Abstract
The Wnt/β‐catenin signalling pathway is pivotal for stem cell function and the control of cellular differentiation, both during embryonic development and tissue homeostasis in adults. Its activity is carefully controlled through the concerted interactions of concentration‐limited pathway components and a wide range of post‐translational modifications, including phosphorylation, ubiquitylation, sumoylation, poly(ADP‐ribosyl)ation (PARylation) and acetylation. Regulation of Wnt/β‐catenin signalling by PARylation was discovered relatively recently. The PARP tankyrase PARylates AXIN1/2, an essential central scaffolding protein in the β‐catenin destruction complex, and targets it for degradation, thereby fine‐tuning the responsiveness of cells to the Wnt signal. The past few years have not only seen much progress in our understanding of the molecular mechanisms by which PARylation controls the pathway but also witnessed the successful development of tankyrase inhibitors as tool compounds and promising agents for the therapy of Wnt‐dependent dysfunctions, including colorectal cancer. Recent work has hinted at more complex roles of tankyrase in Wnt/β‐catenin signalling as well as challenges and opportunities in the development of tankyrase inhibitors. Here we review some of the latest advances in our understanding of tankyrase function in the pathway and efforts to modulate tankyrase activity to re‐tune Wnt/β‐catenin signalling in colorectal cancer cells.
Linked Articles
This article is part of a themed section on WNT Signalling: Mechanisms and Therapeutic Opportunities. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v174.24/issuetoc
Abbreviations
- APC
adenomatous polyposis coli
- ARC
ankyrin repeat cluster
- ARTD
Diphtheria‐toxin‐like ADP‐ribosyltransferase
- AXIN
axis inhibition protein
- B9L
B‐cell CLL/lymphoma 9‐like protein
- CRC
colorectal cancer
- DKK1
Dickkopf‐related protein 1
- DVL
Dishevelled
- FRAP
fluorescence recovery after photobleaching
- GSK3
glycogen synthase kinase 3
- ISC
intestinal stem cell
- LRP5/6
LDL receptor‐related protein 5/6
- NAD+
nicotinamide adenine dinucleotide
- PARdU
PAR‐dependent ubiquitylation
- RING
really interesting new gene
- RNF
RING finger
- SAM
sterile α motif
- TBM
tankyrase‐binding motif
- TNKS/TNKS2
tankyrase
- TNKSi
tankyrase inhibitor
- V5
epitope tag derived from the P and V proteins of paramyxovirus and simian virus 5, respectively
- Wg/WNT
Wingless and its vertebrate orthologue
- WWE
domain named after a motif containing two conserved Trp (W) residues and one conserved Glu (E)
- β‐TRCP
β‐transducin repeats‐containing protein
Regulation of Wnt/β‐catenin signalling by tankyrase‐dependent AXIN poly(ADP‐ribosyl)ation – an overview
The Wnt/β‐catenin signalling pathway plays key roles during embryonic development, tissue homeostasis and regeneration (see Clevers and Nusse, 2012; and Clevers et al., 2014). Central to the pathway is the β‐catenin destruction complex, which tightly controls the levels of nuclear, transcriptionally active β‐catenin (see Stamos and Weis, 2013). Dysregulation of β‐catenin destruction complex function underlies the vast majority of colorectal cancers (CRCs) and other conditions such as fibrosis, neurodegeneration and osteoporosis (see Clevers and Nusse, 2012; and Kahn, 2014). AXIN (AXIN1/AXIN2), the central scaffold of the destruction complex, directly binds all its core components: the scaffolding protein adenomatous polyposis coli (APC), the kinases glycogen synthase kinase 3 (GSK3) and casein kinase 1 and β‐catenin (see Stamos and Weis, 2013) (Figure 1A). (AXIN1 and AXIN2 will be referred to collectively as AXIN where the discussed aspects apply to both.) The complex enables the phosphorylation of β‐catenin at a phosphodegron to prime it for ubiquitylation and subsequent degradation by the proteasome (see Stamos and Weis, 2013). AXIN is thought to be the concentration‐limiting component of the complex (Salic et al., 2000; Lee et al., 2003). Therefore, controlling its abundance is an effective way to regulate β‐catenin destruction. Wnt/β‐catenin signalling is regulated by a wide range of post‐translational modifications (see Gao et al., 2014). The discovery of a regulatory role of tankyrase in Wnt/β‐catenin signalling sparked much excitement given the limited number of known targetable enzymes in the pathway (Huang et al., 2009). Tankyrase, with two human paralogues (TNKS and TNKS2; Tnks and Tnks2 in other species discussed; from here on simply referred to as ‘tankyrase’ where concepts apply to both tankyrases), is a PARP, and as such catalyses the attachment of PAR chains onto its substrates (see Gibson and Kraus, 2012; and Haikarainen et al., 2014a). In addition to its auto‐PARylation activity, tankyrase binds and PARylates AXIN. In turn, PARylation activates the PAR‐dependent E3 ubiquitin ligase really interesting new gene (RING) finger (RNF)146/Iduna, which then ubiquitylates AXIN, tankyrase and itself, targeting the entire complex for proteasomal degradation (Callow et al., 2011; Zhang et al., 2011) (Figure 1A). This process, known as PAR‐dependent ubiquitylation (PARdU) (DaRosa et al., 2015), is thought to occur constitutively and tune the receptiveness of cells to Wnt stimuli by limiting destruction complex formation (Wang et al., 2016b). A tankyrase‐associated ubiquitin‐specific protease (USP25) can de‐ubiquitylate tankyrase, thereby stabilizing it and supporting PARdU of AXIN (Xu et al., 2017a). Recent studies point toward another role of tankyrase, namely in promoting the formation of active, membrane‐localized Wnt signalosomes upon Wnt stimulation (Yang et al., 2016; Wang et al., 2016a) (Figure 1B). Furthermore, additional tankyrase interactors in the Wnt/β‐catenin pathway, other than AXIN, are emerging (Croy et al., 2016). Structure–function studies are providing a detailed picture of the molecular mechanisms by which tankyrase controls Wnt/β‐catenin signalling and are revealing non‐catalytic scaffolding roles of tankyrase (Guettler et al., 2011; Morrone et al., 2012; DaRosa et al., 2015, 2016; Eisemann et al., 2016; Mariotti et al., 2016; Riccio et al., 2016; Xu et al., 2017a). Conserved functions of tankyrase in the Wnt/β‐catenin pathway are being increasingly appreciated from studies in Drosophila and human CRC cell lines (Lau et al., 2013; de la Roche et al., 2014; Yang et al., 2016; Wang et al., 2016b, 2016c). Recently developed tankyrase‐specific catalytic inhibitors are serving as tool compounds and promising preclinical leads for the treatment of CRC and other Wnt‐dependent conditions (Lau et al., 2013; see Haikarainen et al., 2014a). Here, we discuss a selection of recent insights into the roles of tankyrase in the Wnt/β‐catenin signalling pathway. In terms of potential therapeutic applications, we will focus on CRC, given its high incidence and the importance of the Wnt/β‐catenin pathway in its emergence.
Figure 1.
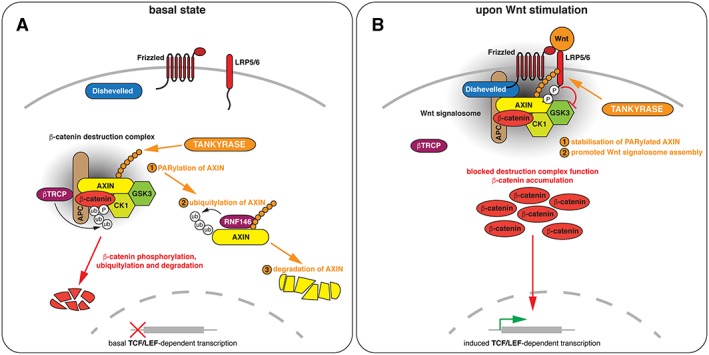
Roles of tankyrase‐dependent poly(ADP‐ribosyl)ation (PARylation) in Wnt/β‐catenin signalling. (A) Under basal Wnt/β‐catenin signalling conditions, PARylation by tankyrase limits the levels of AXIN. Following PARylation, AXIN is ubiquitylated by RNF146 and targeted for proteasomal degradation. (B) Upon Wnt stimulation, PARylated AXIN is stabilized. PARylation facilitates AXIN interaction with LRP5/6 in Wnt signalosomes. Note that AXIN, Dishevelled and tankyrase polymerize and APC dimerizes, and that this is a mechanistically important aspect of the dynamic signalling complexes (Fiedler et al., 2011; Kunttas‐Tatli et al., 2014; Mariotti et al., 2016). For simplicity, proteins are shown as monomers; higher‐order stoichiometry and multivalency are not reflected in the diagrams and nomenclature does not consider multiple paralogues of pathway components.
Tankyrase as a scaffold in Wnt/β‐catenin signalling – a structural perspective
TNKS and TNKS2 share a highly similar multi‐domain organization: five N‐terminal ankyrin repeat clusters (ARCs) for substrate binding are followed by a polymerizing sterile α motif (SAM) domain and a C‐terminal PARP catalytic domain (Figure 2A). With the exception of ARC3, the ARCs act as discrete substrate recognition domains for degenerate 8‐amino‐acid peptides of the consensus R‐[any]‐[any]‐[small hydrophobic or G]‐[D/E]‐G‐[no P]‐[D/E], termed the tankyrase‐binding motif (TBM) (Seimiya, 2002; Seimiya et al., 2004; Guettler et al., 2011). Briefly, an arginine at position 1 and glycine at position 6 of the TBM are essential. A small, hydrophobic residue or glycine is preferred at position 4, and acidic residues are optimal at positions 5 and 8, while proline is disallowed at position 7. Crystal structures of human TNKS2 ARC4 with various TBM peptides (Guettler et al., 2011), of murine Tnks ARC2–3 in complex with Axin1 (Morrone et al., 2012), of human TNKS ARC1–3 bound to TBM peptides derived from leucyl‐cystinyl aminopeptidase (LNPEP), combined with in‐solution structural studies (Eisemann et al., 2016), and of human TNKS ARC5 bound to the TBM of USP25 (Xu et al., 2017a), revealed the architecture of the ARCs and the principles of their substrate recognition. AXIN contains two TBMs in its N‐terminus (Morrone et al., 2012), with the second motif bearing an unusual insertion (Figure 2B–E). The Tnks ARC2–3:Axin1 crystal structure shows a dimeric arrangement of ARC2–3 with each TBM peptide bound to one copy of ARC2 (Morrone et al., 2012). AXIN is also able to contact two ARCs in the same Tnks molecule (Eisemann et al., 2016). TNKS ARCs 1–3 adopt a relatively rigid asymmetric U‐shape, whereas ARCs 4–5 are more dynamic and flexibly linked to ARC1–3 (Eisemann et al., 2016). Multiple AXIN binding sites in the ARCs and two TBMs in AXIN enable their cooperative interaction, but distance and conformational restraints create a preference for bivalent AXIN to either simultaneously bind ARCs 1 and 2, 4 and 5 or 2 and 5, with a preference for combinations involving ARC2, the strongest AXIN binder (Eisemann et al., 2016). When binding to ARCs 2 and 5, AXIN induces a more compact conformation of the ARCs, which might place the PARP domain into closer proximity to ARC‐bound AXIN, in turn promoting AXIN PARylation (Eisemann et al., 2016). Further studies are needed to explore this hypothesis. In the context of polymeric tankyrase, it appears equally likely that AXIN binds separate tankyrase molecules in the same tankyrase filament, with different implications for tankyrase conformation and potentially a further augmentation of cooperativity (Mariotti et al., 2016; Riccio et al., 2016).
Figure 2.
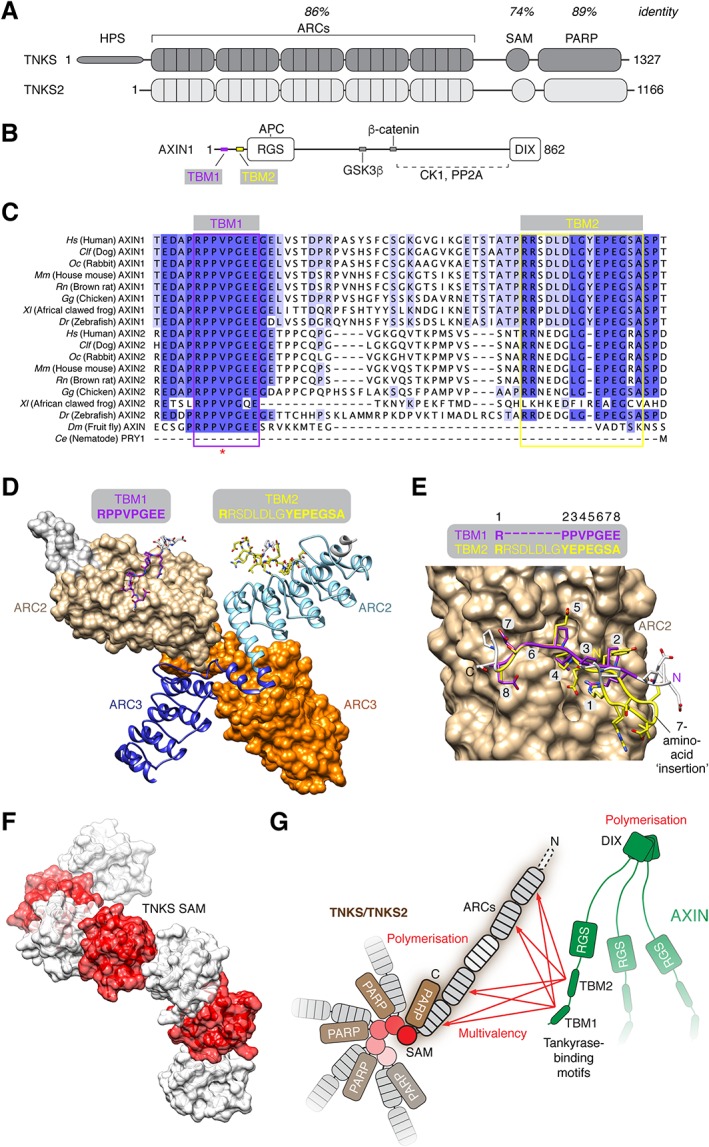
Scaffolding functions of tankyrase. (A) Domain organization of human TNKS and TNKS2. HPS, N‐terminal extension containing homopolymeric stretches of His, Pro and Ser; ARCs, ankyrin repeat clusters; SAM, sterile α motif domain; PARP, catalytic domain. The percentage identity of amino acids between TNKS and TNKS2 is specified for the indicated functional domains. (B) Domain organization of AXIN1. TBM, tankyrase‐binding motif; RGS, regulator of G‐protein signalling domain; DIX, polymerizing domain present in Dishevelled and AXIN (sometimes referred to as DAX domain in AXIN). Binding sites for other β‐catenin destruction complex components are indicated. (C) Multiple sequence alignment of AXIN orthologues/paralogues from the indicated species, coloured by percentage identity. The TBMs are indicated. Note that Drosophila Axin lacks the second TBM. The red asterisk denotes a V26D mutation identified in murine Axin2 (Qian et al., 2011). (D) Structural (surface and cartoon) representation of murine Tnks ARC2–3, bound to the murine Axin1 N‐terminus with two TBMs, shown in stick representation [protein data bank (PDB) code 3UTM] (Morrone et al., 2012). In the crystal, ARC2–3 forms a dimer in which both copies of ARC2 are bound by one of the two TBMs of Axin1, respectively. (E) Detailed structural representation of the Axin1 TBMs (with indicated amino acid positions) on Tnks ARC2. The figure was generated by superimposing both ARC2–3 copies onto each other and displaying ARC2 bound to TBM1. Despite the N‐terminal insertion in TBM2, the arginine (typically at position 1) occupies the same sub‐pocket on the ARC, resulting in a looping out of the intervening residues. (F) Structural (transparent surface and cartoon) representation of a TNKS SAM polymer observed by X‐ray crystallography (PDB code 5JU5) (Mariotti et al., 2016). (G) Avidity model for the interaction of AXIN and tankyrase, modified from Mariotti et al. (2016). Multivalency and polymerization of both tankyrase and AXIN enable avidity contributions in the interaction between both proteins. Note that tankyrase polymerization also promotes its PARP activity (Mariotti et al., 2016; Riccio et al., 2016).
Both TNKS and TNKS2 polymerize through their SAM domains (De Rycker and Price, 2004; Mariotti et al., 2016; Riccio et al., 2016). Recent crystallographic studies of the SAM domains revealed the primarily electrostatic nature of the head‐to‐tail SAM–SAM interfaces within the helical filament (Mariotti et al., 2016; Riccio et al., 2016) (Figure 2F), in agreement with a polymer model guided by NMR studies to identify the residues perturbed upon polymerization (DaRosa et al., 2016). Compatible with the outward‐facing N‐ and C‐termini in the filament (DaRosa et al., 2016; Mariotti et al., 2016; Riccio et al., 2016), full‐length tankyrase indeed polymerizes (De Rycker and Price, 2004; Mariotti et al., 2016; Riccio et al., 2016). TNKS and TNKS2 form cytoplasmic puncta rather than microscopically visible filaments, which may reflect the dynamic nature of the polymers (Mariotti et al., 2016; Riccio et al., 2016). This is consistent with observations made for other proteins containing polymerizing SAM domains (Isono et al., 2013) and for polymerizing AXIN and Dishevelled (DVL)/DVL2 (Schwarz‐Romond et al., 2007; Fiedler et al., 2011; see Bienz, 2014). Supporting this view, polymerization‐deficient TNKS and TNKS2 mutants localize diffusely (Mariotti et al., 2016; Riccio et al., 2016). Luciferase reporter assays revealed that scaffolding through the ARCs and SAM domain is essential for tankyrase function in Wnt/β‐catenin signalling (Mariotti et al., 2016; Riccio et al., 2016). Surprisingly, tankyrase can substantially drive Wnt/β‐catenin activity even in the absence of its catalytic PARP activity, entirely through scaffolding (Huang et al., 2009; Mariotti et al., 2016). Tankyrase polymerization enables productive interactions with the limited pool of AXIN, through avidity effects arising from multivalency and polymerization in both tankyrase and AXIN (Fiedler et al., 2011; Mariotti et al., 2016) (Figure 2G), a requirement that appears overridden by AXIN overexpression (Riccio et al., 2016, and our unpublished observations). The SAM domain and SAM domain‐dependent polymerization are also required for full tankyrase PARP activity (De Rycker and Price, 2004; Levaot et al., 2011; Mariotti et al., 2016; Riccio et al., 2016). Interestingly, while PARP activity is dispensable for tankyrase‐driven Wnt/β‐catenin signalling under basal conditions (Mariotti et al., 2016), it is necessary for tankyrase to potentiate Wnt‐induced β‐catenin activity (Riccio et al., 2016). This may well reflect the recently discovered requirement of AXIN PARylation in the formation of Wnt‐induced signalosomes (see below) (Yang et al., 2016; Wang et al., 2016a) (Figure 1B).
A potential role of tankyrase in the formation of β‐catenin degradasomes
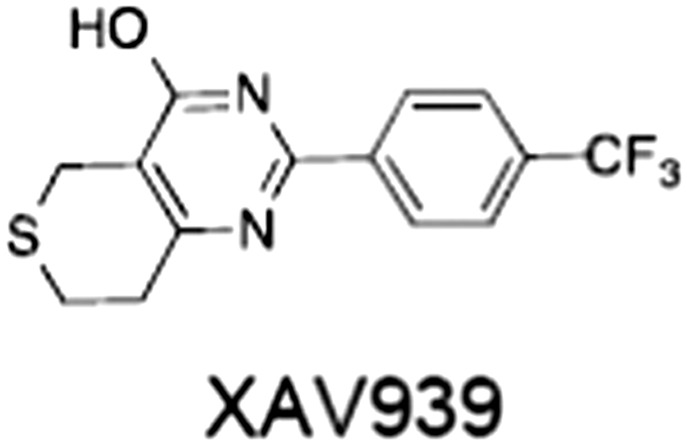
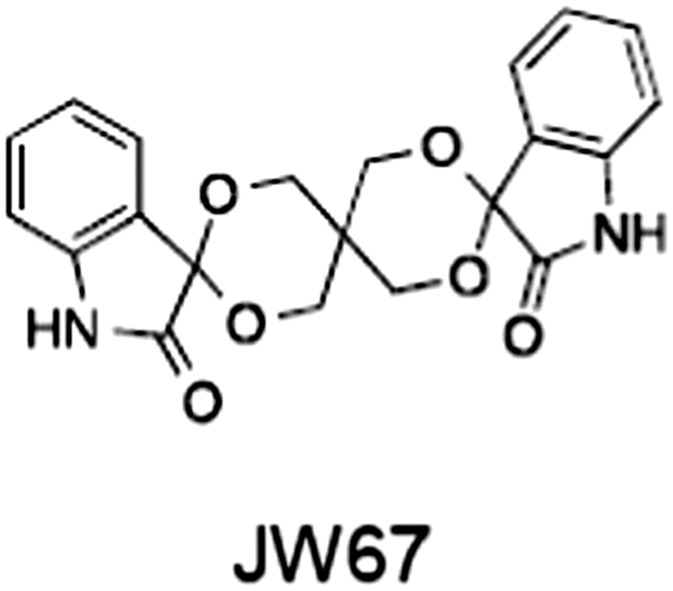
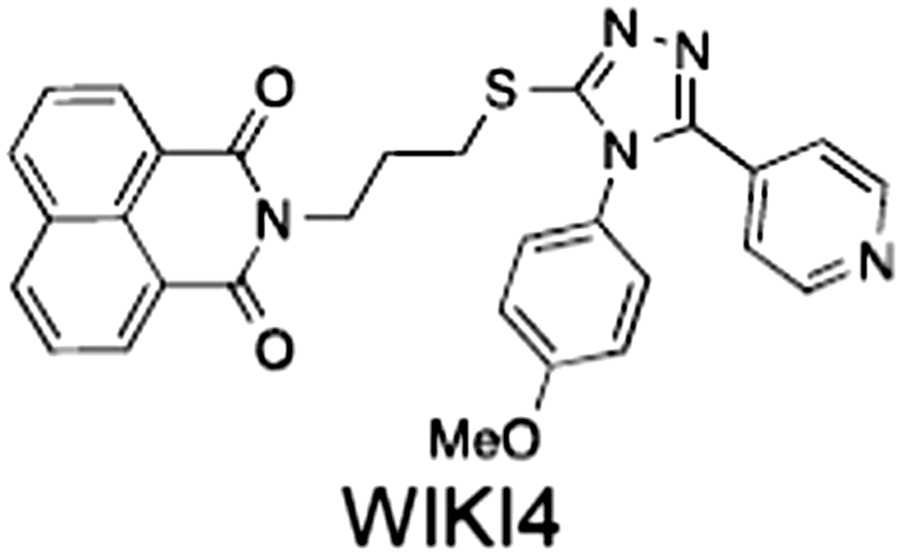
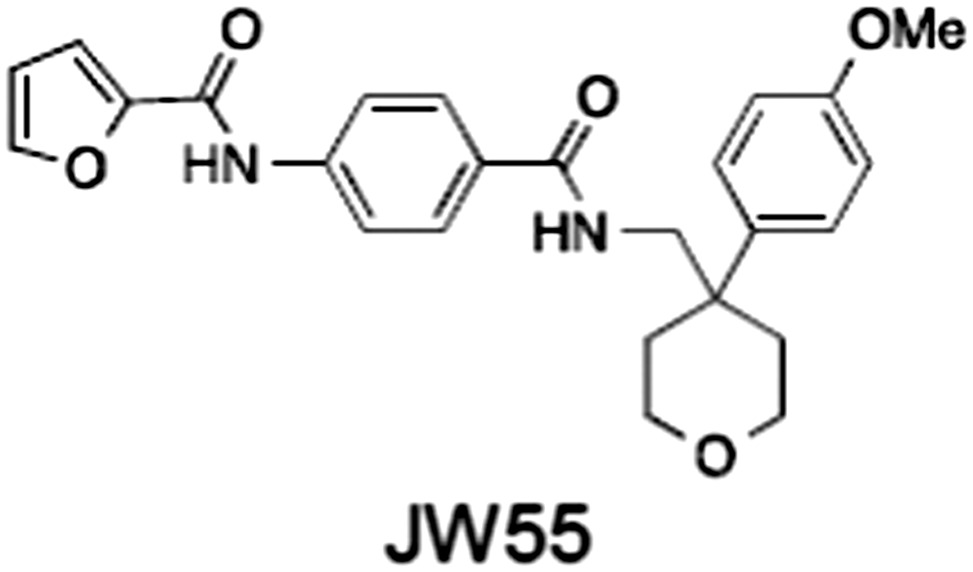
When AXIN is overexpressed or stabilized in APC‐mutant cells by tankyrase inhibition (using the inhibitors JW67, JW74, JW55, XAV939 or G007‐LK; see Table 1 and below for a discussion of inhibitors), it accumulates in cytoplasmic puncta, together with other β‐catenin destruction complex components, including GSK3β, APC, β‐catenin, β‐transducin repeats‐containing protein (β‐TRCP) and tankyrase (Waaler et al., 2011, 2012; de la Roche et al., 2014; Thorvaldsen et al., 2015; Martino‐Echarri et al., 2016) (Figure 3). The puncta gradually disappear upon removal of the tankyrase inhibitor (TNKSi) and re‐establish with subsequent inhibitor treatment (Thorvaldsen et al., 2015). The puncta, referred to as β‐catenin degradasomes, are understood to be ‘morphological correlates’ of β‐catenin destruction complexes, which without tankyrase inhibition are not visible by light microscopy due to the normally low AXIN levels (de la Roche et al., 2014; Thorvaldsen et al., 2015; Pedersen et al., 2016). Several features illustrate functionality of these degradasomes. Firstly, they contain phosphorylated β‐catenin and also colocalize with ubiquitin and β‐TRCP, a component of the E3 ubiquitin ligase responsible for β‐catenin ubiquitylation (Thorvaldsen et al., 2015). Secondly, live‐cell imaging by fluorescence recovery after photobleaching (FRAP) in SW480 cells showed that β‐catenin is rapidly turned over in degradasomes (Thorvaldsen et al., 2015), demonstrating their capacity to degrade β‐catenin.
Table 1.
Tankyrase inhibitors
| No. | Tankyrase inhibitor | TNKS IC50 (nM) | TNKS2 IC50 (nM) | PARP1 [PARP2] IC50 (nM) | IC50 in Wnt reporter assay (nM) | Binding site | References |
|---|---|---|---|---|---|---|---|
| 1 |
(1).
|
11 | 4 | PARP domain: 2194 [114] full‐length:74 [27] | 78 (HEK293, Wnt‐3a) | Nicotinamide | (Huang et al., 2009; Karlberg et al., 2010; Kirby et al., 2012; Shultz et al., 2012; Thorsell et al., 2017) |
| 2 |
(2).
|
6 | 72 | 19 100 [34900] | ‐ | Nicotinamide | (Narwal et al., 2013a, 2013b) |
| 3 |
(3).
|
860 | 52 | >10 000 [>10 000] | ‐ | Nicotinamide | (Larsson et al., 2013) |
| 4 |
(4).
|
‐ | 9 | ‐ | ‐ | Nicotinamide | (Larsson et al., 2013) |
| 5 |
(5).
|
32 | 9 | >5000 | ‐ | Nicotinamide | (Nathubhai et al., 2013, 2016) |
| 6 |
(6).
|
12 | 200 | >10 000 [>10 000] | ‐ | Nicotinamide | (Liscio et al., 2014) |
| 7 |
(7).
|
1.7 | 1.1 | 3400 | ‐ | Nicotinamide | (Kumpan et al., 2015) |
| 8 |
(8).
|
370 | 71 | 5900 [4200] | ‐ | Nicotinamide | (Nkizinkiko et al., 2015) |
| 9 |
(9).
|
12 | ‐ | ‐a | 25 (DLD‐1) | Nicotinamide | (Elliott et al., 2015; Paine et al., 2015) |
| 10 |
(10).
|
32 | ‐ | >1995 | 32 (HEK293, Wnt‐3a) | Nicotinamide | (Thomson et al., 2017) |
| 11 |
(11).
|
8 | 24 | >10 000 | 8 (HEK293, Wnt‐3a) | Nicotinamide | (patent by Feng et al., 2013; Zhong et al., 2016b) |
| 12 |
(12).
|
131 | 56 | >18 750 [>18 750] | 180 (L cells, Wnt‐3a) | Adenosine | (Chen et al., 2009; Huang et al., 2009; Lu et al., 2009; Gunaydin et al., 2012) |
| 13 |
(13).
|
‐ | ‐ | ‐ | 1170 (HEK293, Wnt‐3a) | Unknown | (Waaler et al., 2011) |
| 14 |
(14).
|
2550 | 650 | ‐ | 790 (HEK293, Wnt‐3a) | Adenosine | (Waaler et al., 2011; Shultz et al., 2012; Voronkov et al., 2013) |
| 15 |
(15).
|
‐ | 33 | >19 000 [>19 000] | 215 (HEK293, Wnt‐3a) | Adenosine | (Shultz et al., 2012) |
| 16 |
(16).
|
26 | 15 | >10 000 [>10 000] | ‐ | Adenosine | (James et al., 2012; Haikarainen et al., 2013) |
| 17 |
(17).
|
1800 | 2010 | ‐ | 1230 (HEK293, Wnt‐3a) | Adenosine | (Waaler et al., 2012; Haikarainen et al., 2016) |
| 18 |
(18).
|
46 | 25 | >10 000 [>10 000] | 50 (HEK293, Wnt‐3a) | Adenosine | (Lau et al., 2013; Voronkov et al., 2013) |
| 19 |
(19).
|
1 | ‐ | >85 000 [>170 000] | ‐ | Adenosine | (Bregman et al., 2013a) |
| 20 |
(20).
|
2 | 2 | >85 000 [>170 000] | 12 (DLD‐1) | Adenosine | (Huang et al., 2013) |
| 21 |
(21).
|
31 | 36 | ‐ | 110 (DLD‐1) | Adenosine | (Okada‐Iwasaki et al., 2016) |
| 22 |
(22).
|
8 | 2 | [931] | 36 000 (HEK293, Wnt‐3a) | Dual site | (Bregman et al., 2013b) |
| 23 |
(23).
|
0.2 | 2.5 | ‐ | 1.3 (DLD‐1) | Dual site | (Hua et al., 2013) |
| 24 |
(24).
|
5.1 | 0.1 | 6500 [11600] | 37 (HEK293, Wnt‐3a) | Dual site | (Nathubhai et al., 2017) |
| 25 |
(25).
|
3 | 1 | 2.0 [0.5] | 5 (DLD‐1) | Dual site | (Johannes et al., 2015a) |
| 26 |
(26).
|
‐ | 6 | >19 000 [>30 000] | 3.5 (HEK293, Wnt‐3a) | Dual site | (Shultz et al., 2013) |
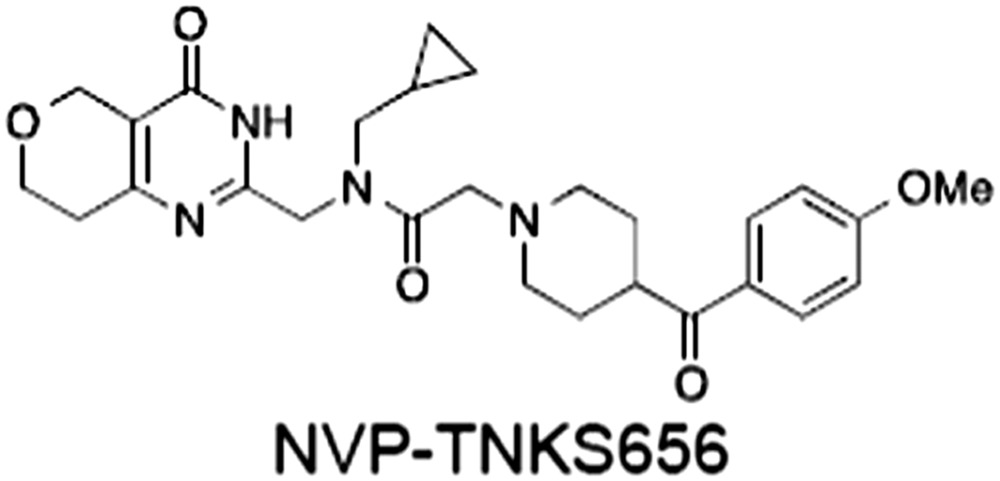
The structures and properties of reported tankyrase‐selective inhibitors are shown, grouped according to binding site. Dual site binders occupy both the nicotinamide and adenosine subsites. One typical structure is given if numerous similar compounds in a series were detailed in the literature. Biochemical IC50 values (i.e. corresponding to inhibition of PARP activity) for TNKS, TNKS2, PARP1 and PARP2 and IC50 values in Wnt/β‐catenin reporter assays are listed if reported. ‘‐’ indicates that values have not been reported. Note that NVP‐TNKS656 is reported as a ‘triple site binder’, also interacting with a hydrophobic ‘nook’ adjacent to the phosphate‐binding groove (Shultz et al., 2012, 2013).
Figure 3.
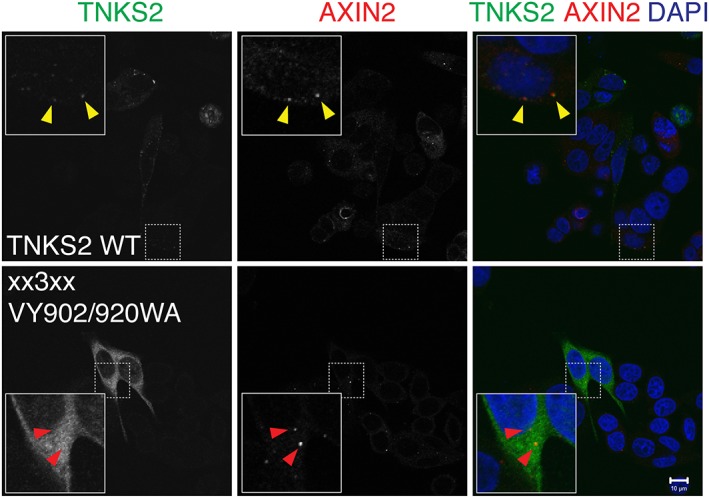
β‐catenin degradasomes induced by TNKSi. SW480 CRC cells were treated with the TNKSi XAV939 and immunostained for AXIN2 (red) and transiently expressed epitope‐tagged wild‐type TNKS2 (TNKS2 WT) or a TNKS2 mutant variant deficient in substrate binding (through site‐directed mutation of ARCs 1, 2, 4 and 5) and polymerization (TNKS2 xx3xx VY903/920WA) (green). Yellow arrowheads indicate colocalization of TNKS2 and AXIN2 in β‐catenin degradasomes; red arrowheads indicate absence of colocalization for the scaffolding‐defective mutant variant of TNKS2. The figure was modified from Mariotti et al. (2016).
Two recent studies suggest that tankyrase plays a structural role in degradasome formation (Thorvaldsen et al., 2015; Martino‐Echarri et al., 2016). Correlative light and electron microscopy suggests that TNKS‐GFP‐containing degradasomes in TNKSi‐treated SW480 cells correspond to electron‐dense and possibly filamentous substructures (Thorvaldsen et al., 2015), perhaps reflecting the polymeric nature of both AXIN and tankyrase. Simultaneous silencing of both tankyrases abolishes degradasome formation (Martino‐Echarri et al., 2016). Other studies have shown that, like tankyrase inhibition, TNKS/TNKS2 RNAi increases the levels of AXIN1/2 (Huang et al., 2009). Strikingly, despite increased AXIN levels, degradasomes are absent under TNKS/TNKS2‐depleted conditions (Martino‐Echarri et al., 2016), supporting a direct, structural role for tankyrase in degradasome formation. FRAP studies have shown that TNKS stably resides in degradasomes, similarly to AXIN (Schwarz‐Romond et al., 2005; Thorvaldsen et al., 2015), although tankyrase and AXIN dynamics have not yet been studied in the same cell. The multivalent interactions of AXIN with tankyrase and the avidity‐enhancing polymerization of both proteins may underlie a scaffolding function of tankyrase in degradasome formation (Mariotti et al., 2016) (Figure 2G). Tankyrase polymerization may be promoted by its catalytic inhibition (De Rycker and Price, 2004), which might offer a potential explanation for the TNKSi‐induced stabilization of degradasomes. APC2, which was recently reported to bind tankyrase (Croy et al., 2016), may also contribute to the avidity‐dependent degradasome assembly, given its numerous AXIN and β‐catenin binding sites.
AXIN2 protein levels rapidly increase upon tankyrase inhibition in SW480 cells, whereas AXIN1 levels do not change until much later (Pedersen et al., 2016; Thorvaldsen et al., 2017). Knockdown of AXIN2 but not AXIN1 prevents degradasome formation (Thorvaldsen et al., 2017), indicating that AXIN2 is the predominant AXIN scaffold in these cells. The TNKSi‐induced accumulation of AXIN2 is dependent on new protein synthesis (Thorvaldsen et al., 2017) and active transcription of the AXIN2 gene (Pedersen et al., 2016). Proteasome inhibition leads to increased levels of phosphorylated β‐catenin in degradasomes (Thorvaldsen et al., 2015), but prolonged proteasome inhibition impairs degradasome formation (Martino‐Echarri et al., 2016; Pedersen et al., 2016). Pedersen et al. (2016) proposed that the transcription factor Forkhead box protein M1 (FoxM1), whose activating phosphorylation is suppressed by proteasome inhibition, controls the AXIN2 gene. AXIN2 is also a Wnt/β‐catenin target gene as part of a negative feedback loop and is highly expressed in APC‐mutated CRC cells (Yan et al., 2001; Jho et al., 2002; Lustig et al., 2002). It is not known why TNKSi induces β‐catenin degradasomes in many CRC cells but far less so in cells with an intact Wnt/β‐catenin pathway (de la Roche et al., 2014). It is possible that tankyrase inhibition strongly represses the AXIN2 gene in Wnt/β‐catenin wild‐type cells. Conversely, APC‐mutant cells might still display residual AXIN2 transcription with AXIN2 protein accumulation arising from continued AXIN2 synthesis and blocked PARdU, leading to the formation of large degradasomes. Degradasome assembly depends on the concentrations of their components (Bienz, 2014), and it is likely that fully functional degradasomes also form in Wnt/β‐catenin wild‐type cells, but these structures remain small.
The structural basis of PARdU
Once PARylated, AXIN is engaged by the PAR‐binding E3 ubiquitin ligase RNF146/Iduna (Callow et al., 2011; Kang et al., 2011; Zhang et al., 2011). RNF146 consists of an RING domain followed by a PAR‐binding WWE domain and an extended C‐terminus, which is predicted to be largely unstructured (Figure 4B). The PAR‐dependency of the enzyme suggested an allosteric activation mechanism; in addition, PAR may serve as a scaffold to enable increased local concentrations of the enzyme (Callow et al., 2011; Kang et al., 2011; Zhang et al., 2011). A crystal structure of RNF146 (RING‐WWE) bound to iso‐ADP‐ribose, an internal unit of PAR (Figure 4A), and an E2 conjugating enzyme is compatible with the allosteric activation of RNF146 (DaRosa et al., 2015) (Figure 4C). Iso‐ADP‐ribose not only binds the WWE but also the RING domain (Wang et al., 2012; DaRosa et al., 2015) and appears to induce restructuring of a loop, which in the apo form of the RING domain extends into the E2‐E3 enzyme contact region, thereby precluding the interaction (Figure 4C). The restructured loop residues become part of an extended central helix in the RING domain, which no longer obstructs E2 binding (Figure 4C), a model supported by NMR spectroscopy and mutagenesis (DaRosa et al., 2015). The extended C‐terminus of RNF146 directly binds tankyrase via five proposed TBMs (DaRosa et al., 2015) (Figure 4B). It is noteworthy that all of these TBMs are atypical in their length or sequence, suggesting that they might be of relatively low individual affinity and may need to act collectively to recruit tankyrase. A model in which the tankyrase:RNF146 complex can still bind other TBM‐containing proteins via the multivalent ARCs implies that the substrate specificity of tankyrase determines RNF146 substrate specificity (DaRosa et al., 2015). Like many PARPs, tankyrase modifies itself, and not all tankyrase binders are also PARylated (Bae, 2002; Guettler et al., 2011; Bisht et al., 2012). This raises the interesting possibility that non‐PARylated tankyrase binders may still be ubiquitylated by RNF146 present in the complex, with RNF146 getting activated by tankyrase auto‐PARylation or PAR attachment to different, simultaneously bound substrates.
Figure 4.
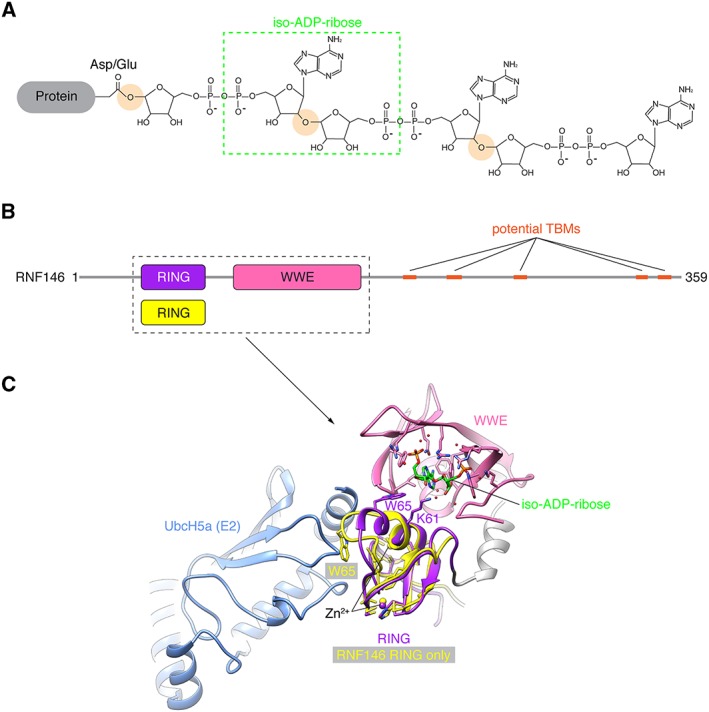
Allosteric regulation of RNF146/Iduna by PAR binding. (A) Structure of a linear PAR chain, here attached to Asp/Glu. The O‐glycosidic bonds linking ADP‐ribose units are highlighted. The green box indicates iso‐ADP‐ribose. Tankyrase is thought to generate linear PAR chains (Rippmann et al., 2002); PAR branches are therefore omitted. (B) Domain organization of human RNF146. Potential TBMs are indicated (DaRosa et al., 2015). The boxed area, which includes the isolated RING domain, corresponds to (C). (C) Structural representation of RNF146 bound to iso‐ADP‐ribose [protein data bank (PDB) code 4QPL] and the E2 enzyme UbcH5a (DaRosa et al., 2015). The isolated RING domain of RNF146 (PDB code 2D8T, one representative of the solution structure ensemble) is superimposed (DaRosa et al., 2015). Domains and corresponding Zn2+ ions are colour‐coded as in (B). Key residues involved in PAR coordination and the allosteric switch are shown in stick representation. Note the clash occurring between the RING domain (yellow) and the E2 enzyme (blue) in the absence of the PAR ligand, and the conformational change upon PAR binding, resulting in a reorientation of Trp65.
While tankyrase RNAi stabilizes AXIN and reduces β‐catenin‐dependent transcription in certain CRC cell lines (Huang et al., 2009; Callow et al., 2011), silencing of RNF146 fails to increase AXIN levels in HCT‐15 or SW480 CRC cells, both of which bear APC truncations, and does not inhibit Wnt/β‐catenin signalling in HCT‐15 cells (Callow et al., 2011). This suggests that there are alternative pathways for the degradation of PARylated AXIN in these cells (Callow et al., 2011). The existence of numerous RING‐type E3 ubiquitin ligases with PAR‐binding WWE domains suggests that functional redundancies may exist (Wang et al., 2012).
Tankyrase and APC set a threshold for Wnt responsiveness by limiting AXIN abundance
To assess the maximum AXIN level still allowing productive Wnt/β‐catenin signalling, Wang et al. (2016b) engineered flies overexpressing C‐terminally V5 epitope‐tagged Axin (Axin‐V5). (Note that Drosophila has a single Axin paralogue; see Figure 2C.) Despite an up to fourfold Axin‐V5 overexpression, flies develop normally with only a mild defect attributable to inhibited Wg signalling (Wang et al., 2016b), in agreement with previous studies (Peterson‐Nedry et al., 2008). This suggested that Axin‐V5 is still subject to physiological regulation at this level. Likewise, loss of tankyrase (of which there is also only a single paralogue in Drosophila) results in a mild (two‐ to threefold) increase in endogenous Axin abundance in larvae, without measurable developmental effects (Feng et al., 2014; Wang et al., 2016b, 2016c). However, developmental defects and loss of Wg/Armadillo target gene expression are observed when Axin‐V5 is expressed in wing imaginal discs in a tankyrase null background, which results in a further threefold increase of Axin‐V5 levels, positioning the inhibitory threshold for Axin three‐ to ninefold above endogenous‐regulated levels (Wang et al., 2016b). This illustrates tankyrase's strong capacity to buffer negative regulation of Wg signalling by Axin, a phenomenon also seen in mammalian cells (Mariotti et al., 2016). These observations are compatible with the previous finding that knockdown of tankyrase in the developing wing only leads to a Wg phenotype if Axin is simultaneously overexpressed (Feng et al., 2014).
Besides their role in promoting β‐catenin/Armadillo degradation, Apc/Apc2 also play a positive role in regulating Wg/Armadillo signalling by post‐transcriptionally limiting Axin levels (Takacs et al., 2008; Wang et al., 2016b). Accordingly, Axin‐V5 accumulates in imaginal discs lacking Apc1 and Apc2 (Wang et al., 2016b). The regulation of Axin levels by Apc is strictly dependent on the Apc‐binding regulator of G‐protein signalling (RGS) domain of Axin, suggesting that a physical interaction between the two proteins is required (Wang et al., 2016b). Surprisingly, while the ability of tankyrase to destabilize Axin is independent of Apc, the TBM of Axin is necessary for Axin destabilization by Apc, suggesting that tankyrase binding is required (Wang et al., 2016b). Alternatively, the Axin N‐terminus may play a tankyrase‐independent role in Axin regulation by Apc. The mechanism by which Apc destabilizes Axin remains unclear, and it will be interesting to decipher whether PARdU is involved. A recent report suggests the existence of a TBM in Apc2 (Croy et al., 2016). Tankyrase may bind to both Apc2 and Axin, thereby providing an additional scaffolding role. The existence of partially separable degradation pathways for Axin, through tankyrase and Apc/Apc2, may explain the mild Tnks null phenotype in flies (Wang et al., 2016b). By analogy, Wang et al. (2016b) propose that if AXIN regulation by APC is lost in APC‐mutant CRC cells, then these cells might be particularly susceptible to tankyrase inhibition, in contrast to APC wild‐type cells, opening the possibility for selectively targeting APC‐mutant cells. This is an interesting idea for further exploration.
Tankyrase mouse models
The physiological role of tankyrase in Wnt/β‐catenin signalling is still far from being fully understood. In mice, loss of both tankyrases gives rise to embryonic lethality, without indication that lethality is attributable to defective Wnt/β‐catenin signalling (Chiang et al., 2008). Individual knockout of either Tnks or Tnks2 results in non‐pathogenic phenotypes, but again, there is no sign of a dysregulated Wnt pathway at the phenotypic level (Chiang et al., 2006; Hsiao et al., 2006; Chiang et al., 2008). One may speculate that embryonic lethality in the double knockout arises from another essential role of tankyrase in development or pleiotropy due to the complex involvement of tankyrase in many biological processes, masking a function in Wnt/β‐catenin signalling. Moreover, these observations point toward a substantial functional redundancy between the two tankyrases. In the absence of conditional Tnks/Tnks2 knockout mice, the role of tankyrase in Wnt‐dependent physiology is difficult to assess. However, an Axin2 V26D mutation, which maps to the stronger N‐terminal TBM of murine Axin2 (Figure 2C), also results in embryonic lethality but with an identifiable Wnt phenotype (Qian et al., 2011). The mutation is expected to abolish tankyrase binding (Guettler et al., 2011), compatible with increased Axin2 levels and reduced Wnt/β‐catenin signalling in most tissues (Qian et al., 2011). Surprisingly, increased rather than decreased Wnt/β‐catenin signalling is seen in the late primitive streak, a structure that marks the beginning of gastrulation and the definition of body axes, and consequential formation of ectopic tails (Qian et al., 2011). This observation points to complex functions of the tankyrase‐Axin interaction. The Axin2 mutation likely exposes a physiological role of tankyrase in Wnt/β‐catenin signalling that may have been masked in the Tnks/Tnks2 double‐knockout mice, although alternative functions of the Axin N‐terminus cannot be excluded. A study otherwise focussing on the role of tankyrase in glucose metabolism corroborates the tankyrase‐Axin‐β‐catenin link in vivo: adipocyte‐specific loss of the Tnks catalytic domain stabilizes Axin1 and reduces the levels of active β‐catenin in adipose tissue (Zhong et al., 2016a).
Tankyrase controls adult intestinal stem cell homeostasis in Drosophila
A more detailed analysis of Tnks mutant flies revealed a sharp drop in viability upon nutrient limitation, paralleled by an accumulation of Axin and hyperproliferation of intestinal stem cells (ISCs) in the midgut (Wang et al., 2016c). The Drosophila midgut can be subdivided into several morphologically and physiologically distinct domains. It displays high levels of Wg target gene transcription close to the inter‐domain boundaries, which may represent source areas for the Wg ligand (Buchon et al., 2013; Tian et al., 2016). Loss of tankyrase distant from the midgut‐hindgut boundary, where β‐catenin/Armadillo activity is low, abolishes the activation of Wg reporters in this region (Wang et al., 2016c). This is not the case in areas close to the midgut‐hindgut boundary, where Wg signalling is high. The authors suggested that by counteracting Axin, tankyrase amplifies β‐catenin/Armadillo activity in compartments of otherwise low pathway activity. They hypothesized that context‐specific roles of tankyrase may explain seemingly contradicting in vivo functions of tankyrase in flies and zebrafish (Huang et al., 2009; Feng et al., 2014) and that similar mechanisms may be in place where Wnt gradients are observed in vertebrates, for example, in the gut (Wang et al., 2016c). (See below for a further discussion of tankyrase's roles in the gastrointestinal tract.) Interestingly, hyperproliferation of ISCs is non‐cell‐autonomous: it does not result from tankyrase loss or inhibited Wg/Armadillo signalling in ISCs but in enterocytes (Tian et al., 2016; Wang et al., 2016c). The authors suggested that decreased Wg signalling in enterocytes upon loss of tankyrase promotes JAK/STAT signalling in ISCs, which in turn drives their proliferation during homeostasis.
A role of AXIN PARylation in Wnt signalosome assembly
While tankyrase's role in PARdU of AXIN is well established, two recent studies point to a novel function in Wnt signalosome assembly (Yang et al., 2016; Wang et al., 2016a) (Figure 1B). During Drosophila embryonic development, the abundance of weakly expressed Axin‐V5 changes in a biphasic manner (Yang et al., 2016). Initially distributed uniformly in the ectoderm, Axin‐V5 first accumulates in segmental stripes marked by Wg induction. Secondly, with progressing development, Axin levels drop specifically in the Wg stripes, an observation analogous to delayed AXIN destabilization in mammalian cells (Li et al., 2012 and references therein). In embryos expressing an Axin‐V5 mutant variant lacking the TBM or in embryos lacking tankyrase, Axin‐V5 levels are uniformly high. Early Axin‐V5 stripes fail to form, with a concomitant loss of normal Wg target gene expression. Importantly, this is not merely due to increased Axin‐V5 levels under these circumstances since strong Axin‐V5 overexpression does not copy this phenotype (Yang et al., 2016). The authors hence proposed a role of tankyrase in regulating Axin function rather than levels (Figure 1B). Studies in Drosophila and work with Drosophila cells and HEK293T cells showed that Wg/Wnt stimulation results in accumulation of PARylated Axin‐V5/AXIN1 with an enhanced formation of AXIN‐ and LDL receptor‐related protein 6 (LRP6)/Arrow‐containing Wnt/Wg signalosomes (Yang et al., 2016). Capitalizing on the ability to detect endogenous Drosophila Axin, a second study suggests that Axin is present in both the cytoplasm and at the membrane, even under basal conditions. Wg stimulation results in Axin accumulation in both compartments, with an enrichment of PARylated Axin at the membrane (Wang et al., 2016a). Forcing Axin to the membrane under basal conditions increases its PARylation and destabilization, presumably by PARdU, suggesting that tankyrase acts at the membrane or Axin's susceptibility to PARylation is augmented upon membrane recruitment (Wang et al., 2016a). Phosphorylation by GSK3 (Kim et al., 2013) or binding of the small molecule HLY78 (Wang et al., 2013) was reported to induce an open AXIN conformation for productive interaction with LRP6. AXIN PARylation may control the assembly of the Wnt signalosome complex in a similar manner. Alternatively, PAR itself may act as a molecular glue to recruit AXIN to Wnt signalosomes. While the intact LRP5 C‐terminus is required for AXIN binding (Mao et al., 2001), it is presently not clear whether AXIN and LRP5/6 interact directly (Mao et al., 2001; Kim et al., 2013; Wang et al., 2013), but there are indications that this may be the case (MacDonald et al., 2011). Future studies will unravel the mechanisms behind the Wnt/Wg‐induced accumulation of PARylated Axin and the role of PARylation in signalosome assembly. Yang et al. (2016) propose that the PAR‐assisted signalosome formation enables a rapid response to Wnt signals while the subsequent down‐regulation of AXIN limits the re‐assembly of β‐catenin destruction complexes, thereby conferring both responsiveness and robustness to the pathway.
Tankyrase inhibitors
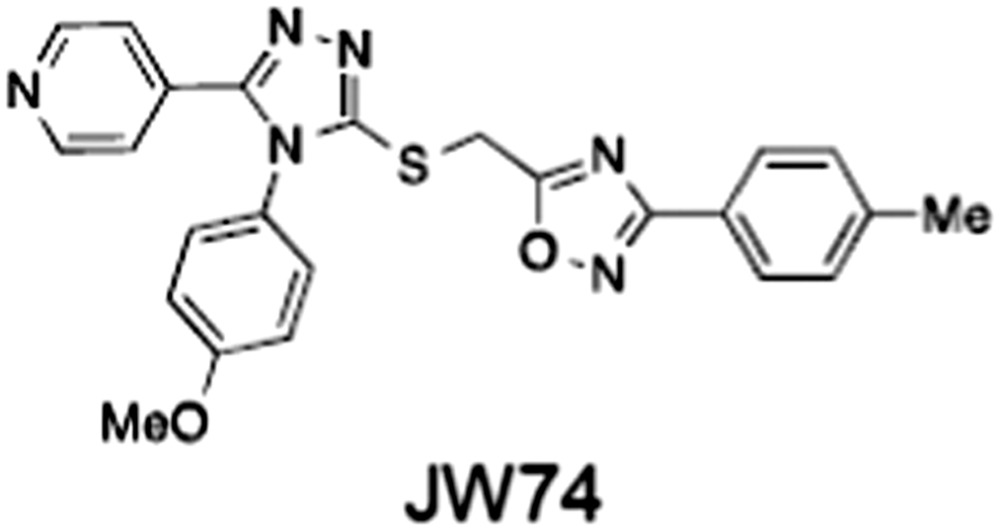
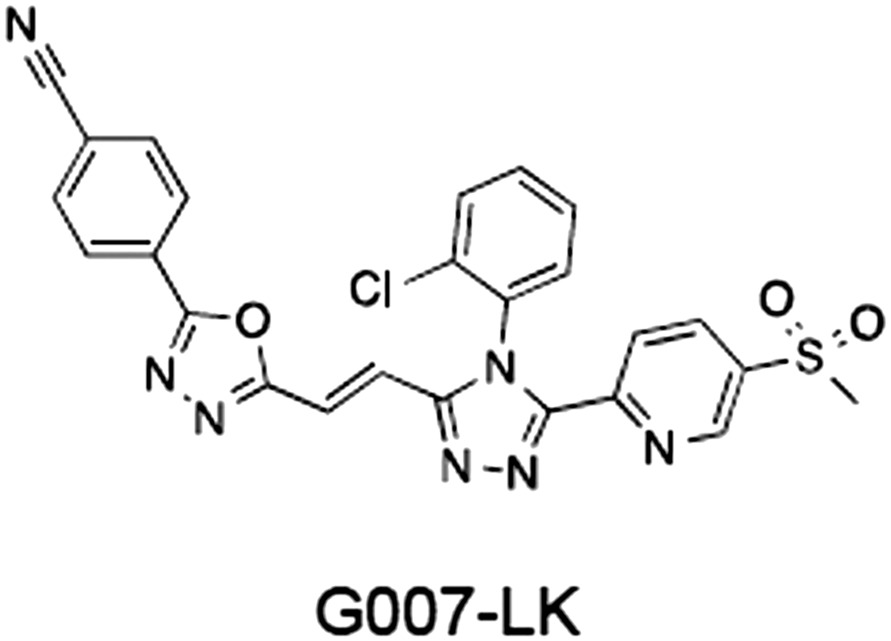
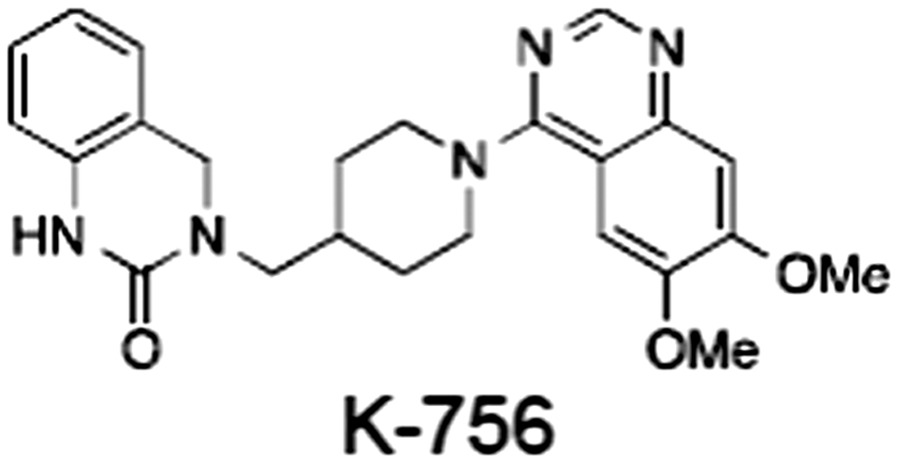
In this section, we provide an overview of TNKSi; for more comprehensive discussions, we refer the reader to other recent reviews (Lehtiö et al., 2013; Steffen et al., 2013; Haikarainen et al., 2014a). All low MW TNKSi developed to date are mimetics of β‐nicotinamide adenine dinucleotide (NAD+) or its adenosine or nicotinamide portions. The first potent toolbox TNKSi, XAV939 (Huang et al., 2009), IWR‐1 and IWR‐2 (Chen et al., 2009; Gunaydin et al., 2012), were discovered in phenotypic screens designed to identify antagonists of the Wnt/β‐catenin pathway, as were the inhibitors JW74 (Waaler et al., 2011), JW55 (Waaler et al., 2012), WIKI4 (James et al., 2012) and K‐756 (Okada‐Iwasaki et al., 2016). Numerous additional inhibitors were established through diverse approaches (see Zhan et al., 2014), including screening for compounds that rescue tankyrase‐induced lethality of yeast cells (Yashiroda et al., 2010) or induce a mitotic spindle defect (Johannes et al., 2015b), fragment screening (Larsson et al., 2013; de Vicente et al., 2015), proteomics (Thomson et al., 2017), in silico screening or substructure searching, followed by compound optimization (Bregman et al., 2013b; Elliott et al., 2015), screening of a DNA‐encoded library (Samain et al., 2015) and extensive structure–activity relationship studies, assisted by the structural analysis of tankyrase/PARP:inhibitor complexes (Hua et al., 2013; Shultz et al., 2013; Voronkov et al., 2013; Narwal et al., 2013a; Liscio et al., 2014; Qiu et al., 2014; Haikarainen et al., 2014b; Kumpan et al., 2015; Nkizinkiko et al., 2015; Paine et al., 2015; Haikarainen et al., 2016; Thomson et al., 2017). Numerous more drug‐like molecules, with optimized pharmacological properties, are now available, for example, G007‐LK (Lau et al., 2013; Voronkov et al., 2013) and NVP‐TNKS656 (Shultz et al., 2013). Table 1 gives examples of published TNKSi.
Inhibitor binding sites on the tankyrase PARP domain
The tankyrase PARP domain shares strong homology with the catalytic domains of the 17 human Diphtheria‐toxin‐like ADP‐ribosyltransferases (ARTDs) (see Hottiger et al., 2010). Among all ARTDs known to synthesize PAR (Vyas et al., 2014), the TNKS/TNKS2 PARP domains are unique in that they coordinate zinc, lack an N‐terminal helical regulatory subdomain and also lack a loop distal to the active site (Lehtiö et al., 2008). In all other respects, they share the typical structural elements with other ARTD catalytic domains (Figure 5A). A so‐called donor site coordinates the co‐substrate NAD+ while the acceptor site accommodates either the peptide for the priming modification or the growing PAR chain poised for extension (Figure 5A). The donor site is lined by three loops: the donor site or D‐loop, a glycine‐rich G‐loop and a phenylalanine‐containing F‐loop (Figure 5A). Tankyrase contains a catalytic H‐Y‐E triad required for PAR synthesis (Figure 5A). All TNKSi developed to date target the NAD+‐binding donor site but can broadly be classified by three binding modes, according to our structural understanding of NAD+ binding to Diphtheria toxin (Bell and Eisenberg, 1996). Inhibitors either primarily engage the nicotinamide subsite (e.g. XAV939) (Karlberg et al., 2010), the adenosine subsite (e.g. IWR‐1 and G007‐LK) (Narwal et al., 2012; Voronkov et al., 2013) or both sites in the case of the more recently developed dual‐site inhibitors (Bregman et al., 2013b) (Figure 5A; Table 1). In some instances, a phosphate site between the two subsites is specified as well (see Steffen et al., 2013). Crystal structures of tankyrase catalytic domains without inhibitor show closed D‐loop conformations, albeit different ones in apo‐TNKS and apo‐TNKS2 with indications of flexibility (Lehtiö et al., 2008; Karlberg et al., 2010). Adenosine site binders appear to induce their own pocket, conferring disorder or conformational changes to the D‐loop (Lehtiö et al., 2008; Gunaydin et al., 2012; Shultz et al., 2012). Hence, the adenosine subsite appears highly adaptable and is sometimes referred to as the ‘induced pocket’.
Figure 5.
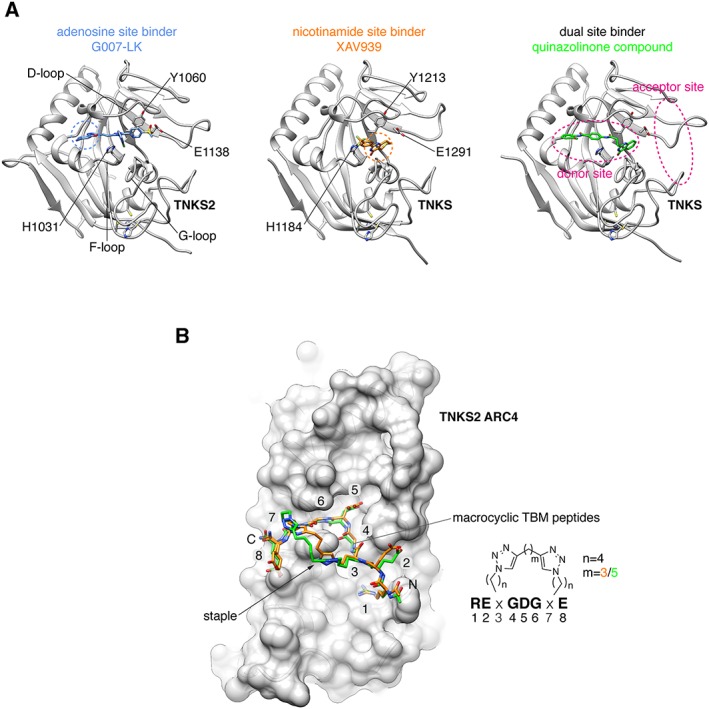
Binding modes of catalytic TNKSi and alternative inhibition strategy. (A) Structural representation of TNKS/TNKS2 PARP domain:inhibitor complexes. Residues of the catalytic H‐Y‐E triad are indicated. Adenosine and nicotinamide sites are highlighted with blue and orange dashed circles, respectively, in the left and central panels, respectively. Left, structure of the TNKS2 PARP domain with the adenosine site binder G007‐LK (shown in blue) [protein data bank (PDB) code 4HYF] (Voronkov et al., 2013). Loops lining the inhibitor binding site are indicated. Centre, structure of the TNKS PARP domain with the nicotinamide site binder XAV939 (shown in orange) (PDB code 3UH4) (Kirby et al., 2012). Right, structure of the TNKS PARP domain with a dual site binder (shown in green) (PDB code 4I9I) (Bregman et al., 2013b). The donor and acceptor sites are highlighted in magenta. (B) Structural representation of TNKS2 ARC4 (in surface representation) bound to two macrocyclized TBM peptides shown in superposition (PDB codes 5BXO and 5BXU) (Xu et al., 2017b). The peptide sequence is shown on the right with the position of the two different peptide staples, whose structures are shown. Amino acid positions of the TBM are indicated.
Achieving selectivity
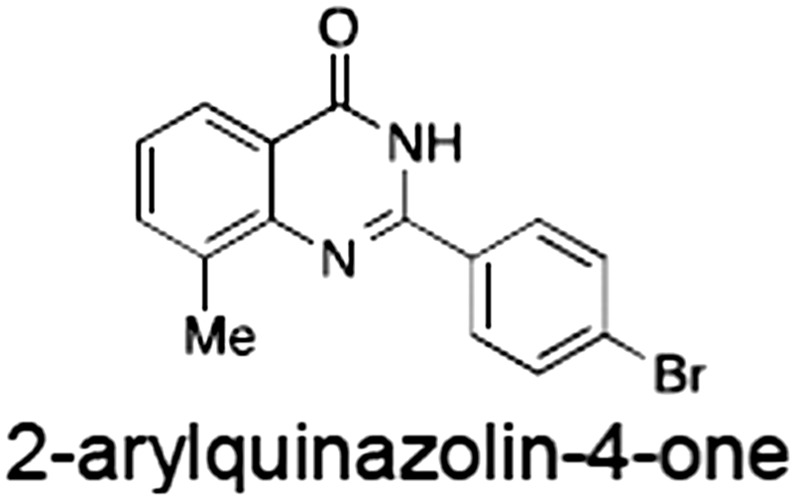
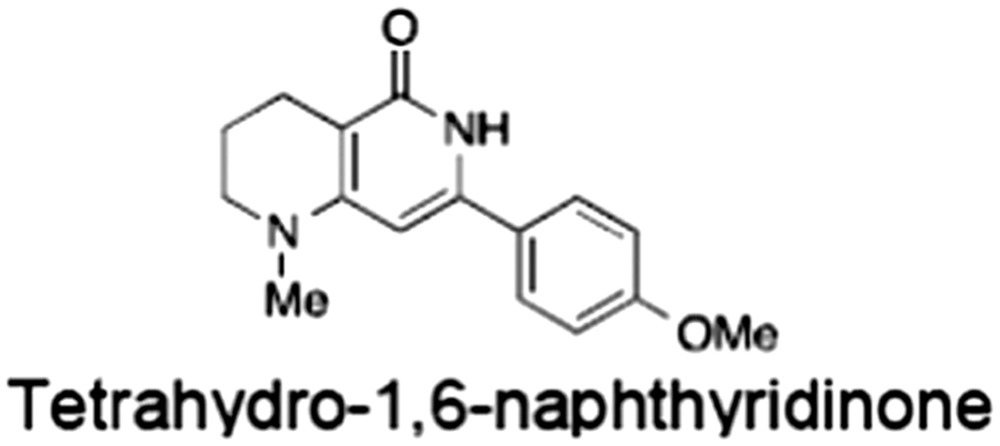
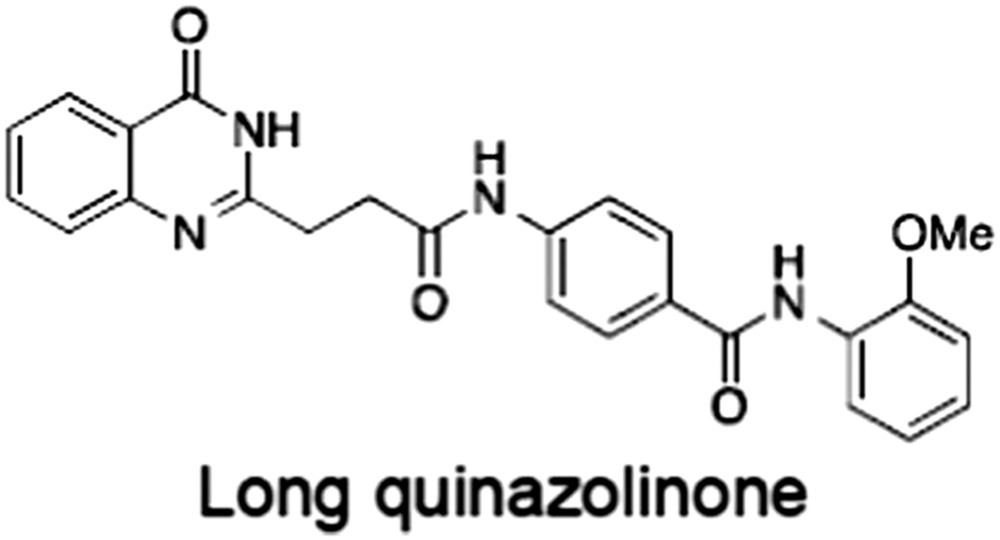
PARP inhibitor profiling against ARTDs revealed a remarkable promiscuity for a number of PARP inhibitors (Wahlberg et al., 2012; Thorsell et al., 2017). For example, XAV939 inhibits TNKS, TNKS2, PARP1 and PARP2 with comparable potency (e.g. IC50 values of 95, 5, 74 and 27 nM in a direct comparison using the catalytic domains of TNKS and TNKS2 and full‐length PARP1 and 2, respectively), while IWR‐1 is more specific for the tankyrases (no measurable IC50 for full‐length PARPs 1 and 2) (Thorsell et al., 2017). High‐resolution PARP domain:TNKSi co‐crystal structures have (i) rationalized TNKSi selectivity and (ii) enabled structure‐based drug design of more selective and potent tankyrase binders (see Lehtiö et al., 2013; Steffen et al., 2013; Haikarainen et al., 2014a). Specific examples for the former include G007‐LK, which was optimized from JW74 (Lau et al., 2013; Voronkov et al., 2013), and WIKI4 (James et al., 2012; Haikarainen et al., 2013). Examples for the latter include NVP‐TNKS656, developed from XAV939 (Shultz et al., 2013), and dual‐site inhibitors (Hua et al., 2013) (Table 1). These studies demonstrated that the unique structural features of the tankyrase catalytic domain can be exploited to gain selectivity. For example, WIKI4 in the adenosine subsite would sterically clash with the helical subdomain in PARPs 1–3 (Haikarainen et al., 2013). More subtle differences can also be harnessed. Compared to other PARPs, the D‐loop of tankyrases is three amino acids shorter, more flexible and often disordered in crystal structures of the domain with inhibitors, due to the absence of three proline residues, and characterized by large hydrophobic amino acids, which confer a narrower, more hydrophobic donor site pocket (Lehtiö et al., 2008; Wahlberg et al., 2012). Selectivity and potency can be gained by ‘growing’ compounds toward this narrow pocket, as seen in the optimization of quinazolinones (Nathubhai et al., 2013), tetrahydro‐1,6‐naphtyridin‐5‐ones (Kumpan et al., 2015) and the XAV939 core to NVP‐TNKS656 (Shultz et al., 2013) (Table 1). Of note, although selectivity over other ARTDs can be achieved, many other enzymes also use NAD+ as a co‐substrate, and so inhibitors designed to target the NAD+ donor site may have unknown off‐target effects at high concentrations. However, the example of sirtuins shows that this potential challenge can be overcome (Ekblad and Schüler, 2016).
Future developments
While further optimization of catalytic TNKSi is progressing, non‐catalytic scaffolding roles of tankyrase in Wnt/β‐catenin signalling are emerging (Mariotti et al., 2016), and these may be augmented when prolonged TNKSi treatment results in tankyrase stabilization by blocked PARdU (Huang et al., 2009). Furthermore, overexpression of tankyrase in several tumour types has been reported (Matsutani et al., 2001; Gelmini et al., 2004, 2006, 2007; Shervington et al., 2007; Shebzukhov et al., 2008; Zhao et al., 2009; Gao et al., 2011; Tang et al., 2012; Busch et al., 2013) and may accentuate tankyrase's concentration‐dependent scaffolding functions, contributing to TNKSi resistance (Mariotti et al., 2016). Therefore, blocking tankyrase's ARC‐ and SAM‐dependent scaffolding functions holds considerable potential. Importantly, the ARCs and SAM domain are highly conserved between the two tankyrases (Figure 2A) but unique among the ARTD family, therefore offering the opportunity for target selectivity of potential compounds over other ARTDs, in addition to the potential benefits of inhibiting non‐catalytic scaffolding functions. Moreover, potential interference with other NAD+‐dependent enzymes could be circumvented by this approach. Whereas blocking SAM‐domain‐dependent polymerization appears challenging due to the relatively shallow polymerization interface, targeting the deeper TBM‐binding pocket on the ARCs is more promising (Guettler et al., 2011; Morrone et al., 2012). This binding pocket is not conserved across ankyrin repeat proteins in general and appears to be unique to tankyrase. Given the presence of four substrate‐binding ARCs, blockage of each of these substrate/ligand binding sites is likely to be required, in both TNKS and TNKS2. However, this appears feasible given the conservation of the TBM‐binding pocket across the TNKS and TNKS2 ARCs (Guettler et al., 2011). Xu et al. (2017b) have recently shown that a stapled TBM peptide, based on a previously reported optimized TBM sequence (Guettler et al., 2011) and fused to a cell‐permeability conferring peptide, can compete with AXIN and block Wnt/β‐catenin signalling (Figure 5B). This proof‐of‐concept study will encourage further development of tankyrase substrate binding antagonists. While a recent tankyrase interactome study is in agreement with the notion that TNKS and TNKS2 are largely functionally redundant (Li et al., 2017), there may be benefit to selectively targeting either TNKS or TNKS2, should unique functions emerge in the future, which may be TNKS/TNKS2‐intrinsic or result from other sources such as differential expression or regulation. One inhibitor study (see Table 1, compound 3) suggests that, in principle, a certain degree of such selectivity can be achieved (Larsson et al., 2013).
Functional and preclinical studies of tankyrase inhibitors in CRC
Differential sensitivity of CRC cell lines to tankyrase inhibition
Most CRC tumours (≈80%) are hemizygous for C‐terminal truncations of APC (see Bodmer, 2006), focussed at a hotspot area known as the mutation cluster region (Miyoshi et al., 1992; Kohler et al., 2008; see Minde et al., 2011) (Figure 6A). Following the N‐terminal Armadillo repeat domain, APC contains four 15‐amino‐acid β‐catenin‐binding repeats, seven 20‐amino‐acid β‐catenin‐binding repeats and three interspersed AXIN‐binding SAMP repeats, among other elements not required for APC's function in Wnt/β‐catenin signalling (see Stamos and Weis, 2013). Dependent on the position of the truncating mutation, a variable number of these motifs is lost (Figure 6A). In APC‐truncated CRC cells, an inability to assemble a functional β‐catenin destruction complex underlies the accumulation of transcriptionally active β‐catenin. A range of studies has investigated the responsiveness of model CRC cell lines to tankyrase inhibition (Lau et al., 2013; de la Roche et al., 2014; Tanaka et al., 2017). While it would have been conceivable that the assembly and function of the β‐catenin destruction complex cannot be sufficiently rescued by tankyrase inhibition in the absence of fully functional APC, there clearly are cases in which dysregulated Wnt/β‐catenin signalling can be curbed. In SW403 and COLO‐320DM cells, both of which bear extensive C‐terminal APC truncations (Figure 6A), tankyrase inhibition (by G007‐LK, IWR‐1 or XAV939) gives rise to AXIN2 stabilization, the formation of β‐catenin degradasomes (in COLO‐320DM cells), a robust reduction in active (non‐phosphorylated) β‐catenin and prominently attenuated β‐catenin‐dependent transcription, both in reporter assays and at the level of endogenous Wnt/β‐catenin target genes (Lau et al., 2013; de la Roche et al., 2014; Tanaka et al., 2017). Importantly, tankyrase inhibition limits the proliferation of these cells in cell culture (for G007‐LK and IWR‐1) and xenograft (for G007‐LK) models (Lau et al., 2013; Tanaka et al., 2017). A similar cell response to tankyrase inhibition is observed in DLD‐1 and HCT‐15 cells, although the levels of active β‐catenin are reduced less robustly and the transcriptional effect is more subtle (Lau et al., 2013; de la Roche et al., 2014; Tanaka et al., 2017). Conversely, in SW480 and SW620 cells, although tankyrase inhibition results in the stabilization of AXIN2, degradasome formation and a strong decrease in active β‐catenin levels, it fails to block Wnt/β‐catenin target genes or the TOPFlash reporter (Lau et al., 2013; de la Roche et al., 2014). In yet another group of CRC cells (COLO‐205, HT‐29, HCC2998 and LS‐411 N), tankyrase inhibition increases AXIN1/2 levels but with only a modest or no decrease in active β‐catenin levels (Lau et al., 2013; Tanaka et al., 2017). For KM12 cells, no effect on the levels of AXIN1/2 or active β‐catenin levels and β‐catenin‐dependent transcription was observed upon tankyrase inhibition (Tanaka et al., 2017). As expected, Wnt/β‐catenin signalling in CRC cells with oncogenic mutations in β‐catenin (LS174T, HCT116) or β‐catenin‐independent CRC cells (RKO) are not sensitive to tankyrase inhibition (Lau et al., 2013; Tanaka et al., 2017).
Figure 6.
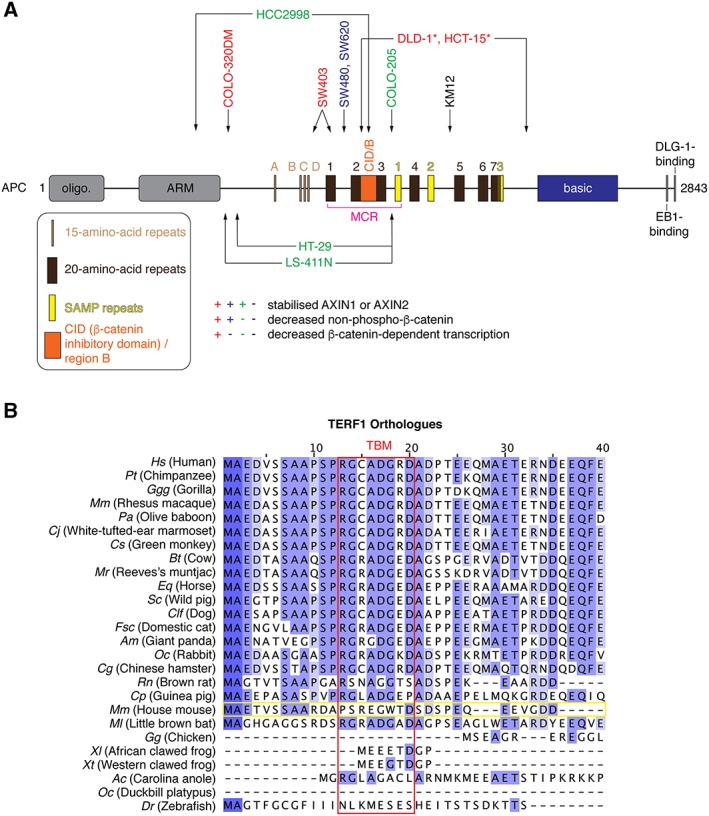
Potential determinants of TNKSi responses: APC mutation status and telomeric roles of tankyrase. (A) Schematic representation of APC with domains and motifs drawn to scale (see Stamos and Weis, 2013). So‐called 15‐ and 20‐amino‐acid‐repeats (15R and 20R) bind β‐catenin (except for 20R2), with the affinity of the 20Rs for β‐catenin being enhanced by their phosphorylation (Eklof Spink et al., 2001; Ha et al., 2004; Xing et al., 2004). SAMP repeats bind to AXIN1/2 (Spink et al., 2000). 20R2 and the catenin interaction domain (CID) / region B are required for β‐catenin ubiquitylation but do not bind β‐catenin; instead, they regulate AXIN1/2 binding to APC, and CID / region B is proposed to bind α‐catenin (Liu et al., 2006; Kohler et al., 2008; Choi et al., 2013; Pronobis et al., 2015). The mutation cluster region (MCR), a mutation hotspot in CRC (Kohler et al., 2008), is indicated in magenta. APC truncations observed in commonly used CRC cell lines are indicated by the arrows (Rowan et al., 2000; Ikediobi et al., 2006); labels are colour‐coded according to the indicated effects of TNKSi on AXIN and non‐phospho (active) β‐catenin levels and β‐catenin‐dependent transcription. *Note that the classification of DLD‐1 and HCT‐15 cells as TNKSi‐sensitive or ‐resistant varies between studies, given an ‘intermediate’ response (Huang et al., 2009; Lau et al., 2013; de la Roche et al., 2014; Tanaka et al., 2017). Very low AXIN1/2 levels in KM12 cells (Tanaka et al., 2017) may be responsible for non‐detectable AXIN accumulation upon tankyrase inhibition. (B) Multiple sequence alignment of the N‐termini of TERF1/TRF1 (telomeric repeat binding factor 1) orthologues from the indicated species, coloured by percentage identity. The amino acid numbering refers to human TERF1. The 8‐amino‐acid TBM is boxed in red. The murine Terf1 orthologue sequence is boxed in yellow and shows no conservation of the TBM.
Tankyrase inhibition can therefore restore, at least partly, β‐catenin destruction complex function in a subset of APC‐mutant CRC cells (Figure 6A). Why another subset of CRC cell lines is TNKSi‐resistant is being investigated. It has been proposed that high levels of β‐catenin in SW480 cells account for TNKSi resistance (Lau et al., 2013), but β‐catenin levels in the TNKSi‐sensitive COLO‐320DM cells appear comparable (de la Roche et al., 2014). Compared to SW480, COLO‐320DM cells appear to have higher levels of AXIN1, TNKS and phospho‐β‐catenin after tankyrase inhibition (de la Roche et al., 2014), suggesting that β‐catenin may be sequestered in stalled destruction complexes, thereby limiting the availability of active β‐catenin.
A potential correlation between the site of APC truncation and TNKSi sensitivity has been explored, both in CRC cell lines and tumour‐derived cells from patients (Tanaka et al., 2017). APC truncations removing all β‐catenin‐binding 20‐amino‐acid repeats (as in COLO‐320DM and SW403 cells; Figure 6A) were proposed to render cells TNKSi‐responsive at the level of cell proliferation and might serve as a predictive biomarker (Tanaka et al., 2017). Another distinguishing feature of these cells is their particularly strong Wnt/β‐catenin pathway activity (Tanaka et al., 2017). The authors suggest that the longer APC variants of other cell lines act as hypomorphs maintaining a higher residual level of β‐catenin regulation: their silencing further stabilizes β‐catenin. TNKSi (G007‐LK, IWR‐1) can reverse this accumulation but not reduce β‐catenin abundance below its cell‐characteristic elevated levels (Tanaka et al., 2017). In turn, cell proliferation remains unresponsive to tankyrase inhibition (Tanaka et al., 2017). The genetic background of additional cell lines and tumour samples will need to be explored to confirm the suitability of APC truncations as predictive biomarkers for TNKSi sensitivity. In line with a requirement of AXIN2 for degradasome formation (see above), TNKSi depend on AXIN2 to reduce active β‐catenin levels (Tanaka et al., 2017). Large APC truncations, eliciting high β‐catenin activity and thus AXIN2 gene transcription, may be required for the TNKSi‐induced accumulation of sufficient amounts of AXIN2. Indeed, absolute AXIN2 mRNA levels are high in COLO‐320DM cells and, upon tankyrase inhibition, remain higher than in many other CRC cell lines (Tanaka et al., 2017).
Upon prolonged Wnt stimulation, sequestration of β‐catenin in nuclear transcriptional complexes may shield β‐catenin from the β‐catenin destruction complex and account for TNKSi (XAV939) resistance (de la Roche et al., 2014). High expression levels of lymphoid enhancer‐binding factor 1 (LEF1) and B9L and a CRC environment providing sustained Wnt levels are potential predictors of TNKSi resistance (de la Roche et al., 2014). Moreover, the acquisition of APC mutations is considered an early event in the emergence of CRC, and secondary mutations in genes such as KRAS, P53 and SMAD4 contribute to driving carcinogenesis (Drost et al., 2015); such mutations may modulate the TNKSi response, although this remains speculative. Of note, DLD‐1 colony formation can be inhibited with XAV939 under low‐ but not high‐serum conditions (Huang et al., 2009; Bao et al., 2012; Lau et al., 2013), and similarly, DLD‐1 and HCT‐15 colony formation does not respond to G007‐LK at high serum, while that of COLO‐320DM and SW403 cells does (Lau et al., 2013). This points to additional sensitivity determinants outside the APC and β‐catenin mutational landscape, although simple compound sequestration by serum components may in some cases also contribute. In support of a more complex determination of sensitivity, Mashima et al. (2017) generated a TNKSi‐resistant COLO‐320DM line, showing decreased Wnt/β‐catenin signalling and up‐regulated mTOR signalling. These cells were generated to tolerate IWR‐1 but also displayed considerable resistance to G007‐LK. The authors showed that the mTOR pathway determines TNKSi resistance in these cells. In conclusion, we need to better understand how the genetic and signalling profile of CRC cells and tumours affects TNKSi responsiveness. Furthermore, a much deeper analysis of the β‐catenin destruction complex and the Wnt signalosome is required to appreciate the mechanisms by which tankyrase inhibition affects the molecular events underlying Wnt/β‐catenin signalling, both in the context of wild‐type and mutant APC.
Tankyrase inhibitors in murine models
In vivo preclinical studies have demonstrated the anti‐tumour activity of various TNKSi (Waaler et al., 2011, 2012; Lau et al., 2013). JW74 was studied in both xenograft and the ApcMin mouse models and reported to be well tolerated while reducing both the total tumour load in the small intestine and the tumour number in the colon (Waaler et al., 2011). Similar observations were made for JW55 in mice with a conditional Apc truncation in the ISC compartment (Waaler et al., 2012). An improved derivative of JW74, G007‐LK, which inhibits tankyrase with double‐digit nanomolar IC50 values and good specificity (Table 1) (Voronkov et al., 2013), decreases the tumour area in the small intestine of these mice by approximately two thirds and shows significant inhibition of tumour growth in various xenograft models with APC‐mutant human cell lines (SW480, COLO‐320DM and SW403) (Lau et al., 2013).
Intestinal toxicity remains a major challenge for many Wnt/β‐catenin pathway inhibitors (see Kahn, 2014). A more careful analysis of the effects of G007‐LK at dose‐limiting levels revealed reduced cell proliferation in crypt bases of the small intestine, inflammation, necrosis, disrupted epithelial architecture, with ensuing weight loss and moribundity (Lau et al., 2013). Whether the observed toxicity is reversible has not been explored. Conversely, in a study investigating the role of tankyrase in glucose metabolism, the long‐term (6 months) treatment of mice with G007‐LK at a lower dose delivered orally, as opposed to intraperitoneally, did not result in detectable toxicity despite the observed stabilization of Axin1 and reduction in active β‐catenin levels (Zhong et al., 2016a). As the expression of the Wnt pathway antagonist Dickkopf‐related protein 1 (Dkk1) in the gut epithelium gives rise to similar toxicity as highly dosed G007‐LK (Pinto et al., 2003; Kuhnert et al., 2004; Lau et al., 2013), it is likely that TNKSi toxicity is an on‐target, Wnt/β‐catenin pathway‐specific effect. A mouse xenograft study evaluating the TNKSi G‐631 (patent by Feng et al., 2013) also revealed considerable intestinal toxicity, even at sub‐therapeutic doses (Zhong et al., 2016b). Importantly, ablation of Wnt signalling by either adenoviral Dkk1 expression or treatment with G‐631 is reversible (Kuhnert et al., 2004; Zhong et al., 2016b).
While toxicity poses challenges in continued efforts to explore the therapeutic potential of TNKSi, it is no reason to be discouraged. Does TNKSi toxicity indeed reflect on‐target or off‐target action of the inhibitors? What is the tissue distribution of the compounds? For example, do they accumulate in the gastrointestinal tract, aggravating toxicity? Do chemically distinct TNKSi display similar signs of toxicity? The fact that SW480 cell xenograft growth can be contained by tankyrase inhibition despite the incomplete penetrance on β‐catenin dependent transcription (see above) suggests that full inhibition of signalling might not be required for a therapeutic effect; so what degree of β‐catenin inhibition translates into a biological effect? Moreover, different dosage and timing regimes could be implemented to manage known toxicities (see Meric‐Bernstam and Mills, 2012). Encouragingly, inhibitors of the acyl transferase Porcupine, which palimitoylates Wnt during its biogenesis, show limited intestinal toxicity at effective doses, suggesting that substantial therapeutic windows can be achieved by targeting the Wnt/β‐catenin pathway (Liu et al., 2013; Proffitt et al., 2013; see also Madan and Virshup, 2015). The point of intervention in the pathway, drug specificity, potency and method of delivery, pharmacokinetics, functional redundancy of targeted pathway components and the genetic background of the tumour cells may all define the therapeutic window. Combination with inhibitors targeting additional cancer dependencies (e.g. EGFR and PI3K‐AKT) provides another possible strategy for increasing the effectiveness of TNKSi while ensuring their safety (Casas‐Selves et al., 2012; Tenbaum et al., 2012; Arques et al., 2016).
A deeper knowledge of the responses and toxicities elicited by Wnt/β‐catenin pathway modulators in different species is much needed to exploit β‐catenin dependencies in cancer. With the vast range of known and putative tankyrase targets (Guettler et al., 2011; Li et al., 2017), it would be surprising if TNKSi effects and toxicities were entirely due to Wnt/β‐catenin pathway inhibition. Another prominent system highly relevant to the human stem cell compartment is telomere length homeostasis, which is also regulated by tankyrase, as is sister telomere resolution in mitosis (Smith et al., 1998; Smith and de Lange, 2000; Canudas et al., 2007; Kulak et al., 2015). Telomeric functions require tankyrase to bind telomeric repeat‐binding factor 1 (TRF1/TERF1). Importantly, telomere regulation by tankyrase is not conserved in mice since murine Trf1/Terf1 lacks the TBM (Figure 6B) and does not bind tankyrase (Muramatsu et al., 2007; see Hsiao and Smith, 2008). Consistent with the absence of a role for telomeric tankyrase functions in mice, Tnks and Tnks2 knockout mice do not display any telomere phenotype (Hsiao et al., 2006; Chiang et al., 2008), but a definitive answer will be obtained from comparing murine and human cells deficient in both tankyrases. Therefore, preclinical studies of TNKSi in mice and other species lacking the Terf1 TBM (e.g. zebrafish; Figure 6B) are unlikely to predict the full extent of biological effects and toxicity in humans. Conversely, rabbits or Chinese hamsters, for example, both display a functional TBM in Terf1 and might be more suitable models for studying the in vivo consequences of tankyrase inhibition, at least with regards to Wnt/β‐catenin signalling and telomere maintenance. In rats, the somewhat stronger deviation of the TBM will require a prior validation of a telomeric role for tankyrase (Muramatsu et al., 2007). Telomere maintenance in Drosophila occurs via a transposon‐mediated mechanism rather than telomerase (Villasante et al., 2008), and a telomeric tankyrase link in flies is therefore unlikely. While Wg/Armadillo pathway regulation by tankyrase is clearly evident in Drosophila, some mechanistic aspects may be different, for example given that Drosophila Axin only bears a single TBM (Figure 2C).
Outstanding questions
The past few years have seen a rapid progress in our understanding of how Wnt/β‐catenin signalling is regulated by PARylation and tankyrase. Tankyrase is now an established core component of the Wnt/β‐catenin network. Nonetheless, we are still far from a full understanding of the complex roles that tankyrase plays in the pathway. How does tankyrase promote Wnt/β‐catenin signalling non‐catalytically, and do these mechanisms contribute to TNKSi resistance? Does scaffolding through tankyrase directly control β‐catenin degradasome assembly, and how is this process regulated? How does AXIN PARylation promote the function of the Wnt signalosome? It will be interesting to explore the consequences of AXIN PARylation on both its conformation and interactions with components of both the signalosome and degradasome complexes. Given early indications of a role for tankyrase in APC‐regulated destabilization of AXIN, additional work is needed to decipher how APC limits AXIN abundance. TNKSi have now reached a remarkable specificity. The continued exploration of their pharmacodynamics, the identification of potential biomarkers for their therapeutic implementation and the in‐depth analysis of emerging resistance and toxicity mechanisms are important avenues of further research. The latter will require a careful choice of model systems. The development of alternative tankyrase inhibition strategies through interfering with the non‐catalytic, scaffolding functions of tankyrase, in particular substrate binding, will undoubtedly offer new and exciting opportunities to understand tankyrase function and explore alternative therapeutic strategies.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Southan et al., 2016), and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 (Alexander et al., 2015a, 2015b).
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank members of the Guettler team and Chris Lord (ICR) for feedback on the manuscript. Work in S.G.'s laboratory is supported by The Institute of Cancer Research (ICR) and by Cancer Research UK through a Career Establishment Award to S.G. (C47521/A16217). L.M. and K.P. are supported by ICR and Wellcome Trust (WT102360/Z/13/Z) PhD studentships, respectively.
Mariotti, L. , Pollock, K. , and Guettler, S. (2017) Regulation of Wnt/β‐catenin signalling by tankyrase‐dependent poly(ADP‐ribosyl)ation and scaffolding. British Journal of Pharmacology, 174: 4611–4636. doi: 10.1111/bph.14038.
References
- Alexander SPH, Davenport AP, Kelly E, Marrion N, Peters JA, Benson HE et al (2015a). The Concise Guide to PHARMACOLOGY 2015/16: G protein‐coupled receptors. Br J Pharmacol 172: 5744–5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE et al (2015b). The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. Br J Pharmacol 172: 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arques O, Chicote I, Puig I, Tenbaum SP, Argiles G, Dienstmann R et al (2016). Tankyrase inhibition blocks Wnt/β‐catenin pathway and reverts resistance to PI3K and AKT inhibitors in the treatment of colorectal cancer. Clin Cancer Res 22: 644–656. [DOI] [PubMed] [Google Scholar]
- Bae J (2002). Tankyrase 1 interacts with Mcl‐1 proteins and inhibits their regulation of apoptosis. J Biol Chem 278: 5195–5204. [DOI] [PubMed] [Google Scholar]
- Bao R, Christova T, Song S, Angers S, Yan X, Attisano L (2012). PLOS ONE: inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS One 7: e48670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CE, Eisenberg D (1996). Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide †. Biochemistry 35: 1137–1149. [DOI] [PubMed] [Google Scholar]
- Bienz M (2014). Signalosome assembly by domains undergoing dynamic head‐to‐tail polymerization. Trends Biochem Sci 39: 487–495. [DOI] [PubMed] [Google Scholar]
- Bisht KK, Dudognon C, Chang WG, Sokol ES, Ramirez A, Smith S (2012). GDP‐mannose‐4,6‐dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP‐ribose) polymerase activity. Mol Cell Biol 32: 3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer WF (2006). Cancer genetics: colorectal cancer as a model. J Hum Genet 51: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman H, Chakka N, Guzman‐Perez A, Gunaydin H, Gu Y, Huang X et al (2013a). Discovery of novel, induced‐pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors. J Med Chem 56: 4320–4342. [DOI] [PubMed] [Google Scholar]
- Bregman H, Gunaydin H, Gu Y, Schneider S, Wilson C, DiMauro EF et al (2013b). Discovery of a class of novel tankyrase inhibitors that bind to both the nicotinamide pocket and the induced pocket. J Med Chem 56: 1341–1345. [DOI] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FPA, Fang HY, Boquete J‐P, Deplancke B et al (2013). Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep 3: 1725–1738. [DOI] [PubMed] [Google Scholar]
- Busch AM, Johnson KC, Stan RV, Sanglikar A, Ahmed Y, Dmitrovsky E et al (2013). Evidence for tankyrases as antineoplastic targets in lung cancer. BMC Cancer 13: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN et al (2011). Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One 6: e22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S (2007). Protein requirements for sister telomere association in human cells. Nat Struct Mol Biol 26: 4867–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas‐Selves M, Kim J, Zhang Z, Helfrich BA, Gao D, Porter CC et al (2012). Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res 72: 4154–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan C‐W et al (2009). Small molecule‐mediated disruption of Wnt‐dependent signaling in tissue regeneration and cancer. Nat Chem Biol 5: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YJ, Hsiao SJ, Yver D, Cushman SW, Tessarollo L, Smith S et al (2008). Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One 3: e2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YJ, Nguyen M‐L, Gurunathan S, Kaminker P, Tessarollo L, Campisi J et al (2006). Generation and characterization of telomere length maintenance in tankyrase 2‐deficient mice. Mol Cell Biol 26: 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Estarás C, Moresco JJ, Yates JR, Jones KA (2013). α‐Catenin interacts with APC to regulate β‐catenin proteolysis and transcriptional repression of Wnt target genes. Genes Dev 27: 2473–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R (2012). Wnt/β‐catenin signaling and disease. Cell 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Clevers H, Loh KM, Nusse R (2014). An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346 1248012–1248012. [DOI] [PubMed] [Google Scholar]
- Croy HE, Fuller CN, Giannotti J, Robinson P, Foley AVA, Yamulla RJ et al (2016). The PARP enzyme tankyrase antagonizes activity of the β‐catenin destruction complex through ADP‐ribosylation of Axin and APC2. J Biol Chem 291: 12747–12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa PA, Ovchinnikov S, Xu W, Klevit RE (2016). Structural insights into SAM domain‐mediated tankyrase oligomerization. Protein Sci 25: 1744–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE et al (2015). Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP‐ribosyl)ation signal. Nature 517: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Roche M, Ibrahim AEK, Mieszczanek J, Bienz M (2014). LEF1 and B9L shield β‐catenin from inactivation by Axin, desensitizing colorectal cancer cells to tankyrase inhibitors. Cancer Res 74: 1495–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rycker M, Price CM (2004). Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP‐ribose) polymerase domains. Mol Cell Biol 24: 9802–9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vicente J, Tivitmahaisoon P, Berry P, Bolin DR, Carvajal D, He W et al (2015). Fragment‐based drug design of novel pyranopyridones as cell active and orally bioavailable tankyrase inhibitors. ACS Med Chem Lett 6: 1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A et al (2015). Sequential cancer mutations in cultured human intestinal stem cells. Nature 521: 43–47. [DOI] [PubMed] [Google Scholar]
- Eisemann T, McCauley M, Langelier M‐F, Gupta K, Roy S, Van Duyne GD et al (2016). Tankyrase‐1 ankyrin repeats form an adaptable binding platform for targets of ADP‐ribose modification. Structure 24: 1679–1692. [DOI] [PubMed] [Google Scholar]
- Ekblad T, Schüler H (2016). Sirtuins are unaffected by PARP inhibitors containing planar nicotinamide bioisosteres. Chem Biol Drug Des 87: 478–482. [DOI] [PubMed] [Google Scholar]
- Eklof Spink K, Fridman SG, Weis WI (2001). Molecular mechanisms of beta‐catenin recognition by adenomatous polyposis coli revealed by the structure of an APC‐beta‐catenin complex. EMBO J 20: 6203–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RJR, Jarvis A, Rajasekaran MB, Menon M, Bowers L, Boffey R et al (2015). Design and discovery of 3‐aryl‐5‐substituted‐isoquinolin‐1‐ones as potent tankyrase inhibitors. Med Chem Commun 6: 1687–1692. [Google Scholar]
- Feng J, Haynes NE, Hermann JC, Kim K, Liu JJ, Scott NR et al (2013). Pyrazolopyrimidone and pyrazolopyridone inhibitors of tankyrase. Patent WO2013182546 A1, filed June 4, 2013, issued December 12, 2013.
- Feng Y, Li X, Ray L, Song H, Qu J, Lin S et al (2014). The Drosophila tankyrase regulates Wg signaling depending on the concentration of Daxin. Cell Signal 26: 1717–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M, Mendoza‐Topaz C, Rutherford TJ, Mieszczanek J, Bienz M (2011). Dishevelled interacts with the DIX domain polymerization interface of Axin to interfere with its function in down‐regulating β‐catenin. Proc Natl Acad Sci 108: 1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Xiao G, Hu J (2014). Regulation of Wnt/β‐catenin signaling by posttranslational modifications. Cell Biosci 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhang J, Long Y, Tian Y, Lu X (2011). Expression of tankyrase 1 in gastric cancer and its correlation with telomerase activity. Pathol Oncol Res 17: 685–690. [DOI] [PubMed] [Google Scholar]
- Gelmini S, Poggesi M, Distante V, Bianchi S, Simi L, Luconi M et al (2004). Tankyrase, a positive regulator of telomere elongation, is over expressed in human breast cancer. Cancer Lett 216: 81–87. [DOI] [PubMed] [Google Scholar]
- Gelmini S, Poggesi M, Pinzani P, Mannurita SC, Cianchi F, Valanzano R et al (2006). Distribution of tankyrase‐1 mRNA expression in colon cancer and its prospective correlation with progression stage. Oncol Rep 16: 1261–1266. [PubMed] [Google Scholar]
- Gelmini S, Quattrone S, Malentacchi F, Villari D, Travaglini F, Giannarini G et al (2007). Tankyrase‐1 mRNA expression in bladder cancer and paired urine sediment: preliminary experience. Clin Chem Lab Med 45: 862–866. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL (2012). New insights into the molecular and cellular functions of poly(ADP‐ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411–424. [DOI] [PubMed] [Google Scholar]
- Guettler S, LaRose J, Petsalaki E, Gish G, Scotter A, Pawson T et al (2011). Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease. Cell 147: 1340–1354. [DOI] [PubMed] [Google Scholar]
- Gunaydin H, Gu Y, Huang X (2012). Novel binding mode of a potent and selective tankyrase inhibitor. PLoS One 7: e33740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha N‐C, Tonozuka T, Stamos JL, Choi H‐J, Weis WI (2004). Mechanism of phosphorylation‐dependent binding of APC to beta‐catenin and its role in beta‐catenin degradation. Mol Cell 15: 511–521. [DOI] [PubMed] [Google Scholar]
- Haikarainen T, Krauss S, Lehtiö L (2014a). Tankyrases: structure, function and therapeutic implications in cancer. Curr Pharm Des 20: 6472–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikarainen T, Narwal M, Joensuu P, Lehtiö L (2014b). Evaluation and structural basis for the inhibition of tankyrases by PARP inhibitors. ACS Med Chem Lett 5: 18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikarainen T, Venkannagari H, Narwal M, Obaji E, Lee H‐W, Nkizinkiko Y et al (2013). Structural basis and selectivity of tankyrase inhibition by a Wnt signaling inhibitor WIKI4. PLoS One 8: e65404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikarainen T, Waaler J, Ignatev A, Nkizinkiko Y, Venkannagari H, Obaji E et al (2016). Development and structural analysis of adenosine site binding tankyrase inhibitors. Bioorg Med Chem Lett 26: 328–333. [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch‐Nolte F (2010). Toward a unified nomenclature for mammalian ADP‐ribosyltransferases. Trends Biochem Sci 35: 208–219. [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S (2008). Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90: 83–92. [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, Poitras MF, Cook BD, Liu Y, Smith S (2006). Tankyrase 2 poly (ADP‐ribose) polymerase domain‐deleted mice exhibit growth defects but have normal telomere length and capping. Mol Cell Biol 26: 2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Bregman H, Buchanan JL, Chakka N, Guzman‐Perez A, Gunaydin H et al (2013). Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors. J Med Chem 56: 10003–10015. [DOI] [PubMed] [Google Scholar]
- Huang H, Guzman‐Perez A, Acquaviva L, Berry V, Bregman H, Dovey J et al (2013). Structure‐based design of 2‐aminopyridine oxazolidinones as potent and selective tankyrase inhibitors. ACS Med Chem Lett 4: 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S‐MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA et al (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461: 614–620. [DOI] [PubMed] [Google Scholar]
- Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S et al (2006). Mutation analysis of 24 known cancer genes in the NCI‐60 cell line set. Mol Cancer Ther 5: 2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Endo TA, Ku M, Yamada D, Suzuki R, Sharif J et al (2013). SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell 26: 565–577. [DOI] [PubMed] [Google Scholar]
- James RG, Davidson KC, Bosch KA, Biechele TL, Robin NC, Taylor RJ et al (2012). WIKI4, a novel inhibitor of tankyrase and Wnt/ß‐catenin signaling. PLoS One 7: e50457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho, E. , Zhang, T. , Domon, C. , and Joo, C.K. (2002). Wnt/β‐catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22: 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes JW, Almeida L, Barlaam B, Boriack‐Sjodin PA, Casella R, Croft RA et al (2015a). Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology. ACS Med Chem Lett 6: 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes JW, Almeida L, Daly K, Ferguson AD, Grosskurth SE, Guan H et al (2015b). Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering. Bioorg Med Chem Lett 25: 5743–5747. [DOI] [PubMed] [Google Scholar]
- Kahn M (2014). Can we safely target the WNT pathway? Nat Rev Drug Disc 13: 513–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HC, Lee Y‐I, Shin J‐H, Andrabi SA, Chi Z, Gagné J‐P et al (2011). Iduna is a poly(ADP‐ribose) (PAR)‐dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci 108: 14103–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg T, Markova N, Johansson I, Hammarström M, Schütz P, Weigelt J et al (2010). Structural basis for the interaction between tankyrase‐2 and a potent Wnt‐signaling inhibitor. J Med Chem 53: 5352–5355. [DOI] [PubMed] [Google Scholar]
- Kim S‐E, Huang H, Zhao M, Zhang X, Zhang A, Semonov MV et al (2013). Wnt stabilization of β‐catenin reveals principles for morphogen receptor‐scaffold assemblies. Science 340: 867–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby CA, Cheung A, Fazal A, Shultz MD, Stams T (2012). Structure of human tankyrase 1 in complex with small‐molecule inhibitors PJ34 and XAV939. Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler EM, Derungs A, Daum G, Behrens J, Schneikert J (2008). Functional definition of the mutation cluster region of adenomatous polyposis coli in colorectal tumours. Hum Mol Genet 17: 1978–1987. [DOI] [PubMed] [Google Scholar]
- Kuhnert F, Davis CR, Wang H‐T, Chu P, Lee M, Yuan J et al (2004). Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf‐1. Proc Natl Acad Sci U S A 101: 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak O, Chen H, Holohan B, Wu X, He H, Borek D et al (2015). Disruption of Wnt/β‐catenin signaling and telomeric shortening are inextricable consequences of tankyrase inhibition in human cells. Mol Cell Biol 35: 2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpan K, Nathubhai A, Zhang C, Wood PJ, Lloyd MD, Thompson AS et al (2015). Structure‐based design, synthesis and evaluation in vitro of arylnaphthyridinones, arylpyridopyrimidinones and their tetrahydro derivatives as inhibitors of the tankyrases. Bioorg Med Chem 23: 3013–3032. [DOI] [PubMed] [Google Scholar]
- Kunttas‐Tatli E, Roberts DM, McCartney BM (2014). Self‐association of the APC tumor suppressor is required for the assembly, stability, and activity of the Wnt signaling destruction complex. Mol Biol Cell 25: 3424–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson EA, Jansson A, Ng FM, Then SW, Panicker R, Liu B et al (2013). Fragment‐based ligand design of novel potent inhibitors of tankyrases. J Med Chem 56: 4497–4508. [DOI] [PubMed] [Google Scholar]
- Lau T, Chan E, Callow M, Waaler J, Boggs J, Blake RA et al (2013). A novel tankyrase small‐molecule inhibitor suppresses APC mutation‐driven colorectal tumor growth. Cancer Res 73: 3132–3144. [DOI] [PubMed] [Google Scholar]
- Lee E, Salic A, Krüger R, Heinrich R, Kirschner MW (2003). The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol 1: E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtiö L, Chi N‐W, Krauss S (2013). Tankyrases as drug targets. FEBS J 280: 3576–3593. [DOI] [PubMed] [Google Scholar]
- Lehtiö L, Collins R, van den Berg S, Johansson A, Dahlgren L‐G, Hammarström M et al (2008). Zinc binding catalytic domain of human tankyrase 1. J Mol Biol 379: 136–145. [DOI] [PubMed] [Google Scholar]
- Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M et al (2011). Loss of tankyrase‐mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell 147: 1324–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li VSW, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP et al (2012). Wnt signaling through inhibition of β‐catenin degradation in an intact Axin1 complex. Cell 149: 1245–1256. [DOI] [PubMed] [Google Scholar]
- Li X, Han H, Zhou M‐T, Yang B, Ta AP, Li N et al (2017). Proteomic analysis of the human tankyrase protein interaction network reveals its role in pexophagy. Cell Rep 20: 737–749. [DOI] [PubMed] [Google Scholar]
- Liscio P, Carotti A, Asciutti S, Karlberg T, Bellocchi D, Llacuna L et al (2014). Design, synthesis, crystallographic studies, and preliminary biological appraisal of new substituted triazolo[4,3‐b]pyridazin‐8‐amine derivatives as tankyrase inhibitors. J Med Chem 57: 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T et al (2013). Targeting Wnt‐driven cancer through the inhibition of porcupine by LGK974. Proc Natl Acad Sci 110: 20224–20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xing Y, Hinds TR, Zheng J, Xu W (2006). The third 20 amino acid repeat is the tightest binding site of APC for beta‐catenin. J Mol Biol 360: 133–144. [DOI] [PubMed] [Google Scholar]
- Lu J, Ma Z, Hsieh J‐C, Fan C‐W, Chen B, Longgood JC et al (2009). Structure‐activity relationship studies of small‐molecule inhibitors of Wnt response. Bioorg Med Chem Lett 19: 3825–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U et al (2002). Negative feedback loop of Wnt signaling through upregulation of conductin/Axin2 in colorectal and liver tumors. Mol Cell Biol 22: 1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Semenov MV, Huang H, He X (2011). Dissecting molecular differences between Wnt coreceptors LRP5 and LRP6. PLoS One 6 e23537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B, Virshup DM (2015). Targeting Wnts at the source – new mechanisms, new biomarkers, new drugs. Mol Cancer Ther 14: 1087–1094. [DOI] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH, Flynn C et al (2001). Low‐density lipoprotein receptor‐related protein‐5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell 7: 801–809. [DOI] [PubMed] [Google Scholar]
- Mariotti L, Templeton CM, Ranes M, Paracuellos P, Cronin N, Beuron F et al (2016). Tankyrase requires SAM domain‐dependent polymerization to support Wnt‐β‐catenin signaling. Mol Cell 63: 498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino‐Echarri E, Brocardo MG, Mills KM, Henderson BR (2016). Tankyrase inhibitors stimulate the ability of tankyrases to bind Axin and drive assembly of β‐catenin degradation‐competent Axin puncta. PLoS One 11 e0150484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima T, Taneda Y, Jang M‐K, Mizutani A, Muramatsu Y, Yoshida H et al (2017). mTOR signaling mediates resistance to tankyrase inhibitors in Wnt‐driven colorectal cancer. Oncotarget 8: 47902–47915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsutani N, Yokozaki H, Tahara E, Tahara H, Kuniyasu H, Haruma K et al (2001). Expression of telomeric repeat binding factor 1 and 2 and TRF1‐interacting nuclear protein 2 in human gastric carcinomas. Int J Oncol 19: 507–512. [PubMed] [Google Scholar]
- Meric‐Bernstam F, Mills GB (2012). Overcoming implementation challenges of personalized cancer therapy. Nat Rev Clin Oncol 9: 542–548. [DOI] [PubMed] [Google Scholar]
- Minde DP, Anvarian Z, Rüdiger SG, Maurice MM (2011). Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer? Mol. Cancer 10: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S et al (1992). Somatic mutations of the APCgene in colorectal tumors: mutation cluster region in the APCgene. Hum Mol Genet 1: 229–233. [DOI] [PubMed] [Google Scholar]
- Morrone S, Cheng Z, Moon RT, Cong F, Xu W (2012). Crystal structure of a tankyrase‐Axin complex and its implications for Axin turnover and tankyrase substrate recruitment. Proc Natl Acad Sci 109: 1500–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu Y, Ohishi T, Sakamoto M, Tsuruo T, Seimiya H (2007). Cross‐species difference in telomeric function of tankyrase 1. Cancer Sci 98: 850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narwal M, Haikarainen T, Fallarero A, Vuorela PM, Lehtiö L (2013a). Screening and structural analysis of flavones inhibiting tankyrases. J Med Chem 56: 3507–3517. [DOI] [PubMed] [Google Scholar]
- Narwal M, Koivunen J, Haikarainen T, Obaji E, Legala OE, Venkannagari H et al (2013b). Discovery of tankyrase inhibiting flavones with increased potency and isoenzyme selectivity. J Med Chem 56: 7880–7889. [DOI] [PubMed] [Google Scholar]
- Narwal M, Venkannagari H, Lehtiö L (2012). Structural basis of selective inhibition of human tankyrases. J Med Chem 55: 1360–1367. [DOI] [PubMed] [Google Scholar]
- Nathubhai A, Haikarainen T, Hayward PC, Muñoz‐Descalzo S, Thompson AS, Lloyd MD et al (2016). Structure‐activity relationships of 2‐arylquinazolin‐4‐ones as highly selective and potent inhibitors of the tankyrases. Eur J Med Chem 118: 316–327. [DOI] [PubMed] [Google Scholar]
- Nathubhai A, Haikarainen T, Koivunen J, Murthy S, Koumanov F, Lloyd MD et al (2017). Highly potent and isoform selective dual site binding tankyrase/Wnt signaling inhibitors that increase cellular glucose uptake and have antiproliferative activity. J Med Chem 60: 814–820. [DOI] [PubMed] [Google Scholar]
- Nathubhai A, Wood PJ, Lloyd MD, Thompson AS, Threadgill MD (2013). Design and discovery of 2‐arylquinazolin‐4‐ones as potent and selective inhibitors of tankyrases. ACS Med Chem Lett 4: 1173–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkizinkiko Y, Suneel Kumar BVS, Jeankumar VU, Haikarainen T, Koivunen J, Madhuri C et al (2015). Discovery of potent and selective nonplanar tankyrase inhibiting nicotinamide mimics. Bioorg Med Chem 23: 4139–4149. [DOI] [PubMed] [Google Scholar]
- Okada‐Iwasaki R, Takahashi Y, Watanabe Y, Ishida H, Saito J‐I, Nakai R et al (2016). The discovery and characterization of K‐756, a novel Wnt/β‐catenin pathway inhibitor targeting tankyrase. Mol Cancer Ther 15: 1525–1534. [DOI] [PubMed] [Google Scholar]
- Paine HA, Nathubhai A, Woon ECY, Sunderland PT, Wood PJ, Mahon MF et al (2015). Exploration of the nicotinamide‐binding site of the tankyrases, identifying 3‐arylisoquinolin‐1‐ones as potent and selective inhibitors in vitro. Bioorg Med Chem 23: 5891–5908. [DOI] [PubMed] [Google Scholar]
- Pedersen NM, Thorvaldsen TE, Schultz SW, Wenzel EM, Stenmark H (2016). Formation of tankyrase inhibitor‐induced degradasomes requires proteasome activity. PLoS One 11 e0160507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson‐Nedry W, Erdeniz N, Kremer S, Yu J, Baig‐Lewis S, Wehrli M (2008). Unexpectedly robust assembly of the Axin destruction complex regulates Wnt/Wg signaling in Drosophila as revealed by analysis in vivo. Dev Biol 320: 226–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, Clevers H (2003). Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev 17: 1709–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt KD, Madan B, Ke Z, Pendharkar V, Ding L, Lee MA et al (2013). Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT‐driven mammary cancer. Cancer Res 73: 502–507. [DOI] [PubMed] [Google Scholar]
- Pronobis MI, Rusan NM, Peifer M (2015). A novel GSK3‐regulated APC:Axin interaction regulates Wnt signaling by driving a catalytic cycle of efficient βcatenin destruction. Elife 4 e08022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Mahaffey JP, Alcorn HL, Anderson KV (2011). Tissue‐specific roles of Axin2 in the inhibition and activation of Wnt signaling in the mouse embryo. Proc Natl Acad Sci 108: 8692–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Lam R, Voytyuk O, Romanov V, Gordon R, Gebremeskel S et al (2014). Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase‐2. Acta Crystallogr D Biol Crystallogr 70: 2740–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio AA, McCauley M, Langelier M‐F, Pascal JM (2016). Tankyrase sterile α motif domain polymerization is required for its role in Wnt signaling. Structure 24: 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippmann JF, Damm K, Schnapp A (2002). Functional characterization of the poly(ADP‐ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J Mol Biol 323: 217–224. [DOI] [PubMed] [Google Scholar]
- Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A et al (2000). APC mutations in sporadic colorectal tumors: a mutational ‘hotspot’ and interdependence of the ‘two hits’. Proc Natl Acad Sci U S A 97: 3352–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Lee E, Mayer L, Kirschner MW (2000). Control of beta‐catenin stability: reconstitution of the cytoplasmic steps of the wnt pathway in Xenopus egg extracts. Mol Cell 5: 523–532. [DOI] [PubMed] [Google Scholar]
- Samain F, Ekblad T, Mikutis G, Zhong N, Zimmermann M, Nauer A et al (2015). Tankyrase 1 inhibitors with drug‐like properties identified by screening a DNA‐encoded chemical library. J Med Chem 58: 5143–5149. [DOI] [PubMed] [Google Scholar]
- Schwarz‐Romond T, Fiedler M, Shibata N, Butler PJG, Kikuchi A, Higuchi Y et al (2007). The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol 14: 484–492. [DOI] [PubMed] [Google Scholar]
- Schwarz‐Romond T, Merrifield C, Nichols BJ, Bienz M (2005). The Wnt signalling effector Dishevelled forms dynamic protein assemblies rather than stable associations with cytoplasmic vesicles. J Cell Sci 118: 5269–5277. [DOI] [PubMed] [Google Scholar]
- Seimiya H (2002). The telomeric poly(ADP‐ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182‐kDa tankyrase‐binding protein (TAB182). J Biol Chem 277: 14116–14126. [DOI] [PubMed] [Google Scholar]
- Seimiya H, Muramatsu Y, Smith S, Tsuruo T (2004). Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP‐ribosyl)ation of TRF1 and telomere elongation. Mol Cell Biol 24: 1944–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebzukhov YV, Lavrik IN, Karbach J, Khlgatian SV, Koroleva EP, Belousov PV et al (2008). Human tankyrases are aberrantly expressed in colon tumors and contain multiple epitopes that induce humoral and cellular immune responses in cancer patients. Cancer Immunol Immunother 57: 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shervington A, Patel R, Lu C, Cruickshanks N, Lea R, Roberts G et al (2007). Telomerase subunits expression variation between biopsy samples and cell lines derived from malignant glioma. Brain Res 1134: 45–52. [DOI] [PubMed] [Google Scholar]
- Shultz MD, Cheung AK, Kirby CA, Firestone B, Fan J, Chen CH‐T et al (2013). Identification of NVP‐TNKS656: the use of structure‐efficiency relationships to generate a highly potent, selective, and orally active tankyrase inhibitor. J Med Chem 56: 6495–6511. [DOI] [PubMed] [Google Scholar]
- Shultz MD, Kirby CA, Stams T, Chin DN, Blank J, Charlat O et al (2012). ([1,2,4]Triazol‐3‐ylsulfanylmethyl)‐3‐phenyl‐[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding. J Med Chem 55: 1127–1136. [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T (2000). Tankyrase promotes telomere elongation in human cells. Curr Biol 10: 1299–1302. [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat S, Schmitt A, de Lange T (1998). Tankyrase, a poly(ADP‐ribose) polymerase at human telomeres. Science 282: 1484–1487. [DOI] [PubMed] [Google Scholar]
- Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SPH et al (2016). The IUPHAR/BPS guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucl Acids Res 44: D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink KE, Polakis P, Weis WI (2000). Structural basis of the Axin‐adenomatous polyposis coli interaction. EMBO J 19: 2270–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos JL, Weis WI (2013). The β‐catenin destruction complex. Cold Spring Harb Perspect Biol 5: a007898–a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen JD, Brody JR, Armen RS, Pascal JM (2013). Structural implications for selective targeting of PARPs. Front Oncol 3: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs CM, Baird JR, Hughes EG, Kent SS, Benchabane H, Paik R et al (2008). Dual positive and negative regulation of wingless signaling by adenomatous polyposis coli. Science 319: 333–336. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Mashima T, Mizutani A, Sato A, Aoyama A, Gong B et al (2017). APC mutations as a potential biomarker for sensitivity to tankyrase inhibitors in colorectal cancer. Mol Cancer Ther 16: 752–762. [DOI] [PubMed] [Google Scholar]
- Tang B, Wang J, Fang J, Jiang B, Zhang M, Wang Y et al (2012). Expression of TNKS1 is correlated with pathologic grade and Wnt/β‐catenin pathway in human astrocytomas. PubMed – NCBI. J Clin Neurosci 19: 139–143. [DOI] [PubMed] [Google Scholar]
- Tenbaum SP, Ordóñez‐Morán P, Puig I, Chicote I, Arques O, Landolfi S et al (2012). β‐catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nat Med 18: 892–901. [DOI] [PubMed] [Google Scholar]
- Thomson DW, Wagner AJ, Bantscheff M, Benson RE, Dittus L, Duempelfeld B et al (2017). Discovery of a highly selective tankyrase inhibitor displaying growth inhibition effects against a diverse range of tumor derived cell lines. J Med Chem 60: 5455–5471. [DOI] [PubMed] [Google Scholar]
- Thorsell A‐G, Ekblad T, Karlberg T, Löw M, Pinto AF, Trésaugues L et al (2017). Structural basis for potency and promiscuity in poly(ADP‐ribose) polymerase (PARP) and tankyrase inhibitors. J Med Chem 60: 1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen TE, Pedersen NM, Wenzel EM, Stenmark H (2017). Differential roles of AXIN1 and AXIN2 in tankyrase inhibitor‐induced formation of degradasomes and β‐catenin degradation. PLoS One 12 e0170508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen TE, Pedersen NM, Wenzel EM, Schultz SW, Brech A, Liestol K et al (2015). Structure, dynamics, and functionality of tankyrase inhibitor‐induced degradasomes. Mol Cancer Res 13: 1487–1501. [DOI] [PubMed] [Google Scholar]
- Tian A, Benchabane H, Wang Z, Ahmed Y (2016). Regulation of stem cell proliferation and cell fate specification by Wingless/Wnt signaling gradients enriched at adult intestinal compartment boundaries. PLoS Genet 12 e1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasante A, de Pablos B, Méndez‐Lago M, Abad JP (2008). Telomere maintenance in Drosophila: rapid transposon evolution at chromosome ends. Cc 7: 2134–2138. [DOI] [PubMed] [Google Scholar]
- Voronkov A, Holsworth DD, Waaler J, Wilson SR, Ekblad B, Perdreau‐Dahl H et al (2013). Structural basis and SAR for G007‐LK, a lead stage 1,2,4‐triazole based specific tankyrase 1/2 inhibitor. J Med Chem 56: 3012–3023. [DOI] [PubMed] [Google Scholar]
- Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT et al (2014). Family‐wide analysis of poly(ADP‐ribose) polymerase activity. Nat Comms 5: 4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaler J, Machon O, von Kries JP, Wilson SR, Lundenes E, Wedlich D et al (2011). Novel synthetic antagonists of canonical Wnt signaling inhibit colorectal cancer cell growth. Cancer Res 71: 197–205. [DOI] [PubMed] [Google Scholar]
- Waaler J, Machon O, Tumova L, Dinh H, Korinek V, Wilson SR et al (2012). A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res 72: 2822–2832. [DOI] [PubMed] [Google Scholar]
- Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell A‐G et al (2012). Family‐wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol 30: 283–288. [DOI] [PubMed] [Google Scholar]
- Wang S, Yin J, Chen D, Nie F, Song X, Fei C et al (2013). Small‐molecule modulation of Wnt signaling via modulating the Axin‐LRP5/6 interaction. Nat Chem Biol 9: 579–585. [DOI] [PubMed] [Google Scholar]
- Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E et al (2012). Recognition of the iso‐ADP‐ribose moiety in poly(ADP‐ribose) by WWE domains suggests a general mechanism for poly(ADP‐ribosyl)ation‐dependent ubiquitination. Genes Dev 26: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tacchelly‐Benites O, Yang E, Ahmed Y (2016a). Dual roles for membrane association of drosophila Axin in Wnt signaling. PLoS Genet 12 e1006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tacchelly‐Benites O, Yang E, Thorne CA, Nojima H, Lee E et al (2016b). Wnt/Wingless pathway activation is promoted by a critical threshold of Axin maintained by the tumor suppressor APC and the ADP‐ribose polymerase tankyrase. Genetics 203: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tian A, Benchabane H, Tacchelly‐Benites O, Yang E, Nojima H et al (2016c). The ADP‐ribose polymerase tankyrase regulates adult intestinal stem cell proliferation during homeostasis in Drosophila. Development 143: 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Le Trong I, Hinds TR, Stenkamp R, Kimelman D et al (2004). Crystal structure of a beta‐catenin/APC complex reveals a critical role for APC phosphorylation in APC function. Mol Cell 15: 523–533. [DOI] [PubMed] [Google Scholar]
- Xu D, Liu J, Fu T, Shan B, Qian L, Pan L et al (2017a). USP25 regulates Wnt signaling by controlling the stability of tankyrases. Genes Dev 31: 1024–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Lau YH, Fischer G, Tan YS, Chattopadhyay A, de la Roche M et al (2017b). Macrocyclized extended peptides: inhibiting the substrate‐recognition domain of tankyrase. J Am Chem Soc 139: 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L et al (2001). Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta‐catenin signaling is activated in human colon tumors. Proc Natl Acad Sci U S A 98: 14973–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, Tacchelly‐Benites O, Wang Z, Randall MP, Tian A, Benchabane H et al (2016). Wnt pathway activation by ADP‐ribosylation. Nat Comms 7: 11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiroda Y, Okamoto R, Hatsugai K, Takemoto Y, Goshima N, Saito T et al (2010). A novel yeast cell‐based screen identifies flavone as a tankyrase inhibitor. Biochem Biophys Res Commun 394: 569–573. [DOI] [PubMed] [Google Scholar]
- Zhan P, Song Y, Itoh Y, Suzuki T, Liu X (2014). Recent advances in the structure‐based rational design of TNKSIs. Mol Biosyst 10: 2783–2799. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA et al (2011). RNF146 is a poly(ADP‐ribose)‐directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol 13: 623–629. [DOI] [PubMed] [Google Scholar]
- Zhao F, Vermeer B, Lehmann U, Kreipe H, Manns MP, Korangy F et al (2009). Identification of a novel murine pancreatic tumour antigen, which elicits antibody responses in patients with pancreatic carcinoma. Immunology 128: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Ding Y, Bandyopadhyay G, Waaler J, Börgeson E, Smith S et al (2016a). The PARsylation activity of tankyrase in adipose tissue modulates systemic glucose metabolism in mice. Diabetologia 59: 582–591. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Katavolos P, Nguyen T, Lau T, Boggs J, Sambrone A et al (2016b). Tankyrase inhibition causes reversible intestinal toxicity in mice with a therapeutic index < 1. Toxicol Pathol 44: 267–278. [DOI] [PubMed] [Google Scholar]