewers or clinician-administered DSM-IV checklists. Controls were screened for absence of MDD. Nine of these cohorts also provide phenotyping of exposure to environmental factors as risk for psychiatric disorders including childhood trauma (CT). Five cohorts used the most widely applied Childhood Trauma Questionnaire (CTQ) that distinguishes between different dimensions of childhood abuse and neglect (Bernstein et al., 2003) (table S1): Cognition and Function in Mood Disorders Study (COFAMS) from Australia (Baune and Air, 2016), the Netherlands Study of Depression and Anxiety (NESDA) (Penninx et al., 2008), Radiant-UK from the United Kingdom (Lewis et al., 2010), and two independent samples from the Study of Health in Pomerania (SHIP) from Germany (Völzke et al., 2011). Six cohorts used study-specific versions of childhood trauma assessment that do not capture all five dimensions of abuse and neglect from the CTQ or used different types of questions (table S1): Depression Gene Network (DGN) from the USA (Mostafavi et al., 2014), the Genetics of Recurrent Early-Onset Depression (GenRED) from the USA (Holmans et al., 2007), two independent samples from the Queensland Institute of Medical Research (QIMR) from Australia (Nelson et al., 2002; Wray et al., 2012), the psychiatric arm of the population-based CoLaus study (PsyCoLaus) from Switzerland (Preisig et al., 2009) and the Bonn/Mannheim study from Germany (BOMA) (Cichon et al., 2011). To reduce the heterogeneity in our samples, we only included the five studies that measured childhood trauma with the same standardized instrument (CTQ). In addition, COFAMS was excluded from the analysis due to the low number (N=56) of MDD cases with available CTQ data.
Childhood Trauma Questionnaire
The CTQ assesses childhood trauma, defined as trauma before the age of 16 (CTQ, table S1) (Bernstein et al., 2003), which covers three sub-scales of abuse, sexual abuse, physical abuse and emotional abuse, as well as two sub-scales of neglect, emotional neglect, and physical neglect, all covered by five questions (range 1 to 5). This results in a score per domain ranging from 5 to 25, and an overall CTQ continuous score ranging from 25–125. Per domain, cutoffs from the CTQ manual (Bernstein et al., 2003) were applied to get a broad definition of childhood trauma separating no trauma from mild, moderatel, or severe trauma. CT was transformed to a dichotomous abuse variable separating childhood abuse in any of the three domains (1=Yes) from no abuse in all domains (0=No) to address the skewness of the CTQ score.
Genotyping, quality control and imputation
The cohorts were genotyped following their local protocols, after which quality control and imputation to the 1000 genomes reference panel (Abecasis, 2010) was conducted through the standardized PGC pipeline (see Schizophrenia Working Group of the PGC, 2014) per cohort (PGC MDD: wave 2 GWAS results, In preparation). For details see supplemental material.
Statistical Analysis
We performed these analyses in three steps: 1. Power calculation and selection of candidate SNPs/genes for GxE analyses, 2. Analysis of the candidate variants and genes in GxE, and 3. Analysis of the genome-wide GxE GWAS results.
Different methodological approaches
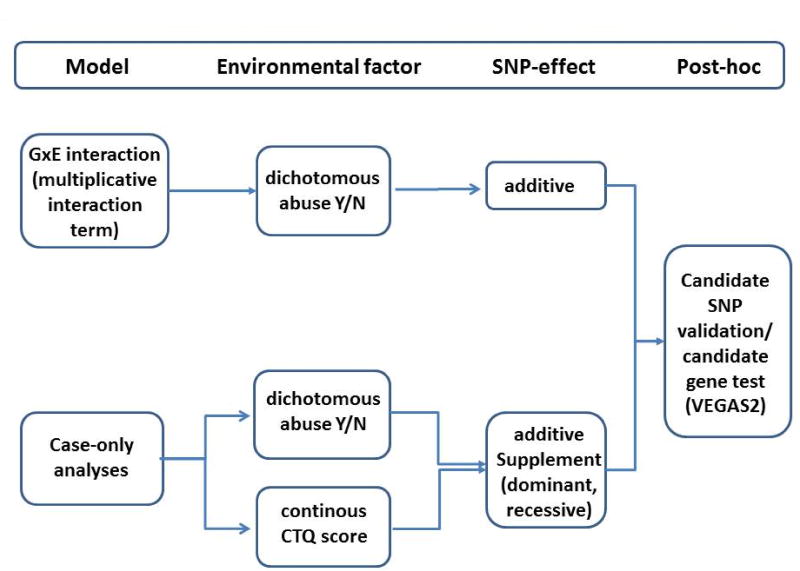
Our main analytic models assuming additive genetic effects include (i) a standard GxE analysis with a multiplicative interaction term and a dichotomous environmental exposure (abuse 0/1) and (ii) case-only analyses, recommended by VanderWeele (VanderWeele et al., 2010; Explanation in Causal Inference, 2015), with the dichotomous abuse 0/1 variable as well as the continuous CTQ score as outcome (for overview see figure 1). Case-only analyses have a higher statistical power to detect GxE effects (Gauderman et al., 2013) than case-control GxE models with a multiplicative interaction term and circumvent the statistical difficulties of the low robustness of interaction terms. But these models require that no gene-environment (G~E) correlation is present (VanderWeele et al., 2010; VanderWeele, 2015). For all case-only models G~E correlation in MDD negative controls was analyzed. We used MDD negative controls because these constitute of roughly 85% of the population and can thus be used as an approximation for the full population.
- GxE case-control (CC) interaction analyses included a multiplicative interaction term between the SNPs and abuse 0/1 assessing the p-value for the interaction term. Analyses were controlled for sex, the first three genetic principal components as well as all SNPxCov and ABUSExCov interaction terms as recommended by Keller (Keller, 2014).
- MDD case-only (CO) analyses with abuse 0/1 as dependent variable assessed the SNP p-value:
MDD CO analyses with CTQ score as dependent variable assessed the SNP p-value:
-
These three approaches enable the comparisons between CC GxE and COy analysis as well as between dichotomous and continuous measurement of childhood trauma (abuse 0/1 vs. CTQ-score).As a sensitivity analysis, the CO analyses were also performed assuming a dominant and recessive SNP effect (see supplement). This analysis was empirically driven by the fact that in many candidate studies for GxE in MDD dominant or recessive effects were found (Mandelli and Serretti, 2013).
Figure 1.
Overview of the seven different models and approaches for GxE
GWAS and meta-analyses
For each of the four cohorts (SHIP-0, SHIP-TREND, NESDA, Radiant-UK) GWAS have been performed as described above. A logistic regression model was used for a binary outcome and a linear regression model for a continuous outcome using PLINK (Chang et al., 2015). Quantile-quantile (QQ) and Manhattan (MH) plots were generated using R (https://cran.r-project.org/). The final meta-analysis comprised N=3944 individuals (N=1891 MDD cases and N=2053 controls). The results were combined using an inverse variance-weighted fixed effects meta-analysis in METAL (Willer et al., 2010) with the following QC parameters; MAF>0.05, info score>0.6, HWE>0.001 and including only SNP present in at least 3 of the 4 cohorts. The genomic inflation factor λ for each study was calculated, and genomic-control (GC) correction was applied when λ>1. The I2 statistic was used to evaluate between-study heterogeneity.
RESULTS
The number of MDD cases and controls with and without CT are summarized in table 1. In each of the four cohorts the CTQ total score as well as the abuse 0/1 variable were highly associated with MDD, adjusted for sex and age.
Table 1.
Sample description and descriptive statistic of the four samples included in the meta-analysis
| N | MDD cases | Controls | MDD~abuse | Abuse prevalence |
||||
|---|---|---|---|---|---|---|---|---|
| No abuse |
abuse | No abuse |
abuse | odds ratio |
p-value | |||
| SHIP-0 | 1505 | 239 | 117 | 921 | 171 | 2.55 | 7.6E-11 | 19% |
| SHIP-TREND-0 | 665 | 114 | 46 | 428 | 59 | 3.06 | 1.5E-6 | 16% |
| NESDA/NTR | 1396 | 500 | 627 | 208 | 61 | 4.19 | 1.1E-19 | 49% |
| Radiant UK | 525 | 85 | 175 | 199 | 66 | 6.09 | 2.6E-20 | 46% |
| Total | 4091 | 938 | 965 | 1756 | 357 | − | − | − |
1. Power calculation and selection of candidate genes/SNPs
The software Quanto (vs. 1.2.4) was used to determine the interaction effect size we would be able to detect as genome-wide significant given our sample (see table S2). In the case-control GxE model with N≈1900 MDD cases, we would only be able to detect large effects (OR≥2.5) with a power of 80% for SNPs with high MAF (MAF≥0.2). In the case-only model with N≈2000 MDD cases, we would be able to detect large effects (OR≥2.4) for SNPs with low MAF (5%), and medium effects (OR>1.5) with SNPs with large MAF (50%). Because in GWAS small effects (OR<1.5) are observed, we assumed that with our current sample size we were underpowered to detect genome-wide interaction signals. Thus, we will focus on candidate SNPs and genes for GxE in MDD.
Since the literature on candidate genes for MDD and GxE interaction in MDD is very broad, we focused on papers that reviewed the previous work in the field. The candidate list comprises SNPs/genes that were taken from two major reviews (Mandelli and Serretti, 2013 for GxE interaction in MDD; Luo et al., 2016 for candidate genes/SNPs in MDD). These candidates cover genes from central monoaminergic systems such as serotonin, dopamine or noradrenalin, from the glutamatergic system, corticotrophin system, neurotropic system or from inflammatory processes (e.g. SLC6A4, DRD2, COMT, NR1, CRHR1, BDNF, FKBP5, NR3C1). We also included SNPs from recent GWAS results for MDD or GxE interaction (Dunn et al., 2016; Otowa et al., 2016; Hyde et al., 2016). The final list included 268 different candidate SNPs (table S4) and 27 candidate genes (table S6). From the candidate SNP list, 184 SNPs were available in the meta-analyses after QC (most of these candidate SNPs were excluded based on MAF<0.05).
2. Analysis of candidate SNPs/genes
The candidate genes/variants were analyzed using different methodological approaches. In the CO analyses the candidate SNPs revealed no excess of G~E correlation (supplemental material, table S7), justifying continuation into case-only analyses. A full list of the results from the candidate SNPs in all models is provided in table S4.
Case-control GxE analysis with a multiplicative interaction term
In the fully adjusted GxE model no candidate SNP reached statistical significance after correcting for multiple testing (pcorrected set to 0.05/184≈0.0003) and five SNPs showed nominal significance (p<0.05), which was fewer than expected by chance (expected N=9): rs2433320 (PDLIM5), rs1656369 (RSRC1), rs1539243 (IKBKE), rs900144 (ARNTL) and rs6582078 (TPH2).
Case-only approach on abuse 0/1 and CTQ score
Abuse 0/1: From the candidate list, eight SNPs were at least nominally significant in the additive SNP model: rs1656369 (RSRC1), rs41423247/rs6191/rs33388 (NR3C1), rs1801262 (NEUROD1), rs4763327 (EMP1), rs2433320 (PDLIM5). CTQ-score: Seven SNPs from the candidate list were nominally significant in the additive SNP model: rs909486 (CSF2RB), rs3754674 (NPAS2), rs9450282 (NT5E), rs4244813/rs2279861 (SLC29A2), rs6191 (NR3C1), rs737865 (COMT). Results for the dominant/recessive case-only models can be found in supplementary table S4.
Exploratory comparison of all three approaches
Taking the CC GxE and CO GxE approaches, only 16 (≈9%) of the 184 SNPs showed nominal significance in at least one of the approaches (table S4), 13 of them with consistent directions of effects in all three approaches. Some of them showed consistently significant associations across approaches. Two SNPs (rs33388 and rs6191) of NR3C1 (glucocorticoid receptor) which acts as a transcription factor and player in the hypothalamic-pituitary-adrenal (HPA) axis could be supported (Keller et al., 2016) in both case-only models but not in the direct GxE interaction. Rs2433320 in PDLIM5 (PDZ and LIM domain containing 5) and rs16566369 in RSRC1 (arginine and serine rich coiled-coil 1) showed nominal significant results in at least two different approaches.
3. GWAS to identify GxE interaction loci
Meta-analysis of all four cohorts included nearly 4.3 million variants. An overview of the top loci in all three models, assuming an additive SNP-effect, is given in table 2. In all three meta-analysis no SNP achieved genome-wide significance (all p>5E-8). The top SNPs from the genome-wide CO approaches revealed no excess of gene-environment correlation (supplemental material, table S7).
Table 2.
List of top independent signals from the 3 meta-analyses (case-control GxE, case-only with abuse 0/1 and case-only with CTQ-score) assuming an additive SNP effect (p<E-5);
| RSID | MAF | p- value |
Effect | Genes nearby |
Alleles | CHR | I2 | direction | Gene information from NCBI resource |
|---|---|---|---|---|---|---|---|---|---|
| Case-only analysis (dichotomous childhood abuse 0/1)* | |||||||||
| rs17578476 | 0.27 | 3.3E-6 | −0.368 | LRRIQ3 | AC | chr1 | 0.0 | − − − − | A SNP at the LRRIQ3 locus has been associated with SCZ |
| rs6553019 | 0.40 | 5.1E-6 | 0.321 | FAT1 | AG | chr4 | 46 | + + + + | Probable function as an adhesion molecule or signaling receptor, and is likely to be important in developmental processes and cell communication; cadherine gene |
| rs10846719 | 0.49 | 5.6E-6 | 0.335 | SCARB1, NCOR2 | TC | chr12 | 0.0 | + + + ? | plasma membrane receptor for high density lipoprotein cholesterol (HDL) |
| rs10504765 | 0.39 | 9.5E-6 | −0.547 | - | AG | chr8 | 0.0 | − − ? − | |
| Case-only analysis (dimensional CTQ score)** | |||||||||
| rs3214187 | 0.13 | 7.4E-7 | −3.443 | NPY | D_I3 | chr7 | 48 | − − − − | widely expressed in the central nervous system and influences many physiological processes, including cortical excitability, stress response, food intake, circadian rhythms, and cardiovascular function |
| rs199719135 | 0.09 | 1.5E-6 | 6.044 | SH3BP4 | D_I11 | chr2 | 62 | + + ? + | involved in cargo-specific control of clathrin-mediated endocytosis |
| rs75184661 | 0.06 | 4.1E-6 | 5.479 | CACNA1C (intronic) | TC | chr12 | 0.0 | ? + + + | mediate the influx of calcium ions into the cell upon membrane polarization |
| rs6822352 | 0.34 | 6.8E-6 | 2.291 | KDR | AG | chr4 | 16 | + + + ? | VEGF receptor |
| rs6997589 | 0.26 | 7.7E-6 | −3.575 | SH2D4A (intronic) | AG | chr8 | 0.0 | − − ? − | |
| rs200510841 | 0.23 | 9.9E-6 | 3.787 | SEMA6D | D_I2 | chr15 | 0.0 | + + ? + | mediates transport to and from the nucleus |
| GxE analysis (dichotomous childhood abuse 0/1)* | |||||||||
| rs7128637 | 0.26 | 4.4E-6 | −0.880 | ARHGAP20 | CG | chr11 | 0.0 | − − ? − | |
| rs10772578 | 0.06 | 6.0E-6 | 1.268 | CREBL2 | TC | chr12 | 45 | + + + + | suggestions that CREBL2 encodes a protein with DNA binding capabilities and has tumor-suppressor properties |
| rs10808504 | 0.44 | 7.9E-6 | 0.755 | ENPP2 | TC | chr8 | 0.0 | + + ? + | regulating myelin formation |
log(OR);
beta.
I2 as measurement of heterogeneity between studies. Direction reporting the direction of effects in SHIP-0, TREND, NESDA, Radiant-UK.
Manhattan-plots for all three models are given in figure S1. The quantile-quantile plots showed a deflation of the observed results to those expected by chance (figure S2) and the λs were between 0.96 and 0.99. A full list of SNPs with p<1E-5 in all different approaches is given in table S3.
The top-hit in the case-control GxE approach on abuse 0/1 assuming an additive SNP effect was the variant rs7128637 near the ARHGAP20 gene, a Rho GTPase activating protein, with p=4.4E-6. The top hit in the case-only analysis on abuse 0/1 was rs17578476 (p=3.3E-6) harboring at LRRIQ3 locus, a locus previously implicated as one of the 108 loci associated with schizophrenia (Schizophrenia Working Group of the PGC, 2014). In the case-only analysis for CTQ score the minimum p-value was achieved for rs3214187 (p=7.4E-7) near the NPY (neuropeptide Y) gene. Another top variant from this analysis was rs75184661 (p=4.1E-6), intronic of CACNA1C (subunit of calcium voltage-gated channel). Results for the dominant/recessive case-only models can be found in supplementary table S3.
Exploratory comparison of all three approaches
The correlation between effect estimates (betas and log(OR)) of all three genome-wide approaches assuming an additive SNP effect was highest between both CO approaches on abuse 0/1 and CTQ-score (r=0.58), medium between both analyses on abuse 0/1 (CC and CO) (r=0.54) and lowest between the dimensional CO approach using CTQ score and the GxE interaction approach using abuse 0/1 (r=0.33). The overlap between SNPs with a notable p-value <0.001 is given in the Venn diagram (figure S3). Although in theory all approaches were applied to measure GxE interaction for CT in MDD, the overlap between all three analyses with p<0.001 was only one SNP (rs10504767) on chromosome 8 with no known gene nearby.
Lookup of candidate genes
We performed a gene-based test using VEGAS2 (Mishra and Macgregor, 2015) on the genome-wide summary statistics of the three main analyses assuming additive SNP-effects. The gene definition was set to ±10kb and all SNPs per gene were used for analysis. No gene was significant after correction for multiple testing and the top results contained no gene previously associated with a psychiatric phenotype (table S5). Also our list of candidate genes revealed not even a nominally significant (p<0.05) hit in the VEGAS2 results (tables S6).
DISCUSSION
This is the first genome-wide GxE interaction GWAS for depression and childhood trauma in subjects of European ancestry. The aims of this study were to validate candidate SNPs and genes for GxE in MDD while applying different model assumptions to acknowledge the variety of GxE models in previous candidate gene studies. Our methods included standard case-control GxE analysis with a multiplicative interaction term as well as case-only analyses with two different parametrizations of childhood trauma (dichotomous childhood abuse 0/1 and a continuous childhood trauma score) assuming an additive SNP-effect.
Two published GxE studies on depressive symptoms reported genome-wide significant SNPs in African American women (rs4652467) (Dunn et al., 2016) and in a Japanese population (rs1051057) (Otowa et al., 2016). We were not able to replicate these findings as rs4652467 only has a MAF <0.001 in populations of European ancestry and was therefore excluded from our analyses. The association between rs1051057 and MDD in this sample was non-significant, even at a nominal level. One explanation for our failure to replicate this finding could be due to the different phenotype definition, as we used lifetime MDD and not current depressive symptoms. As expected, due to the limited number of subjects, we were underpowered to identify robust genome-wide significant interaction effects in 3944 individuals. None of the genome-wide approaches suggested an inflation of the p-values in the QQ-plots (figure S2).
Overall, the candidate variants could not sufficiently be supported by our analyses; only 9% of the SNPs revealed a nominally significant effect in at least one of the three main approaches. Also the introduction of dominant and recessive SNP models led to no association with p<0.0003. Subsequent gene-based analyses on the summary statistics using VEGAS2 allowed for no biologically meaningful interpretation. These findings are also consistent with recent large-scale efforts to validate candidate genes, especially the 5-HTTLPR variant (Culverhouse et al., 2017). With such limited validation of candidate variants it seems questionable if the current approaches of candidate gene studies are the right tool to gain insights into the biology of gene-environment interactions in MDD. Our results suggest that published studies on candidate variants in GxE for MDD are in part likely subject to publication bias.
Methodological limitations and challenges
GxE studies face even larger methodological challenges than genetic association studies looking for main effects of SNPs.
Power: Our main limitation was the lack of power due to the limited sample size in our analysis which only allows for the robust identification of huge genetic effects which are not expected when analyzing common variants. – We tried to circumvent this limitation by focusing on previously reported candidate variants.
Assumptions behind GxE models: The methodological approaches that have been performed are based on different assumptions. In case-only analysis, independence between the genetic signal and the environmental factor is required. This might be a problem as studies suggest a significant heritability of childhood adversity through inherited ways of behavior. – Nevertheless, as previously shown for the MDD PGC wave2 data, SNP heritability for childhood trauma was estimated to be not significantly different from 0.00 in GRM based analyses (Peyrot et al., in preparation). Because of limited sample size, estimating the proportion of variance attributable to the interaction between CT and genome-wide genetic effects was not possible. But we also found no evidence for a gene-environment correlation for the top hits of our meta-analyses.
Heterogeneity across samples: The samples used in these analyses were taken from different settings, general population and clinical patients. – Although this might have biased the results, all subjects were of European ancestry, all subjects were screened for MDD with the same instrument, all controls were also screened for absence of MDD and childhood trauma was assessed using the same instrument (CTQ).
Childhood trauma measurement: As with many other measures for childhood trauma, the CTQ is a retrospective self-report measure and thus reports are likely to be influenced by recall bias and particularly depressive state. One solution would be to control for mood at the time of reporting in the analyses (Fisher et al., 2013). But other groups have found that depressive symptoms do not result in exaggeration of retrospectively recalled stressful events (Brewin et al., 1993; Fisher et al., 2011) and thus the use of retrospective self-reports is likely to have had only a minimal impact on these results. Nevertheless, this could be a source of bias leading to false positive results in our analyses.
GxE models: Although a number of different model assumptions were tested in this study, there are other models that need further investigation like the distinction between additive and multiplicative interaction models which is circumvented in the case-only approach as this method is assuming a multiplicative interaction. Also environmental factors different from childhood trauma such as social status or BMI could exhibit a GxE interaction as these are also risk factors for depression (Mansur et al., 2015; Schlossberg et al., 2010).
Lack of replication samples: We had no independent replication samples but we used different models to assess the robustness of the results. Unfortunately, no consistency between models was found.
Coverage of genetic variants: A GWAS approach does not cover all possible genetic variants that could contribute to depression, e.g. insertions, deletions or rare genetic variants. One of the most prominent examples is the serotonin-transporter polymorphism (Caspi et al., 2003).
Nevertheless, our analyses may provide some insights into GxE analyses and the genetic underpinning of gene-environment interactions in MDD as some of the top signals in the three main analyses involved genes previously implicated in psychiatric phenotypes (table 2) like ENPP2 (Aston et al., 2005), the LRRIQ3 locus identified in the latest GWAS for schizophrenia (Schizophrenia Working Group of the PGC, 2014), FAT1 (Light et al., 2007; Abou Jamra et al., 2008), NPY (Nakhate et al., 2016; Soleimani et al., 2015) and CACNA1C, a candidate gene in depression that also shows pleiotropic effects on other major psychiatric disorders (Rao et al., 2016; Cross-Disorder Group of the PGC, 2013). Although some of these genes are well known in psychiatric research, the significant SNPs from the analyses showed no overlap with previously identified candidate SNPs of these genes.
Finally, we can say that with the current sample size we were not able to detect a robust genome-wide significant interaction with childhood trauma and depression and most of the candidate variants and genes could not be supported when utilizing different methodological approaches. An important point of this analysis is the lack of replication, even if only nominal p-values are considered. Moreover, the analyses showed a lack of stability of findings in the different methodological approaches.
It will be necessary to collect more data on childhood trauma in MDD samples to validate our top hits and achieve a higher power to detect robust genome-wide significant findings. Also it might be prudent to at least partially question some of the former candidate SNP results for MDD as these could be attributed to publication bias and inconsistent models. One approach could be to harmonize the different CT measures throughout all of the PGC cohorts and perform the analyses with a much larger sample size. We also recommend reconsideration of the different models currently performed in GxE analyses for MDD and to perform consistent analyses in samples large enough to identify robust GxE interaction signals. Our next steps will be to perform the GxE analyses on single sub-dimensions of abuse where we have more data within the PGC as well as performing the analysis under the assumption of an additive interaction effect.
Supplementary Material
Acknowledgments
We thank all people participating in the studies.
Role of Funding Sources
The Netherlands Study of Depression and Anxiety (NESDA) and the Netherlands Twin Register (NTR): funding was obtained from the Netherlands Organization for Scientific Research (NWO) and MagW/ZonMW grants Middelgroot-911- 09-032, Spinozapremie 56-464-14192, Center for Medical Systems Biology (CSMB, NWO Genomics), Genetic influences on stability and change in psychopathology from childhood to young adulthood (ZonMW 912-10-020), NBIC/BioAssist/RK (2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI–NL, 184.021.007), VU University’s Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); the European Science Council (ERC Advanced, 230374). The infrastructure for the NESDA study (www.nesda.nl) is funded through the Geestkracht program of the Netherlands Organization for Health Research and Development (ZonMw, grant number 10-000-1002) and is supported by participating universities and mental health care organizations (VU University Medical Centre, GGZ inGeest, Arkin, Leiden University Medical Centre, GGZ Rivierduinen, University Medical Centre Groningen, Lentis, GGZ Friesland, GGZ Drenthe, Institute for Quality of Health Care (IQ Healthcare), Netherlands Institute for Health Services Research (NIVEL) and Netherlands Institute of Mental Health and Addiction (Trimbos)). Part of the genotyping of NESDA and NTR was funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health, Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH R01 HD042157-01A1, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995). Computing was supported by BiG Grid, the Dutch e- Science Grid, which is financially supported by NWO.
SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research Grant Nos. 01ZZ9603, 01ZZ0103, and 01ZZ0403; the Ministry of Cultural Affairs; and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide data were supported by the Federal Ministry of Education and Research Grant No. 03ZIK012 and a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG. SHIP-LEGEND is funded by the German Research Foundation (DFG: GR 1912/5-1). S. V. was supported by the German Federal Ministry of Education and Research within the framework of the e:Med research and funding concept (IntegraMent) Grant No. 01ZX1314E.
This report represents independent research part-funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health. The RADIANT studies were funded by a joint grant from the UK Medical Research Council, GlaxoSmithKline (G0701420) and by the National Institute for Health Research (NIHR) Biomedical Research Centre for Mental Health at South London and Maudsley NHS Foundation Trust and Institute of Psychiatry, Psychology and Neuroscience, King's College London. N.M. and C.M.L. have received funding from the European Community's Seventh Framework Programme under the Marie Curie Industry-Academia Partnership and Pathways (grant 286213). H.L.F. is supported by an MQ Fellows Award (MQ14F40). We thank all individuals who participated in the RADIANT study and all involved with data collection and management.
NRW acknowledges funding from the Australian National Health and Medical Research Council 1078901, 1087889.
The CoLaus|PsyCoLaus study was and is supported by research grants from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (grants 3200B0–105993, 3200B0-118308, 33CSCO-122661, 33CS30-139468 and 33CS30-148401).
Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (NWO 480-05-003 PI: Posthuma) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam.
Footnotes
Conflicts of interest
The authors declare that no conflicts of interest exist.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Jamra R, Becker T, Georgi A, Feulner T, Schumacher J, Stromaier J, Schirmbeck F, Schulze TG, Propping P, Rietschel M, Nöthen MM, Cichon S. Genetic variation of the FAT gene at 4q35 is associated with bipolar affective disorder. Mol Psychiatry. 2008;13(3):277–84. doi: 10.1038/sj.mp.4002111. [DOI] [PubMed] [Google Scholar]
- Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah M-H, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F. Accelerating Novel Candidate Gene Discovery in Neurogenetic Disorders via Whole-Exome Sequencing of Prescreened Multiplex Consanguineous Families. Cell Rep. 2015;10(2):148–61. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Appel K, Schwahn C, Mahler J, Schulz A, Spitzer C, Fenske K, Stender J, Barnow S, John U, Teumer A, Biffar R, Nauck M, Völzke H, Freyberger HJ, Grabe HJ. Moderation of adult depression by a polymorphism in the FKBP5 gene and childhood physical abuse in the general population. Neuropsychopharmacology. 2011;36(10):1982–91. doi: 10.1038/npp.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. FAT and bipolar affective disorder. Mol Psychiatry. 2007;12(10):899–900. doi: 10.1038/sj.mp.4002040. [DOI] [PubMed] [Google Scholar]
- Baune BT, Air T. Clinical, Functional, and Biological Correlates of Cognitive Dimensions in Major Depressive Disorder - Rationale, Design, and Characteristics of the Cognitive Function and Mood Study (CoFaM-Study) Front Psychiatry. 2016;26(7):150. doi: 10.3389/fpsyt.2016.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DP, Stein JA, Newcomb MD, Walker E, Pogge D, Ahluvalia T, Strokes J, Handelsman L, Medrano M, Desmond D, Zule W. Development and validation of a brief screening version of the childhood trauma questionnaire. Child Abuse Negl. 2003;27(2):169–190. doi: 10.1016/s0145-2134(02)00541-0. [DOI] [PubMed] [Google Scholar]
- Brewin C, Andrews B, Gotlib IH. Psychopathology and early experience: a reappraisal of retrospective reports. Psychological Bulletin. 1993;113:82–98. doi: 10.1037/0033-2909.113.1.82. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–9. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Cerdá M, Sagdeo A, Johnson J, Galea S. Genetic and environmental influences on psychiatric comorbidity: A systematic review. J Affect Disord. 2010;126(1–2):14–38. doi: 10.1016/j.jad.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miro X, Strohmaier J, Steffens M, Meesters C, Herms S, Weingarten M, Priebe L, Haenisch B, Alexander M, Vollmer J, Breuer R, Schmäl C, Tessmann P, Moebus S, Wichmann HE, Schreiber S, Müller-Myhsok B, Lucae S, Jamain S, Leboyer M, Bellivier F, Etain B, Henry C, Kahn JP, Heath S, Bipolar Disorder Genome Study (BiGS) Consortium. Hamshere M, O'Donovan MC, Owen MJ, Craddock N, Schwarz M, Vedder H, Kammerer-Ciernioch J, Reif A, Sasse J, Bauer M, Hautzinger M, Wright A, Mitchell PB, Schofield PR, Montgomery GW, Medland SE, Gordon SD, Martin NG, Gustafsson O, Andreassen O, Djurovic S, Sigurdsson E, Steinberg S, Stefansson H, Stefansson K, Kapur-Pojskic L, Oruc L, Rivas F, Mayoral F, Chuchalin A, Babadjanova G, Tiganov AS, Pantelejeva G, Abramova LI, Grigoroiu-Serbanescu M, Diaconu CC, Czerski PM, Hauser J, Zimmer A, Lathrop M, Schulze TG, Wienker TF, Schumacher J, Maier W, Propping P, Rietschel M, Nöthen MM. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–381. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523(7562):588–91. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse RC, Saccone NL, Horton AC, Ma Y, Anstey KJ, Banaschewski T, Burmeister M, Cohen-Woods S, Etain B, Fisher HL, Goldman N, Guillaume S, Horwood J, Juhasz G, Lester KJ, Mandelli L, Middeldorp CM, Olié E, Villafuerte S, Air TM, Araya R, Bowes L, Burns R, Byrne EM, Coffey C, Coventry WL, Gawronski KA, Glei D, Hatzimanolis A, Hottenga JJ, Jaussent I, Jawahar C, Jennen-Steinmetz C, Kramer JR, Lajnef M, Little K, Zu Schwabedissen HM, Nauck M, Nederhof E, Petschner P, Peyrot WJ, Schwahn C, Sinnamon G, Stacey D, Tian Y, Toben C, Van der Auwera S, Wainwright N, Wang JC, Willemsen G, Anderson IM, Arolt V, Åslund C, Bagdy G, Baune BT, Bellivier F, Boomsma DI, Courtet P, Dannlowski U, de Geus EJ, Deakin JF, Easteal S, Eley T, Fergusson DM, Goate AM, Gonda X, Grabe HJ, Holzman C, Johnson EO, Kennedy M, Laucht M, Martin NG, Munafò MR, Nilsson KW, Oldehinkel AJ, Olsson CA, Ormel J, Otte C, Patton GC, Penninx BW, Ritchie K, Sarchiapone M, Scheid JM, Serretti A, Smit JH, Stefanis NC, Surtees PG, Völzke H, Weinstein M, Whooley M, Nurnberger JI, Jr, Breslau N, Bierut LJ. Collaborative meta-analysis finds no evidence of a strong interaction between stress and 5-HTTLPR genotype contributing to the development of depression. Mol Psychiatry. 2017 Apr 4; doi: 10.1038/mp.2017.44. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168:1041–9. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EC, Uddin M, Subramanian SV, Smoller JW, Galea S, Koenen KC. Gene-environment interaction (GxE) research in youth depression: A systematic review with recommendations for future research. Journal of Child Psychology and Psychiatry. 2011;52(12):1223–1238. doi: 10.1111/j.1469-7610.2011.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EC, Brown RC, Dai Y, Rosand J, Nugent NR, Amstadter AB, Smoller JW. Genetic determinants of depression: Recent findings and future directions. Harv Rev Psychiatry. 2015;23(1):1–18. doi: 10.1097/HRP.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EC, Wiste A, Radmanesh F, Almli LM, Gogarten SM, Sofer T, Faul JD, Kardia SL, Smith JA, Weir DR, Zhao W, Soare TW, Mirza SS, Hek K, Tiemeier H, Goveas JS, Sarto GE, Snively BM, Cornelis M, Koenen KC, Kraft P, Purcell S, Ressler KJ, Rosand J, Wassertheil-Smoller S, Smoller JW. Genome-wide Association Study (GWAS) and Genome-wide by Environment Interaction Study (GWEIS) of Depressive Symptoms in African American and Hispanic/Latina Woman. Depress Anxiety. 2016;33(4):265–80. doi: 10.1002/da.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher HL, Cohen-Woods S, Hosang GM, Korszun A, Owen M, Craddock N, Craig IW, Farmer AE, McGuffin P, Uher R. Interaction between specific forms of childhood maltreatment and the serotonin transporter gene (5-HTT) in recurrent depressive disorder. J Affect Disord. 2013;145(1):136–41. doi: 10.1016/j.jad.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher HL, Craig TK, Fearon P, Morgan K, Dazzan P, Lappin J, Hutchinson G, Doody GA, Jones PB, McGuffin P, Murray RM, Leff J, Morgan C. Reliability and comparability of psychosis patients’ retrospective reports of childhood abuse. Schizophrenia Bulletin. 2011;37(3):546–553. doi: 10.1093/schbul/sbp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Kendler KS. The Genetics of Major Depression. Neuron. 2014;81(3):484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Zhang P, Morrison JL, Lewinger JP. Finding novel genes by testing G × E interactions in a genome-wide association study. Genet Epidemiol. 2013;37(6):603–13. doi: 10.1002/gepi.21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe HJ, Schwahn C, Mahler J, Appel K, Schulz A, Spitzer C, Fenske K, Barnow S, Freyberger HJ, Teumer A, Petersmann A, Biffar R, Rosskopf D, John U, Völzke H. Genetic epistasis between the brain-derived neurotrophic factor Val66Met polymorphism and the 5-HTT promoter polymorphism moderates the susceptibility to depressive disorders after childhood abuse. Prog Neuropsychopharmacol Biol Psychiatry. 2012;36(2):264–70. doi: 10.1016/j.pnpbp.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC, Amin N, Bakshis E, Baumert J, Ding J, Liu Y, Marciante K, Meirelles O, Nalls MA, Sun YV, Vogelzangs N, Yu L, Bandinelli S, Benjamin EJ, Bennett DA, Boomsma D, Cannas A, Coker LH, de Geus E, De Jager PL, Diez-Roux AV, Purcell S, Hu FB, Rimm EB, Hunter DJ, Jensen MK, Curhan G, Rice K, Penman AD, Rotter JI, Sotoodehnia N, Emeny R, Eriksson JG, Evans DA, Ferrucci L, Fornage M, Gudnason V, Hofman A, Illig T, Kardia S, Kelly-Hayes M, Koenen K, Kraft P, Kuningas M, Massaro JM, Melzer D, Mulas A, Mulder CL, Murray A, Oostra BA, Palotie A, Penninx B, Petersmann A, Pilling LC, Psaty B, Rawal R, Reiman EM, Schulz A, Shulman JM, Singleton AB, Smith AV, Sutin AR, Uitterlinden AG, Völzke H, Widen E, Yaffe K, Zonderman AB, Cucca F, Harris T, Ladwig KH, Llewellyn DJ, Räikkönen K, Tanaka T, van Duijn CM, Grabe HJ, Launer LJ, Lunetta KL, Mosley TH, Jr, Newman AB, Tiemeier H, Murabito J. A genome-wide association study of depressive symptoms. Biol Psychiatry. 2012;73(7):667–78. doi: 10.1016/j.biopsych.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, Depaulo JR, Jr, Knowles JA, Zubenko WN, Murphy-Eberenz K, Marta DH, Boutelle S, McInnis MG, Adams P, Gladis M, Steele J, Miller EB, Potash JB, Mackinnon DF, Levinson DF. Genetics of recurrent early-onset major depression (GenRED): final genome scan report. Am J Psychiatry. 2007;164(2):248–58. doi: 10.1176/ajp.2007.164.2.248. [DOI] [PubMed] [Google Scholar]
- Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, Tung JY, Hinds DA, Perlis RH, Winslow AR. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016 doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry. 2011;68(5):444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J, Gomez R, Williams G, Lembke A, Lazzeroni L, Murphy GM, Jr, Schatzberg AF. HPA axis in major depression: cortisol, clinical symptomatology and genetic variation predict cognition. Mol Psychiatry doi. 2016 doi: 10.1038/mp.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC. Gene × environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol Psychiatry. 2014;75(1):18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, Witte JS, Absher D, Agartz I, Akil H, Amin F, Andreassen OA, Anjorin A, Anney R, Anttila V, Arking DE, Asherson P, Azevedo MH, Backlund L, Badner JA, Bailey AJ, Banaschewski T, Barchas JD, Barnes MR, Barrett TB, Bass N, Battaglia A, Bauer M, Bayés M, Bellivier F, Bergen SE, Berrettini W, Betancur C, Bettecken T, Biederman J, Binder EB, Black DW, Blackwood DH, Bloss CS, Boehnke M, Boomsma DI, Breen G, Breuer R, Bruggeman R, Cormican P, Buccola NG, Buitelaar JK, Bunney WE, Buxbaum JD, Byerley WF, Byrne EM, Caesar S, Cahn W, Cantor RM, Casas M, Chakravarti A, Chambert K, Choudhury K, Cichon S, Cloninger CR, Collier DA, Cook EH, Coon H, Cormand B, Corvin A, Coryell WH, Craig DW, Craig IW, Crosbie J, Cuccaro ML, Curtis D, Czamara D, Datta S, Dawson G, Day R, De Geus EJ, Degenhardt F, Djurovic S, Donohoe GJ, Doyle AE, Duan J, Dudbridge F, Duketis E, Ebstein RP, Edenberg HJ, Elia J, Ennis S, Etain B, Fanous A, Farmer AE, Ferrier IN, Flickinger M, Fombonne E, Foroud T, Frank J, Franke B, Fraser C, Freedman R, Freimer NB, Freitag CM, Friedl M, Frisén L, Gallagher L, Gejman PV, Georgieva L, Gershon ES, Geschwind DH, Giegling I, Gill M, Gordon SD, Gordon-Smith K, Green EK, Greenwood TA, Grice DE, Gross M, Grozeva D, Guan W, Gurling H, De Haan L, Haines JL, Hakonarson H, Hallmayer J, Hamilton SP, Hamshere ML, Hansen TF, Hartmann AM, Hautzinger M, Heath AC, Henders AK, Herms S, Hickie IB, Hipolito M, Hoefels S, Holmans PA, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hultman CM, Hus V, Ingason A, Ising M, Jamain S, Jones EG, Jones I, Jones L, Tzeng JY, Kähler AK, Kahn RS, Kandaswamy R, Keller MC, Kennedy JL, Kenny E, Kent L, Kim Y, Kirov GK, Klauck SM, Klei L, Knowles JA, Kohli MA, Koller DL, Konte B, Korszun A, Krabbendam L, Krasucki R, Kuntsi J, Kwan P, Landén M, Långström N, Lathrop M, Lawrence J, Lawson WB, Leboyer M, Ledbetter DH, Lee PH, Lencz T, Lesch KP, Levinson DF, Lewis CM, Li J, Lichtenstein P, Lieberman JA, Lin DY, Linszen DH, Liu C, Lohoff FW, Loo SK, Lord C, Lowe JK, Lucae S, MacIntyre DJ, Madden PA, Maestrini E, Magnusson PK, Mahon PB, Maier W, Malhotra AK, Mane SM, Martin CL, Martin NG, Mattheisen M, Matthews K, Mattingsdal M, McCarroll SA, McGhee KA, McGough JJ, McGrath PJ, McGuffin P, McInnis MG, McIntosh A, McKinney R, McLean AW, McMahon FJ, McMahon WM, McQuillin A, Medeiros H, Medland SE, Meier S, Melle I, Meng F, Meyer J, Middeldorp CM, Middleton L, Milanova V, Miranda A, Monaco AP, Montgomery GW, Moran JL, Moreno-De-Luca D, Morken G, Morris DW, Morrow EM, Moskvina V, Muglia P, Mühleisen TW, Muir WJ, Müller-Myhsok B, Murtha M, Myers RM, Myin-Germeys I, Neale MC, Nelson SF, Nievergelt CM, Nikolov I, Nimgaonkar V, Nolen WA, Nöthen MM, Nurnberger JI, Nwulia EA, Nyholt DR, O'Dushlaine C, Oades RD, Olincy A, Oliveira G, Olsen L, Ophoff RA, Osby U, Owen MJ, Palotie A, Parr JR, Paterson AD, Pato CN, Pato MT, Penninx BW, Pergadia ML, Pericak-Vance MA, Pickard BS, Pimm J, Piven J, Posthuma D, Potash JB, Poustka F, Propping P, Puri V, Quested DJ, Quinn EM, Ramos-Quiroga JA, Rasmussen HB, Raychaudhuri S, Rehnström K, Reif A, Ribasés M, Rice JP, Rietschel M, Roeder K, Roeyers H, Rossin L, Rothenberger A, Rouleau G, Ruderfer D, Rujescu D, Sanders AR, Sanders SJ, Santangelo SL, Sergeant JA, Schachar R, Schalling M, Schatzberg AF, Scheftner WA, Schellenberg GD, Scherer SW, Schork NJ, Schulze TG, Schumacher J, Schwarz M, Scolnick E, Scott LJ, Shi J, Shilling PD, Shyn SI, Silverman JM, Slager SL, Smalley SL, Smit JH, Smith EN, Sonuga-Barke EJ, St Clair D, State M, Steffens M, Steinhausen HC, Strauss JS, Strohmaier J, Stroup TS, Sutcliffe JS, Szatmari P, Szelinger S, Thirumalai S, Thompson RC, Todorov AA, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Van Os J, Vicente AM, Vieland VJ, Vincent JB, Visscher PM, Walsh CA, Wassink TH, Watson SJ, Weissman MM, Werge T, Wienker TF, Wijsman EM, Willemsen G, Williams N, Willsey AJ, Witt SH, Xu W, Young AH, Yu TW, Zammit S, Zandi PP, Zhang P, Zitman FG, Zöllner S, Devlin B, Kelsoe JR, Sklar P, Daly MJ, O'Donovan MC, Craddock N, Sullivan PF, Smoller JW, Kendler KS, Wray NR, et al. International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekman M, Hössjer O, Andrews P, Källberg H, Uvehag D, Charney D, Manji H, Rush JA, McMahon FJ, Moore JH, Kockum I. The genetic interacting landscape of 63 candidate genes in Major Depressive Disorder: an explorative study. BioData Mining. 2014;7:19. doi: 10.1186/1756-0381-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, Weale ME, Schosser A, Paredes UM, Rivera M, Craddock N, Owen MJ, Jones L, Jones I, Korszun A, Aitchison KJ, Shi J, Quinn JP, Mackenzie A, Vollenweider P, Waeber G, Heath S, Lathrop M, Muglia P, Barnes MR, Whittaker JC, Tozzi F, Holsboer F, Preisig M, Farmer AE, Breen G, Craig IW, McGuffin P. Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry. 2010;167:949–57. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- Light KJ, Miller AL, Doughty CJ, Joyce PR, Olds RJ, Kennedy MA. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10(3):309–22. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- Lubke GH, Hottenga JJ, Walters R, Laurin C, de Geus EJ, Willemsen G, Smit JH, Middeldorp CM, Penninx BW, Vink JM, Boomsma DI. Estimating the genetic variance of major depressive disorder due to all single nucleotide polymorphisms. Biol Psychiatry. 2012;15;72(8):707–9. doi: 10.1016/j.biopsych.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Stavrakakis N, Penninx BW, Bosker FJ, Nolen WA, Boomsma DI, de Geus EJ, Smit JH, Snieder H, Nolte IM, Hartman C. Does refining the phenotype improve replication rates? A review and replication of candidate gene studies on Major Depressive Disorder and Chronic Major Depressive Disorder. Am J Med Genet B Neuropsychiatr Genet. 2016;171B(2):215–36. doi: 10.1002/ajmg.b.32396. [DOI] [PubMed] [Google Scholar]
- Mandelli L, Petrelli C, Serretti A. The role of specific early trauma in adult depression: A meta-analysis of published literature. Childhood trauma and adult depression. Eur Psychiatry. 2015;30(6):665–80. doi: 10.1016/j.eurpsy.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Mandelli L, Serretti A. Gene environment interaction studies in depression and suicidal behavior: An update. Neurosci Biobehav Rev. 2013;37(10 Pt 1):2375–97. doi: 10.1016/j.neubiorev.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Mansur RB1, Brietzke E2, McIntyre RS. Is there a "metabolic-mood syndrome"? A review of the relationship between obesity and mood disorders. Neurosci Biobehav Rev. 2015;52:89–104. doi: 10.1016/j.neubiorev.2014.12.017. [DOI] [PubMed] [Google Scholar]
- Mishra A, Macgregor S. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Research and Human Genetics. 2015:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Battle A, Zhu X, Potash JB, Weissman MM, Shi J, Beckman K, Haudenschild C, McCormick C, Mei R, Gameroff MJ, Gindes H, Adams P, Goes FS, Mondimore FM, MacKinnon DF, Notes L, Schweizer B, Furman D, Montgomery SB, Urban AE, Koller D, Levinson DF. Type I Interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing. Mol Psychiatry. 2014;19:1267–74. doi: 10.1038/mp.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhate KT, Yedke SU, Bharne AP, Subhedar NK, Kokare DM. Evidence for the involvement of neuropeptide Y in the antidepressant effect of imipramine in type 2 diabetes. Brain Res. 2016 doi: 10.1016/j.brainres.2016.05.035. pii: S0006-8993(16)30390-0. [DOI] [PubMed] [Google Scholar]
- Nelson EC, Heath AC, Madden PA, Cooper ML, Dinwiddie SH, Bucholz KK, Glowinski A, McLaughlin T, Dunne MP, Statham DJ, Martin NG. Association between self-reported childhood sexual abuse and adverse psychosocial outcomes: results from a twin study. Arch Gen Psychiatry. 2002;59(2):139–45. doi: 10.1001/archpsyc.59.2.139. [DOI] [PubMed] [Google Scholar]
- Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, Meddens SF, Linnér RK, Rietveld CA, Derringer J, Gratten J, Lee JJ, Liu JZ, de Vlaming R, Ahluwalia TS, Buchwald J, Cavadino A, Frazier-Wood AC, Furlotte NA, Garfield V, Geisel MH, Gonzalez JR, Haitjema S, Karlsson R, van der Laan SW, Ladwig KH, Lahti J, van der Lee SJ, Lind PA, Liu T, Matteson L, Mihailov E, Miller MB, Minica CC, Nolte IM, Mook-Kanamori D, van der Most PJ, Oldmeadow C, Qian Y, Raitakari O, Rawal R, Realo A, Rueedi R, Schmidt B, Smith AV, Stergiakouli E, Tanaka T, Taylor K, Wedenoja J, Wellmann J, Westra HJ, Willems SM, Zhao W, LifeLines Cohort Study. Amin N, Bakshi A, Boyle PA, Cherney S, Cox SR, Davies G, Davis OS, Ding J, Direk N, Eibich P, Emeny RT, Fatemifar G, Faul JD, Ferrucci L, Forstner A, Gieger C, Gupta R, Harris TB, Harris JM, Holliday EG, Hottenga JJ, De Jager PL, Kaakinen MA, Kajantie E, Karhunen V, Kolcic I, Kumari M, Launer LJ, Franke L, Li-Gao R, Koini M, Loukola A, Marques-Vidal P, Montgomery GW, Mosing MA, Paternoster L, Pattie A, Petrovic KE, Pulkki-Råback L, Quaye L, Räikkönen K, Rudan I, Scott RJ, Smith JA, Sutin AR, Trzaskowski M, Vinkhuyzen AE, Yu L, Zabaneh D, Attia JR, Bennett DA, Berger K, Bertram L, Boomsma DI, Snieder H, Chang SC, Cucca F, Deary IJ, van Duijn CM, Eriksson JG, Bültmann U, de Geus EJ, Groenen PJ, Gudnason V, Hansen T, Hartman CA, Haworth CM, Hayward C, Heath AC, Hinds DA, Hyppönen E, Iacono WG, Järvelin MR, Jöckel KH, Kaprio J, Kardia SL, Keltikangas-Järvinen L, Kraft P, Kubzansky LD, Lehtimäki T, Magnusson PK, Martin NG, McGue M, Metspalu A, Mills M, de Mutsert R, Oldehinkel AJ, Pasterkamp G, Pedersen NL, Plomin R, Polasek O, Power C, Rich SS, Rosendaal FR, den Ruijter HM, Schlessinger D, Schmidt H, Svento R, Schmidt R, Alizadeh BZ, Sørensen TI, Spector TD, Steptoe A, Terracciano A, Thurik AR, Timpson NJ, Tiemeier H, Uitterlinden AG, Vollenweider P, Wagner GG, Weir DR, Yang J, Conley DC, Smith GD, Hofman A, Johannesson M, Laibson DI, Medland SE, Meyer MN, Pickrell JK, Esko T, Krueger RF, Beauchamp JP, Koellinger PD, Benjamin DJ, Bartels M, Cesarini D. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48(6):624–33. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otowa T, Kawamura Y, Tsutsumi A, Kawakami N, Kan C, Shimada T, Umekage T, Kasai K, Tokunaga K, Sasaki T. The First Pilot Genome-Wide Gene-Environment Study of Depression in the Japanese Population. PLoS One. 2016;11(8):e0160823. doi: 10.1371/journal.pone.0160823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninx BWJH, Beekman ATF, Smit JH, Zitman FG, Nolen WA, Spinhoven P, Cuijpers P, De Jong PJ, Van Marwijk HW, Assendelft WJ, Van Der Meer K, Verhaak P, Wensing M, De Graaf R, Hoogendijk WJ, Ormel J, Van Dyck R NESDA Research Consortium. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Methods Psychiatr Res. 2008;17:121–40. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrot WJ, Van der Auwera S, Milaneshi Y, The childhood trauma working-group of PGC-MDD wave 2. Dolan CV, Boomsma DI, Wray NR, Grabe HJ, Penninx BWJH. Evaluating childhood trauma and polygenic risk for major depressive disorder in approximately 5,765 subjects. In preparation. [Google Scholar]
- Preisig M, Waeber G, Vollenweider P, Bovet P, Rothen S, Vandeleur C, Guex P, Middleton L, Waterworth D, Mooser V, Tozzi F, Muglia P. The PsyCoLaus study: methodology and characteristics of the sample of a population-based survey on psychiatric disorders and their association with genetic and cardiovascular risk factors. BMC Psychiatry. 2009;17(9):9. doi: 10.1186/1471-244X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric Genomics Consortium. Major Depressive Disorder: wave 2 GWAS results. In preperation. [Google Scholar]
- Psychiatric GWAS Consortium Steering Committee. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14(1):10–7. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- Rao S, Yao Y, Zheng C, Ryan J, Mao C, Zhang F, Meyre D, Xu Q. Common variants in CACNA1C and MDD susceptibility: A comprehensive meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2016 doi: 10.1002/ajmg.b.32466. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani L, Oquendo MA, Sullivan GM, Mathé AA, Mann JJ. Cerebrospinal fluid neuropeptide Y levels in major depression and reported childhood trauma. Int J Neuropsychopharmacol. 2014;18(1) doi: 10.1093/ijnp/pyu023. pii: pyu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera S, Janowitz D, Schulz A, Homuth G, Nauck M, Völzke H, Rose M, Meyer zu Schwabedissen H, Freyberger HJ, Grabe HJ. Interaction among childhood trauma and functional polymorphisms in the serotonin pathway moderate the risk of depressive disorders. Eur Arch Psychiatry Clin Neurosci. 2014;264(Suppl 1):S45–54. doi: 10.1007/s00406-014-0536-2. [DOI] [PubMed] [Google Scholar]
- VanderWeele TJ. Explanation in Causal Inference: Methods for Mediation and Interaction. Oxford University Press; 2015. [Google Scholar]
- VanderWeele TJ, Hernández-Díaz S, Hernán MA. Case-only gene- environment interaction studies: when does association imply mechanistic interaction? Genet Epidemiol. 2010;34(4):327–34. doi: 10.1002/gepi.20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, Friedrich N, Aumann N, Lau K, Piontek M, Born G, Havemann C, Ittermann T, Schipf S, Haring R, Baumeister SE, Wallaschofski H, Nauck M, Frick S, Arnold A, Jünger M, Mayerle J, Kraft M, Lerch MM, Dörr M, Reffelmann T, Empen K, Felix SB, Obst A, Koch B, Gläser S, Ewert R, Fietze I, Penzel T, Dören M, Rathmann W, Haerting J, Hannemann M, Röpcke J, Schminke U, Jürgens C, Tost F, Rettig R, Kors JA, Ungerer S, Hegenscheid K, Kühn JP, Kühn J, Hosten N, Puls R, Henke J, Gloger O, Teumer A, Homuth G, Völker U, Schwahn C, Holtfreter B, Polzer I, Kohlmann T, Grabe HJ, Rosskopf D, Kroemer HK, Kocher T, Biffar R, John U, Hoffmann W. Cohort profile: the study of health in Pomerania. Int J Epidemiol. 2011;40:294–307. doi: 10.1093/ije/dyp394. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DHR, Penninx BWJH, Gordon SD, Nyholt DR, Ripke S, MacIntyre DJ, McGhee KA, Maclean AW, Smit JH, Hottenga JJ, Willemsen G, Middeldorp CM, de Geus EJ, Lewis CM, McGuffin P, Hickie IB, van den Oord EJ, Liu JZ, Macgregor S, McEvoy BP, Byrne EM, Medland SE, Statham DJ, Henders AK, Heath AC, Montgomery GW, Martin NG, Boomsma DI, Madden PA, Sullivan PF. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.