Abstract
The present work reveals the potential of biosurfactant producing P. aeruginosa PBS for microbial enhanced oil recovery (MEOR). The biosurfactant production medium and culture conditions were optimized using response surface methodology. The optimization of media components and process parameters was consecutively executed in two sets of experimental runs designed by central composite rotatable design (CCRD). The maximum biosurfactant yield was attained with 2% fresh inoculum of P. aeruginosa PBS in minimal salt medium (pH 7), possessing 2.17% sodium citrate as C-source and 0.5% yeast extract as N-source, after 48 h upon incubation at 30 °C/150 rpm. Under optimum conditions, biosurfactant yield was increased more than threefold and turned out to be 2.65 g/L as compared to 0.82 g/L under previous conditions. The biosurfactant was characterized as a glycolipid comprising of four rhamnolipid homologs (RhaRhaC10C10, RhaRhaC8C10, RhaRhaC12C10/RhaRhaC10C12, RhaC10C10) by thin layer chromatography, fourier transform infrared spectroscopy, nuclear magnetic resonance and mass spectrometry. The produced biosurfactant was highly efficient for oil recovery application showing extreme reduction in surface tension of medium (71.80 to 23.76 mN/m), immense hydrocarbons emulsification capacity (50–60%) and greater stability at wide range of temperature (4–100 °C) and pH (4–10) along with an excellent (56.18 ± 1.59%) additional oil recovery in sand-pack column lab test.
Keywords: Rhamnolipids, Biosurfactant, MEOR, Pseudomonas, RSM, Oil recovery
Introduction
SURFace ACTive AgeNTS, renowned as surfactants, are amphiphilic molecules displaying relevance to enormous fields such as pharmaceuticals, agriculture, remediation, textile, food and cosmetics (Ferhat et al. 2017; Mouafi et al. 2016). These molecules also play a significant role in the recovery of oil from the rocks (Yela et al. 2016). These are competent to reduce the surface tension/interfacial tension, viscosity as well as capillary forces retaining the oil inside the reservoir. Consequently, the surfactants improve the oil mobilization and force it to pump out of the reservoir (Al-Wahaibi et al. 2014). These chemical surfactants can be superseded by surfactants of biological origin due to their biodegradability and non-toxicity in addition to high specific activity at extreme pH, temperature and salinity. Biosurfactants are produced by wide range of bacteria, yeast and fungi (Ferhat et al. 2017). Despite the remarkable features, the commercial production of the biosurfactant is small owing to low yield and high production cost (Dobler et al. 2016).
The economics of biosurfactant production can be improved using cost-effective raw materials, high yielding strains or optimizing the process conditions (Mouafi et al. 2016; Pereira et al. 2013). Large scale biosurfactant production is possible from fermentation of cheap and waste substrates. Waste cooking oil, frying rice bran oil, molasses and corn steep liquors were utilized to produce biosurfactants at high yield (Lan et al. 2015; Venkatesh and Vedaraman 2012). The optimization of process parameters and media components was also useful in enhancing the biosurfactant yield (Pereira et al. 2013). Response surface methodology (RSM) is the most commonly employed technique for the optimization which is used to study the influence of multiple parameters for a single response and to obtain the best operating conditions with reduced experimental runs (Najafi et al. 2010).
The type of biosurfactant produced by the microbes also depends upon the media and environmental parameters (Sepahy et al. 2005). The chemical diversity of biosurfactants incorporates glycolipids, lipopeptides/lipoproteins, phospholipids, fatty acids and polymers. Glycolipids and lipopeptides are the most comprehensively studied biosurfactants (Zou et al. 2014). Pseudomonas aeruginosa strains are well-known producers of rhamnolipids, a type of glycolipids (Dobler et al. 2016). These are considered to be involved in both survival and virulence, though their accurate role remains ambiguous. The rhamnolipids consist of one or two rhamnose rings associated with one or two β-hydroxy fatty acids of variable chain length (C8–C22) (Yin et al. 2009). The most common homolog structures include di-rhamnolipids, α-l-rhamnopyranosyl-α-l-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate (RhaRhaC10C10) and RhaRhaC10, whereas mono-rhamnolipid homolog structures comprise of RhaC10 and RhaC10C10. Despite these frequently known structures, various other homologues were found to be present ranging m/z value of 475–677 depending upon the number of rhamnose rings and fatty acid chains as well as on the number of carbons in the fatty acid chains (Dobler et al. 2016; Yin et al. 2009).
The rhamnolipids are eminent for anti-microbial activity and bioremediation of recalcitrant organic compounds and heavy metal contaminants (Dobler et al. 2016). Besides this, application for oil recovery is of utmost interest due to their efficacy at stringent environment (Amani 2015). It is amazing to mention that three to four trillion barrels of oil remain ambushed in the rocks. The oil inefficiency to move out of the reservoir is due to its high viscosity and interfacial tension (Amani et al. 2013). Biosurfactants are highly efficient to mobilize oil by reducing viscosity, surface/interfacial tension and capillary forces (Amani 2015; Pathak and Keharia 2014a).
The objectives of present study included the optimization, characterization and application of biosurfactant produced by an isolate, namely P. aeruginosa PBS. Different nutritional and environmental parameters including nature and concentration of carbon and nitrogen sources, pH of media, inoculum size, incubation time and temperature were optimized using RSM for good biosurfactant yield. Thereafter, the produced biosurfactant was characterized by various techniques such as TLC, FTIR, NMR and MS. The emulsification and stability of the biosurfactant were compared with synthetic surfactants. Ultimately, the efficiency of biosurfactant was evaluated for oil recovery in sand-pack column test.
Materials and methods
Materials
The analytical grade chemicals were used throughout the study. Soil samples were collected from oil contaminated sites (bus stands, petrol pumps and automobiles shops) of different regions of Punjab. Crude oil (CAS No. 8002-05-9) was kindly provided by Guru Gobind Singh Refinery, Bathinda, Punjab (India).
Isolation and screening of biosurfactant producers
One gram of each oil contaminated soil sample was dispensed into 100 mL of enrichment medium (Bushnell Hass Broth, BHB) to augment biosurfactant producing microbes. The filter sterilized kerosene (3%) was used as a sole source of carbon while olive oil (20 µL) as an inducer (Chandran and Das 2010). Following incubation at 37 °C for 72 h, 1 mL aliquot was further transferred to fresh BHB medium. Three cycles of subculturing were followed by serial dilution, spreading and streak plate method to isolate pure colonies of distinct morphology. The isolated microbes were maintained on the nutrient agar slants at 4 °C (Varadavenkatesan and Murty 2013). To screen biosurfactant producers, oil spreading and CTAB (cetyltrimethylammonium bromide) agar tests (Satpute et al. 2008) were performed with cell free supernatants of 48–72 h grown cultures in BHB media. The selected microbial isolate PBS was identified by 16S rRNA gene sequencing and phylogenetic analysis (Dobler et al. 2016; Varadavenkatesan and Murty 2013).
Optimization of biosurfactant production
Glucose minimum salt (GMS) medium (pH 7) comprised of (g/L) 50.0 glucose, 5.0 KNO3, 1.0 KH2PO4·2H2O, 1.0 K2HPO4·2H2O, 0.2 MgSO4·7H2O and 0.02 CaCl2·2H2O was used as basic biosurfactant production media (Pathak and Keharia 2014a). 2% fresh culture of isolate (6.0–7.0 × 108 cells/mL) was inoculated in the medium and incubated at 37 °C on orbital shaker (150 rpm) for 72 h. For the optimum biosurfactant production, production medium composition and environmental parameters were optimized. First of all, the optimum carbon and nitrogen sources were selected. Twelve different C-sources (glucose, fructose, sucrose, sodium succinate, sorbitol, sodium acetate, sodium citrate, glycerol, maltose, lactose, tryptone and yeast extract) were analyzed in production medium keeping KNO3 as N-source. Subsequently, optimum N-source was chosen out of seven sources (ammonium citrate, ammonium sulphate, potassium nitrate, sodium nitrate, yeast extract, tryptone, meat peptone and urea) using optimized C-source (Pereira et al. 2013) in production medium.
Following the optimization of nature of C- and N-sources manually, other media components (C-source%, N-source%, pH) and process parameters (inoculum size, temperature and incubation time) were optimized by response surface methodology (RSM) (Najafi et al. 2010) using Design Expert 10 statistical software (Stat-Ease Inc., Minneapolis, MN). Table 1 presents the actual and coded values of different independent variables. The ranges selected for these parameters were as follow: 3–7% C-source, 0.3–0.7% N-source, pH 4–7, inoculum size 2–6%, temperature 30–45 °C and incubation time 48–96 h. A set of 15 experimental runs were designed individually for the optimization of media components and process parameters using centre composite rotatable design (CCRD) (Table 2). Following incubation at specified conditions, the culture was centrifuged (11,730×g, 20 min, 4 °C) to remove biomass and cell free supernatant was analyzed for biosurfactant production by surface tension (ST) measurement (Pathak and Keharia 2014a). All measurements were performed in triplicates and average was used for analysis of variance (ANOVA).
Table 1.
Coded values of independent variables
| No. | Variables | Symbols | Coded values | ||||
|---|---|---|---|---|---|---|---|
| − 1.68 | − 1 | 0 | 1 | 1.68 | |||
| Media components | |||||||
| 1 | C-conc. (%) | X | 2.17 | 3 | 5 | 7 | 7.83 |
| 2 | N-conc. (%) | Y | 0.217 | 0.3 | 0.5 | 0.7 | 0.783 |
| 3 | pH | Z | 2.76 | 4 | 7 | 10 | 11.24 |
| Process parameters | |||||||
| 4 | Temperature (°C) | A | 26.9 | 30 | 37.5 | 45 | 48.1 |
| 5 | Incubation time (h) | B | 38.06 | 48 | 72 | 96 | 105.9 |
| 6 | Inoculum size (%) | C | 1.17 | 2 | 4 | 6 | 6.83 |
Table 2.
Central composite rotatable design (CCRD) matrix of independent variables (for media components and process parameters) with their experimental and predicted response (surface tension)
| Run | Variables | Surface tension (mN/m) | |||
|---|---|---|---|---|---|
| X (C-conc. %) | Y (N-conc. %) | Z (pH) | Experimental | Predicted | |
| Media components | |||||
| 1 | 7 | 0.3 | 10 | 54.43 | 54.94 |
| 2 | 5 | 0.783 | 7 | 29.93 | 29.43 |
| 3 | 5 | 0.5 | 7 | 30.59 | 30.84 |
| 4 | 5 | 0.5 | 7 | 31.59 | 30.84 |
| 5 | 5 | 0.5 | 2.76 | 58.40 | 57.89 |
| 6 | 5 | 0.5 | 7 | 30.64 | 30.84 |
| 7 | 3 | 0.7 | 10 | 27.61 | 28.12 |
| 8 | 5 | 0.5 | 11.24 | 62.12 | 61.61 |
| 9 | 5 | 0.5 | 7 | 29.67 | 30.84 |
| 10 | 7 | 0.7 | 4 | 32.78 | 33.29 |
| 11 | 7.83 | 0.5 | 7 | 38.02 | 37.51 |
| 12 | 2.17 | 0.5 | 7 | 26.32 | 25.81 |
| 13 | 5 | 0.5 | 7 | 30.67 | 30.84 |
| 14 | 3 | 0.3 | 4 | 67.76 | 68.26 |
| 15 | 5 | 0.217 | 7 | 34.54 | 34.03 |
| Run | Variables | Surface tension (mN/m) | |||
|---|---|---|---|---|---|
| A (temperature, °C) | B (incubation time, h) | C (inoculum size, %) | Experimental | Predicted | |
| Process parameters | |||||
| 1 | 37.5 | 72 | 4 | 26.734 | 25.78 |
| 2 | 37.5 | 72 | 6.83 | 29.813 | 30.29 |
| 3 | 30 | 96 | 6 | 24.917 | 24.44 |
| 4 | 37.5 | 72 | 4 | 26.172 | 25.78 |
| 5 | 37.5 | 72 | 4 | 25.814 | 25.78 |
| 6 | 45 | 48 | 6 | 64.13 | 63.65 |
| 7 | 37.5 | 38.06 | 4 | 30.692 | 31.17 |
| 8 | 45 | 96 | 2 | 50.106 | 49.63 |
| 9 | 30 | 48 | 2 | 23.755 | 23.28 |
| 10 | 48.1 | 72 | 4 | 54.961 | 55.44 |
| 11 | 37.5 | 105.9 | 4 | 32.655 | 33.13 |
| 12 | 37.5 | 72 | 4 | 25.128 | 25.78 |
| 13 | 26.9 | 72 | 4 | 25.757 | 26.23 |
| 14 | 37.5 | 72 | 1.17 | 35.826 | 36.30 |
| 15 | 37.5 | 72 | 4 | 25.984 | 25.78 |
The bold values indicate the optimized parameters corresponding to minimal surface tension
Emulsification and stability of biosurfactant
The emulsification ability of biosurfactant was evaluated for various hydrocarbons (crude oil, petrol, kerosene, hexane, heptane and hexadecane) and compared with chemical surfactants (1% SDS, 1% CTAB and 0.1% Tween-100) (Chandran and Das 2010). The emulsification was evaluated by adding 2 mL of hydrocarbon to 2 mL of cell free supernatant/surfactant solution taken in graduated tube which was vortexed for 2 min at high speed. The tubes were kept undisturbed for 24 h and emulsification index (E 24) was determined by measuring percentage of emulsion height (mm) with respect to the total height of liquid column (mm) (Ferhat et al. 2017) as:
Stability of biosurfactant was monitored at different pH and temperature conditions. Cell free broth was adjusted to different pH (2, 4, 7 and 10) to study the effect of pH on biosurfactant and incubated at 30 °C for 2 h. On the other hand, to evaluate the influence of temperature, 20–20 mL of cell free broth was kept for 2 h at different temperatures (4, 25, 37, 70 and 100 °C). Both types of samples were then equilibrated by keeping at room temperature for 6 h and then ST of each sample was measured (Pathak and Keharia 2014a).
Analytical techniques
The biomass was removed from fermented broth by centrifugation at 11,730×g (10,000 rpm) for 20 min at 4 °C (Pathak and Keharia 2014b). The surface tension of cell free broth was measured at room temperature with Digital Surface/Interfacial Tensiometer (Culture instrument lab, Bangalore) according to ring method (Zou et al. 2014). The instrument was calibrated beforehand with MilliQ water taking ST of water equal to 72 mN/m (25 °C). The measurements were repeated until three concordant readings. The biosurfactant was quantified by Orcinol Method (Bharali and Konwar 2011) in which amount of rhamnolipid was estimated from the standard curve of l-rhamnose. The obtained rhamnose value was multiplied by correction factor 3.4 for rhamnolipid/rhamnose precise quantification.
Extraction and purification of the biosurfactant
The biosurfactant was extracted from cell free broth by acid precipitation and solvent extraction method (Pathak and Keharia 2014a). The biosurfactant was precipitated by adjusting the pH of broth to 2.0 with 6 N HCl following incubation at 4 °C overnight. The precipitates were harvested from acidified broth by centrifugation at 11,730×g for 20 min (4 °C). Thereafter, precipitates were solubilized in pure methanol leaving behind the undissolved fraction which was subsequently removed by centrifugation. The methanolic extract of biosurfactant was then subjected to vacuum dry with the aid of rotary evaporator (Buchi, Switzerland) to obtain purified biosurfactant.
Characterization of biosurfactant
Thin layer chromatography
Methanolic extract of biosurfactant was preliminary characterized by thin layer chromatography (TLC). The sample (2 µL) was spotted on pre-coated silica gel plates (Merck, USA) with the help of automated Linomat-5 spotting device of HPTLC system (CAMAG, Switzerland) (Das et al. 2008). Simultaneously three plates were developed with solvent system comprised chloroform:methanol:water (60:20:0.5) in air-tight saturated chamber. The qualitative biochemical analysis of biosurfactant was completed by applying different color developing reagents on three plates. For the detection of protein, lipid and sugar moieties, ninhydrin reagent (0.5% ninhydrin in anhydrous acetone) (Lan et al. 2015), iodine vapors (Mukherjee et al. 2008) and diphenylamine (DPA) reagent (Anderson et al. 2002), respectively, were employed on air-dried plates. The plates were then incubated at 105 °C for the appearance of colored spots.
Fourier transform infrared spectrophotometry
The infrared absorption spectrum of homogenized KBr pellet of biosurfactant was studied using FTIR spectrophotometer (Perkin Elmer Spectrum Spotlight 400 imaging, Germany) to reveal the overall chemical nature. IR spectra were scanned over the range of 400–4000 cm−1 with a spectral resolution of 4 cm−1 and final spectrum was presented as average of 5 scans (Lan et al. 2015).
Nuclear magnetic resonance
The purified biosurfactant was re-dissolved in deuterated dimethyl sulphoxide (DMSO). The 1H-NMR spectrum was recorded at 25 °C using Bruker II Avance 400 NMR spectrometer operating at 400 mHz (SAIF, Panjab University, Chandigarh). The chemical shifts were presented on the ppm scale with respect to tetramethyl silane, TMS (Pereira et al. 2013).
Mass spectrometry
The absolute characterization of purified biosurfactant was achieved by mass spectrometry. Electro spray ionization–mass spectrum (ESI–MS) was recorded on Waters, Q-TOF Micromass Mass Spectrometer (SAIF, Panjab University, Chandigarh). A sample aliquot of 20 µL was infused with the flow rate of 0.4 mL/min maintaining nitrogen and auxiliary gas flows at 35 and 550 L/h, respectively, in ESI. The heated capillary temperature was maintained at 250 °C and capillary voltage was set at 3000 V. Scanning was done at 50–2000 m/z range in negative ion mode (Lan et al. 2015).
Enhanced oil recovery with biosurfactant
The efficacy of biosurfactant for enhancement of oil recovery was estimated by sand-pack column technique (Al-Wahaibi et al. 2014). 100 g of acid washed sand (HIMEDIA) was packed in a glass column (25 cm height, 3 cm internal diameter). The brine solution (10% sodium chloride) was flooded through the column and pore volume (PV) was estimated by determining the volume required to wet the whole sand matrix with brine solution. To confirm 100% saturation, more brine solution was passed approximately 3 PVs. Thereafter, column was pumped with crude oil under pressure which discharged the brine solution. The discharged volume indicated the initial oil saturation (IOS). Further, 5–6 PVs of brine solution were passed to wash the column until no more discharging of oil. The actual oil retained (ROS, residual oil saturation) was estimated by subtracting the discharged oil after water flooding (SOR, secondary oil recovery) from initial oil saturation. Finally, 48 h grown cell-free broth (20 mL) possessing biosurfactant was loaded in the crude oil saturated column and the system was kept closed for 24–48 h. After incubation, the fraction of released oil (AOR, additional oil recovery) was collected and measured to estimate the percentage of oil recovery (Pathak and Keharia 2014a).
Results
Selection and identification of biosurfactant producer
Forty-two morphologically distinct bacterial colonies were isolated from twelve different soil samples using kerosene as a sole source of carbon in enrichment media. Out of these, only three were able to produce biosurfactant as indicated from oil spreading techniques. The isolate PBS was selected for study which allowed the crude oil displacement above a significant level (> 0.5 cm) while other two were showing the lesser displacement (0.2–0.3 cm). Oil spreading is a semi-quantitative method for the screening as higher the content of biosurfactant in solution, the greater will be the reduction in ST of water with respect to oil which will lead to more spreading (Lan et al. 2015). Further, in CTAB–methylene agar plate test, isolate PBS indicated the production of anionic biosurfactant with bluish-green halos around the wells. Halos are formed due to the precipitation of CTAB with extracellularly secreted anionic biosurfactant which predominates in the presence of the methylene blue (Satpute et al. 2008).
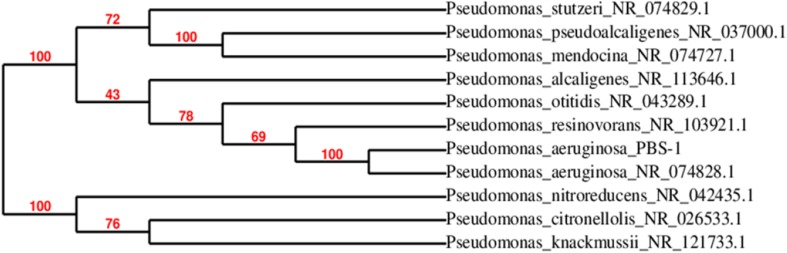
The isolate PBS was identified as P. aeruginosa based on 16S rDNA sequencing showing 99% homology with P. aeruginosa NR 0748281. The phylogenetic tree of isolate PBS was constructed depending upon the sequence similarity with closely related microorganisms (Fig. 1) and gene sequence of isolate PBS was submitted in GenBank of NCBI with accession no. KX180920.
Fig. 1.
Phylogenetic tree of the isolate PBS based on 16S rDNA sequences of closely related microorganisms
Optimization of biosurfactant production
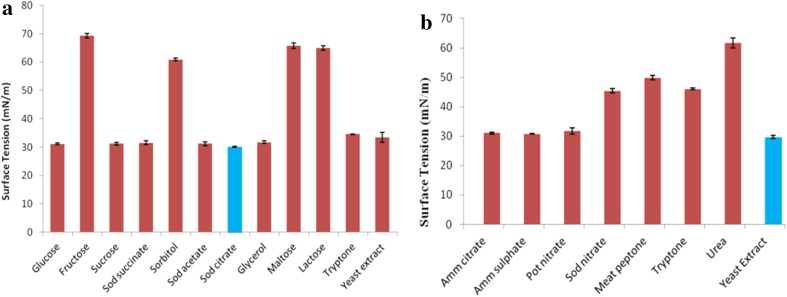
For the optimization of biosurfactant production medium, firstly, carbon and nitrogen sources were optimized. Out of twelve carbon sources examined in the production medium, most of the carbon sources led to ST reduction up to 30–34 mN/m with respect to 71.80 ± 0.96 mN/m (ST of uninoculated media) except fructose, sorbitol, maltose and lactose (Fig. 2a). Greater ST reduction of medium has given the indication of large amount of biosurfactant produced by microbe. From the previous reports it is clear that more and more ST reduction takes place with increase in the concentration of biosurfactant until critical micelle concentration (CMC) is achieved (Najafi et al. 2010). Hence, sodium citrate was selected as the best C-source resulting in the maximum surface tension reduction (30.060 ± 0.214 mN/m) followed by glucose (31.142 ± 0.219 mN/m), sodium acetate (31.180 ± 0.650 mN/m), sucrose (31.246 ± 0.492 mN/m), sodium succinate (31.530 ± 0.681 mN/m), glycerol (31.731 ± 0.551 mN/m), yeast extract (33.440 ± 1.811 mN/m) and tryptone (34.590 ± 0.116 mN/m). Regarding the N-sources, maximum surface tension reduction was observed with yeast extract (29.661 ± 0.494 mN/m) which is followed by ammonium sulphate (30.792 ± 0.032 mN/m), ammonium citrate (31.019 ± 0.316 mN/m), potassium nitrate (31.705 ± 1.012 mN/m) (Fig. 2b). On the whole, sodium citrate and yeast extract were selected as the optimum C- and N-sources, respectively, for biosurfactant production with P. aeruginosa PBS.
Fig. 2.
Surface tension of biosurfactant production medium supplemented with different a carbon sources and b nitrogen sources
Afterwards, media components including pH of medium, concentration of carbon and nitrogen source (%) and process parameters such as temperature (°C), incubation time (h) and inoculum size (%) were optimized in two sets of 15 experiments designed by CCRD. The experimental design, predicted and the experimental results are described in Table 2. The predicted values were in agreement with the experimental values. Table 3 recapitulate the ANOVA summary for response surface quadratic models obtained with these designed experiments. The models were found significant with Fisher’s statistical test for ANOVA. Significance of the model was assured from model F values 331.48 and 323.30 and there were merely 0.01% chances that these large F values could arise due to noise. F value is the ratio of the mean square due to regression to the mean square due to error and specifies the significance of each controlled factor on the tested model. The values of ‘Probability > F’ less than 0.05 indicated model terms are significant. Henceforth, the model terms X, Y, Z, XY, XZ, YZ, Z 2 are significant while studying the optimization of media components. The fitted model equation proposed by regression analysis is given by
where X is the concentration of C-source (%), Y is the concentration of N-source (%) and Z is the pH of the medium. The model determination coefficient R 2 = 0.9986 recommended that the fitted model could explain 99.86% of the total variation. This indicated a reasonable depiction of the process by the model. The predicted R 2 of 0.9159 is in logical agreement with the adjusted R 2 of 0.9960. On the other hand, the model terms A, C, AB, AC, BC, A 2, B 2, C 2 were found to be significant with values of ‘Probability > F’ less than 0.05 in case of optimization of process parameters. The fitted model equation for the response (surface tension) for second set experiments is
where A is the temperature (°C), B is the incubation time (h) and C is the inoculum size (%). The model determination coefficient R 2 = 0.9983 indicated that 99.86% of the total variation in biosurfactant production is attributed to the selected variables and only 0.14% variation was not justified by the model. The predicted R 2 of 0.8799 is in reasonable agreement with the adjusted R 2 of 0.9952.
Table 3.
Results of analysis of variance (ANOVA) for response surface quadratic model obtained from experimental design (for media components and process parameters)
| Source of variation | Sum of squares | Degree of freedom | Mean square | F value | Probability > F |
|---|---|---|---|---|---|
| Media components | |||||
| Model | 2755.41 | 9 | 306.16 | 331.48 | <0.0001 |
| X—C-conc. | 68.43 | 1 | 68.43 | 74.09 | 0.0003 |
| Y—N-conc. | 10.60 | 1 | 10.60 | 11.47 | 0.0195 |
| Z—pH | 6.92 | 1 | 6.92 | 7.50 | 0.0409 |
| XY | 70.53 | 1 | 70.53 | 76.37 | 0.0003 |
| XZ | 382.11 | 1 | 383.11 | 413.71 | <0.0001 |
| YZ | 76.25 | 1 | 76.25 | 82.56 | 0.0003 |
| X 2 | 1.32 | 1 | 1.32 | 1.42 | 0.2862 |
| Y 2 | 1.54 | 1 | 1.54 | 1.67 | 0.2532 |
| Z 2 | 1612.42 | 1 | 1612.42 | 1745.76 | < 0.0001 |
| Residual | 4.62 | 5 | 0.92 | ||
| Lack of fit | 2.78 | 1 | 2.78 | 6.06 | 0.0695 |
| Pure error | 1.84 | 4 | 0.46 | ||
| Core total | 2760.3 | 14 | |||
| Process parameters | |||||
| Model | 2212.05 | 9 | 245.78 | 323.30 | < 0.0001 |
| A—temperature | 426.44 | 1 | 426.44 | 560.93 | < 0.0001 |
| B—incubation time | 1.93 | 1 | 1.93 | 2.53 | 0.1723 |
| C—inoculum size | 18.08 | 1 | 18.08 | 23.78 | 0.0046 |
| AB | 70.15 | 1 | 70.15 | 92.27 | 0.0002 |
| AC | 30.57 | 1 | 30.57 | 40.21 | 0.0014 |
| BC | 73.59 | 1 | 73.59 | 96.80 | 0.0002 |
| AC | 30.57 | 1 | 30.57 | 40.21 | 0.0014 |
| BC | 73.59 | 1 | 73.59 | 96.80 | 0.0002 |
| A 2 | 437.32 | 1 | 437.32 | 575.25 | < 0.0001 |
| B 2 | 78.33 | 1 | 78.33 | 103.03 | 0.0002 |
| C 2 | 109.03 | 1 | 109.03 | 143.42 | < 0.0001 |
| Residual | 3.80 | 5 | 0.76 | ||
| Lack of fit | 2.44 | 1 | 2.44 | 7.20 | 0.0551 |
| Pure error | 1.36 | 4 | 0.34 | ||
| Cor total | 2215.85 | 14 | |||
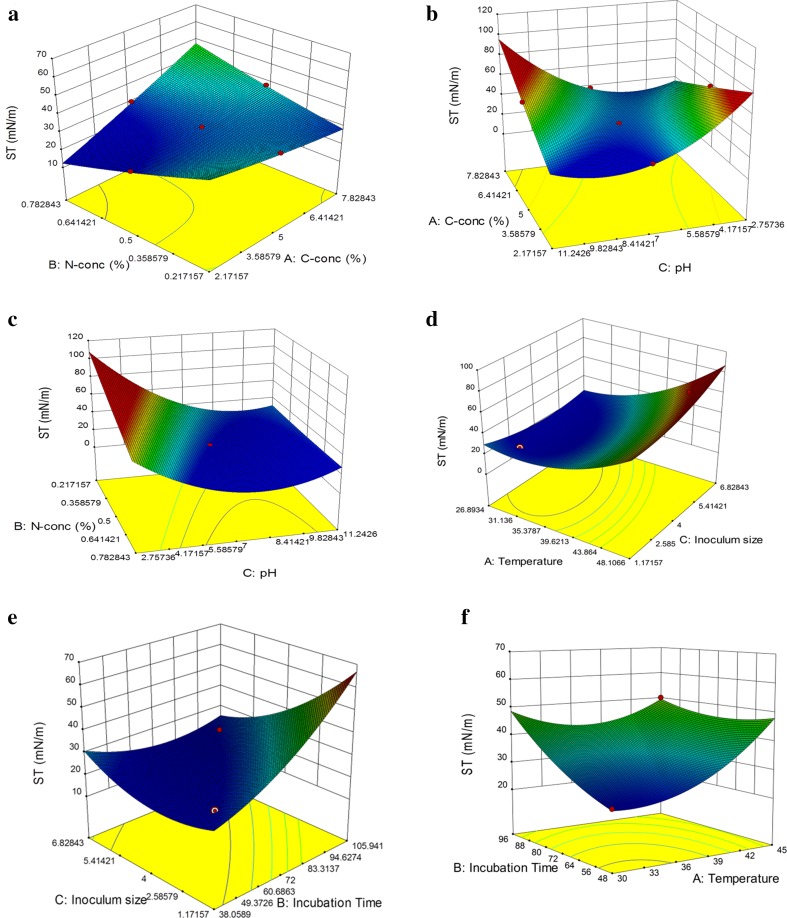
The presented models were employed to construct the fitted 3D-response surfaces for biosurfactant production. Figure 3 demonstrates the response surfaces due to interactive effects of two variables at a time on surface tension of the media fixing the third at the middle level. Figure 3a shows the interactive effect of concentrations of carbon and nitrogen sources on the biosurfactant production fixing the pH value at 7. The maximum biosurfactant production and the least ST of medium appeared at lower concentration of C-source (sodium citrate) and intermediate concentration of N-source (yeast extract). It is supported by the finding that yeast extract used as a nitrogen source also fulfill the carbon requirement (Pereira et al. 2013), therefore it supplemented the primary C-source and resulted in good production even at lower concentration of sodium citrate. Further, higher concentration of yeast extract may inhibit the growth due to its complex nature. Figure 3b presents the effect of concentration of C-source and pH on the ST of media at the fixed mid-value of N-source (0.5%). The low biosurfactant production was observed in acidic medium which increased on moving towards alkaline medium at lower concentration of carbon source. Whilst studying the interactive effect of concentration of N-source and pH on the biosurfactant production, higher biosurfactant production was observed at low N-concentration and neutral pH of the medium at fixed concentration (5%) of C-source (Fig. 3c). On a whole, the highest ST reduction and hence maximum biosurfactant production was observed with 2.17% C-source and 0.5% N-source at pH 7 of medium.
Fig. 3.
Response surface plot showing the interactive effect of two factors on surface tension of media. a C-source% and N-source%, b C-source% and pH of media, c C:N-source% and pH of media, d temperature and inoculum size, e inoculum size and incubation time, f incubation time and temperature. The plots generated using the data obtained from Table 2
Further, the temperature and inoculum size are found to influence the biosurfactant production effectively (Fig. 3d). At a fixed incubation time of 72 h, the low biosurfactant production was observed at high temperature and larger inoculum size due to the poor microbial growth in thermophilic range (Najafi et al. 2010). However, reduction in the surface tension was increased while moving towards lower temperature as well as low inoculum size, indicating the mesophilic nature of microbe. Figure 3(e–f) represents the change in ST of media due to the interactive effect of incubation time with inoculum size and temperature, respectively. The biosurfactant production was found to commence after 30 h of inoculation as the biosurfactants are secondary metabolites excreted in the stationary phase (Xia et al. 2013). High biosurfactant production was observed with smaller inoculum size at lower incubation time while studying their interactive effect fixing temperature at 37 °C (Fig. 3e). Higher biosurfactant production at low incubation time is beneficial on account of less energy consumption. Similarly, the combined effect of temperature and incubation time on biosurfactant production presented the low ST of media at mesophilic range of temperature and low incubation time at fixed inoculum (4%) value (Fig. 3f). The maximum biosurfactant production was observed with 2% inoculum (6.0–7.0 × 108 cells/mL), incubation temperature of 30 °C at incubation time of 48 h after the optimization of growth conditions.
Ultimately, after the optimization of composition of media and growth conditions, the highest ST reduction and hence maximum biosurfactant production was observed in MSM media (pH 7) comprised of (per 100 mL) 2.17 g sodium citrate, 0.5 g yeast extract, 0.1 g KH2PO4·2H2O, 0.1 g K2HPO4·2H2O, 0.02 g MgSO4·7H2O and 0.002 g CaCl2·2H2O (pH 7) incubated at 30 °C/150 rpm for 48 h with 2% fresh inoculum of P. aeruginosa PBS. Under these conditions, 2.65 g/L biosurfactant was produced which was more than thrice as compared to the previous conditions (0.82 g/L).
Emulsification and stability of biosurfactant production
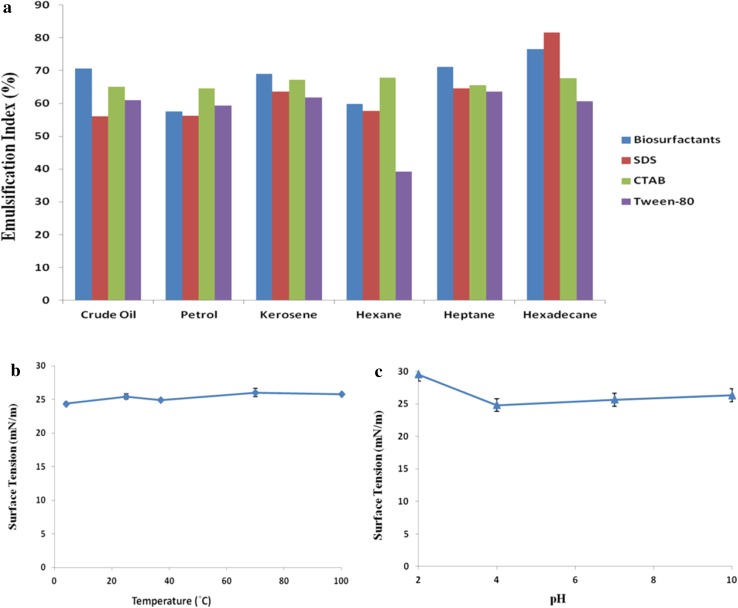
The produced biosurfactant exhibited good emulsification (E 24 50–60%) with heptane, hexadecane, kerosene, petrol as well as with crude oil, whereas low E 24 value was observed with hexane (34.79%). The higher emulsification index value (E 24%) for hexadecane as compared to hexane and heptane was justified by higher emulsification with increase in length of alkyl chains (Pathak and Keharia 2014a). Emulsification value obtained with kerosene, petrol and crude oil depended upon the composition of hydrocarbons. The biosurfactant produced by other P. aeruginosa also exhibited emulsification index of 52–70% with hydrocarbons (Anyanwu and Chukwudi 2010; Kumar et al. 2010). The comparison of E 24 value of biosurfactant (approximately 0.1%) and other chemical surfactants such as anionic SDS (1%), cationic CTAB (1%) and neutral Tween-80 (0.1%) with different hydrocarbons is presented in Fig. 4a. The emulsification index of the biosurfactant was found comparable (or slightly lower) to the other chemical surfactants. More and above, the emulsions formed with biosurfactant were found to be stable for more than 60 days as compared to 15–20 days with chemical surfactants.
Fig. 4.
Emulsification and stability of biosurfactant. a Emulsification indices of biosurfactant and chemical surfactants with various hydrocarbons, b stability study at different temperature, c stability study at different pH
The surface tension of the cell free supernatant possessing biosurfactant remained unchanged over pH 4–10 and temperature even up to 100 °C (Fig. 4b, c). Henceforth, the biosurfactant was stable at wide pH (4–10) and temperature range (4–100 °C) making it suitable for oil recovery application. Many workers also demonstrated the stability of the biosurfactant at wide temperature and pH range (Pathak and Keharia 2014a; Varadavenkatesan and Murty 2013).
Characterization of biosurfactant
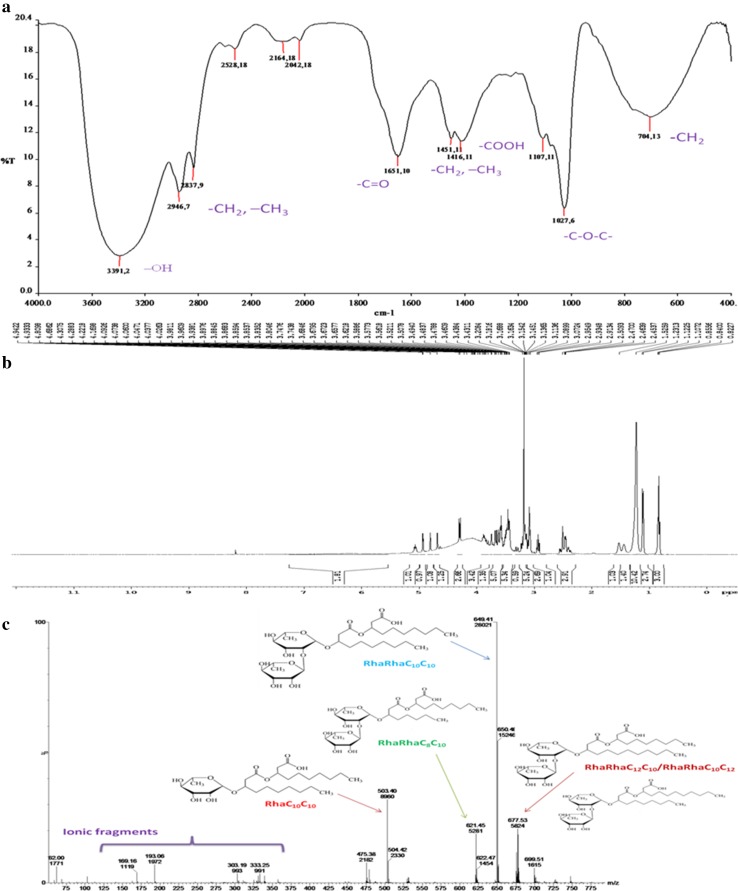
The structural characterization of the extracted biosurfactant was performed by TLC, FTIR, NMR and MS techniques. Two spots appeared on the TLC plates visualized with iodine vapors and DPA reagent which confirmed the presence of lipids and sugar moieties, respectively. No color developed with ninhydrin indicated the absence of protein moiety. TLC technique was generally used for qualitative characterization of extracted biosurfactant as reported earlier (Mukherjee et al. 2008). The spots of glycolipid with retardation factor (R f) of 0.43 and 0.76 directly corresponded to mono-rhamnolipids and di-rhamnolipids, respectively, as supported by the literature (Lan et al. 2015; Antonlou et al. 2015). FTIR absorption spectrum (Fig. 5a) of the biosurfactant also revealed the involvement of rhamnolipids. The strong absorption band in the region of 3394 cm−1 indicated the stretching vibrations of –OH and/or –NH bonds. The absorption peaks at 2946 and 2837 cm−1 confirmed the asymmetrical and symmetrical stretching of methylene group, respectively. The appearance of absorption peak at 1416 cm−1 due to the in plane bending of C–O–H corresponded to carboxylic (–COOH) group (Antonlou et al. 2015). Further the absorption at 1451 and 704 cm−1 indicated the presence of alkyl (–CH2 and –CH3) groups (Pereira et al. 2013). An intense absorption peak at 1651 cm−1 confirmed the presence of carbonyl (–C=O) group. The absorption peak at 1107 cm−1 was assigned to the stretching vibrations of –C–O–C– group of ether linkage of rhamnose (Lan et al. 2015).
Fig. 5.
Characterization of biosurfactant. a The FTIR spectra, b the NMR spectra, c ESI–MS of biosurfactant produced by P. aeruginosa PBS
1H-NMR spectrum (Fig. 5b) of biosurfactant produced by P. aeruginosa PBS also presented the peaks corresponding to rhamnose ring and long hydrocarbon chain. The presence of resonance signals at 0.822–0.856 ppm and 1.23–1.31 ppm corresponded to hydrogen atoms of –CH3 and –(CH2)5– groups of long fatty acid chain, respectively (Raza et al. 2009). The chemical shift of 2.454–2.509 ppm reported the presence of –COOCH2– group (Lan et al. 2015). The rhamnose ring of RhaC10C10 was confirmed by the presence of signal for proton situated at particular positions of the ring, viz. 4.686 ppm for C-1′, 3.621 ppm for C-2′, 3.589 ppm for C-3′ and C-5′, 3.479 ppm for C-4′ and 1.231 ppm for methyl group of ring. Similarly, peaks ranging from 3.438 to 4.686 ppm assigned to proton associated with ring carbons of RhaRhaC10C10 (Raza et al. 2009). Mass spectrometric observations of biosurfactant (Fig. 5c) validated the existence of four homolog structures of rhamnolipid with molecular mass ranging from m/z 503 to 677 (Heyd et al. 2008). The major peaks with m/z 649 corresponded to di-rhamnolipid, RhaRhaC10C10 (Antonlou et al. 2015). Other di-rhamnolipid structures reported with m/z 621 (RhaRhaC8C10) and 677 (RhaRhaC12C10/RhaRhaC10C12). The signal at m/z 503 confirmed the presence of mono-rhamnolipids, RhaC10C10, respectively. The additional minor peaks were detected due to ion fragments of alkyl chains and ester links (Raza et al. 2009). Consequently, it was concluded that the isolated strain P. aeruginosa PBS produced a mixture of four isoforms of rhamnolipids. The rhamnolipids are the most widely studied type of biosurfactant in the members of genus Pseudomonas (Dobler et al. 2016; Heyd et al. 2008).
Application of biosurfactant for enhanced oil recovery
The produced biosurfactant resulted in 56.18 ± 1.59% additional oil recovery (AOR) in laboratory scale sand-pack column test (SPC). Table 4 presents the data of several parameters measured in SPC test including PV, IOS, IOS %, SOR, SOR%, ROS, ROS%, AOR and AOR%. Pore volume (PV) of the column was recorded about 30 mL by flooding with 5% brine solution (w/v). Approximately 18.0–18.5 mL (ROS) of crude oil was entrapped within the sand matrix simulating the natural reservoirs after flooding with crude oil under pressure. Washing of oil-saturated column with a brine solution displaced crude oil. However, water flooding was not sufficient to remove more than 13.76 ± 1.41% of oil from the saturated column which can be considered equivalent to secondary oil recovery phase employed in the oil well. Hence, 86.24 ± 1.41% of initially saturated oil remaining entrapped in the column was attempted to release by the application of biosurfactant and 56.18 ± 1.59% was recovered successfully. The present strain P. aeruginosa PBS producing rhamnolipids appeared as a potential candidate for the oil recovery enhancement.
Table 4.
Sand-pack column test for biosurfactant produced by P. aeruginosa PBS
| Parameters | Sand-pack column test | Mean ± SD | ||
|---|---|---|---|---|
| Trial 1 | Trial 2 | Trial 3 | ||
| PV (mL) | 30.0 | 29.5 | 30.0 | 29.83 ± 0.29 |
| IOS (mL) | 18.5 | 18.0 | 18.0 | 18.17 ± 0.29 |
| IOS (%) | 61.67 | 61.02 | 60.00 | 60.90 ± 0.84 |
| SOR (mL) | 2.6 | 2.2 | 2.7 | 2.50 ± 0.26 |
| SOR (%) | 14.05 | 12.22 | 15.00 | 13.76 ± 1.41 |
| ROS (mL) | 15.9 | 15.8 | 15.3 | 15.67 ± 0.32 |
| ROS (%) | 85.95 | 87.78 | 85.00 | 86.24 ± 1.41 |
| AOR (mL) | 9 | 8.6 | 8.8 | 8.80 ± 0.20 |
| AOR (%) | 56.6 | 54.43 | 57.52 | 56.18 ± 1.59 |
PV pore volume, IOS initial oil saturation, SOR secondary oil recovery after water flooding, ROS residual oil saturation, AOR additional oil recovery, IOS% percent initial oil saturation, SOR% percent secondary oil recovery, ROS% percent residual oil saturation, AOR% percent additional oil recovery
Discussion
Being more beneficial than their chemical counterparts, the use of biosurfactant is an astounding approach in the industrial market (Mouafi et al. 2016). The major obstacles in the economic production are, however, low yielding strains and expensive recovery. Henceforth, it is highly appreciated to explore the new strain capable of producing characteristics biosurfactant at high yield (Pereira et al. 2013). The yield of biosurfactant by native strain can also be enhanced by the optimization of media composition and growth conditions (Sepahy et al. 2005).
Many researchers reported the enormous increase in the yield after the optimization of different parameters using RSM (Najafi et al. 2010) which is an imperative tool to study the influence of multiple factors on a single response. Box–Behnken design (BBD) and central composite rotatable design (CCRD) were the most commonly employed designs for optimization of multiple parameters in RSM ((Najafi et al. 2010). CCRD was used in the present work for the optimization by RSM. Temperature, pH, glucose and salinity were selected as critical factors influencing the biosurfactant production by Najafi et al. (2010). According to Lan et al. (2015), inoculum size, nitrogen source, carbon nitrogen ratio (C/N), concentration of carbon source and concentration of the mineral elements influenced the biosurfactant production effectively.
In the current work, the nature of carbon and nitrogen sources, their concentrations, temperature, pH, inoculum size and incubation time are considered for the optimization of biosurfactant production. Sodium citrate was selected as the best C-source in the biosurfactant production media. This was on the contrary to many works in which sucrose or glucose were reported as the best C-source (Pathak and Keharia 2014a; Pereira et al. 2013). Among the various nitrogen sources, yeast extract was selected as the best nitrogen source. Many other workers also reported the yeast extract as the best nitrogen source in biosurfactant production media (Pathak and Keharia 2014b) whereas others presented sodium nitrate as the best N-source (Venkatesh and Vedaraman 2012).
It is well known that P. aeruginosa strains usually produce rhamnolipids, a type of glycolipids (Dobler et al. 2016). The different strains were found to form different homolog structures of rhamnolipids ranging from m/z 475 to 703 depending upon the number of rhamnose rings and fatty acid chains as well as on the number of carbons in the fatty acid chains (Heyd et al. 2008; Raza et al. 2009). Nineteen homologs of rhamnolipids were produced by P. aeruginosa CPCL as reported by Arutchelvi and Doble (2010), whereas four types of rhamnolipids with m/z values of 621, 649, 650 and 677 were produced by P. aeruginosa PBS.
The biosurfactant produced by isolate PBS reduced the ST of media as low as 23.76 mN/m which was cited first time for Pseudomonas strain in the literature as best of our knowledge. Thorough comparison of existing work with previously studied Pseudomonas strains with respect to surface tension, yield, emulsification index and stability of the biosurfactant is displayed in Table 5. As clear from the data (Table 5), the biosurfactant yield was quite higher as compared to the most of the strains. Though some workers (Kanna et al. 2014) reported slightly higher yield than the presented work, nevertheless, the surface tension reduction was lesser (34.0 mN/m), and moreover, nothing was explained for biosurfactant stability.
Table 5.
Properties of biosurfactant produced by different Pseudomonas strains
| Microbial strain | ST (mN/m) | Yield (g/L) | E 24 (%) | Stability (pH/temperature °C) | References |
|---|---|---|---|---|---|
| P. chlororaphis pBS29-P2-rhlC | – | 0.242 | – | – | Cabrera-Valladares et al. (2006) |
| P. aeruginosa CPCL | 44.0 | – | – | – | Arutchelvi and Doble (2010) |
| P. aeruginosa SP4 | 29.0 | – | – | – | Pornsunthoontawae et al. (2008) |
| P. aeruginosa S6 | 33.9 | – | – | – | Yin et al. (2009) |
| Pseudomonas sp. | – | 0.122 | – | 5–9, 20–50 | Anandaraj and Thivakaran (2010) |
| P. aeruginosa DS10-129 | 27.9 | 0.106 | – | – | Rahman et al. (2010) |
| P. aeruginosa BS-161R | 26.5 | 1.50 | 60–70 | – | Kumar et al. (2010) |
| P. aeruginosa | 34.5 | – | 52.85 | 4–12, 100 | Anyanwu and Chukwudi (2010) |
| P. aeruginosa | – | 0.70 | – | – | Amani et al. (2013) |
| P. stutzeri | 0.68 | – | Joshi and Shekhawat (2014) | ||
| P. aeruginosa TMN | 34.0 | – | 46 | – | Moussa et al. (2014) |
| P. putida MTCC 2497 | 34.0 | 2.80 | – | 11, 100 | Kanna et al. (2014) |
| P. aeruginosa PBS | 23.76 | 2.65 | 50–60 | 4–10, 100 | The present study |
Most of the rhamnolipids produced by Pseudomonas strains were used for the bioremediation and anti-microbial activities (Moussa et al. 2014; Lan et al. 2015). Only a few reports are available for the use of rhamnolipids for the oil recovery (Amani et al. 2013). The rhamnolipids produced by P. aeruginosa MM1011 exhibited only 27% enhanced oil recovery (Amani et al. 2013). Recently, homogeneous glass micromodel was employed to estimate the potential of P. aeruginosa HATH rhamnolipids in MEOR and only 5% oil recovery was reported (Amani 2015). Besides Pseudomonas rhamnolipids, other types of biosurfactants were also used for the oil recovery applications at lab scale and field trials. The strain Acinetobacter baylyi ZJ2 reported 28% displacement of residual oil (Zou et al. 2014), whereas 31 and 43% AOR were attained with Bacillus subtilis strains B30 and K1, respectively (Al-Wahaibi et al. 2014; Pathak and Keharia 2014a). The rhamnolipid produced by P. aeruginosa in the present study was found highly efficient in oil recovery application exhibiting 56.18 ± 1.59% additional oil recovery. Therefore, the present strain P. aeruginosa PBS producing rhamnolipids appeared as a potential candidate for the oil recovery enhancement.
Conclusion
The current work presents the optimization, characterization and application of biosurfactant produced by P. aeruginosa PBS. The biosurfactant production medium and culture conditions were optimized in two sets of experimental runs designed by CCRD under response surface methodology. The optimized parameters were: 2% fresh inoculum, minimal salt medium at pH 7 with 2.17% sodium citrate as C-source and 0.5% yeast extract as N-source, incubation at 30 °C/150 rpm for 48 h. Multiple correlation coefficients (R 2) for two sets 0.9986 and 0.9983, respectively, were justifying the excellent correlation between the experimental and predicted values, whereas the model F values 331.48 and 323.30 assured the significance of the models. The optimization of different parameters displayed about three times increase in the yield (2.65 g/L) of biosurfactant. The composition analysis by thin layer chromatography (TLC), Fourier transform infrared (FTIR) spectroscopy, nuclear magnetic resonance (NMR) and mass spectrometry (MS) characterized the produced biosurfactant as a glycolipid comprising of four homolog structures of rhamnolipid. The biosurfactant was efficient to reduce the surface tension of media up to 23.76 mN/m and allowed high (50–60%) emulsification with heptane, hexadecane, kerosene, petrol as well as with crude oil. The produced biosurfactant appeared as an efficient tool for oil recovery showing stability at wide range of pH (4–10) and temperature (4–100 °C) and 56.18 ± 1.59% additional oil recovery (AOR) in the lab scale sand-pack column experiment.
Acknowledgements
RS acknowledges University Grant Commission (UGC), New Delhi, India for financial support in the form of Senior Research Fellowship (SRF).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Al-Wahaibi Y, Joshi S, Al-Bahry S, Elshafie A, Al-Bemani A, Shibulal B. Biosurfactant production by Bacillus subtilis B30 and its application in enhancing oil recovery. Coll Surf B Biointerf. 2014;114:324–333. doi: 10.1016/j.colsurfb.2013.09.022. [DOI] [PubMed] [Google Scholar]
- Amani H. Study of enhanced oil recovery by rhamnolipids in a homogeneous 2D micromodel. J Pet Sci Eng. 2015;128:212–219. doi: 10.1016/j.petrol.2015.02.030. [DOI] [Google Scholar]
- Amani H, Muller MM, Syldatk CX, Hausmann R. Production of microbial rhamnolipid by Pseudomonas aeruginosa MM1011 for ex situ enhanced oil recovery. Appl Biochem Biotechnol. 2013;170:1080–1093. doi: 10.1007/s12010-013-0249-4. [DOI] [PubMed] [Google Scholar]
- Anandaraj B, Thivakaran P. Isolation and production of biosurfactant producing organism from oil spilled soil. Biosci Tech. 2010;1:120–126. [Google Scholar]
- Anderson K, Li S-C, Li Y-T. Diphenylamine–aniline–phosphoric acid reagent, a versatile spray reagent for revealing glycoconjugates on thin-layer chromatography plates. Anal Biochem. 2002;287:337–339. doi: 10.1006/abio.2000.4829. [DOI] [PubMed] [Google Scholar]
- Antonlou E, Fodelianakis S, Korkakaki E, Kalogerakis N. Biosurfactant production from marine hydrocarbon-degrading consortia and pure bacterial strains using crude oil as carbon source. Front Microbiol. 2015;6:1–14. doi: 10.3389/fmicb.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyanwu, Chukwudi U. Surface activity of extracellular products of a Pseudomonas aeruginosa isolated from petroleum contaminated soil. Int J Environ Sci. 2010;1:225–235. [Google Scholar]
- Arutchelvi J, Doble M. Characterization of glycolipid biosurfactant from Pseudomonas aeruginosa CPCL isolated from petroleum contaminated soil. Lett Appl Microbiol. 2010;51:75–82. doi: 10.1111/j.1472-765X.2010.02858.x. [DOI] [PubMed] [Google Scholar]
- Bharali P, Konwar K. Production and physico-chemical characterization of a biosurfactant produced by Pseudomonas aeruginosa OBP1 isolated from petroleum sludge. Appl Biochem Biotechnol. 2011;164:1444–1460. doi: 10.1007/s12010-011-9225-z. [DOI] [PubMed] [Google Scholar]
- Cabrera-Valladares N, Richardson AP, Olvera C, Trevino LG, Deziel E, Lepine F. Monorhamnolipids and 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs) production using Escherichia coli as a heterologous host. Appl Microbiol Biotechnol. 2006;73:187–194. doi: 10.1007/s00253-006-0468-5. [DOI] [PubMed] [Google Scholar]
- Chandran P, Das N. Biosurfactant production and diesel oil degradation by yeast species Trichosporon asahii isolated from petroleum hydrocarbon contaminated soil. Int J Eng Sci Technol. 2010;2:6942–6953. [Google Scholar]
- Das P, Mukherjee S, Sen R. Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J Appl Microbiol. 2008;104:1675–1684. doi: 10.1111/j.1365-2672.2007.03701.x. [DOI] [PubMed] [Google Scholar]
- Dobler L, Vilela LF, Almeida RV, Neves BC. Rhamnolipids in perspective: gene regulatory pathways, metabolic engineering, production and technological forecasting. New Biotechnol. 2016;33:123–135. doi: 10.1016/j.nbt.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Ferhat S, Alouaoui R, Badis A, Moulai-Mostefa N. Production and characterization of biosurfactant by free and immobilized cells from Ochrobactrum intermedium isolated from the soil of southern Algeria with a view to environmental application. Biotechnol Biotechnol Equip. 2017 [Google Scholar]
- Heyd M, Kohnert A, Tan T-H, Nusser M, Kirschhofer F, Brenner-Weiss G, Franzreb M, Berensmeier S. Development and trends of biosurfactant analysis and purification using rhamnolipids as an example. Anal Bioanal Chem. 2008;391:1579–1590. doi: 10.1007/s00216-007-1828-4. [DOI] [PubMed] [Google Scholar]
- Joshi PA, Shekhawat DB. Screening and isolation of biosurfactant producing bacteria from petroleum contaminated soil. Eur J Exp Biol. 2014;4:164–169. [Google Scholar]
- Kanna R, Sathyanarayan N, Gummadi N, Kumar GS. Production and characterization of biosurfactant by Pseudomonas putida MTCC 2467. J Biol Sci. 2014;14:436–445. doi: 10.3923/jbs.2014.436.445. [DOI] [Google Scholar]
- Kumar CG, Mamidyala SK, Das B, Sridhar B, Devi GS, Karuna MS. Synthesis of biosurfactant-based silver nanoparticles with purified Rhamnolipids isolated from Pseudomonas aeruginosa BS-161R. Microbiol Biotechnol. 2010;20:1061–1068. doi: 10.4014/jmb.1001.01018. [DOI] [PubMed] [Google Scholar]
- Lan G, Fan Q, Liu Y, Chen C, Li G, Liu Y, Yu X. Rhamnolipid production from waste cooking oil using Pseudomonas SWP-4. Biochem Eng J. 2015;101:44–54. doi: 10.1016/j.bej.2015.05.001. [DOI] [Google Scholar]
- Mouafi FE, Elsouda MMA, Moharamb ME. Optimization of biosurfactant production by Bacillus brevis using response surface methodology. Biotechnol Rep. 2016;9:31–37. doi: 10.1016/j.btre.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa TAA, Mohamed MS, Samak N. Production and characterization of di-rhamnolipid produced by Pseudomonas aeruginosa TMN. Braz J Chem Eng. 2014;31:867–880. doi: 10.1590/0104-6632.20140314s00002473. [DOI] [Google Scholar]
- Mukherjee S, Das P, Sivapathasekaran C, Sen R. Enhanced production of biosurfactant by a marine bacterium on statistical screening of nutritional parameters. Biochem Eng J. 2008;42:254–260. doi: 10.1016/j.bej.2008.07.003. [DOI] [Google Scholar]
- Najafi AR, Rahimpoura MR, Jahanmiria AH, Roostaazad R, Arabianb D, Ghobadi Z. Enhancing biosurfactant production from an indigenous strain of Bacillus mycoides by optimizing the growth conditions using a response surface methodology. Chem Eng J. 2010;163:188–194. doi: 10.1016/j.cej.2010.06.044. [DOI] [Google Scholar]
- Pathak KV, Keharia H. Application of extracellular lipopeptide biosurfactant produced by endophytic Bacillus subtilis K1 isolated from aerial roots of banyan (Ficus benghalensis) in microbially enhanced oil recovery (MEOR). 3. Biotech. 2014;4:41–48. doi: 10.1007/s13205-013-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak KV, Keharia H. Identification of surfactins and iturins produced by potent fungal antagonist, Bacillus subtilis K1 isolated from aerial roots of banyan (Ficus benghalensis) tree using mass spectrometry. 3. Biotech. 2014;4:283–295. doi: 10.1007/s13205-013-0151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JFB, Gudina E, Costa R, Vitorino R, Teixeira J, Coutinho J. Optimization and characterization of biosurfactant production by Bacillus subtilis isolates towards microbial enhanced oil recovery applications. Fuel. 2013;111:259–268. doi: 10.1016/j.fuel.2013.04.040. [DOI] [Google Scholar]
- Pornsunthoontawae O, Wongpanit P, Chavadej S, Abe M, Rujiravanit R. Structural and physicochemical characterization of crude biosurfactant produced by Pseudomonas aeruginosa SP4 isolated from petroleum-contaminated soil. Biores Technol. 2008;99:1589–1595. doi: 10.1016/j.biortech.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Rahman PKSM, Pasirayi G, Auger V, Ali Z. Production of rhamnolipid biosurfactant by Pseudomonas aeruginosa DS10-129 in a microfluidic bioreactor. Biotechnol Appl Biochem. 2010 doi: 10.1042/BA20090277. [DOI] [PubMed] [Google Scholar]
- Raza ZA, Khalid ZM, Banat IM. Characterization of rhamnolipids produced by a Pseudomonas aeruginosa mutant strain grown on waste oils. J Environ Sci Health Part A Toxic Hazard Subst Environ Eng. 2009;44:1367–1373. doi: 10.1080/10934520903217138. [DOI] [PubMed] [Google Scholar]
- Satpute SK, Bhawsar BD, Dhakephalkar PK, Chopade BA. Assessment of different screening methods for selecting different biosurfactant producing marine bacteria. Ind J Mar Sci. 2008;37:243–250. [Google Scholar]
- Sepahy A, Assadi M, Saggadian V, Noohi A. Production of biosurfactant from Iranian oil fields by isolated Bacilli. Int J Environ Sci Technol. 2005;1:287–293. doi: 10.1007/BF03325844. [DOI] [Google Scholar]
- Varadavenkatesan T, Murty VR. Production of a lipopeptide biosurfactant by a novel Bacillus sp. and its applicability to enhanced oil recovery. ISRN Microbiol. 2013 doi: 10.1155/2013/621519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh NM, Vedaraman N. Remediation of soil contaminated with copper using Rhamnolipids produced from Pseudomonas aeruginosa MTCC 2297 using waste frying rice bran oil. Ann Microbiol. 2012;62:85–91. doi: 10.1007/s13213-011-0230-9. [DOI] [Google Scholar]
- Xia WJ, Luo ZB, Dong HP, Yu L. Studies of biosurfactant for microbial enhanced oil recovery by using bacteria isolated from the formation water of a petroleum reservoir. Pet Sci Technol. 2013;31:2311–2317. doi: 10.1080/10916466.2011.569812. [DOI] [Google Scholar]
- Yela ACA, Tibaquir MA, Martínez Pineros GAR, Lopez VC, Villamizar SH, Veleza VLN, Abraham WR, Florez MJV, Barrios AFG. A comparison between conventional Pseudomonas aeruginosa rhamnolipids and Escherichia coli transmembrane proteins for oil recovery enhancing. Int Biodeter Biodegrad. 2016;112:59–65. doi: 10.1016/j.ibiod.2016.04.033. [DOI] [Google Scholar]
- Yin H, Qiang J, Jia Y, Ye J, Peng H, Qin H, Zhang N, He B. Characteristics of biosurfactant produced by Pseudomonas aeruginosa S6 isolated from oil-containing wastewater. Process Biochem. 2009;44:302–308. doi: 10.1016/j.procbio.2008.11.003. [DOI] [Google Scholar]
- Zou C, Wang M, Xing Y, Lan G, Ge T, Yan X, Gu T. Characterization and optimization of biosurfactant produced by Acinetobacter baylyi ZJ2 isolated from crude oil-contaminated soil sample toward microbial enhanced oil recovery applications. Biochem Eng J. 2014;90:49–58. doi: 10.1016/j.bej.2014.05.007. [DOI] [Google Scholar]