Abstract
Basic domain-leucine zipper (bZIP) transcription factor, one type of conserved gene family, plays an important role in plant development and stress responses. Although 77 MebZIPs have been genome-wide identified in cassava, their in vivo roles remain unknown. In this study, we analyzed the expression pattern and the function of two MebZIPs (MebZIP3 and MebZIP5) in response to pathogen infection. Gene expression analysis indicated that MebZIP3 and MebZIP5 were commonly regulated by flg22, Xanthomonas axonopodis pv. manihotis (Xam), salicylic acid (SA), and hydrogen peroxide (H2O2). Subcellular localization analysis showed that MebZIP3 and MebZIP5 are specifically located in cell nucleus. Through overexpression in tobacco, we found that MebZIP3 and MebZIP5 conferred improved disease resistance against cassava bacterial blight, with more callose depositions. On the contrary, MebZIP3- and MebZIP5-silenced plants by virus-induced gene silencing (VIGS) showed disease sensitive phenotype, lower transcript levels of defense-related genes and less callose depositions. Taken together, this study highlights the positive role of MebZIP3 and MebZIP5 in disease resistance against cassava bacterial blight for further utilization in genetic improvement of cassava disease resistance.
Keywords: basic leucine zipper (bZIP) transcription factor, cassava (Manihot esculenta), cassava bacterial blight, disease resistance, virus-induced gene silencing (VIGS)
Introduction
The genus Xanthomonas is a kind of plant pathogen that infects a wide range of plant species, including rice, pepper, tomato, citrus, and Nicotiana benthamiana. Xam is the causal pathogen of cassava bacterial blight, resulting in leaf wilting, shoot dieback, and stem vascular necrosis (McCallum et al., 2017). Cassava is one major tropical crop; however, its yield is seriously affected by cassava bacterial blight (Pereiral et al., 2003; Camilo et al., 2005; Quintero et al., 2013; Muñoz-Bodnar et al., 2014). To date, the molecular mechanism underlying cassava in response to bacterial blight is largely unknown, and the identification and utilization of disease-related genes are very limited. With the public available cassava genome sequence (Wang et al., 2014), more and more researches start to isolate cassava genes and investigate their role in stress response, starch metabolism, and postharvest physiological deterioration of cassava storage roots (Okogbenin et al., 2013; Xu et al., 2013; Zeng et al., 2014; Zhao et al., 2015; Wei et al., 2016). Although some disease resistant cassava varieties have been identified (Boher and Verdier, 1994; Wydra et al., 2007), functional characterization of disease-related genes remains limited.
The bZIP transcription factor, one type of conserved gene family, plays an important role in plant growth, development, abiotic and biotic stress responses (Alves et al., 2013; E et al., 2014). With the conserved bZIP domain, bZIP family is one of the largest transcription factors in plants. The bZIP domain contains two structural features, a basic region and a leucine zipper (Alagarasan et al., 2017; Zha et al., 2017). The basic region consists of about 16 amino acid residues and an invariant N-x7-R/K motif, which are responsible for nuclear localization and DNA binding, respectively. The leucine zipper includes a heptad repeat of leucines or other bulky hydrophobic amino acids that are positioned exactly nine amino acids toward the C-terminus, forming a superimposing coiled-coil structure (Alves et al., 2013; E et al., 2014). So far, plant bZIP transcription factors preferentially bind to DNA sequences with a core motif of ACGT, such as A-box (TACGTA), C-box (GACGTC), and G-box (CACGTG) (Foster et al., 1994; Sibéril et al., 2001; Jakoby et al., 2002; Schütze et al., 2008).
Through genome-wide analysis, bZIP gene family has been identified in numerous plant species, including Arabidopsis (Jakoby et al., 2002), pepper (Capsicum annum) (Hwang et al., 2005), rice (Oryza sativa L.) (Nijhawan et al., 2008), maize (Zea mays L.) (Wei et al., 2012), Populus (Ji et al., 2013), Phaseolus vulgaris (Astudillo et al., 2013), castor bean (Ricinus communis L.) (Jin et al., 2014), grapevine (Vitis vinifera) (Liu et al., 2014), cucumber (Cucumis sativus) (Baloglu et al., 2014), Brassica rapa (Hwang et al., 2014), barley (Hordeum vulgare L.) (Pourabed et al., 2015), Brachypodium distachyon (Liu and Chu, 2015), tomato (Solanum lycopersicum L.) (Li et al., 2015), legume (Lablab purpureus L.) (Wang et al., 2015), cassava (Manihot esculenta) (Hu et al., 2016), apple (Malus sieversii L.) (Zhao et al., 2016), and cabbage (Brassica oleracea) (Bai et al., 2016). Functional analysis found that plant bZIPs are widely involved in metabolism (Hartmann et al., 2015; Zhang et al., 2015; Sagor et al., 2016), abiotic stress (salt, drought) (Inaba et al., 2015; Moon et al., 2015; Sornaraj et al., 2016; Xu et al., 2016; Zong et al., 2016; Banerjee and Roychoudhury, 2017) and plant–pathogen interaction (Kim and Delaney, 2002; Shearer et al., 2012; Alves et al., 2013, 2015; Lim et al., 2015).
As transcription factors, plant bZIPs regulate down-stream genes through directly binding to their promoter regions (Foster et al., 1994; Sibéril et al., 2001; Jakoby et al., 2002; Schütze et al., 2008). TGA is widely known in plant defense responses. In Arabidopsis, TGAs interact with NPR1, and binding to the promoters of SA-responsive genes such as PR1 (Alves et al., 2013). Moreover, plant bZIPs regulate disease resistance through interacting with other proteins in defense responses, including the interaction of AtbZIP10 and LSD1 (Kaminaka et al., 2006), NtTGAs and NtWRKY12 (van Verk et al., 2011). Although 77 MebZIPs have been genome-wide identified in cassava (Hu et al., 2016), their in vivo role remains unknown. In this study, the expression pattern and gene function of two MebZIPs (MebZIP3 and MebZIP5) in response to pathogen infection were analyzed. We highlight the positive role of MebZIP3 and MebZIP5 in disease resistance against cassava bacterial blight for further utilization in genetic improvement of cassava resistance to disease.
Materials and Methods
Plant Materials and Growth Conditions
South China 124 variety of Manihot esculenta was used. SC124 cassava and tobacco plants were grown in soil with Hoagland’s solution, at 26–28°C, with 12 h light at 120–150 μmol quanta m-2 s-1 irradiance and 12 h dark cycles.
RNA Isolation and Quantitative Real-Time PCR
Total RNA extraction and cDNA synthesis were performed from plant leaves using RNAprep Pure Plant Kit (TIANGEN, DP441, Beijing, China) and RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, K1622, Waltham, MA, United States), according to the manufacturer’s instruction. The quantitative real-time PCR was performed using cDNA and FastStart Essential DNA Green Master (Roche, 06924204001, Basel, Switzerland) and analyzed using the comparative ΔΔCT method as Wei et al. (2016) described. NtEF1a and MeEF1a were used as internal references for analysis. The primers used for real-time PCR were listed in Supplementary Table S1.
Vector Construction and Transient Expression in Nicotiana benthamiana Leaves
For the vector construction, the coding regions of MebZIP3 and MebZIP5 were first amplified by PCR from plant leave samples. Thereafter, the PCR products were cloned into SpeI and NcoI/SpeI digested modified pCAMBIA1302 (Liu et al., 2015) by restriction enzyme digestion and T4 ligase ligation, respectively. The primers responsible for vector constructs were listed in Supplementary Table S2, and the restriction enzymes and their cutting sites were marked. The vector cassettes were illustrated in Supplementary Figure S1. After DNA sequencing for confirmation, the recombinant plasmids as well as P19 were transformed into Agrobacterium tumefaciens strain GV3101. After syringe infiltrating into Nicotiana benthamiana leaves as Sparkes et al. (2006) described for 2 dpi, the green fluorescent and DAPI-stained cell nuclei in the infiltrated leaf areas were examined using a confocal laser-scanning microscope (TCS SP8, Leica, Heidelberg, Germany).
Generation of Transgenic Tobacco Plants
The transgenic MebZIP3 and MebZIP5 tobacco plants were generated through Agrobacterium-mediated transformation of MebZIP3-pCAMBIA1302 and MebZIP5-pCAMBIA1302 as Horsch et al. (1985) described. Briefly, 14-day-old sterilized tobacco leaves were incubated in Agrobacterium cell suspension for 10 min. Subsequently, the treated leaves were dried with sterilized tissue paper and placed on full MS-Agar medium for co-cultivation. After 2 days, the leaves were transferred to shoot initiation medium with cephalosporin (250 mg L-1) and hygromycin (50 mg L-1) and the surviving seedlings were grown in a greenhouse to produce seeds for further analysis. The T2 transgenic seedlings were selected on MS medium with 50 mg L-1 hygromycin, and the green seedlings with long roots were transferred to soil for further semi-quantitative reverse transcriptase-PCR and seed harvest. The transgenic T3 seedlings were further selected on MS medium with 50 mg L-1 hygromycin to obtain homozygous lines with no segregation on hygromycin resistance, and the independent transgenic T3 lines were used for phenotype analysis.
Virus-Induced Gene Silencing (VIGS) in Cassava
For the vector construction, the partial coding regions of MebZIP3 and MebZIP5 were first amplified by PCR from plat leave samples. Thereafter, the PCR products of these genes were cloned into EcoRI/BamHI digested pTRV2 vector (Liu et al., 2002) by restriction enzyme digestion and T4 ligase ligation. The primers that are responsible for vector constructs were listed in Supplementary Table S2, the restriction enzymes and their cutting sites were marked. The vector cassettes were illustrated in Supplementary Figure S1. After DNA sequencing for confirmation, the recombinant plasmid (pTRV2-MebZIP3 and pTRV2-MebZIP5) as well as pTRV1 were transformed into Agrobacterium tumefaciens strain GV3101. The GV3101 strains were first cultured in 10 ml of LB liquid medium at 28°C for 12 h, and shaken in the new LB liquid culture to reached OD600 at about 2. After diluted to OD600 of 1 by 10 mM MgCl2, 10 mM MES, and 20 mM acetosyringone, the GV3010 strain with pTRV1 and the strain with pTRV2 or MebZIP3-pTRV2 or MebZIP5-pTRV2 were mixed with ratio of 1:1 and co-infiltrated into cassava leaves as Wei et al. (2017) described. After 14 days, the corresponding gene expression assay and disease resistance assay were performed in plant leaves.
Xam Infection
The bacterial pathogen of Xam was first cultured in 10 ml of LB liquid medium at 28°C for 12 h, and shaken in the new LB liquid culture to reach OD600 at about 0.6. After diluted to 108 cfu ml-1 by 10 mM MgCl2 and 0.05% silwet L-77, the Xam was syringe infiltrated into abaxial side of plant leaves. Then the plants with pathogen infection were grown in the green house. At indicated time-points, at least 20 leaves were harvested in every biological repeat. Plant leaves were gently washed by sterile distilled water for 1 min, then the bacterial populations in plant leaves were quantified using 10 μl five 10-fold dilutions of homogenate in LB medium.
Callose Staining
Callose deposition in plant leaves was visualized by callose staining, using alcoholic lactophenol solution, 0.01% (w/v) aniline blue solution, 50% (v/v) glycerol and fluorescence microscope (DM6000B, Leica, Heidelberg, Germany) as Hauck et al. (2003) described.
Reactive Oxygen Species (ROS) Quantification
The endogenous levels of H2O2 in plant leaves were extracted and determined using the peroxide-titanium buffer as Wei et al. (2016) previously described.
Transcriptional Activation Assay in Yeast Cells
For the vector construction, the coding regions of MebZIP3 and MebZIP5 were first amplified by PCR from plant leave samples. Thereafter, the PCR products of these were cloned into NdeI/BamHI and NcoI/BamHI digested pGBKT7 by restriction enzyme digestion and T4 ligase ligation, respectively. The primers that are responsible for vector constructs were listed in Supplementary Table S2, the restriction enzymes and their cutting sites were marked. After DNA sequencing for confirmation, the recombinant plasmids were transformed into yeast strain AH109, according to the manufacturer’s protocol (Clontech, United States). The transformed clones were screened on the SD/-Trp and SD/-His mediums, respectively. The transcriptional activation was evidenced by the growth of yeast cells on SD/-His medium with 5 mM X-α-gal at 30°C for 3 days.
Accession Numbers
The accession numbers and CDS length of all genes are shown as following: MebZIP3 (KU160294, 1,788 bp), MebZIP5 (KU160296, 1,488 bp), MePR1 (Me07G050300, 492 bp), MePR2 (Me10G089800, 492 bp), MePR3 (Me07G050700, 486 bp), MePR4 (Me07G050400, 492 bp), MeEF1a (AF041463, 1,035 bp), NtEF1a (AY206004, 661 bp).
Statistical Analysis
All results in this study were obtained from at least three biological repeats, and the average values and SDs of these biological repeats were shown. In the meanwhile, asterisk symbols (∗) indicting the significant differences at p < 0.05 were also shown after ANOVA analysis.
Results
Expression Profiles of MebZIP3 and MebZIP5 in Response to Stress Treatments
In the previous study (Hu et al., 2016), 77 MebZIPs have been identified in Manihot esculenta Phytozome database v10.31. Herein, a phylogenetic tree between MebIP3/MebZIP5 and their homologs from other plant species were constructed (Supplementary Figure S2A), and the results implied the functional similarities among the bZIP proteins in different plants. Moreover, the conserved bZIP domain of MebZIP3 and MebZIP5 was identified (Supplementary Figure S2B), further indicating that bZIPs are conserved during evolution.
Using quantitative real-time PCR, we found that the transcript levels of MebZIP3 and MebZIP5 were significantly regulated after flg22, Xam, SA and H2O2 treatments (Figures 1A,B). After flg22 treatment, the transcript levels of MebZIP3 and MebZIP5 were down-regulated at 3 h, but largely up-regulated at 6 h. After Xam treatment, MebZIP3 and MebZIP5 transcripts were significantly induced at 6 h. MebZIP3 transcript was largely increased after SA treatment for 3 and 6 h, while MebZIP5 expression was decreased after SA treatment for 1 h. Moreover, MebZIP3 and MebZIP5 transcripts were largely induced after H2O2 treatment for 3 and 6 h (Figures 1A,B). Generally, the transcripts of MebZIP3 and MebZIP5 displayed common expression patterns in response to these treatments, indicating the possible involvement of them in plant disease response. Moreover, we found that MebZIP3 and MebZIP5 were expressed in all assayed organs, with higher transcript levels in cassava stem and storage root than in leaf (Figure 1C).
FIGURE 1.
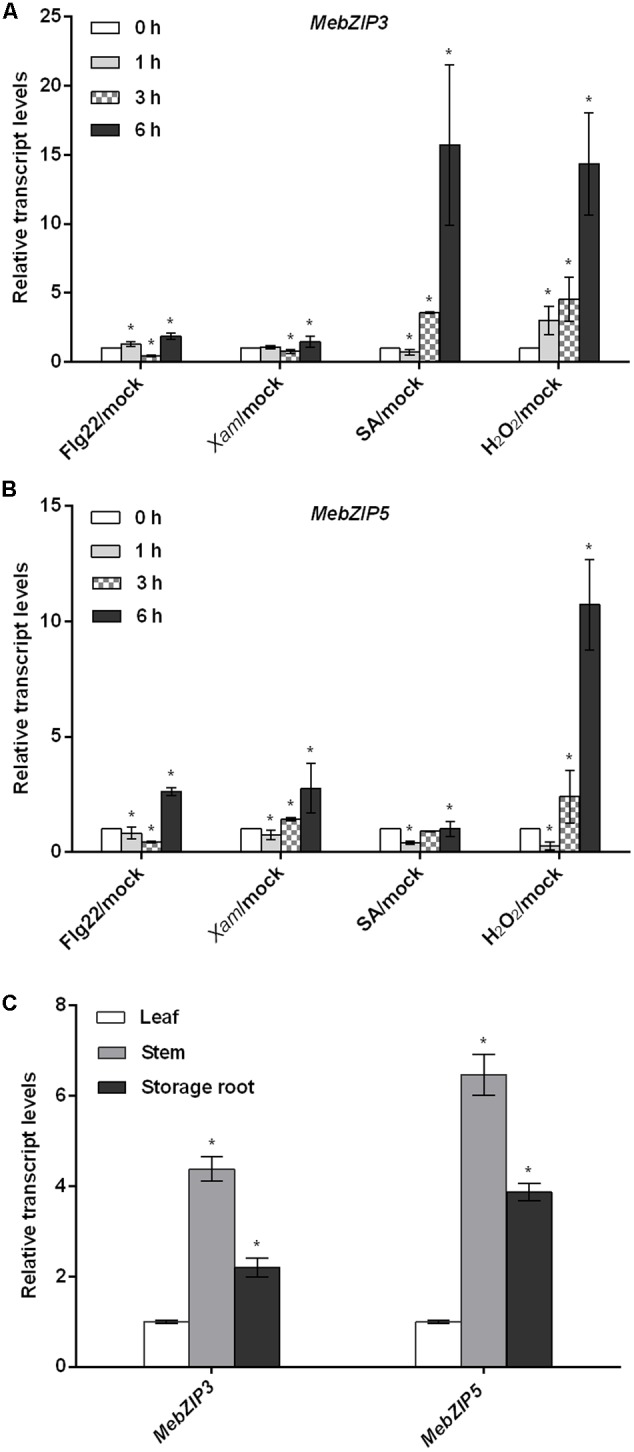
The expression patterns of MebZIP3 and MebZIP5. (A,B) The transcript levels of MebZIP3 (A) and MebZIP5 (B) in response to different treatments. For the assay, cassava leaves were sprayed with water (mock), or 10 μM flg22, or 100 μM SA, or 10 mM H2O2, or syringe infiltrated with 10 mM MgCl2 (mock) or 108 cfu ml-1 of Xam for 0, 1, 3, and 6 h for sample harvest. (C) The transcript levels of MebZIP3 and MebZIP5 in cassava leaf, stem and storage root. Asterisk symbols (∗) indicting the significant differences at p < 0.05.
Subcellular Localization of MebZIP3 and MebZIP5
To investigate the subcellular location of MebZIP3 and MebZIP5, the coding regions of these genes were fused with GFP and transiently expressed in Nicotiana benthamiana leaves. The control vector (35S::GFP)-transformed leaves displayed GFP in both cell nuclei and membrane, consistent with many previous studies (Wei et al., 2017). The GFP signals of MebZIP3-GFP and MebZIP5-GFP were co-localized with DAPI-stained cell nuclei in the infiltrated leaf areas, as marked by the arrow, suggesting that MebZIP3 and MebZIP5 are specifically located in cell nucleus (Figure 2).
FIGURE 2.
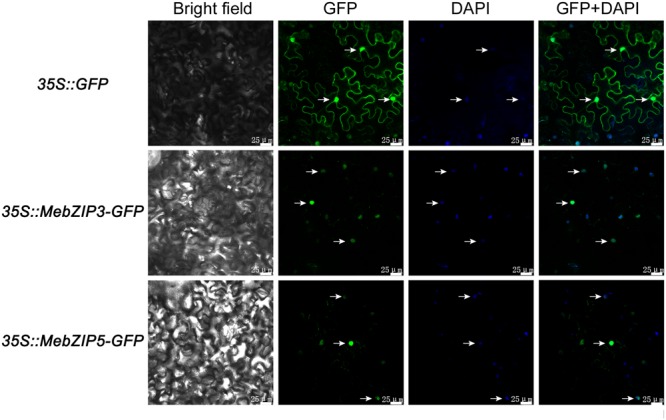
Subcellular localization of MebZIP3 and MebZIP5 in Nicotiana benthamiana leaves. After 2 dpi in Nicotiana benthamiana leaves, green fluorescent with green color and DAPI-stained cell nuclei with blue color were examined using a confocal laser-scanning microscope. The co-localization of GFP with DAPI was marked by arrow. Bars = 25 μm.
Transcriptional Activation Assays of MebZIP3 and MebZIP5
Since bZIPs belongs to transcription factor family, transcription activation assays of MebZIP3 and MebZIP5 were performed in yeast cells. The coding regions of MebZIP3 and MebZIP5 were fused to the GAL4 DNA binding domain in pGBKT7, and the constructs were transformed into yeast strain AH109. As evidenced by the growth of yeast cells and LacZ staining on SD/-His medium with 5 mM X-α-gal, the yeast cells transformed with MebZIP3-pGBKT7 and MebZIP5-pGBKT7 had transcriptional activity (Supplementary Figure S3), suggesting the transcriptional activities of MebZIP3 and MebZIP5 in yeast cells.
Isolation of MebZIP3 and MebZIP5 Overexpressing Plants in Nicotiana benthamiana
To further reveal the in vivo roles of MebZIP3 and MebZIP5, the transgenic plants overexpressing MebZIP3 or MebZIP5 were generated in tobacco. After selection on MS medium with hygromycin, the resistant T1 transgenic seedlings were transferred to soil, and the gene expressions in the overexpressing lines were confirmed by semi-quantitative reverse transcriptase-PCR (Figures 3A,B and Supplementary Figure S4). The corresponding MebZIP3 or MebZIP5 could be examined in the transgenic MebZIP3 or MebZIP5 overexpressing tobacco lines, but could not be amplified in the WT tobacco leaves (Figures 3A,B and Supplementary Figure S4). The transgenic T2 and T3 seedlings were further selected on MS medium with 50 mg L-1 hygromycin to obtain homozygous lines with no segregation on hygromycin resistance. Because no PCR product was detected in the WT sample by semi-quantitative reverse transcriptase-PCR (Figures 3A,B), quantitative real-time PCR was performed to show the relative transcript level in different transgenic T3 lines (Figures 3C,D). Based on the gene transcript level, three independent transgenic T3 lines were used for the phenotype analysis of MebZIP3 (OE1, OE2, and OE3) and MebZIP5 (OE2, OE5, and OE7).
FIGURE 3.
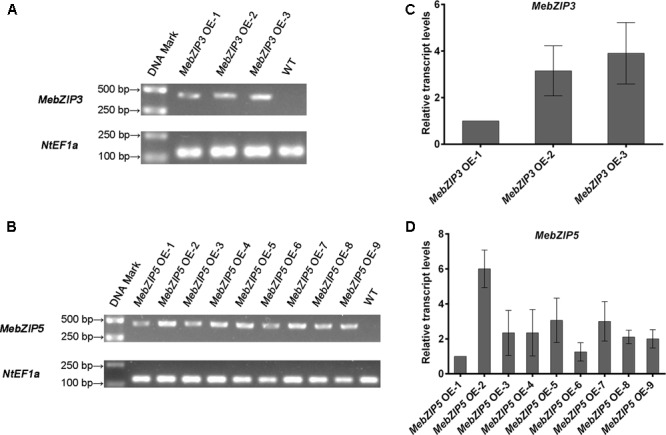
Isolation of MebZIP3 and MebZIP5 overexpressing plants in Nicotiana benthamiana. (A,B) Confirmation of the gene expression in overexpressing lines by semi-quantitative reverse transcriptase-PCR. The expression of NtEF1a was used as the internal control. (C,D) The relative transcript levels in overexpressing lines by quantitative real-time PCR. Asterisk symbols (∗) indicting the significant differences in comparison to mock treatment at p < 0.05.
MebZIP3 and MebZIP5 Confer Improved Disease Resistance against Cassava Bacterial Blight
Although Nicotiana benthamiana is non-host of Xam, its leaves can be infected by Xam with disease symptom and pathogen growth. To investigate the function of MebZIP3 and MebZIP5 in plant disease resistance, the leaf surfaces of WT, MebZIP3, and MebZIP5 transgenic lines were infected with 108 cfu ml-1 of Xam. At 2, 4, and 6 dpi, three MebZIP3 (OE1, OE2, and OE3) and three MebZIP5 (OE2, OE5, and OE7) overexpressing lines exhibited significant less bacterial number in the leaves in comparison to that of WT (Figures 4A–C). Moreover, when Xam was infected, the H2O2 and callose depositions were substantially higher in the overexpressing plant leaves than those in WT (Figures 4D,E). These results suggested that overexpression of MebZIP3 and MebZIP5 conferred improved disease resistance.
FIGURE 4.
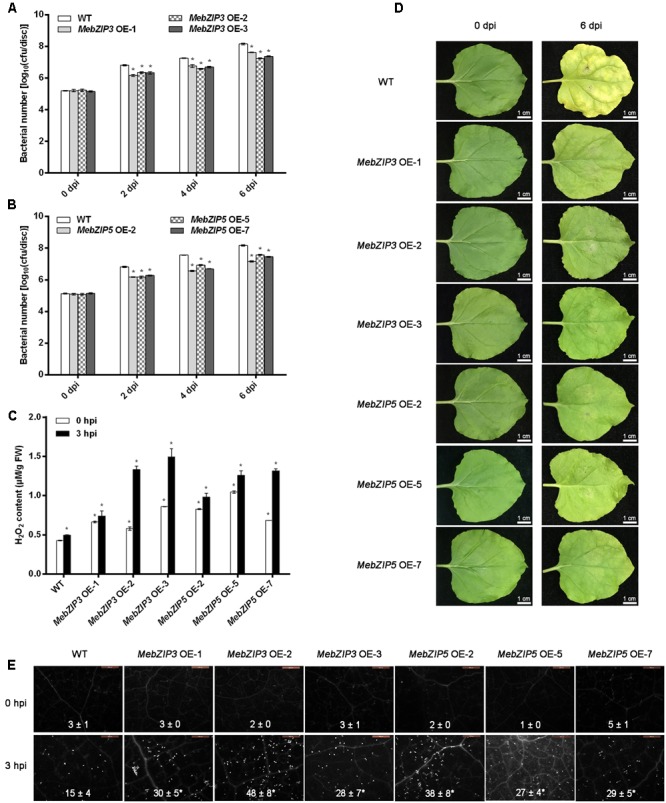
Overexpression of MebZIP3 and MebZIP5 confers improved disease resistance against cassava bacterial blight. (A,B) The bacterial number of Xam in WT, MebZIP3 (A) and MebZIP5 (B) transgenic plant leaves at 0, 2, 4, and 6 dpi of 108 cfu ml-1 of Xam. (C) The pictures showing the plant leaves at 0 and 6 dpi of Xam. Bars = 1 cm. (D) Quantification of H2O2 in plant leaves. (E) The callose depositions in plant leaves. White dots in the figures indicated callose depositions in plant leaves, and the average data was shown. At least 15 cassava leaves were assayed for every biological repeat, and at least three biological repeats were performed. Asterisk symbols (∗) indicting the significant differences in comparison to WT at p < 0.05.
To further confirm the in vivo roles of MebZIP3 and MebZIP5 in cassava defense resistance, we construct the MebZIP3- and MebZIP5-silenced plants through VIGS. As evidenced by the lower transcript of MebZIP3 or MebZIP5, the VIGS plants (pTRV-MebZIP3 and pTRV-MebZIP5) were successfully acquired (Figure 5A). In comparison to mock plants, the VIGS plant (pTRV-MebZIP3 and pTRV-MebZIP5) leaves showed more bacterial number (Figure 5B), lower transcripts of defense-related genes (PR1, PR2, PR3, and PR4) (Figure 6), less callose depositions and lower levels of H2O2 in plant leaves upon Xam infection (Figures 7A,B). Thus, MebZIP3 and MebZIP5 are essential for plant disease resistance in cassava.
FIGURE 5.
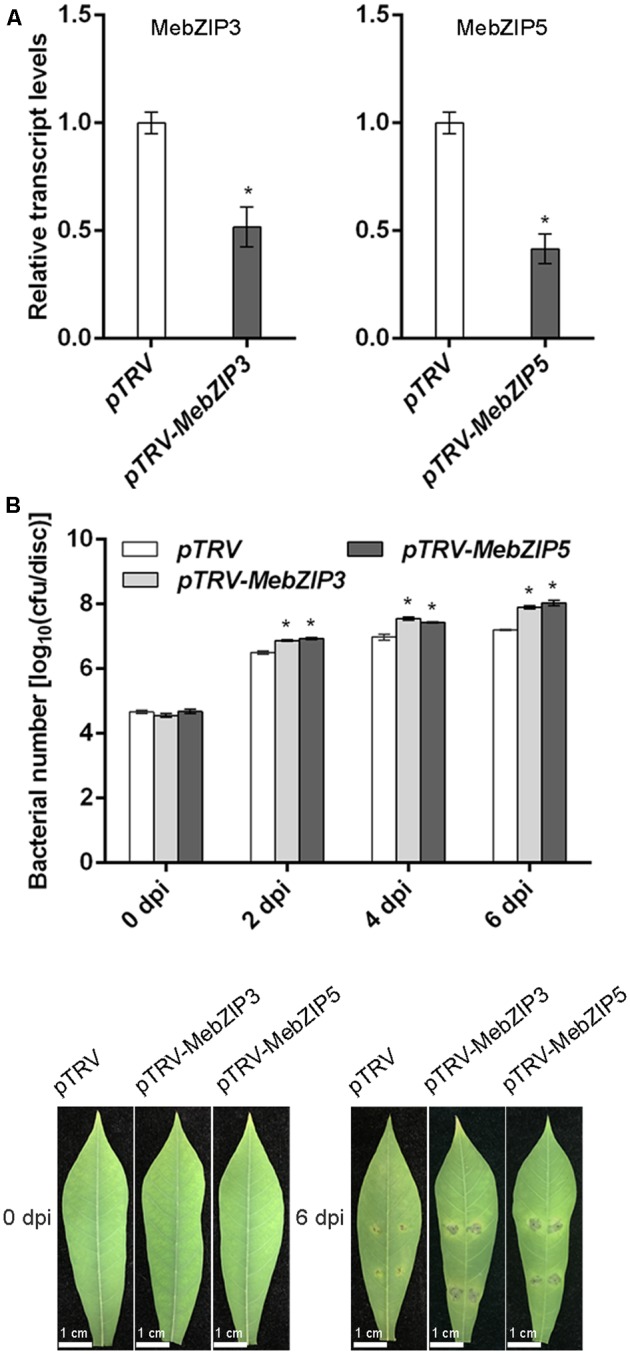
MebZIP3- and MebZIP5-silenced plants result in disease sensitive. (A) The gene transcript levels of MebZIP3 and MebZIP5 in the VIGS plants. (B) The bacterial number of Xam in WT, MebZIP3- and MebZIP5-silenced plant leaves at 0, 2, 4, and 6 dpi of 108 cfu ml-1 of Xam. For the assay, Agrobacterium tumefaciens strains with the recombinant plasmid (pTRV2-MebZIP3 and pTRV2-MebZIP5) as well as pTRV1 were syringe infiltrated into cassava leaves for 14 days, thereafter the corresponding gene expression assay and disease resistance assay were performed. The cassava leaves were syringe infiltrated by 108 cfu ml-1 of Xam for another 0, 2, 4, 6 days, and the bacterial number in the cassava leaves were quantified. At least 15 cassava leaves were assayed for every biological repeat, and at least three biological repeats were performed. Asterisk symbols (∗) indicting the significant differences in comparison to vector transformation at p < 0.05.
FIGURE 6.
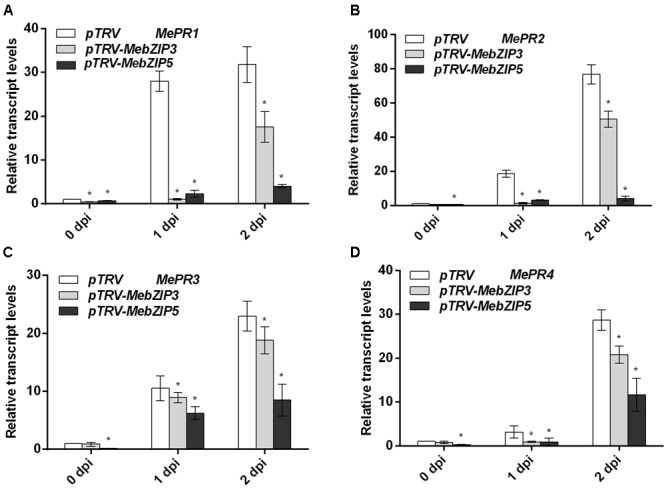
Silencing of MebZIP3- and MebZIP5 regulates the transcripts of defense-related genes. (A–D) The relative transcript levels of MePR1 (A), MePR2 (B), MePR3 (C), and MePR4 (D) in the gene silenced plants. At least 15 cassava leaves were assayed for every biological repeat, and at least three biological repeats were performed. Asterisk symbols (∗) indicting the significant differences in comparison to vector transformation at p < 0.05.
FIGURE 7.
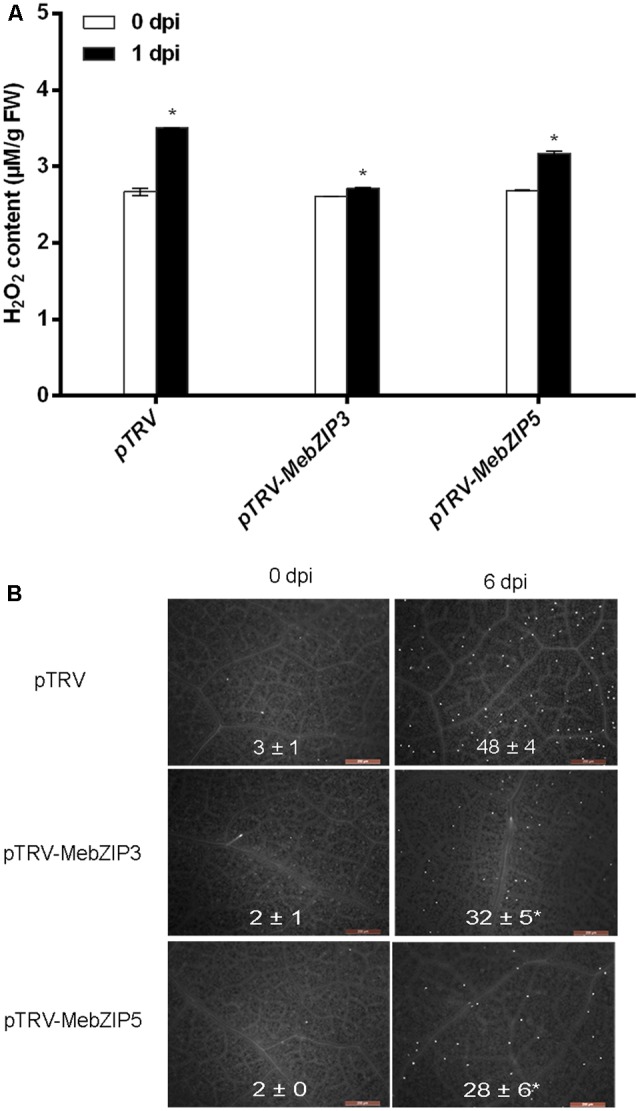
Silencing of MebZIP3- and MebZIP5 regulates callose depositions and ROS level. (A) Quantification of H2O2 in plant leaves. (B) The visualization and quantification of calloses in plant leaves. White dots in the figures indicate callose depositions in plant leaves, and the average data was shown. At least 15 cassava leaves were assayed for every biological repeat, and at least three biological repeats were performed. Asterisk symbols (∗) indicting the significant differences in comparison to vector transformation at p < 0.05.
Discussion
Although some aquatic plants can move, most plants live as sessile organisms. When subjected to abiotic stress (soil salinity, drought, and extreme temperature) and pathogen infection, plants have to response and cope with these stressors. In the long time of evolution, plants have developed several stress-signaling pathways, including signal receptor, protein kinase, transcription factor, and downstream genes. In the core stress-signaling pathways, transcription factors (including bHLHs, bZIPs, ERFs, ZFPs, WRKYs, MYBs, MYCs) play important roles in linking upstream protein kinase with downstream gene expression (Alves et al., 2013; Okogbenin et al., 2013; E et al., 2014; Hu et al., 2016).
Although 77 MebZIPs have been identified in cassava recently (Hu et al., 2016), their in vivo roles remain unknown so far. In this study, gene expression analysis showed that MebZIP3 and MebZIP5 were commonly regulated by flg22, Xam, SA, and H2O2. With the conserved bZIP domain, transcription activity and specific localization in cell nucleus, MebZIP3 and MebZIP5 are confirmed to be transcription factors. Through overexpression in tobacco, we found that MebZIP3 and MebZIP5 conferred improved disease resistance against cassava bacterial blight and more callose depositions. Through VIGS, MebZIP3- and MebZIP5-silenced plants resulted in disease sensitive, lower transcripts of defense-related genes and less callose depositions. These results are consistent with previous studies that plant bZIP transcription factors are widely involved in plant–pathogen interaction (Kim and Delaney, 2002; Shearer et al., 2012; Alves et al., 2013, 2015; Lim et al., 2015). Thus, we highlight the positive role of MebZIP3 and MebZIP5 in disease resistance against cassava bacterial blight for further utilization in genetic improvement of cassava resistance to disease. As reviewed by Alves et al. (2013), TGA is an important bZIP gene in SA signaling. Under control conditions, NPR1 is retained in the cytoplasm as oligomer through S-nitrosylation of NPR1 by NO. When the pathogen is infected, SA induces monomeric NPR1 translocates to the nucleus, and NPR1 interacts with TGA family members (bZIPs), and binds to the promoters of SA-responsive genes such as PR1 (Alves et al., 2013). Although the molecular mechanism of MebZIP-mediated defense response remains elusive, the present study provided strong evidence that MebZIP3 and MebZIP5 are positive regulators of disease resistance against cassava bacterial blight. Plant bZIPs serve as important regulators of defense resistance through two ways. On one hand, plant bZIPs interact with other proteins in defense responses, including the interaction of AtbZIP10 and AtLSD1 (Kaminaka et al., 2006), AtTGAs and AtNPR1 (Alves et al., 2013), and NtTGAs and NtWRKY12 (van Verk et al., 2011). On the other hand, plant bZIPs preferentially bind to DNA sequences with A-box (TACGTA), C-box (GACGTC), and G-box (CACGTG) (Foster et al., 1994; Sibéril et al., 2001; Jakoby et al., 2002; Schütze et al., 2008). Herein, MebZIP3- and MebZIP5-silenced plants had significant effects on the transcripts of other MePRs, the clone and analysis of MePRs promoters will display whether A-box, C-box, and G-box are distributed in these regions. If one of these motifs is distributed in MePRs promoters, the underlying MePRs may be the direct target of MebZIP3 and MebZIP5. Otherwise, the transcripts of MePRs may be affected by MebZIP3 and MebZIP5 indirectly. In further study, the identification of direct targets and interacting proteins of MebZIPs will provide more clues to the underlying mechanism in MebZIPs-mediated defense response in cassava. MebZIP3 and MebZIP5 may interact with other transcription factors to regulate their directly binding to MePRs. As a kind of glucan and plant polysaccharide, callose is directly related with callose-associated cell wall and papillae-associated defense (Hauck et al., 2003). Although the underlying mechanism remains unclear, MebZIP3 and MebZIP5-mediated callose accumulation may also contribute to their effects on disease resistance. Taken together, this is the first study showing the positive effects of MebZIP3 and MebZIP5 in plant disease resistance against cassava bacterial blight.
Author Contributions
HS conceived and directed this study, analyzed the data, wrote and revised the manuscript. XL, SF, WH, GL, and YW performed the experiments, analyzed the data, wrote and revised the manuscript. CH provided suggestions and revised the manuscript. All authors approved the manuscript and the version to be published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Chris R. Somerville, Dr. Jie Zhou, Dr. Yanru Hu, and Dr. Jian Tian for sharing their vector plasmids.
Abbreviations
- bHLH
basic helix-loop-helix
- bZIP
basic domain-leucine zipper
- DAPI
4′,6-diamidino-2-phenylindole
- dpi
days post infiltration
- ERFs
ethylene-responsive element-binding factors
- GFP
green fluorescent protein
- H2O2
hydrogen peroxide
- LSD1
lesions simulating disease resistance 1
- NO
nitric oxide
- NPR1
non-expresser of PR genes
- POX
peroxidase
- PR1
pathogenesis-related gene 1
- ROS
reactive oxygen species
- SA
salicylic acid
- SC124
South China 124
- TGA
TGACGTCA cis-element-binding protein
- VIGS
virus-induced gene silencing
- WT
wild type
- Xam
Xanthomonas axonopodis pv. manihotis
- ZFPs
zinc finger proteins
Funding. This research was supported by the National Natural Science Foundation of China (No. 31760067), the startup funding and the Scientific Research Foundation of Hainan University (No. kyqd1531) to HS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02110/full#supplementary-material
The vector cassettes in this study.
The phylogenetic tree of MebIP3/5 homologs from other plant species. (B) The conserved bZIP domain of MebZIP3 and MebZIP5. Multiple sequence alignment and phylogenetic tress were performed by Clustalx 1.83 and MEGA5.05.
Transcriptional activation assays of MebZIP3 and MebZIP5 in yeast cells.
The original gel images of gene expressions in MebZIP3 and MebZIP5 overexpressing lines by semi-quantitative reverse transcriptase-PCR.
References
- Alagarasan G., Dubey M., Aswathy K. S., Chandel G. (2017). Genome wide identification of orthologous ZIP genes associated with zinc and iron translocation in Setaria italica. Front. Plant Sci. 8:775. 10.3389/fpls.2017.00775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves M. S., Dadalto S. P., Gonçalves A. B., De Souza G. B., Barros V. A., Fietto L. G. (2013). Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14 7815–7828. 10.3390/ijms14047815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves M. S., Soares Z. G., Vidigal P. M., Barros E. G., Poddanosqui A. M., Aoyagi L. N., et al. (2015). Differential expression of four soybean bZIP genes during Phakopsora pachyrhizi infection. Funct. Integr. Genomics 15 685–696. 10.1007/s10142-015-0445-440 [DOI] [PubMed] [Google Scholar]
- Astudillo C., Fernandez A. C., Blair M. W., Cichy K. A. (2013). The Phaseolus vulgaris ZIP gene family: identification, characterization, mapping, and gene expression. Front. Plant Sci. 4:286. 10.3389/fpls.2013.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Zhu W., Hu X., Sun C., Li Y., Wang D., et al. (2016). Genome-wide analysis of the bZIP gene family identifies two ABI5-like bZIP transcription factors, BrABI5a and BrABI5b, as positive modulators of ABA signalling in Chinese cabbage. PLOS ONE 11:e0158966. 10.1371/journal.pone.0158966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloglu M. C., Eldem V., Hajyzadeh M., Unver T. (2014). Genome-wide analysis of the bZIP transcription factors in cucumber. PLOS ONE 9:e96014. 10.1371/journal.pone.0096014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Roychoudhury A. (2017). Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254 3–16. 10.1007/s00709-015-0920-924 [DOI] [PubMed] [Google Scholar]
- Boher B., Verdier V. (1994). Cassava bacterial blight in Africa: the state of knowledge and implication for designing control strategies. Afr. Crop Sci. J. 4 505–509. [Google Scholar]
- Camilo L., Soto M., Restrepo S. (2005). Gene expression profile in response to Xanthomonas axonopodis pv manihotis infection in cassava using a cDNA microarray. Plant Mol. Biol. 57 393–410. 10.1007/s11103-004-7819-3 [DOI] [PubMed] [Google Scholar]
- E Z. G., Zhang Y. P., Zhou J. H., Wang L. (2014). Mini review roles of the bZIP gene family in rice. Genet. Mol. Res. 13 3025–3036. 10.4238/2014.April.16.11 [DOI] [PubMed] [Google Scholar]
- Foster R., Izawa T., Chua N. H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J. 8 192–200. [DOI] [PubMed] [Google Scholar]
- Hartmann L., Pedrotti L., Weiste C., Fekete A., Schierstaedt J., Göttler J., et al. (2015). Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. Plant Cell 27 2244–2260. 10.1105/tpc.15.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck P., Thilmony R., He S. Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. U.S.A. 100 8577–8582. 10.1073/pnas.1431173100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R. B., Fry J. E., Hoffmann N. L., Eichholtz D., Rogers S. C., Fraley R. T. (1985). A simple and general method for transferring genes into plants. Science 227 1229–1231. 10.1126/science.227.4691.1229 [DOI] [PubMed] [Google Scholar]
- Hu W., Yang H., Yan Y., Wei Y., Tie W., Ding Z., et al. (2016). Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 6:22783. 10.1038/srep22783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E. W., Kim K. A., Park S. C., Jeong M. J., Byun M. O., Kwon H. B. (2005). Expression profiles of hot pepper (Capsicum annuum) genes under cold stress conditions. J. Biosci. 30 657–667. 10.1007/BF02703566 [DOI] [PubMed] [Google Scholar]
- Hwang I., Jung H. J., Park J. I., Yang T. J., Nou I. S. (2014). Transcriptome analysis of newly classified bZIP transcription factors of Brassica rapa in cold stress response. Genomics 104 194–202. 10.1016/j.ygeno.2014.07.008 [DOI] [PubMed] [Google Scholar]
- Inaba S., Kurata R., Kobayashi M., Yamagishi Y., Mori I., Ogata Y., et al. (2015). Identification of putative target genes of bZIP19, a transcription factor essential for Arabidopsis adaptation to Zn deficiency in roots. Plant J. 84 323–334. 10.1111/tpj.12996 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Droge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. 10.1016/S1360-1385(01)02223-3 [DOI] [PubMed] [Google Scholar]
- Ji L., Wang J., Ye M., Li Y., Guo B., Chen Z., et al. (2013). Identification and characterization of the Populus AREB/ABF subfamily. J. Integr. Plant Biol. 55 177–186. 10.1111/j.1744-7909.2012.01183.x [DOI] [PubMed] [Google Scholar]
- Jin Z., Xu W., Liu A. (2014). Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 239 299–312. 10.1007/s00425-013-1979-1979 [DOI] [PubMed] [Google Scholar]
- Kaminaka H., Näke C., Epple P., Dittgen J., Schütze K., Chaban C., et al. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25 4400–4411. 10.1038/sj.emboj.7601312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Delaney T. P. (2002). Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica. Plant J. 32 151–163. 10.1046/j.1365-313X.2001.01411.x [DOI] [PubMed] [Google Scholar]
- Li D., Fu F., Zhang H., Song F. (2015). Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genomics 16:771. 10.1186/s12864-015-1990-1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. W., Baek W., Lim S., Han S. W., Lee S. C. (2015). Expression and functional roles of the pepper pathogen-induced bZIP transcription factor CabZIP2 in enhanced disease resistance to bacterial pathogen infection. Mol. Plant Microbe Interact. 28 825–833. 10.1094/MPMI-10-14-0313-R [DOI] [PubMed] [Google Scholar]
- Liu J., Chen N., Chen F., Cai B., Dal Santo S., Tornielli G. B., et al. (2014). Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15:281. 10.1186/1471-2164-15-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu L., Li Y., Jia C., Zhang J., Miao H., et al. (2015). Role for the banana AGAMOUS-like gene MaMADS7 in regulation of fruit ripening and quality. Physiol. Plant 155 217–231. 10.1111/ppl.12348 [DOI] [PubMed] [Google Scholar]
- Liu X., Chu Z. (2015). Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics 16:227. 10.1186/s12864-015-1457-1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 131 777–786. 10.1111/j.1365-313X.2005.02441.x [DOI] [PubMed] [Google Scholar]
- McCallum E. J., Anjanappa R. B., Gruissem W. (2017). Tackling agriculturally relevant diseases in the staple crop cassava (Manihot esculenta). Curr. Opin. Plant Biol. 38 50–58. 10.1016/j.pbi.2017.04.008 [DOI] [PubMed] [Google Scholar]
- Moon S. J., Han S. Y., Kim D. Y., Yoon I. S., Shin D., Byun M. O., et al. (2015). Ectopic expression of a hot pepper bZIP-like transcription factor in potato enhances drought tolerance without decreasing tuber yield. Plant Mol. Biol. 89 421–431. 10.1007/s11103-015-0378-y [DOI] [PubMed] [Google Scholar]
- Muñoz-Bodnar A., Perez-Quintero A. L., Gomez-Cano F., Gil J., Michelmore R., Bernal A., et al. (2014). RNAseq analysis of cassava reveals similar plant responses upon infection with pathogenic and non-pathogenic strains of Xanthomonas axonopodis pv. manihotis. Plant Cell Rep. 33 1901–1912. 10.1007/s00299-014-1667-7 [DOI] [PubMed] [Google Scholar]
- Nijhawan A., Jain M., Tyagi A. K., Khurana J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146 333–350. 10.1104/pp.107.112821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okogbenin E., Setter T., Ferguson M., Mutegi R., Ceballos H., Olasanmi B., et al. (2013). Phenotypic approaches to drought in cassava: review. Front. Physiol. 4:93. 10.3389/fphys.2013.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereiral L. P., Goodwin P. H., Erickson L. (2003). Cloning of a peroxidase gene from cassava with potential as a molecular marker for resistance to bacterial blight. Braz. Arch. Biol. Technol. 46 149–154. 10.1590/S1516-89132003000200002 [DOI] [Google Scholar]
- Pourabed E., Ghane Golmohamadi F., Soleymani Monfared P., Razavi S. M., Shobbar Z. S. (2015). Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis. Mol. Biotechnol. 57 12–26. 10.1007/s12033-014-9797-9792 [DOI] [PubMed] [Google Scholar]
- Quintero A., Pérez-Quintero A. L., López C. (2013). Identification of ta-siRNAs and cis-nat-siRNAs in cassava and their roles in response tocassava bacterial blight. Genomics Proteomics Bioinformatics 11 172–181. 10.1016/j.gpb.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagor G. H., Berberich T., Tanaka S., Nishiyama M., Kanayama Y., Kojima S., et al. (2016). A novel strategy to produce sweeter tomato fruits with high sugar contents by fruit-specific expression of a single bZIP transcription factor gene. Plant Biotechnol. J. 14 1116–1126. 10.1111/pbi.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze K., Harter K., Chaban C. (2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13 247–255. 10.1016/j.tplants.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Shearer H. L., Cheng Y. T., Wang L., Liu J., Boyle P., Després C., et al. (2012). Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol. Plant Microbe Interact. 25 1459–1468. 10.1094/MPMI-09-11-0256 [DOI] [PubMed] [Google Scholar]
- Sibéril Y., Doireau P., Gantet P. (2001). Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 268 5655–5666. 10.1046/j.0014-2956.2001.02552.x [DOI] [PubMed] [Google Scholar]
- Sornaraj P., Luang S., Lopato S., Hrmova M. (2016). Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta 1860 46–56. 10.1016/j.bbagen.2015.10.014 [DOI] [PubMed] [Google Scholar]
- Sparkes I. A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1 2019–2025. 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- van Verk M. C., Neeleman L., Bol J. F., Linthorst H. J. (2011). Tobacco transcription factor NtWRKY12 interacts with TGA2.2 in vitro and in vivo. Front. Plant Sci. 2:32. 10.3389/fpls.2011.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Feng B., Xiao J., Xia Z., Zhou X., Li P., et al. (2014). Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 5:5110. 10.1038/ncomms6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cheng K., Wan L., Yan L., Jiang H., Liu S., et al. (2015). Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics 16:1053. 10.1186/s12864-015-2258-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K., Chen J., Wang Y., Chen Y., Chen S., Lin Y., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19 463–476. 10.1093/dnares/dss026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Hu W., Wang Q., Liu W., Wu C., Zeng H., et al. (2016). Comprehensive transcriptional and functional analyses of melatonin synthesis genes in cassava reveal their novel role in hypersensitive-like cell death. Sci. Rep. 6:35029. 10.1038/srep35029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Liu G., Bai Y., Xia F., He C., Shi H. (2017). Two transcriptional activators of N-acetylseronin O-methyltransferase 2 and melatonin in cassava. J. Exp. Bot. 68 4997–5006. 10.1093/jxb/erx305 [DOI] [PubMed] [Google Scholar]
- Wydra K., Banito A., Kpemoua K. E. (2007). Characterization of resistance of cassava genotypes to bacterial blight by evaluation of leaf and systemic symptoms in relation to yield in different ecozones. Euphytiea 155 337–348. 10.1007/s10681-006-9335-9 [DOI] [Google Scholar]
- Xu J., Duan X., Yang J., Beeching J. R., Zhang P. (2013). Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 161 1517–1528. 10.1104/pp.112.212803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Ali Z., Xu L., He X., Huang Y., Yi J., et al. (2016). The nuclear protein GmbZIP110 has transcription activation activity and plays important roles in the response to salinity stress in soybean. Sci. Rep. 6:20366. 10.1038/srep20366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Chen Z., Xia J., Zhang K., Chen X., Zhou Y., et al. (2014). Chilling acclimation provides immunity to stress by altering regulatory networks and inducing genes with protective functions in cassava. BMC Plant Biol. 14:207. 10.1186/s12870-014-0207-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha L., Liu S., Liu J., Jiang C., Yu S., Yuan Y., et al. (2017). DNA methylation influences chlorogenic acid biosynthesis in Lonicera japonica by mediating LjbZIP8 to regulate phenylalanine ammonia-lyase 2 expression. Front. Plant Sci. 8:1178. 10.3389/fpls.2017.01178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Fu X., Lv Z., Lu X., Shen Q., Zhang L., et al. (2015). A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol. Plant 8 163–175. 10.1016/j.molp.2014.12.004 [DOI] [PubMed] [Google Scholar]
- Zhao J., Guo R., Guo C., Hou H., Wang X., Gao H. (2016). Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family. Front. Plant Sci. 7:376. 10.3389/fpls.2016.00376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Liu P., Shao J., Li C., Wang B., Guo X., et al. (2015). Analysis of different strategies adapted by two cassava cultivars in response to drought stress: ensuring survival or continuing growth. J. Exp. Bot. 66 1477–1488. 10.1093/jxb/eru507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W., Tang N., Yang J., Peng L., Ma S., Xu Y., et al. (2016). Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 171 2810–2825. 10.1104/pp.16.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The vector cassettes in this study.
The phylogenetic tree of MebIP3/5 homologs from other plant species. (B) The conserved bZIP domain of MebZIP3 and MebZIP5. Multiple sequence alignment and phylogenetic tress were performed by Clustalx 1.83 and MEGA5.05.
Transcriptional activation assays of MebZIP3 and MebZIP5 in yeast cells.
The original gel images of gene expressions in MebZIP3 and MebZIP5 overexpressing lines by semi-quantitative reverse transcriptase-PCR.


