Abstract
Background
Atazanavir causes plasma indirect bilirubin to increase. We evaluated associations between Gilbert’s polymorphism and bilirubin-related atazanavir discontinuation stratified by race/ethnicity.
Patients and methods
Patients had initiated atazanavir/ritonavir-containing regimens at an HIV primary care clinic in the southeastern USA, and had at least 12 months of follow-up data. Metabolizer group was defined by UGT1A1 rs887829 C→T. Genome-wide genotype data were used to adjust for genetic ancestry in combined population analyses.
Results
Among 321 evaluable patients, 15 (4.6%) had bilirubin-related atazanavir discontinuation within 12 months. Homozygosity for rs887829 T/T was present in 28.1% of Black, 21.4% of Hispanic, and 8.6% of White patients. Among all patients the hazard ratio (HR) for bilirubin-related discontinuation with T/T versus C/C genotype was 7.3 [95% confidence interval (CI): 1.7–31.5; P=0.007]. Among 152 White patients the HR was 14.4 (95% CI: 2.6–78.7; P=0.002), but among 153 Black patients the HR was 0.8 (95% CI: 0.05–12.7; P=0.87).
Conclusion
Among patients who initiated atazanavir/ritonavir-containing regimens, UGT1A1 slow metabolizer genotype rs887829 T/T was associated with increased bilirubin-related discontinuation of atazanavir in White but not in Black patients, this despite T/T genotype being more frequent in Black patients.
Keywords: atazanavir, HIV, jaundice, pharmacogenomics, UGT1A1
Introduction
The HIV-1 protease inhibitor atazanavir, with pharmacokinetic enhancement by either ritonavir (atazanavir/r) or cobicistat (atazanavir/c), is generally safe and effective as a first-line regimen for HIV-1 infection 1–5. Atazanavir inhibits bilirubin glucuronidation by hepatic uridine diphosphate glucuronosyltransferase (UGT) 1A1, which causes plasma indirect bilirubin concentrations to increase 6. Although this does not reflect hepatic injury 4,7–9, some patients discontinue atazanavir because of cosmetic jaundice 3,10,11. In AIDS Clinical Trials Group (ACTG) protocol A5257, patients who were randomized to the atazanavir/r-containing arm experienced more toxicity-related drug discontinuations than patients who were randomized to the darunavir/r-containing arm, with the difference driven by jaundice or increased blood bilirubin 12. It was mainly the results from protocol A5257 that led to an update to US prescribing guidelines in 2015, with atazanavir/r-containing regimens changed from recommended to alternative status as an initial therapy for HIV-1 infection, whereas darunavir/r-containing regimens remained recommended 4.
Polymorphisms in UGT1A1 are associated with indirect bilirubin concentrations in the general population (i.e. Gilbert’s syndrome). A promoter tandem TA repeat, UGT1A1*28 (TA)7, is associated with reduced UGT1A1 transcription versus UGT1A1*1 (TA)6, as is the far less frequent UGT1A1*37 (TA)8 13,14. Among atazanavir recipients, UGT1A1*28 has been associated with unconjugated hyperbilirubinemia 15,16, as has a C→T polymorphism (rs887829) that is in almost complete linkage disequilibrium with UGT1A1*28 17. In the latter study, the UGT1A1*37 allele, which was present in 19 individuals, was perfectly tagged by the rs887829 T allele (D.W.H., personal communication, 10 June 2017). In a genome-wide study involving individuals who had been randomized to atazanavir-containing regimens in ACTG protocol A5202, rs887829 T/T genotype and baseline indirect bilirubin were independently associated with peak on-treatment total bilirubin concentration 17. An analysis from the Swiss HIV Cohort Study found an association between Gilbert’s polymorphism and atazanavir discontinuation 18, although a follow-up study based on data from ACTG protocol A5202 did not replicate this finding 16. A subsequent study based on ACTG protocol A5257 showed that, among 481 participants who were randomized to initiate atazanavir/r, the likelihood of bilirubin-related discontinuation of atazanavir/r was low with UGT1A1 rs887829 non-T/T genotypes (i.e. C/C or C/T), but substantially higher with rs887829 T/T, particularly among White participants 19. In 2015, informed largely by the genetic association analysis of protocol A5257, the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommended that providers consider prescribing an agent other than atazanavir among UGT1A1 poor metabolizers, particularly where jaundice would be of a concern to the patient, but no need to avoid atazanavir with other genotypes 20.
The present study examined associations between UGT1A1 rs887829 genotype and bilirubin-related atazanavir/r discontinuation among clients of a large HIV primary care clinic in the Southeastern USA, and whether associations differed by race/ethnicity. Genome-wide genotype data were used to minimize confounding by population stratification.
Patients and methods
Study participants
This is an observational cohort study of patients who had initiated atazanavir/r-containing regimens for HIV-1 infection at the Vanderbilt Comprehensive Care Clinic in Nashville, Tennessee, USA. Eligible participants had at least 12-month of follow-up data, had provided informed consent for genetic research, and had stored DNA available for analysis. Clinical and laboratory data were extracted from de-identified electronic medical records and reviewed independently by two study investigators with no knowledge of genotype data (P.L. and D.W.H.) to determine causes of atazanavir discontinuation. Of 349 patients who initiated atazanavir-containing regimens, no clear reason for atazanavir discontinuation was found for seven individuals, and three discontinued atazanavir for elevated liver function tests but not clearly related to bilirubin. An additional 18 individuals were lost to follow-up within the first 12 months while receiving atazanavir and were censored. This study was approved by the Vanderbilt University Institutional Review Board.
Genotyping
Stored DNA from a total of 837 patients who had initiated either atazanavir-containing regimens (including patients in the present association analysis) or efavirenz-containing regimens (from a previous analysis 21) was genotyped for 535 543 single nucleotide polymorphisms, including rs887829 C→T, by Illumina HumanCore Exome assay (San Diego, California, USA). Genotype call rates exceeded 99% for 807 samples, including 349 individuals for the current analysis. Laboratory personnel with no knowledge of clinical data performed genotyping. The rs887829 polymorphisms, genotyped as G→A, is represented as C→T hereafter for consistency with past publications.
Multifactor dimensionality scaling
To account for possible population stratification, whole genome data were used to generate multidimensional scaling (MDS) coordinates in PLINK 22. Over 500 000 polymorphisms available from 224 individuals in the current study as well as 582 additional participants from a previous efavirenz study were used to generate MDS coordinates. The QC process was reported elsewhere 21.
Statistical analyses
Baseline characteristics of study participants are presented as median and interquartile ranges. Log-rank test (χ2) was used to assess the correlation between genotype and binomial variables. Cox proportional hazard regression model, adjusted for population stratification based on MDS coordinates, was used to examine associations between genotype and bilirubin-related atazanavir discontinuation. Analyses considered three metabolizer groups, slow, intermediate and extensive based on rs887829 T/T, C/T, and C/C genotypes, respectively. All analyses used a 5% two-sided significance level and were performed using Stata/IC, version 14.1 (StataCorp LLC, College Station, Texas, USA).
Results
Study cohort
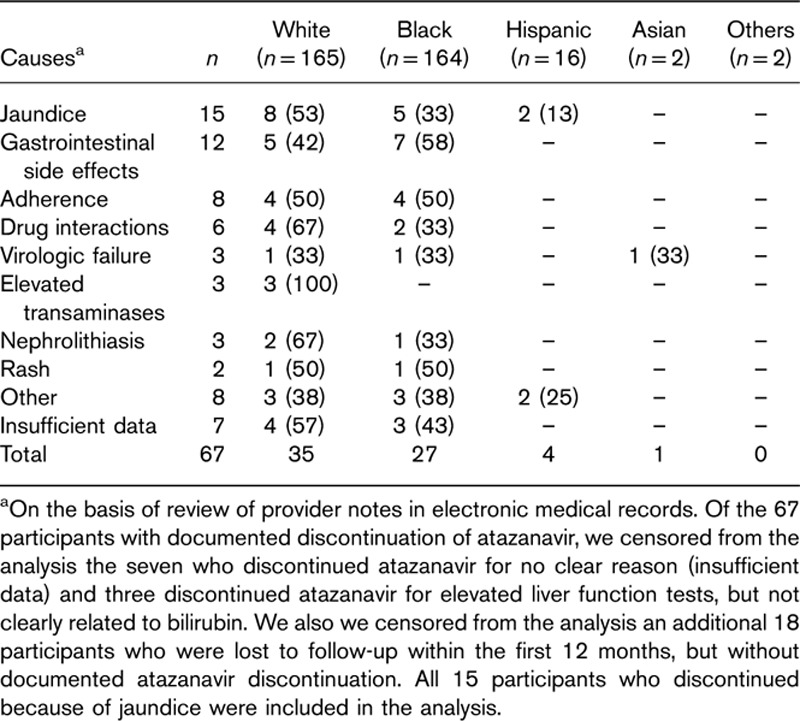
From 2003 to 2012, a total of 349 patients initiated atazanavir-containing regimens at the Vanderbilt Comprehensive Care Clinic. Among them, seven had discontinued atazanavir treatment for no clear reason, three discontinued atazanavir for elevated liver function tests but not clearly related to bilirubin, and 18 were lost to follow-up within the first 12 months while receiving atazanavir. These 28 patients were censored from the analysis. A total of 321 eligible patients had genotype data that passed quality control. Of these 321 patients, 57 (17%) permanently discontinued atazanavir within the first year of therapy, whereas 264 continued to receive atazanavir for at least 12 months. Baseline demographics of study participants are shown in Table 1, and generally reflect the demographics of the Vanderbilt Comprehensive Care Clinic during the study period. Of the 321 patients with MDS-derived genetic ancestry data, 152 (47%) were White, 153 (48%) were Black, 14 (4%) were Hispanic, and two (1%) were Asian. Scatter plots of MDS coordinates were reported elsewhere 21. Of the initial 67 patients who discontinued atazanavir, the primary cause was bilirubin-related in 15 (22.4%) patients. Other causes of atazanavir discontinuation are reported in Table 2.
Table 1.
Baseline demographics of study participants
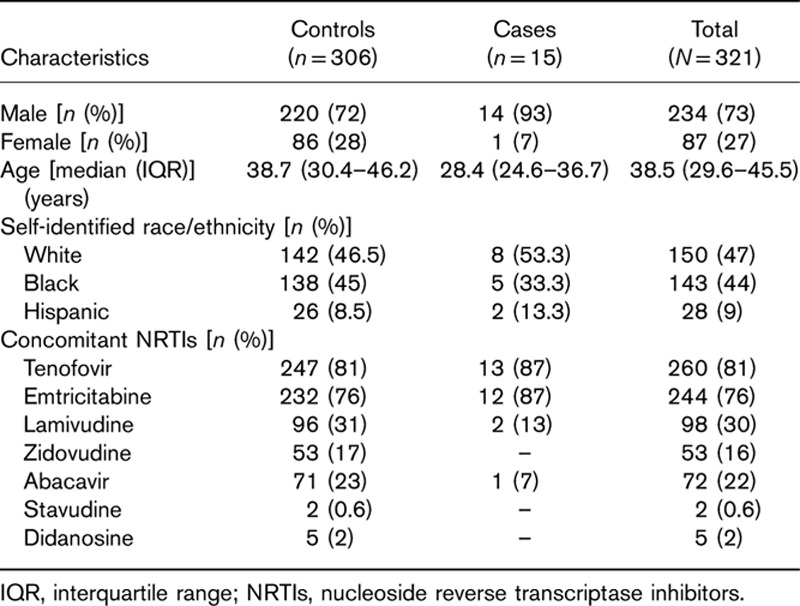
Table 2.
Causes of atazanavir discontinuation stratified by race/ethnicity
UGT1A1 genotypes
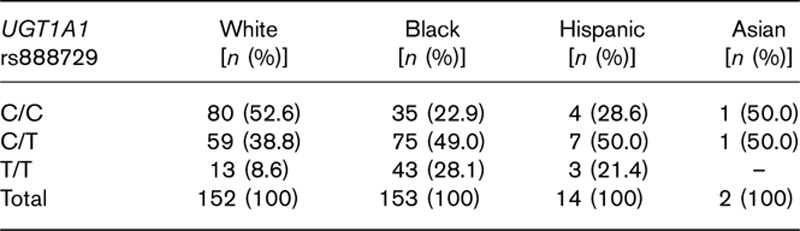
Among the 321 evaluable patients, 120 (37.5%) had rs887829 C/C, 142 (44%) had C/T, and 59 (18.5%) had T/T genotype. Among 59 patients homozygous for rs887829 T/T, 43 (73%) were Black, 13 (22%) were White, and three (5%) were Hispanic. Homozygosity for rs887829 T/T was present in 28.1% of Black, 21.4% of Hispanic, and 8.6% of White patients (Table 3).
Table 3.
Genotype frequencies stratified by race/ethnicity
Associations with atazanavir discontinuation
Among the 321 evaluable patients, in analyses adjusted for the first two MDS coordinates, the hazard ratio (HR) for bilirubin-related atazanavir discontinuation with T/T genotype was 7.4 [95% confidence interval (CI): 1.7–31.5; P=0.007], and with C/T genotype was 1.5 (95% CI: 0.5–8.4; P=0.30). Time to bilirubin-related atazanavir discontinuation stratified by genotype is shown in Fig. 1a. Among 152 White patients, unadjusted HRs for discontinuation with T/T and C/T genotypes were 14.4 (95% CI: 2.6–78.7; P=0.002) and 1.3 (95% CI: 0.2–9.4; P=0.78), respectively (Fig. 1b). Among 153 Black patients, unadjusted HRs with T/T and C/T genotypes were 0.8 (95% CI: 0.1–12.7; P=0.87) and 1.4 (95% CI: 0.1–13.6; P=0.77), respectively (Fig. 1). Among 14 Hispanic patients, two had bilirubin-related atazanavir discontinuation, including one of three with T/T, and one of seven with C/T genotypes (Fig. 1d).
Fig. 1.
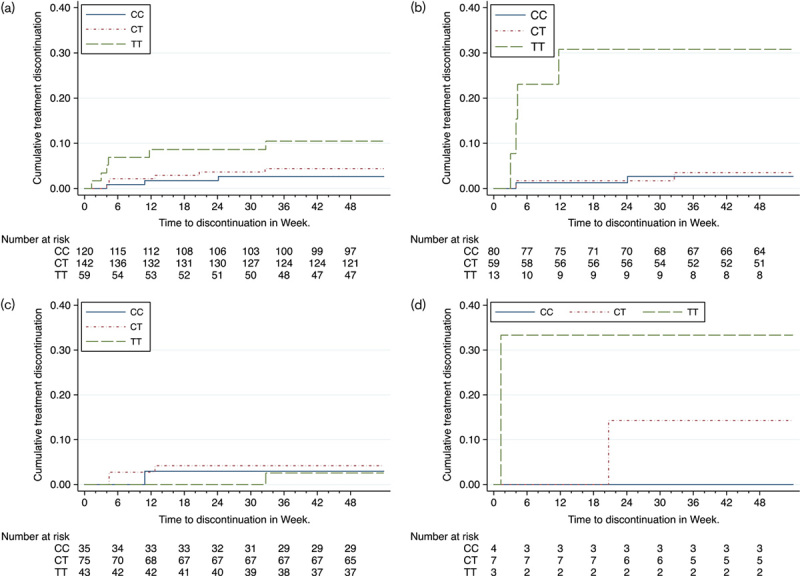
Time to bilirubin-related atazanavir/r discontinuation, stratified by UGT1A1 genotype. (a) All patients stratified by UGT1A1 rs887829 genotype; (b) White patients stratified by UGT1A1 rs887829 genotype; (c) Black patients stratified by UGT1A1 rs887829 genotype; (d) Hispanic patients stratified by UGT1A1 rs887829 genotype.
The above analyses did not consider pretreatment baseline bilirubin as a covariate. At baseline, six patients had bilirubin concentrations above the normal range, including five with bilirubin between 1.1 and 1.3 mg/dl, and one with a bilirubin of 2.3 mg/dl. Among the 321 evaluable patients, in analyses adjusted for the first two MDS coordinates, the HR of baseline bilirubin concentration for bilirubin-related atazanavir discontinuation was 7.3 (95% CI=2.6–20.7; P<0.001). In a multivariable model that included rs887829 genotype, baseline bilirubin concentration, and 2 MDS coordinates, the HR for discontinuation was significant for T/T genotype (HR=5.2; 95% CI: 1.1–24.5; P=0.038) but not for baseline bilirubin (HR=2.0; 95% CI: 0.8–4.9; P=0.14). With this same multivariable model but just including White patients, the HR for discontinuation was significant for T/T genotype (HR=9.4; 95% CI: 1.3–66.9; P=0.026) but not for baseline bilirubin (HR=5.2; 95% CI: 0.7–40.4; P=0.11). With this model but just among Black patients, the HR for discontinuation was not significant for T/T genotype (HR=0.30; 95% CI: 0.01–6.7; P=0.45) but was significant for baseline bilirubin, with an extremely wide confidence interval (HR=140.6; 95% CI: 3.6–5481; P=0.008).
Discussion
Atazanavir with pharmacokinetic enhancement by either ritonavir or cobicistat is generally safe and effective as a first-line regimen for HIV-1 infection 1–5, but some patients discontinue atazanavir due to cosmetic jaundice 3,10,11, and many are not prescribed atazanavir to avoid this possibility. The present study showed that, among 321 evaluable patients who had initiated atazanavir/r-containing regimens at an HIV primary care clinic in the Southeastern USA, and adjusting for genetic ancestry, UGT1A1 rs887829 T/T genotype was associated with increased bilirubin-related atazanavir discontinuation during the first 12 months of therapy (HR=7.3; P=0.007). In analyses stratified by race, the HR in White patients was 14.4 (P=0.002), but in Black patients it was 0.8 (P=0.87), despite the T/T genotype being considerably more frequent in Black patients. The nearly identical number of Black patients and White patients in the analysis, which was by chance, strengthens our findings. Results were consistent in additional analyses that controlled for baseline bilirubin.
This study replicates a difference by race that was first reported in analyses on the basis of data from ACTG clinical trial A5257 19. In that study there was a much stronger association between UGT1A1 slow metabolizer genotype and increased bilirubin-related atazanavir discontinuation among 183 White patients (HR=24, P=1.3×10−4) than among 211 Black patients (HR=10, P=0.03). A similar association in a largely Caucasian cohort was seen in an observational study involving 121 Swiss HIV Cohort Study participants (80% Whites) who had received atazanavir/r, in which carriage of UGT1A1 low expresser alleles (*28/*28 or *28/*37) was associated with increased risk of atazanavir/r discontinuation, with cumulative rates of 63% among 18 participants carrying two alleles, 24% among 48 participants carrying one allele, and 15% among 55 participants carrying no allele 19.
In patients who are prescribed atazanavir, Gilbert’s polymorphism predicts higher peak plasma bilirubin concentration regardless of ancestry 16. A possible explanation for the lack of association between UGT1A1 T/T homozygosity and bilirubin-related atazanavir/r discontinuation in Black patients compared with White patients is that jaundice may be less visible among individuals with darker skin. If so, then this is an example of a known functional pharmacogenetic variant having a different impact on an antiretroviral toxicity outcome depending on race/ethnicity context. A pharmacogenetic difference in reported antiretroviral toxicity depending on race/ethnicity has also been described for the antiretroviral drug efavirenz, for which there is a strong association between CYP2B6 genotype and central nervous system toxicity including suicidality among European Americans, but this association is markedly attenuated or absent among African Americans 21,23,24, despite a greater frequency of CYP2B6 risk genotype with African ancestry 25.
It is possible that providers queried Black patients less than White patients regarding reasons for discontinuing atazanavir. However, this is unlikely as all patients received care at a single HIV primary care clinic where, since 1997 every antiretroviral treatment change is presented and thoroughly discussed at a multidisciplinary Antiretroviral Therapy Conference. There is strong evidence that bilirubin concentrations with atazanavir/r are similar among Black patients and White patients with UGT1A1 slow metabolizer genotypes 16, so this is unlikely to explain the difference by race/ethnicity.
In the present analyses we did not genotype for the UGT1A1 TA tandem repeat polymorphism (rs8175347) because a previous analysis based on data from AIDS Clinical Trial Group Protocol A5202 showed that rs887829 was in almost complete linkage with *28 in White, Black, and Hispanic participants 17. In A5202, every participant with UGT1A1*37 (TA8) slow metabolizer genotype also carried the rs887829 T allele (D.W.H., personal communication). Specifically, 549 participants from A5202 had genotype data for rs887829, UGT1A1*1, *28, *36, and *37, including 241 White, 160 Black, and 122 Hispanic participants. Of the 549 participants, 19 (3.5%) were heterozygous for UGT1A1*37, including 18 Black and one White participant. All 14 with *1/*37 were also rs887829 C/T, and all five with *28/*37 were also rs887829 T/T. In the present study, any participants with UGT1A1*37 slow metabolizer genotypes were therefore almost certainly correctly classified by rs887829 genotyping.
United States prescribing guidelines were recently updated to change atazanavir/r-containing regimens from recommended to alternative status as an initial therapy for HIV-1 infection 4, informed largely by results from ACTG protocol A5257 that showed increased discontinuation of atazanavir versus darunavir, and with decreased tolerability due primarily by participant-driven regimen change for jaundice in the ritonavir-boosted atazanavir arm 12. The present study showed that, among patients who had initiated atazanavir/ritonavir-containing regimens at a clinic in the southeastern USA, increased likelihood of bilirubin-related atazanavir/r discontinuation was driven by UGT1A1 slow metabolizer genotype, and that this association was present in White patients but not in Black patients.
Acknowledgements
This work was supported in part by the National Institute of Allergy and Infectious Diseases grants R01 AI077505, P30 AI110527, and UL1 TR002243 (DWH), and by Fogarty International Center and National Institute of Mental Health award D43 TW009608 (J.N.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Gatell J, Salmon-Ceron D, Lazzarin A, Van Wijngaerden E, Antunes F, Leen C, et al. Efficacy and safety of atazanavir-based highly active antiretroviral therapy in patients with virologic suppression switched from a stable, boosted or unboosted protease inhibitor treatment regimen: the SWAN Study (AI424-097) 48-week results. Clin Infect Dis 2007; 44:1484–1492. [DOI] [PubMed] [Google Scholar]
- 2.Molina JM, Andrade-Villanueva J, Echevarria J, Chetchotisakd P, Corral J, David N, et al. Once-daily atazanavir/ritonavir versus twice-daily lopinavir/ritonavir, each in combination with tenofovir and emtricitabine, for management of antiretroviral-naive HIV-1-infected patients: 48 week efficacy and safety results of the CASTLE study. Lancet 2008; 372:646–655. [DOI] [PubMed] [Google Scholar]
- 3.Daar ES, Tierney C, Fischl MA, Sax PE, Mollan K, Budhathoki C, et al. Atazanavir plus ritonavir or efavirenz as part of a 3-drug regimen for initial treatment of HIV-1. Ann Intern Med 2011; 154:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Panel on Clinical Practices for Treatment of HIV Infection. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. 2016. Available at: http://www.aidsinfo.nih.gov/contentfiles/lvguidelines/adultandadolescentgl.pdf [Accessed 14 July 2016]. [DOI] [PubMed]
- 5.Gallant JE, Koenig E, Andrade-Villanueva J, Chetchotisakd P, DeJesus E, Antunes F, et al. Cobicistat versus ritonavir as a pharmacoenhancer of atazanavir plus emtricitabine/tenofovir disoproxil fumarate in treatment-naive HIV type 1-infected patients: week 48 results. J Infect Dis 2013; 208:32–39. [DOI] [PubMed] [Google Scholar]
- 6.Zhang D, Chando TJ, Everett DW, Patten CJ, Dehal SS, Humphreys WG. In vitro inhibition of UDP glucuronosyltransferases by atazanavir and other HIV protease inhibitors and the relationship of this property to in vivo bilirubin glucuronidation. Drug Metab Dispos 2005; 33:1729–1739. [DOI] [PubMed] [Google Scholar]
- 7.Malan DR, Krantz E, David N, Wirtz V, Hammond J, McGrath D, et al. Efficacy and safety of atazanavir, with or without ritonavir, as part of once-daily highly active antiretroviral therapy regimens in antiretroviral-naive patients. J Acquir Immune Defic Syndr 2008; 47:161–167. [DOI] [PubMed] [Google Scholar]
- 8.Cleijsen RM, van de Ende ME, Kroon FP, Lunel FV, Koopmans PP, Gras L, et al. Therapeutic drug monitoring of the HIV protease inhibitor atazanavir in clinical practice. Journal Antimicrob Chemo 2007; 60:897–900. [DOI] [PubMed] [Google Scholar]
- 9.Torti C, Lapadula G, Antinori A, Quirino T, Maserati R, Castelnuovo F, et al. Hyperbilirubinemia during atazanavir treatment in 2404 patients in the Italian atazanavir expanded access program and MASTER Cohorts. Infection 2009; 37:244–249. [DOI] [PubMed] [Google Scholar]
- 10.Puls RL, Srasuebkul P, Petoumenos K, Boesecke C, Duncombe C, Belloso WH, et al. Efavirenz versus boosted atazanavir or zidovudine and abacavir in antiretroviral treatment-naive, HIV-infected subjects: week 48 data from the Altair study. Clin Infect Dis 2010; 51:855–864. [DOI] [PubMed] [Google Scholar]
- 11.McDonald C, Uy J, Hu W, Wirtz V, Juethner S, Butcher D, et al. Clinical significance of hyperbilirubinemia among HIV-1-infected patients treated with atazanavir/ritonavir through 96 weeks in the CASTLE study. AIDS Patient Care STDs 2012; 26:259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lennox JL, Landovitz RJ, Ribaudo HJ, Ofotokun I, Na LH, Godfrey C, et al. Efficacy and tolerability of 3 nonnucleoside reverse transcriptase inhibitor-sparing antiretroviral regimens for treatment-naive volunteers infected with HIV-1: a randomized, controlled equivalence trial. Ann Intern Med 2014; 161:461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome. New Engl J Med 1995; 333:1171–1175. [DOI] [PubMed] [Google Scholar]
- 14.Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet 1996; 347:578–581. [DOI] [PubMed] [Google Scholar]
- 15.Rotger M, Taffe P, Bleiber G, Gunthard HF, Furrer H, Vernazza P, et al. Gilbert syndrome and the development of antiretroviral therapy-associated hyperbilirubinemia. J Infect Dis 2005; 192:1381–1386. [DOI] [PubMed] [Google Scholar]
- 16.Ribaudo HJ, Daar ES, Tierney C, Morse GD, Mollan K, Sax PE, et al. Impact of UGT1A1 Gilbert variant on discontinuation of ritonavir-boosted atazanavir in AIDS clinical trials group study A5202. J Infect Dis 2013; 207:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson DH, Venuto C, Ritchie MD, Morse GD, Daar ES, McLaren PJ, et al. Genomewide association study of atazanavir pharmacokinetics and hyperbilirubinemia in AIDS Clinical Trials Group protocol A5202. Pharmacogenet Genom 2014; 24:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubomirov R, Colombo S, di Iulio J, Ledergerber B, Martinez R, Cavassini M, et al. Association of pharmacogenetic markers with premature discontinuation of first-line anti-HIV therapy: an observational cohort study. J Infect Dis 2011; 203:246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vardhanabhuti S, Ribaudo HJ, Landovitz RJ, Ofotokun I, Lennox JL, Currier JS, et al. Screening for UGT1A1 genotype in study A5257 would have markedly reduced premature discontinuation of atazanavir for hyperbilirubinemia. Open Forum Infect Dis 2015; 2:ofv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gammal RS, Court MH, Haidar CE, Iwuchukwu OF, Gaur AH, Alvarellos M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for UGT1A1 and atazanavir prescribing. Clin Pharmacol Ther 2015; 99:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leger P, Chirwa S, Turner M, Richardson DM, Baker P, Leonard M, et al. Pharmacogenetics of efavirenz discontinuation for reported central nervous system symptoms appears to differ by race. Pharmacogenet Genomics 2016; 26:473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribaudo HJ, Liu H, Schwab M, Schaeffeler E, Eichelbaum M, Motsinger-Reif AA, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis 2010; 202:717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mollan KRTC, Hellwege JN, Eron JJ, Hudgens M, Gulick RM, Haubrich R, et al. The Pharmacogenetics of reported suicidality with efavirenz varied by race/ethnicity among clinical trials participants. J Infect Dis in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holzinger ER, Grady B, Ritchie MD, Ribaudo HJ, Acosta EP, Morse GD, et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genom 2012; 22:858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]


